Abstract
The de-methylesterification of the pectic polysaccharide homogalacturonan (HG) by pectin methylesterases (PMEs) is a critical step in the control of plant cell expansion and morphogenesis. Plants have large gene families encoding PMEs but also PME inhibitors (PMEIs) with differ in their biochemical properties. The Arabidopsis thaliana PECTIN METHYLESTERASE INHIBITOR 3 (PMEI3) gene is frequently used as a tool to manipulate pectin methylesterase activity in studies assessing its role in the control of morphogenesis. One limitation of these studies is that the exact biochemical activity of this protein has not yet been determined. In this manuscript we produced the protein in Pichia pastoris and characterized its activity in vitro. Like other PMEIs, PMEI3 inhibits PME activity at acidic pH in a variety of cell wall extracts and in purified PME preparations, but does not affect the much stronger PME activity at neutral pH. The protein is remarkable heat stable and shows higher activity against PME3 than against PME2, illustrating how different members of the large PMEI family can differ in their specificities towards PME targets. Finally, growing Arabidopsis thaliana seedlings in the presence of purified PMEI3 caused a dose-dependent inhibition of root growth associated with the overall inhibition of HG de-methylesterification of the root surface. This suggests an essential in vivo role for PME activity at acidic pH in HG de-methylesterification and growth control. These results show that purified recombinant PMEI3 is a powerful tool to study the connection between pectin de-methylesterification and cell expansion.
Keywords: Pectin, Cell expansion, Root, Pectin methylesterase (PME), PME inhibitor, Pichia pastoris
Abbreviations: HG, homogalacturonan; PME, pectin methylesterase; PMEI, PME inhibitor; GalA, galacturonic acid
Introduction
A central question in plant biology is how growth can occur despite the presence of strong cell walls (Cosgrove, 2005). Cell walls of growing cells (i.e. primary cell walls) are highly dynamic polymer assemblies primarily consisting of cellulose, small amounts of structural proteins and the matrix polysaccharides hemicelluloses and pectins. Pectins are galacturonic acid (GalA) - containing polysaccharides among which the linear polymer HG is the most abundant matrix polysaccharide, for instance representing 20% of the total cell wall polysaccharides, in Arabidopsis thaliana leaves (Zablackis et al., 1995) or even 50% in onion epidermis cells (Wilson et al., 2021). In vivo studies show that the metabolism of HG plays a critical role in the control of cell expansion and plant morphogenesis (Andres-Robin et al., 2018, Haas et al., 2020a, Haas et al., 2020b, Jobert et al., 2021, Peaucelle et al., 2015, Peaucelle et al., 2008, Phyo et al., 2017, Qi et al., 2017, Stefanowicz et al., 2021, Wachsman et al., 2020), but the underlying mechanisms are still poorly understood.
HG is synthesized in a highly methylesterified form in Golgi bodies and delivered to the surface via secretory vesicles. In the cell wall, de-methylesterification by PMEs (EC 3.1.1.11) uncovers the negatively charged carboxylic acid groups, which dramatically changes the physical properties of the individual polymers and polymer assemblies (Atmodjo et al., 2013, Haas et al., 2021). In addition, depending on the methylesterification pattern, HG is more or less sensitive to enzymatic degradation by polygalacturonases and pectate lyases (Hocq et al., 2017a, Hocq et al., 2017b). PMEs are part of large gene families (66 members in A. thaliana (Wolf et al., 2009)), with different expression patterns, but also different biochemical characteristics. For instance, most plant PMEs have processive activity, generating blockwise de-methylesterified HG, in contrast to characterized fungal PMEs, which are non-processive, generating more randomly de-methylesterified HG (Wolf et al., 2009). PME activity is also strongly pH dependent, with all plant PMEs studied so far showing neutral or alkaline pH optima (Hocq et al., 2021). Interestingly, the pH also influences the mode of action of the enzymes. For instance, citrus PME and Arabidopsis thaliana PME2 activities are processive at pH 8 and non-processive at pH 5 (Hocq et al., 2021). The biological significance of this behavior, in particular in relation to growth control is not known. PME activity is also regulated by PMEIs, which are also encoded by large gene families in vascular plants (76 in A. thaliana (Wolf et al., 2009)). PMEIs inhibit PME activity by forming 1:1 complexes with them. A critical role for PME activity in the control of cell expansion was shown by the inducible overexpression of PME5 and PMEI3, which promoted and inhibited cell wall expansion respectively, where both treatments caused the loss of growth anisotropy (Peaucelle et al., 2015). PMEI3 overexpression also caused the inhibition of organ formation in the inflorescence meristem (Peaucelle et al., 2011). Similar observations were made in which PME5 and PMEI3 overexpression respectively caused an increased and reduced gynoecium length (Andres-Robin et al., 2018), or reduced or increased cell expansion and loss of organ asymmetry in A. thaliana leaves (Qi et al., 2017). Finally, PMEI3 overexpression also led to the inhibition of lateral root formation (Wachsman et al., 2020). Surprisingly, some studies revealed opposite effects of changing the degree of HG methylesterification on cell wall mechanics and expansion rate. Indeed, depending on the context, de-methylesterification caused an increase or a decrease in wall elasticity and growth rate (Peaucelle et al., 2015, Phyo et al., 2017). It has been proposed that these opposite effects may be related to the pH-dependent, processive or non-processive nature of certain PMEs (Hocq et al., 2017a, Hocq et al., 2017b). In this view, processive activity generates stretches of acidic GalA, which can form cooperative Ca2+ crosslinks (the so-called egg boxes) and which would cause cell wall stiffening and growth inhibition. In contrast, randomly demethylated HG would form a more elastic Ca2+ cross-linked network (Vincent et al., 2013). An alternative view explaining PME-dependent growth was recently proposed based on the observation, using super-resolution optical microscopy, of oriented HG filaments in anticlinal cell walls of leaf pavement cells (Haas et al., 2020a, Haas et al., 2020b). In this view the de-methylesterification of HG leads to a volume increase of HG filaments, which would be a driver for cell expansion.
The above-mentioned studies used the inducible expression of the putative PMEI inhibitor PMEI3 (At5G20740) to manipulate pectin methylesterification. Despite its frequent use in vivo, the biochemical activity of PMEI3 has not been characterized so far. This knowledge is essential however, for the correct interpretation of the induced mechanical, cellular and macroscopic phenotypes. A few other PMEIs (PMEI4, 7 and 9) have been heterologously expressed and biochemically characterized (Hocq et al., 2017b, Sénéchal et al., 2015). This showed, for different PMEIs, pH-dependent (PMEI4, 7) or pH-independent (PMEI9) ineractions with target PMEs. In this report, we produced the PMEI3 protein in Pichia pastoris and characterized its activity and specificity. We show a pH-dependent inhibition of PME activity. Interestingly the inhibitory activity varied against different PME enzymes, showing the potential biochemical specialization of PME-PMEI pairs. Using purified PMEI3, we show that PME activity at acidic pH is critical for in vivo HG de-methylesterification and cell expansion in Arabidopsis roots.
Results
Expression in Pichia pastoris and purification of PMEI3
A codon-usage-optimized sequence of Arabidopsis thaliana PMEI3 (At5G20740), fused at its N-terminus to a secretion signal peptide and at its C-terminus to 6×His and c-MYC tags, was expressed in Pichia pastoris X33 (Fig. 1A). The mature protein could be detected in the culture supernatant using immunoblotting with anti-His antibodies (Fig. 1B). Upon His-tag affinity purification (Fig. S2A), a band of the expected molecular weight (∼25kDa) was observed on Coomassie-stained SDS PAGE (Fig. 1C). The identification of the PMEI3 protein was verified using immunoblotting with anti-His antibodies (Fig. 1C) and LC-MS/MS on the band extracted from the gel, which detected in the tryptic digest 4 peptides matching PMEI3 (Fig. 1D).
Fig. 1.
Expression in Pichia pastoris and purification of PMEI3. A. Scheme of the expression construct, B. Immunoblot with anti-His antibodies of 2 culture supernatants (#1 and #2) and positive control (PC) of purified PMEI3 . C. Coomassie stained gel (left) anti-His immunoblot (right) of purified protein. D. PMEI3-matching tryptic fragments (in red) from the purified protein identified by MS-MS.
pH-dependent inhibition of PME activities
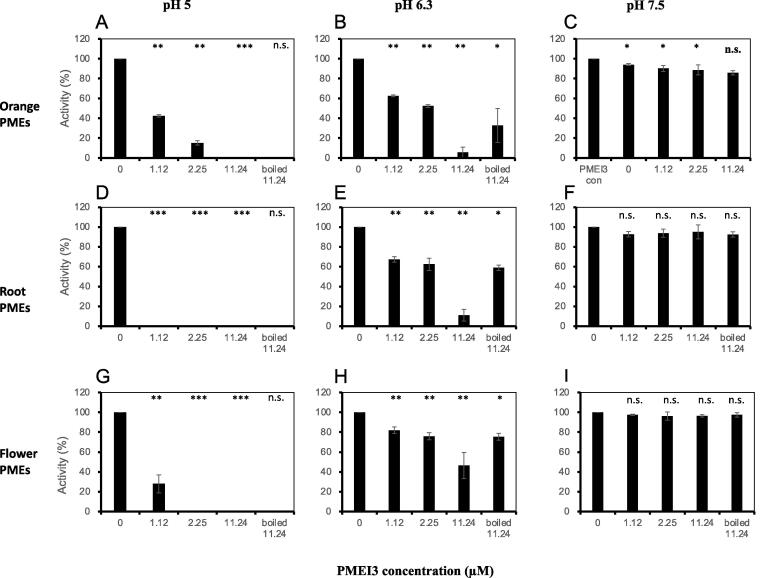
We next characterized the inhibitory activity of PMEI3 on a number of PME preparations. The PME activity was first measured on commercial citrus pectin (Degree of Methylesterification [DM] 85%) at pH 7.5 using a colorimetric assay for methanol production on dilutions of the enzyme preparation (Fig. S1). Non-saturating dilutions were chosen for the PMEI assays (1/100 dilution for Orange PME and 1/10 dilution for flower and root PME preparations). The activity of purified PMEI3 was assayed using a gel diffusion assay at pH 5, 6.3, and 7.5, respectively. The activity was quantified by measuring the radius of the red halo (Fig. S2). We first tested the inhibitory activity of PMEI3 against orange PME. The results showed strong inhibition at pH 5 and decreasing inhibition from pH 6.3 to 7.5, respectively (Fig. 2A-C). Surprisingly, boiling the sample did not (at pH 5) or only slightly (at pH 6.3) reduce the PMEI3 activity. This indicates that this inhibitor is able to correctly refold upon heat denaturation. A similar pH-dependent inhibitory pattern was observed against PME preparations from roots (Fig. 2D-F) and inflorescences (Fig. 2G-I) respectively. Interestingly, the inhibitory activity was stronger on the extracts from roots than from flowers at both pH 5 and 6.3. In conclusion, PMEI3 showed a pH-dependent inhibitory activity against one or more PME isoforms that are expressed in roots and flowers. The lower activity on flower extracts suggests that different PME isoforms may vary in their sensitivity to PMEI3 inhibition.
Fig. 2.
pH-sensitive inhibition of PME activity by PMEI3. Relative PME activity measured at pH 5 (A,D,G); 6.3 (B,E,H) and 7.5 (CF,I) in the presence of 0; 1.12 µM; 2.25 µM; 11.24 µM and boiled 11.24 µM PMEI3 in PME preparations from orange (A-C), Arabidopsis thaliana roots (D-F) or flowers (G-I). PME activity is expressed as % of the control without PMEI3. Error bars are SD (n=3) and stars refer to the P-value of the t-tests on the comparison with the 0 control or for the boiled samples for the comparison with non-boiled 11.24 µM samples. *:<0.05, **:<0.01, ***:<0.001 , ns : not significant.
Inhibitory activity and pH dependence of PMEI3 varies for different PME isoforms
To further investigate the specificity of PMEI3, we compared its activity against two heterologously expressed PME isoforms PME2 (At1g53830) (Hocq et al., 2021) and PME3 (At3g14310) (Guenin et al., 2011) (Figure 3). Both PME2 and PME3 are closely related isoforms with distinct expression patterns. PME3 is broadly expressed in roots and leaves (Fig. S3). Based on mutant analysis, PME3 represented around 30% of the PME activity in root extracts (Guenin et al., 2011). PME2 instead, has a more restricted expression pattern, with higher transcript levels in roots and flower petals (Fig. S4). In addition, mutant analysis showed that the contribution of PME2 to the total PME activity was only marginal at least in the organ investigated (dark-grown hypocotyl) (Hocq et al., 2021). The latter may reflect the epidermis-specific expression pattern of this gene as suggested by the cell type-specific RNA expression data (Klepikova et al., 2016, Wendrich et al., 2020) (Fig. S5). PME2 was produced in Pichia pastoris and purified using ion exchange chromatography (Hocq et al., 2021). PME3 fused to a C-terminal 6xHis-tag was transiently expressed in tobacco leaves and affinity purified (Senechal et al., 2015). Again, at pH 7.5, no PME inhibitory activity was observed against either one of the PMEs, with for the lower PMEI3 concentrations even a small but significant (t-test, P value = 0.008) PME3 promoting activity. Interestingly however, at pH 5, PMEI3 was at least 10-fold more active against PME3 than against PME2 (PME3 inhibition with 1.12 µM PMEI3 was comparable to PME2 inhibition with 11.2 µM PMEI3). In addition, at pH 6.3, PMEI3 was still active against PME3 but not against PME2. Together, these observations illustrate the high pH optimum for PMEI3 and its preference for PME3 over PME2.
Fig. 3.
Specific PMEI3 activity against purified PMEs. Relative PME activity measured in purified PME3 (A-C) and PME2 (D-F) preparations. at pH 5 (A-D); 6.3 (B-E) and 7.5 (C-F) in the presence of 0; 1.12 µM; 2.25 µM; 11.24 µM and boiled 11.24 µM PMEI3. PME activity is expressed as % of the control without PMEI3. Error bars are standard deviations (n=3) and stars refer to the P-value of the t-tests on the comparison with the 0 control or for the boiled samples for the comparison with non-boiled 11.24 µM samples. *:<0.05, **:<0.01, ***:<0.001, ns : not significant.
In vivo effect of PME inhibition
We next assessed the in vivo effect of the inhibition of PME activity by supplementing purified PMEI3 to the growth medium of A. thaliana seedlings. The small molecule PME inhibitor epigallocatechin-3-gallate (EGCG) was used as a positive control. EGCG caused a dose-dependent reduction in the root length of 4-day-old seedlings, relative to the mock-treated control, with an IC50 of around 55 µM (Fig. S6). Similarly, the root length of 3-day-old seedlings grown in the presence of PMEI3 was strongly reduced in a dose-dependent fashion (Fig. 4A,B) with an IC50 of 0.16 µM. We next measured the amount of blockwise de-methylesterified HG using the fluorescent probe COS647 (Mravec et al., 2014). Interestingly the labeling was strongly reduced in roots grown in the presence of 0.2 or 0.6 µM PMEI3 relative to control roots (Fig. 4C,D). Increasing the sensitivity of the detection also showed a dose responsiveness for the inhibition of the fluorescent signal, with a stronger inhibition at 0.6 µM relative to 0.2 µM PMEI3. (0.6 µM sat and 0.2 µM sat, respectively in Fig. 4C,D). In summary, these results show that external supply of PMEI3, like the PME inhibitor EGCG, strongly inhibited processive PME activity and root growth. PMEI3 treatment thus mimics the effects of the inducible overexpression of the PMEI3 gene (Peaucelle et al., 2015).
Fig. 4.
In vivo activity of purified PMEI3. A,B: Representative photographs (A) and primary root lengths (B) of 3-day-old Arabidopsis thaliana seedlings. Curve is a second degree polynomal fit (y = 3,3692x2 - 3,6899x + 1,007; R2 = 0,9947); IC50 = 0.16 µM. C,D: Representative photographs (C) and fluorescence quantification (D) of COS647-labeled roots. 0.2 µM sat and 0.6 µM sat refer to the same samples as for 0.2 µM and 0.6 µM but with an increased detection sensitivity. Two-day-old seedlings were transferred to 0; 0.2 µM or 0.63 µM PMEI3 and incubated for 24h before analysis. Error bars are SD (n > 30 for B and n=5 for D). *** t-test, P-value<0.001.
Discussion
Heterologously-expressed PMEI3 inhibited the activity of a variety of PME preparations. Interestingly, PMEI3 showed quantitative differences in its inhibitory activity, with higher activity against root PMEs and PME3 relative to flower and orange PMEs and PME2. PME3 is the major PME activity in vegetative tissues including roots, whereas PME2 transcripts are more abundant in flower petals. This may explain the higher sensitivity of root PMEs relative to flower PMEs for PMEI3. The comparison of the closely related 3D structures of PME2 and PME3 (Hocq et al., 2021) may provide valuable information on the structural basis for these differences in specificity.
In vitro PMEI3 activity was strongly pH-dependent, with the highest inhibition at pH 5.5 and almost no or no inhibition at pH 7.5. Similar observations were made for PMEI4 and PMEI6 but not PMEI9, which also inhibited PME activity at pH 7.5 (Hocq et al., 2017b). PMEI3, like PMEI4 and 6, is therefore expected not to totally prevent PME activity but only to sharpen its pH optimum in vivo. Interestingly, the pH was recently shown to also affect the de-methylesterification patterns at least for orange PME and Arabidopsis thaliana PME2 (Hocq et al., 2021). Indeed, whereas both enzymes were processive at pH 7.5, generating blockwise demethylated HG, their activity became non-processive at pH 5.5. This suggests that the strong effect of PMEI3 overexpression on growth and development (Andres-Robin et al., 2018, Peaucelle et al., 2015, Peaucelle et al., 2011, Qi et al., 2017, Wachsman et al., 2020) might be related to a change in the HG methylation patterns, rather than the quantitative inhibition of PME activity. This is surprising given the strong overall reduction of COS647 staining in PMEI3-treated seedling roots, and the fact that COS647 selectively binds blockwise de-methylesterified HG, the product of processive PME activity (Fig. 4C,D) (Mravec et al., 2014). To explain this, it is possible that the activity in intact cell walls does not reflect the in vitro specificity on isolated wall extracts or purified PMEs. In this context it is interesting to note that the specificity of PME activity was shown to change when the enzyme was incorporated in pectin or pectin/cellulose gels (Bonnin et al., 2019, Vincent et al., 2013). Alternatively, the apparent inhibition of both non-processive and, the much stronger, processive PME activity in root cell walls by the external application of PMEI3 may suggest that non-processive PME activity at low pH might be an essential step to prime subsequent processive activity at a higher pH. It will be interesting to see whether PME activity at high pH is unable to attack fully methylesterified HG and, if so, to what extent such behavior may contribute to acid growth. It will also be interesting to investigate the effect on in vivo pectin methylesterification and plant development of PMEI9 overexpression, which is expected to suppress PME activity irrespective of the pH, and to what extent HG populations with different methylation patterns can have distinct in vivo functions. A striking example of a HG methylesterification pattern with a specific in vivo function was recently reported for the external cell wall of seed coat epidermis cells (Francoz et al., 2019). Indeed, a specific pattern, recognized by antibody LM20, and requiring the presence of PMEI6, appeared to recruit a peroxidase (PRX36) to a specific location, thus generating a brittle zone in the external cell wall facilitating cell wall rupture and mucilage release at this location during seed hydration.
Finally, we observed that the external supply of PMEI3 to the growth medium inhibited the growth of seedlings in a concentration dependent way paralleling the PME inhibitory activity, corroborating the observations that PMEI3 overexpression inhibits cell wall expansion (Andres-Robin et al., 2018, Peaucelle et al., 2015, Peaucelle et al., 2011, Qi et al., 2017, Wachsman et al., 2020). The purified protein will be a valuable tool to study the in vivo dynamics of pectin metabolism and the developmental consequences of the manipulation of pectin methylesterification. Finally, it will be interesting to further explore the biochemical characteristics of other members of the large PME and PMEI families in plants.
Materials and methods
PMEI3 heterologous expression and purification
pPICZα B is a 3.6 kb vector used to express and secrete recombinant proteins in Pichia pastoris (ThermoFischer, Ref V19520). Recombinant proteins are expressed as fusions to an N-terminal peptide encoding the Saccharomyces cerevisiae α-factor secretion signal. The vector allows high-level, methanol-inducible expression of the gene of interest in Pichia, and can be used in Pichia strain X-33. A plant codon-optimized PMEI3 sequence (see supplementary data) was inserted into plasmid pPICZαB and transformed into E. coli strain TOP10. Transformants was selected on Low Salt LB plates containing 50 μg/mL Zeocin. Eight transformants for each were analyzed by restriction mapping or sequencing to confirm in-frame fusion of PMEI3 with the α-factor secretion signal and the C-terminal tag. The appropriate transformants were cultivated to collect more plasmid. Plasmids were purified using a DNA purification kit (NucleoBond PC 100/BAC kit, obtained from Machery-Nagel). The recombinant plasmids were purified and then linearized using the Pme1 restriction enzyme. Recombinant plasmids (at a concentration > 500 ng/μL) were transformed into Pichia pastoris strain X33. Transformed Pichia was plated onto YPDS medium containing 50 μg/mL of Zeocin.
Transformants were selected and then cultivated in BMGY medium. After 12-h incubation at 37°C with 250 rpm shaking, the OD600 of the culture was measured. The recombinant Pichia was transferred into BMMY medium in a bigger volume (Table 7.2) for large-scale expression. The OD600 was measured every 24 h to follow Pichia growth. In the meantime, 100% methanol was added to the medium to keep its final concentration at 0.5% (v/v). To make sure the recombinant proteins expressed correctly, the concentrated supernatant of the culture was tested using SDS-PAGE and/or Immunoblotting (details see below).
After 4-d expression, Pichia cells were removed by centrifugation at 5000 rpm for 5 min. To the protein-containing supernatant was added PBS buffer 10x (Eurobio, GAUPBS00-01), 400 mM NaCl, 5 mM imidazole and 500 μL cOmplete Hig-tag purification Resin (obtained from Roche, ref 05893682001). The mixture was gently shaken at 4°C for 4 h in dark. The supernatant was removed by centrifugation at 8000 rpm for 5 min at 4°C. The resin was washed by 5 ml of wash buffer 1 (PBS 1x with 400 mM NaCl and 5 mM imidazole), 5 ml of wash buffer 2 (PBS 1x with 400 mM NaCl and 30 mM imidazole) and eluted by elution buffer (PBS 1x with 400 mM NaCl and 400 mM imidazole) for 3 times (800 μL for each). The 3 elutes were collected and centrifuged at 8000 rpm for 30 min by using 3-kDa filter (Amicon Ultra-2 mL, UFC200324). The buffer change was performed using PD SpinTrap G-25 column. Final protein concentration was measured with a Bradford assay. The purified PMEI3 was frozen in liquid nitrogen and stored at -80°C until use.
Cell lysis and immunoblotting
Pichia cells were pelleted, rapidly frozen by liquid nitrogen and then stored at -80°C before use. Cells were resuspended in phosphate buffer pH 7.0 (2 mL for 1 mL pellet) containing 300 μl of protease inhibitor 1x (Constant systems Ltd. United Kingdom). Proteins were resolved using a 15% acrylamide/bisacrylamide SDS-PAGE (Table 7.3), with a running buffer composed of 25 mM Tris-base, 192 mM glycine, and 0.1% SDS at pH 8.7 (for Fig. 1B) or NuPage Bis/Tris 4-12 % acrylamide using MOPS running buffer (Fig. 1C). 13 μL of protein was added to 15 μL Laemmli buffer 2x and 2 μL of 1 M imidazole. The protein mix was heated at 95°C for 20 min. 15 μL of protein mix was loaded into a well of a SDS-PAGE gel. After electrophoresis at 100 mA for 100 min, the gel was stained with Pageblue protein staining solution (Thermo Scientific, catalog no. 24620) and destained with distilled water for 6 h. After migration of proteins, the gel SDS-PAGE was immerged in cathode buffer (25 mM of Tris pH 9.4, 40mM of Glycine, 10% of ethanol) for 15 min. A piece of Hybond-P PVDF membrane (GE Healthcare, catalog no. RPN303F) was placed in 100% ethanol for 15 seconds and rinsed by water for 2 min. Then the membrane was incubated in anode buffer II (25 mM of Tris pH 10.4, 10% of ethanol) for 5 min. The membrane, gel, and 7 same-sized Whatmann papers (previously immerged in anode I, anode II and cathode buffers) were placed. The transfer “sandwich” was exposed to 25 V and 1 A for 30 mins by a Trans-Blot TURBO transfer system (Bio-Rad, catalog no. 170- 4155). The membrane was incubated in 4.0% dry milk in PBS buffer, 0,1% Tween20 for 30 min and then incubated for 1 h at room temperature under shaking (50 rpm) in the same buffer containing 1/5000 of primary antibody raised against poly-His tag directly coupled with peroxidase (Sigma, catalog no. A7058). After washing 3 times by PBS 1x, 10 mins for each, 500 μL of DAB peroxidase substrate was added (Thermo Scientific, catalog no. 34002). Imaging was performed with a LAS-4000 according to the user manual. The concentration of proteins was determined according to Bradford using a BSA standard (1976).
Cell wall-enriched protein extraction
Fifty mg of frozen material of 10-day-old roots or 4-week-old flowers were ground with liquid nitrogen. One ml buffer (containing 50 mM Na2HPO4, 20 mM citric acid, 1 M NaCl, 0.01% Tween 20 at pH 7.0) was added. The solution was shaken at 250 rpm for 1 h. The extracts were clarified by centrifugation at 10000 rpm for 30 min at 4°C. The supernatant was filtered using an Amicon ultracentrifuge filter 0.5 ml/10000 MWCO (Millipore, catalog no. UFC5010BK) in order to remove salts. Protein concentration was determined by the Bradford method (details see below).
Methanol colorimetric assay
Activity of AtPME2, AtPME3, citrus PME, and PMEs from flowers and roots were determined using a colorimetric microassay adapted from Klavons and Bennett (1986). The reaction solution contained 5 μL of purified protein, 85% methylesterified (DM85) Citrus pectin (Sigma P9561) at 1 mg/ml, 0.008 U of Pichia pastoris alcohol oxidase (Sigma A2404) and 50 mM sodium phosphate buffer (pH = 7.5) to a final volume of 100 μl. AtPME2, AtPME3, and citrus PME were prepared at 3 concentrations (1, 1/10 and 1/100 of purified proteins) and PMEs from flowers and roots at 2 concentrations (1 and 1/10). The mixture was incubated at 28°C for 30 min. A first OD reading at 420 nm was done on a BioTek PowerWaveXS2 spectrometer. 100 μL of developing solution (2 mM ammonium acetate, 0.02 M pentane-2,4-dione, 0.05 M glacial acetic acid) was added and the solution was incubated at 68°C for 15 min after which a second reading was done. A standard dilution series from 0 to 10 nmol of methanol was included in each batch. Results were expressed as nmol MeOH/min-1.μL-1 of protein using the methanol standard curve.
Gel diffusion assay
DM85 Citrus pectin (Sigma P9561) was dissolved in 0.1 M citric acid and 0.2 M Na2HPO4 buffer at pH 5.0, 6.3 and 7.5 (Hocq et al., 2017b, Sénéchal et al., 2015 and 2017; Downie et al., 1998). One percent (w/v) agarose was added to the pectin buffer and heated until the agarose had dissolved. The solution was cooled to 60°C and transferred to a Petri dish (50 ml per dish) and allowed to solidify. Wells were punched in the gel with a 2-mm-diameter plastic sucker and the gel plug removed by aspiration. The gel was nicked and the orientation of the nick marked on the outside of the Petri dish with a marker. Ten μL of enzyme mixture was pipetted into each well. the wells were incubated at 37°C for 16 h. After incubation, the gels were rinsed briefly with water and stained with 0.02% Ruthenium Red (Sigma) for 1h at room temperature. The dye was then poured off and the gel was rinsed with water again. Diameters of the red halos around each well were measured using the Fiji software.
Mass spectrometry
Each SDS-PAGE band was manually excised from the gels to be hydrolysed according to (Shevchenko et al., 1996). Each sample was destained in a 40% ethanol-10% acetic acid solution (v/v) for 15 min and progressively dehydrated with 15 min incubations in first a 50% acetonitrile-25 mM ammonium bicarbonate solution and then in 100% acetonitrile. The dehydrated gel pieces were then reduced for 30 min with a 10 mM solution of DTT at 56°C and alkylated for 45 min with 55 mM iodoacetamide. The samples were then dehydrated as above. For digestion, the samples were incubated overnight at 37°C with 0.01 μg/μL of trypsin (Promega, Madison, WI) and dissolved in 50 mM ammonium bicarbonate, pH 8. The tryptic fragments were extracted from the gel by centrifugation after incubation of 15 min in a 0.5% trifluoroacetic acid - 50% acetonitrile (v/v) and in 100% acetonitrile solution, dried in a speed-vac, and reconstituted with 25 μL of a 2% acetonitrile - 0.1% formic acid (v/v) buffer. HPLC was performed on a NanoLC-Ultra system (Eksigent). A 4 μL sample of the peptide solution was loaded at 7.5 μL min−1 on a precolumn cartridge (stationary phase: C18 Biosphere, 5 μm; column: 100 μm inner diameter, 2 cm; Nanoseparations) and desalted with 0.1% fluorhydric acid and 2% acetonitrile. After 3 min, the precolumn cartridge was connected to the separating PepMap C18 column (stationary phase: C18 Biosphere, 3 μm; column: 75 μm inner diameter, 150 mm; Nanoseparations). Buffers A and B respectively were prepared with 0.1% HCOOH in water, and with 0.1% HCOOH in acetonitrile. The peptide separation was achieved at 300 nL min−1 with a linear gradient: from minute 0 to 10, at 5 to 30% of B; from minute 10 to 12, at 30 to 95% of B; from minute 12 to 18, at 95% of B (the regeneration step); from minute 18 to 19, at 95 to 5% of B; from minute 19 to 20, at 5% of B (the equilibration step). Remaining percentages are those of buffer A. One run took 45 min. Eluted peptides were online analyzed with an LTQ XL ion trap (Thermo Electron) using a nanoelectrospray interface. Ionization (1.5 kV ionization potential) was performed with liquid junction and a non-coated capillary probe (10 μm inner diameter; New Objective). Peptide ions were analyzed using Xcalibur 2.07 with the following data- dependent acquisition steps: (1) full MS scan (mass-to-charge ratio (m/z) 350-1 400, centroid mode) and (2) MS/MS (isolation width 1m/z, qz = 0.25, activation time = 30 ms, and normalized collision energy = 35%; centroid mode). Step 2 was repeated for the three major ions detected in step 1. Dynamic exclusion was set to 45s.
A database search was performed with XTandem (version 10-12-01-1) (http://www.thegpm.org/TANDEM/). Enzymatic cleavage was defined as a trypsin digestion with one possible miscleavage. Regarding protein modifications, Cys carboxyamidomethylation was set to static. As potential modifications, Met oxidation, Nter acetylation, Nter deamidation on Gln, Nter deamidation on carbamidomethylated Cys, Nter neutral loss on Glu were set. Precursor mass and fragment mass tolerance were 2.0 and 0.8, respectively. A refinement search was added with similar parameters. The Araport11_genes.201606 database and a contaminant database (trypsin, keratins) were used for peptides identification. Identified peptides were filtered and grouped using Xtandem according to: (1) A minimum of two different peptides required with an E-value smaller than 0.05, (2) a protein log(E-value) (calculated as the product of unique peptide E-values) smaller than -4.
Growth inhibition assays
Arabidopsis seedlings were grown for three days on vertical petridishes on ½ MS medium (Duchefa, reference M0221) with 1.2% phytoagar (Duchefa, reference: P1003) at 22 °C, 60% relative humidity, 50 µmol.m–2.s–1 white light (light source LED) with 18h-8h day-night cycle, before transfer to microtiter plates (4 seedlings per well) containing 100 µl of liquid ½ MS medium supplemented or not with PMEI3 or the PMEI inhibitor epigallocatechin-3-gallate (EGCG, Sigma-Aldrich) as a positive control. The root length was scored 14h later. Statistics were performed using the R statistics platforms. Significance (α = 0.05) was assessed by analysis of variance (ANOVA ; parametric) using linear mixed-effect models followed by Tukey HSD.
COS647 labeling of roots
Seedlings were fixed for 10 min in 10% acetic acid and 3.8% formaldehyde in “2F4 buffer” (Haas et al., 2020a, Haas et al., 2020b) and neutralized overnight in 10 mM NH4Cl in “2F4 buffer”. After washing with 25 mM MES buffer pH5.7, the samples were incubated in the same buffer for 10 min with 1/500 COS647 (Mravec et al., 2014) and, following washing, imaged with a confocal (Zeiss 710). COS647 was kindly provided by Jozef Mravec (University of Copenhagen, DK).
CRediT authorship contribution statement
Fan Xu: Conceptualization, Methodology, Investigation, Formal analysis. Martine Gonneau: Conceptualization, Methodology, Investigation. Elvina Faucher: Investigation, Formal analysis. Olivier Habrylo: Supervision, Methodology, Resources. Valérie Lefebvre: Supervision, Resources. Jean-Marc Domon: Resources. Marjolaine Martin: Investigation. Fabien Sénéchal: Resources. Alexis Peaucelle: Conceptualization, Investigation, Formal analysis. Jérôme Pelloux: Conceptualization, Supervision, Funding acquisition. Herman Höfte: Conceptualization, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Gladys Cloarec is thanked for assistance in the use of the confocal microscope. Jozef Mravec is thanked for kindly providing the COS647 probe. The work was financed by ANR projects “Pectosign” and “Homeowall” and the CSC PhD fellowship Nr 201506300059 to FX. The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007). This work has benefited from the support of IJPB's Plant Observatory technological platforms.
Footnotes
Given his/her/their role as Editor, Herman Hofte had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Prof Neil Gow.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tcsw.2022.100080.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Andres-Robin A., Reymond M.C., Dupire A., Battu V., Dubrulle N., Mouille G., Lefebvre V., Pelloux J., Boudaoud A., Traas J., Scutt C.P., Monéger F. Evidence for the regulation of gynoecium morphogenesis by ETTIN via cell wall dynamics. Plant Physiol. 2018;178:1222–1232. doi: 10.1104/pp.18.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo M.A., Hao Z., Mohnen D. Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 2013;64:747–779. doi: 10.1146/annurev-arplant-042811-105534. [DOI] [PubMed] [Google Scholar]
- Bonnin E., Alvarado C., Crépeau M.J., Bouchet B., Garnier C., Jamme F., Devaux M.F. Mobility of pectin methylesterase in pectin/cellulose gels is enhanced by the presence of cellulose and by its catalytic capacity. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-49108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005;6(11):850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Downie B., Dirk L.M., Hadfield K.A., Bennett A.B., Bradford K.J. A gel diffusion assay for quantification of pectin methylesterase activity. Anal. Biochem. 1998;264:149–157. doi: 10.1006/abio.1998.2847. [DOI] [PubMed] [Google Scholar]
- Francoz E., Ranocha P., Le Ru A., Martinez Y., Fourquaux I., Jauneau A., Dunand C., Burlat V. Pectin Demethylesterification Generates Platforms that Anchor Peroxidases to Remodel Plant Cell Wall Domains. Dev. Cell. 2019;48(2):261–276.e8. doi: 10.1016/j.devcel.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Guenin S., Mareck A., Rayon C., Lamour R., Assoumou Ndong Y., Domon J.M., Senechal F., Fournet F., Jamet E., Canut H., Percoco G., Mouille G., Rolland A., Rusterucci C., Guerineau F., Van Wuytswinkel O., Gillet F., Driouich A., Lerouge P., Gutierrez L., Pelloux J., Guénin S., Mareck A., Rayon C., Lamour R., Assoumou Ndong Y., Domon J.M., Sénéchal F., Fournet F., Jamet E., Canut H., Percoco G., Mouille G., Rolland A., Rustérucci C., Guerineau F., Van Wuytswinkel O., Gillet F., Driouich A., Lerouge P., Gutierrez L., Pelloux J. Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytol. 2011;192:114–126. doi: 10.1111/j.1469-8137.2011.03797.x. [DOI] [PubMed] [Google Scholar]
- Haas K., Rivière M., Wightman R., Peaucelle A. Multitarget Immunohistochemistry for Confocal and Super-resolution Imaging of Plant Cell Wall Polysaccharides. Bio-Protocol. https:// 2020;10(19) doi: 10.21769/bioprotoc.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas K.T., Wightman R., Meyerowitz E.M., Peaucelle A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science (80-. 2020;). 367:1003–1007. doi: 10.1126/science.aaz5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas K.T., Wightman R., Peaucelle A., Höfte H. The role of pectin phase separation in plant cell wall assembly and growth. Cell Surf. 2021;7:100054. doi: 10.1016/j.tcsw.2021.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocq L., Habrylo O., Voxeur A., Pau-Roblot C., Safran J., Senechal F., Fournet F., Bassard S., Hattu V., Demailly H., Tovar J., Pilard S., Marcelo P., Savary B.K., Marcadante D., Fransiska Njo M., Beeckman T., Boudaoud A., Pelloux J., Lefebvre V. The pH-dependent processivity of Arabidopsis AtPME2 can control cell wall mechanical properties. BioRxiv. 2021 doi: 10.1101/2021.03.03.433777. [DOI] [Google Scholar]
- Hocq L., Pelloux J., Lefebvre V. Connecting Homogalacturonan-Type Pectin Remodeling to Acid Growth. Trends Plant Sci. 2017;22:20–29. doi: 10.1016/j.tplants.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Hocq L., Sénéchal F., Lefebvre V., Lehner A., Domon J.-M., Mollet J.-C., Dehors J., Pageau K., Marcelo P., Guérineau F., Kolšek K., Mercadante D., Pelloux J. Combined Experimental and computational approaches reveal distinct pH-dependence of pectin methyl esterase inhibitors. Plant Physiol. 2017;173(2):1075–1093. doi: 10.1104/pp.16.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobert, F., Soriano, A., Brottier, L., Casset, C., Divol, F., Safran, J., Lefebvre, V., Pelloux, J., Robert, S., Péret, B., 2021. Auxin and pectin remodeling interplay during rootlet emergence in white lupin. bioRxiv 03420, 2021.07.19.452882. [DOI] [PubMed]
- Klepikova A.V., Kasianov A.S., Gerasimov E.S., Logacheva M.D., Penin A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016;88:1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- Mravec J., Kračun S.K., Rydahl M.G., Westereng B., Miart F., Clausen M.H., Fangel J.U., Daugaard M., Van Cutsem P., De Fine Licht H.H., Höfte H., Malinovsky F.G., Domozych D.S., Willats W.G.T. Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Dev. 2014;141(24):4841–4850. doi: 10.1242/dev.113365. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Braybrook S.A.A., Le Guillou L., Bron E., Kuhlemeier C., Hofte H., Höfte H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol. 2011;21:1720–1726. doi: 10.1016/j.cub.2011.08.057. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Louvet R., Johansen J.N., Höfte H., Laufs P., Pelloux J., Mouille G. Arabidopsis Phyllotaxis Is Controlled by the Methyl-Esterification Status of Cell-Wall Pectins. Curr. Biol. 2008;18(24):1943–1948. doi: 10.1016/j.cub.2008.10.065. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Wightman R., Höfte H., Hofte H. The Control of Growth Symmetry Breaking in the Arabidopsis Hypocotyl. Curr Biol. 2015;25:1746–1752. doi: 10.1016/j.cub.2015.05.022. [DOI] [PubMed] [Google Scholar]
- Phyo P., Wang T., Kiemle S.N., O’Neill H., Pingali S.V., Hong M., Cosgrove D.J. Gradients in wall mechanics and polysaccharides along growing inflorescence stems. Plant Physiol. 2017;175(4):1593–1607. doi: 10.1104/pp.17.01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Wu B., Feng S., Lü S., Guan C., Zhang X., Qiu D., Hu Y., Zhou Y., Li C., Long M., Jiao Y. Mechanical regulation of organ asymmetry in leaves. Nat. Plants. 2017;3:724–733. doi: 10.1038/s41477-017-0008-6. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sénéchal F., L'Enfant M., Domon J.-M., Rosiau E., Crépeau M.-J., Surcouf O., Esquivel-Rodriguez J., Marcelo P., Mareck A., Guérineau F., Kim H.-R., Mravec J., Bonnin E., Jamet E., Kihara D., Lerouge P., Ralet M.-C., Pelloux J., Rayon C. Tuning of pectin methylesterification: PECTIN METHYLESTERASE INHIBITOR 7 modulates the processive activity of co-expressed PECTIN METHYLESTERASE 3 in a pH-dependent manner. J Biol Chem. 2015;290(38):23320–23335. doi: 10.1074/jbc.M115.639534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanowicz K., Szymanska-Chargot M., Truman W., Walerowski P., Olszak M., Augustyniak A., Kosmala A., Zdunek A., Malinowski R. Plasmodiophora brassicae-Triggered Cell Enlargement and Loss of Cellular Integrity in Root Systems Are Mediated by Pectin Demethylation. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.711838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent R.R.R., Mansel B.W., Kramer A., Kroy K., Williams M.A.K. Micro-rheological behaviour and nonlinear rheology of networks assembled from polysaccharides from the plant cell wall. New J. Phys. 2013;15:35002. doi: 10.1088/1367-2630/15/3/035002. [DOI] [Google Scholar]
- Wachsman, G., Zhang, J., Moreno-Risueno, M.A., Anderson, C.T., Benfey, P.N., 2020. Cell wall remodeling and vesicle trafficking mediate the root clock in <em>Arabidopsis</em> Science (80-.). 370, 819 LP – 823. https://doi.org/10.1126/science.abb7250. [DOI] [PMC free article] [PubMed]
- Wendrich J.R., Yang B.J., Vandamme N., Verstaen K., Smet W., Van de Velde C., Minne M., Wybouw B., Mor E., Arents H.E., Nolf J., Van Duyse J., Van Isterdael G., Maere S., Saeys Y., De Rybel B. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science. 2020;370(6518) doi: 10.1126/science.aay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L.A., Deligey F., Wang T., Cosgrove D.J. Saccharide analysis of onion outer epidermal walls. Biotechnol. Biofuels. 2021;14:1–14. doi: 10.1186/s13068-021-01923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Mouille G., Pelloux J. Homogalacturonan Methyl-Esterification and Plant Development. Molecular Plant. 2009;2(5):851–860. doi: 10.1093/mp/ssp066. [DOI] [PubMed] [Google Scholar]
- Zablackis, E., Huang, J., Muller, B., Darvill, A.G., Albersheim, P., 1995. Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107, 1129–1138. https://doi.org/107/4/1129 [pii]. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.