Abstract
Purinostat Mesylate (PM) is a novel highly selective and active HDAC I/IIb inhibitor, and the injectable formulation of PM (PMF) based on the compound prescription containing cyclodextrin completely can overcome PM's poor solubility and improves its stability and pharmacokinetic properties. Here, we showed that PM effectively repressed the survival of Ph+ leukemia cells and CD34+ leukemia cells from CML patients in vitro. In vivo studies demonstrated that PMF significantly prevented BCR-ABL(T315I) induced CML progression by restraining leukemia stem cells (LSCs), which are insensitive to chemotherapy and responsible for CML relapse. Mechanism studies revealed that targeting HDAC I/IIb repressed several important factors for LSCs survival including c-Myc, β-Catenin, E2f, Ezh2, Alox5, and mTOR, as well as interrupted some critical biologic processes. Additionally, PMF increased glutamate metabolism in LSCs by increasing GLS1. The combination of PMF and glutaminase inhibitor BPTES synergistically eradicated LSCs by altering multiple key proteins and signaling pathways which are critical for LSC survival and self-renewal. Overall, our findings represent a new therapeutic strategy for eliminating LSCs by targeting HDAC I/IIb and glutaminolysis, which potentially provides a guidance for PMF clinical trials in the future for TKI resistance CML patients.
Keywords: Chronic myelogenous leukemia, Leukemia stem cell, Selective HDAC I/IIb inhibitor, GLS1, Mouse model
Graphical abstract
The presence of leukemia stem cells and TKI treatment resistance are the root causes why CML disease cannot be cured. PM, the novel highly selective HDAC I/IIb inhibitor. We prepared the free PM into the injectable formulation of PM (PMF) based on the compound prescription containing cyclodextrin, which can improve the solubility, stability, and pharmacokinetic properties of free PM, and reduces toxicity. Using BCR-ABL(T315I) induced TKI resistance CML mouse model, we demonstrated that PMF could significantly prevent CML disease and diminish survival and maintenance of LSCs. However, PMF treatment alone also increased glutaminolysis of LSCs by increasing GLS1 expression. Treatment of PMF combination with targeting GLS1 can synergistically impede disease and eradicate LSCs of CML. Our findings provide a guidance for PMF clinical trials in the future for TKI resistance CML patients.
Highlights
-
•
PM is a novel HDACI/IIb inhibitor with better selectivity and inhibitory activity than currently marketed HDAC inhibitors.
-
•
PMF completely overcomes the problem of PM's poor solubility, and improved PM stability and pharmacokinetic properties.
-
•
PMF effectively inhibits disease progression and abrogates leukemia stem cells survival in TKI-resistant CML mouse model.
-
•
Simultaneous targeting of I/IIb HDACs and glutaminolysis could sufficiently eradicated LSCs in the mouse model.
1. Introduction
Previously, we discovered a novel highly selective HDAC I/IIb inhibitor, Purinostat Mesylate (PM), which has a higher selectivity and inhibitory activity on HDAC I/IIb compared with the five approved HDAC inhibitors (Vorinostat, Romidepsin, Belinostat, Panobinostat, and Chidamide) [1]. However, the clinical application of free PM was greatly limited by its poor solubility in water and low bioavailability. Here, we developed a new PM formulation (PMF) via hydroxypropyl-β-cyclodextrin (HP-β-CD), arginine, meglumine, and mannitol to make it suitable for intravenous injection. PMF improves the solubility, stability, and pharmacokinetic properties of free PM, and reduces its toxicity. Preclinical studies have demonstrated that PM has good anti-tumor activity and low toxicity in treating multiple hematological malignancies [1,2]. Currently, PMF has been approved by the Food and Drug Administration (FDA; IND143467) and the Chinese National Medical Products Administration (NMPA; CXHL1800174) for clinical trials in relapsed and refractory B-cell malignancies. In order to further expand its applications, we deeply studied the ability of PMF to treat chronic myeloid leukemia (CML).
CML is a disease of hematological malignancy arising from hematopoietic stem cells (HSCs) [3], which is majorly caused by a fusion oncogene, BCR-ABL. BCR-ABL owns continuous tyrosine kinase activity resulting in activating multiple intracellular signaling pathways, including PI3K/AKT1, WNT/CTNNB1, SRC family kinases, and STATs [4]. Accordingly, BCR-ABL kinase activity inhibitors (TKIs; such as Imatinib, Nilotinib, Dasatinib and Ponatinib, etc.) have achieved great remission rates for CML patients [5]. However, some patients develop resistance to TKIs due to accumulation of BCR-ABL variant mutations, including T315I, G250E, G350R, Q252H, E352K, and Y253H [6]. The most common mutation, T315I, accounts for at least 15% of these variants [7]. Therefore, it is urgent to explore new treatment strategies for these patients hosting TKI resistance mutations. In addition, there are also BCR-ABL independent mechanisms involved in TKI resistance [8]. It is speculated that certain genetic evolutionary events have conferred an increased susceptibility to additional oncogenic hits, including genomic stability, epigenetic changes and microenvironmental shifts [9]. These changes could prompt leukemia cells to be promoted by (co-)drivers other than BCR-ABL. TKI response well is restricted to dividing CML progenitors and granulocytes, but not the case for leukemia stem cells (LSCs). It is now broadly accepted that a radical cure for CML depends upon the complete elimination of LSCs [10]. Therefore, it is important to identify and develop new treatments against key LSC targets for CML cure.
Targeting epigenetic regulators has shown promise for eliminating LSCs [11]. Histone deacetylases inhibitor (HDACi) has displayed excellent anti-tumor activity both in CML and AML [12,13]. Previous studies have demonstrated that pan-HDACi (Panobinostat, the most active HDACi) alone partially removed quiescent primitive LSCs while the combination of TKI and HDACi markedly diminished CML stem cells [14]. However, several studies have revealed that inhibition of HDAC IIa and IV has more side effects [15,16], for example, Panobinostat has a boxed warning in “Highlights of prescribing information” due to severe diarrhea, fatal cardiac ischemic events, arrhythmias, and ECG changes [17]. More importantly, previous studies have highlighted that selectively targeting some HDACs is more effective to remove LSCs [18,19]. Therefore, developing a higher affinity and selectivity of HDACi and identifying other relevant pathways are required for developing curative CML therapies. It is recognized that “Glutamine addiction” is a hallmark of cancer cell metabolism and targeting glutaminolysis is an attractive cancer therapeutic strategy [20]. As the essential enzyme in glutaminolysis, glutaminase1 (GLS1) catalyzes the conversion of glutamine to glutamate and ammonia. Inhibiting GLS1 activity with a specific inhibitor or reducing its product with glutamine analogue L-DON has been shown to be effective in cancer therapeutic strategies [21].
By using TKI resistance BCR-ABL(T315I) induced CML mouse model, it was shown that PMF effectively prevented CML progression and significantly suppressed LSCs. Nevertheless, PMF increased glutamate metabolism in LSCs associated with a concomitantly high level of GLS1. Fortunately, we found simultaneous administration of PMF and inhibitor of glutaminolysis significantly prolonged survival of disease mice and markedly diminished LSCs compared with monotherapy. Overall, our findings provide a new therapeutic strategy for eliminating CML LSCs by simultaneous targeting HDAC I/IIb and glutaminolysis, which potentially provides a new option for CML patients hosting BCR-ABL TKI resistant mutations.
2. Materials and methods
2.1. Cell lines and compounds
LAMA84 and K562 were cultured in RPMI1640 medium with 10% FBS (Gibco, Thermo Fisher Scientific) and 100 units/ml penicillin/streptomycin (Gibco). Imatinib (MB1122) and GLS1 inhibitor BPTES (MB2348) were purchased from Meilunbio, China. Purinostat Mesylate was synthesized in our lab. In vitro studies, IM, free PM and BPTES were dissolved in DMSO; In vivo, the IM was dissolved in 0.9% NaCl solution and BPTES was dissolved in PBS with 10% DMSO. For mice treatment, PMF lyophilized powder was dissolved in 0.9% NaCl solution and the Placebo group was given the same amount of PM-free lyophilized powder.
2.2. Preparation of the injectable formulation of PM lyophilized powder
The injectable formulation of PM was prepared by a simple dissolution method. First, 10% HP-β-CD was added to deionized water and stirred at 37 °C for 2 h. Next, PM was added and stirred for 24 h. After filtering with a 0.22 μm membrane to remove excess PM, the filtrate was mixed with arginine, meglumine, and mannitol in a specific molar ratio to form the compound prescription formulation, and the formulation lyophilized in a penicillin bottle, and stored at 2–8 °C. The detailed preparation method was shown in the patent “CN202110815779”. Injectable formulation of PM (PMF) lyophilized powder can be re-dissolved in 0.9% NaCl solution to obtain a clear yellow solution. The doses of PMF were all quantified by the mass of the free PM.
2.3. Western blot analysis
Whole cells were lysed with RIPA buffer plus protease and phosphatase inhibitors. Proteins were separated using PAGE gel electrophoresis, transferred onto membranes, and immunoblotted with primary antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibodies. The antibodies against Ac-α-tubulin (Cat # Sc-23950), α-tubulin (Cat # sc-134237), Ac-H3 (Cat # sc-56616), Abl (Cat # sc-131), P53 (Cat # sc-47698), c-MYC (Cat # sc-70468), Alox5 (Cat # sc-136195), Alox15 (Cat # sc-133085), β-Catenin (Cat # sc-59737), PARP-1 (Cat # sc-8007), P-Erk (Cat # sc-101760), Fak (Cat # sc-558) and P-Fak (Cat # sc-81493) were purchased from Santa Cruz Biotechnology, USA. The antibodies against GAPDH (Cat # 5174), H3 (Cat # 4499), Ezh2 (Cat # 4095), P-Stat5 (Cat # 9359), Stat5 (Cat # 9363), Caspase9 (Cat # 9508), P-Raf (Cat # 9427), Raf (Cat # 9422), P-Mek (Cat # 9121), Mek (Cat # 4694), Erk (Cat # 4695), P-Akt (Cat # 4060) and P21 (Cat # 2946) were purchased from Cell Signaling Technology, USA. The antibodies against Gls1 (Cat # ab262716), Caspase3 (Cat # ab44976), Bad (Cat # ab32245) and Akt (Cat # ab32038) were purchased from Abcam. The antibodies against Caspase8 (Cat #c2976) and Bcl-xl (Cat #L7543) were purchased from Sigma, USA. The antibody against HoxA9 (Cat # EM-1706-21) was purchased from Huabio, China. The final signal was detected with enhanced chemiluminescence (ECL).
2.4. In vivo pharmacokinetics and treatment of T315I-CML mouse model
All animal studies were performed in accordance with the guidelines approved by the Institutional Animal Care and Use Committees of Sichuan University (Chengdu, China).T315I-CML mouse model was established as described previously [22]. Briefly, bone marrow (BM) cells from 5-Fu treated (200 mg/kg) donor mice were transduced twice with BCR-ABL(T315I) retrovirus by co-sedimentation in the presence of IL-3, IL-6, and SCF. Recipient mice received two doses of 500 cGy γ-ray irradiation separated by 3 h and a dose of 5 × 105 cells transplanted via tail vein injection.
For pharmacokinetic studies, 12 days after BMT, the proportions of tumor cells (GFP+Gr-1+, n = 6) in peripheral blood (PB), spleen and BM of model mice were 44.18 ± 5.46%, 21.4 ± 2.82% and 40.72 ± 10.03%, respectively. Free PM was dissolved in 2.0% DMSO, 5.0% castor oil and 2.5% ethanol (v/v), and diluted with 0.9% NaCl solution to 1 mg/mL, and the PMF lyophilized powder was dissolved in 0.9% NaCl solution to 1 mg/mL. Then, the model mice were divided into 2 groups, and received a single intravenous injection of 10 mg/kg free PM and PMF, respectively. Plasma and BM cell samples in free PM and PMF administration groups were collected after 0.083, 0.5, 1, 2, 4, 8, 24, and 48 h, and the PM concentrations were determined by an internal standard UFLC-MS/MS method using protein precipitation prepared in non-treated mouse plasma and BM cells. 1 μL sample was injected into ultrafast liquid chromatography (UFLC) system (SIL-30AC auto-sampler, LC-30AD chromatograph CBM-20A communications bus module, CTO-20AC prominence column oven, Shimadzu, Japan), equipped with Acquity UPLC BEH C18 column (100 mm × 2.1 mm I.D., 1.7 mm; Waters). The concentrations were analyzed in AB SCIEX Qtrap 5500 mass spectrometer, which has electrospray positive mode and analyst 1.6.2 software. Reported concentrations are mean ± SD values from 3 mice/time point/group, and the pharmacokinetic parameters were fitted with DAS2.0 software.
For in vivo therapy, 14 days after BMT, CML mice were randomly grouped and treated with IM (100 mg/kg, orally, twice daily), PMF (5 mg/kg and 10 mg/kg, intravenous, three times a week), BPTES (10 mg/kg, intra-peritoneally, three times a week), or a combination of the aforementioned. Placebo was used as the control.
2.5. Flow cytometry
Cells from PB, spleen and BM content of femurs and tibiae were obtained. The GFP-expressing myeloid cells and stem cell populations were stained with anti-mouse APC-Gr-1 (Cat # 17–5931, eBioscience), anti-mouse Hematopoietic lineage eFluor450® Cocktail (Cat # 88–7772, eBioscience), anti-mouse PE-Sca-1 (Cat # 12–5981, eBioscience), anti-mouse APC-c-Kit (Cat # 17–1171, eBioscience), anti-mouse PE-Cy7-CD16/32 (Cat # 25-0161-82, eBioscience), anti-mouse-Alexa Fluor700-CD34 (Cat # 56-0341-82, eBioscience) and measured by flow cytometry on a BD LSR Fortessa machine. All the above antibodies were purchased from Thermo Fisher Scientific. The FACS data was analyzed and presented using the Flowjo10 software.
2.6. RNA-seq and analysis of gene expression changes induced by PMF and BPTES treatment
T315I-CML induced by BCR-ABL(T315I), at day 14, treated with PMF and BPTES (10 mg/kg). After 24 h, LSCs (GFP + LinlowSca1+c-Kit+) [23] were sorted from the BM using a FACS Aria III (Becton Dickinson). Total RNA was extracted from LSCs using the RNeasy Mini kit (Qiagen). RNA samples were reverse transcribed and amplified using the Ovation Pico WTA System V2 (NuGEN Technologies), and biotin labelled with the Encore Biotin Module (NuGEN Technologies). RNA-seq was performed using the Illumina MiSeq system. Gene Set Enrichment Analysis (GSEA) was performed with GSEA software version 3.0 (http://www.broadinstitute.org/gsea/) for detection of enrichment of predetermined gene sets in hallmark gene sets (H), curated gene sets (C2), and GO gene sets (C5) category from the Molecular Signature Database (MsigDB). Gene sets representing common functional categories were categorized and grouped.
2.7. Cell viability and apoptosis assay of human CD34+ CML cells
Leukemia cells from primary and relapse CML patients PB were incubated with human CD34 Microbeads (130-046-702, Miltenyi Biotec), and CD34+ CML cells (including LSCs and leukemia progenitor cells) were purified in sterile conditions. Indicated numbers cells were cultured in SFM containing SCF, TPO, Flt-3 ligand, and IL3 at 50 ng/mL and treated with various dose of free PM (2.5–20 nM) and BPTES (5 μM or 10 μM) for 48 h, then alive CD34+cells numbers were counted with trypan blue and FACS. Relative viable cells were normalized with that in control treatment group. For cell apoptosis assay, cells were stained with CD34 monoclonal antibody (Cat#12-0349-42, Biosciences) initially and then washed again with binding buffer before staining with Annexin-V and PI. The patient samples were obtained from West China Hospital which were approved by the clinical ethics committee in West China Hospital, Sichuan University. All patients signed the informed consent form, and met all requirements of the Declaration of Helsinki.
2.8. Leukemia stem cell culture assay
CML mice were induced by BCR-ABL(T315I). On day 14 post BMT, 5 × 106 cells isolated from BM were cultured in 6 cm dish containing Stemspan SFEM (Stem Cell Technologies), SCF (Peprotech), IGF-2 (R&D Systems), TPO (R&D Systems), Heparin (Sigma) and α-FGF (Gibco) as described previously for the culture of hematopoietic stem cells [24].
2.9. Colony formation assay
CML mice were induced by BCR-ABL(T315I). On day 14 post BMT, BM cells were collected for analysis with FACS. 1 × 104 GFP + cells were plated into a 6 well plate containing 2 mL M3234 medium with or without indicated concentration of drugs (n = 3 for each treatment group). The colonies were counted 7 days after drug treatment.
2.10. UFLC-MS/MS determination of glutamate, α-ketoglutarate, and succinyl-CoA
K562 cells (1 × 107) were plated in 10 cm dish and treated with free PM at 0, 10, 20 and 40 nM for 24 h, then cells were collected and alive cells numbers were counted. Each cell sample was sonicated after adding chloroform/methanol (2:1, v/v). BCR-ABL(T315I)-induced CML mice were treated with a single dose of PMF at a dose of 10 mg/kg for 24 h, then the spleens were collected. The freshly spleens were homogenized with chloroform/methanol (2:1, v/v). After centrifuging the K562 cells debris or spleen homogenate suspension, and the supernatant was collected and dried with N2. Dried lipids were dissolved with 90 μL 90% methanol (in H2O), then sonicated for 3–5 min, and spiked with 10 μL of 500 ng/ml deuterated internal standard solution. Then, the samples were then assayed with UFLC-MS/MS. For each run, a standard curve was generated with different concentration of glutamate, α-ketoglutarate (α-KG) or succinyl-CoA mixed with IS (50 ng/mL final concentration).
2.11. Histopathology analysis
The lungs and spleens from the Placebo or drug treated group mice were fixed in 4% PFA for 24 h at room temperature, followed by an overnight rinse in water. Then processed, sectioned, and stained with hematoxylin and eosin (H&E) with routine methods.
2.12. Statistical analysis
Statistical analyses were performed using t-test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001) and GraphPad Prism software, version 7.01 for Windows (GraphPad Software). A p value of less than 0.05 was considered statistically significant. Survival data was analyzed by a log-rank test (Mantel-Cox).
3. Results
3.1. Purinostat Mesylate shows potent suppression activity on Ph + cells
Previously, we designed a highly selective and active HDAC I/IIb inhibitor, Purinostat Mesylate (Fig. 1A and Fig. S1A), which shows excellent activity in hematological malignancies [2]. Here, we further investigated the therapeutic effect of free PM upon CML. Firstly, we found that PM significantly reduced the viability of Ph + cell lines with an IC50 of less than 10 nM (Fig. 1B). Additionally, PM obviously arrested the cell cycle at G0/G1 phase and induced apoptosis with a dose- and time-dependent manners (Fig. 1D–E and Figs. S1B–C). Furthermore, PM also induced BaF3-BCR-ABL(T315I) cell apoptosis in a dose-dependent manner (Fig. 1C). Importantly, we also found that PM showed excellent anti-tumor activity on human CD34+ cells (including LSCs and leukemia progenitor cells) isolated from CML patients (including newly diagnosed and TKI treatment relapse patients, Supplementary Table 1). The results indicated that the free PM markedly reduced cell survival and induced apoptosis of CML CD34+ cells in a dose dependent manner (Figs. S2A–B).
Fig. 1.
The free PM shows potent suppression activity on Ph +cell lines. (A) LAMA84 cells were cultured in presence of 20 nM free PM for indicated time, then cells were harvested for westren blotting analysis using Ac-TUBULIN,TUBULIN, Ac-H3, and H3 antibodies. GAPDH served total protein loading control. (B) LAMA84 and K562 cells were transfected with the mCherry-luciferase-puromycin virus to generate stable cell lines, and luciferase-tagged cells were cultured in presence of DMSO or the indicated concentration of free PM. Luminescence activity was measured every 12 h. Relative cell vitality was normalized with cells treated with DMSO. (C) BCR-ABL(T315I) or MIG transducted BaF3 cells were treated with indicated concentration of free PM. Cell apoptosis was measured with Annexin-V/PI staining and analyzed with FACS. (D) LAMA84 and K562 cells were cultured in presence of DMSO or free PM at the indicated concentrations for 24 h, the harvested cells were digested with RNAase and stained with PI for cell cycle analysis. (E) LAMA84 and K562 cells were cultured with DMSO or the free PM at the indicated concentrations for 36 h. Cell apoptosis was measured with Annexin-V/PI staining and analyzed with FACS. (F) LAMA84 cells were treated with 20 nM free PM for the indicated time, and western blotting analysis were performed to detect endogenous and exogenous apoptotic proteins including CASPASE9, CASPASE8, CASPASE3, PARP and their associate proteins P53, BCL-XL and BAD; (G) and proteins involved in cell proliferation pathway including c-RAF, p-c-RAF, MEK, p-MEK, ERK, p-ERK, AKT, p-AKT; and (H) BCR-ABL, c-MYC, ALOX5, ALOX15, β-CATENIN, EZH2, p-STAT5, STAT5, p-FAK, and FAK. GAPDH served as total protein loading control.
Subsequently, to investigate the potential signaling pathways involved in PM induced cell apoptosis, several node factors in apoptosis processes were examined. As shown in Fig. 1F, free PM significantly induced CASPASE8 self-cleavage, and subsequently activated CASPASE3 and 9. As its consequent result, PARP was also cleaved. Simultaneously, the levels of BAD and TP53 were increased and the anti-apoptotic protein BCL-XL was decreased. Secondly, PM substantially suppressed the phosphorylation of “RAF-MEK-ERK-AKT” cell growth signaling axis (Fig. 1G). More importantly, free PM significantly reduced the protein levels of BCR-ABL and its downstream targets STAT5 and FAK (Fig. 1H and Figs. S1D–F). Additionally, many important factors for LSCs survival including c-MYC, ALOX5 [25], and EZH2 [26] were substantively suppressed both in a time- and dose-dependent manner. It was noteworthy that ALOX15 [27] and β-CATENIN [28] were significantly increased (Fig. 1H and Figs. S1D–F). Together, these results suggested that PM effectively prevented CML by suppressing multiple important signaling pathways and key factors for LSCs survival.
3.2. The injectable formulation of PM has excellent solubility
Although free PM exhibits good anti-tumor activity in vitro, the solubility of free PM is only 9.18 μg/mL in water and its oral bioavailability is 6.71%, which greatly limits its clinical application. As previously reported, we prepared free PM into an injectable formulation that meets clinical needs [29]. Through screening solubilizing excipients, we found that hydroxypropyl-β-cyclodextrin (HP-β-CD) could significantly improve the apparent solubility of PM in water (from 9.18 μg/mL to 2.02 mg/mL). The interaction mechanism between PM and HP-β-CD was confirmed by computer simulation, fourier transform infrared spectroscopy (FT-IR), nuclear magnetic resonance (1H NMR), and scanning electron microscopy (SEM) (Figs. S3A–D). Unlike a simple physical mixing of free PM and HP-β-CD, one side of the aminobenzene ring in PM structure is deeply embedded in the hydrophobic cavity of HP-β-CD, while the side of the pyridine ring is slightly or not embedded in the cavity. Although the PM/HP-β-CD inclusion complex improves the solubility of free PM, we found that it was not stable enough when dissolved in 0.9% NaCl solution, and many impurities were formed. Through further extensive screening of co-solvents, we found that a certain proportion of arginine and meglumine could interact with the hydroxamic acid groups of PM in PM/HP-β-CD to form basic in situ salts, which not only further improves the solubility of the free PM but also greatly improves the stability of PM in 0.9% NaCl solution (the solubility of PMF in 0.9% NaCl >10 mg/mL, in terms of the free PM mass). Overall, the PMF lyophilized powder completely overcomes the problems of poor solubility and instability of the free PM in preclinical and clinical administration.
3.3. PMF improves the pharmacokinetics and reduces the toxicity of the free PM in vivo
We further conducted the pharmacokinetic studies of free PM and PMF through a BCR-ABL(T315I) induced CML mouse model to explore whether PMF could improve the pharmacokinetics properties of free PM. Model mice were treated with a single dose of free PM or PMF at 10 mg/kg. The concentrations of free PM in plasma and BM cells were measured at multiple time points. As shown in Table 1 and Figs. S4A–B, the half-lives (t1/2) of free PM in plasma and BM cells in the PMF administration group were 18.99 ± 6.22 h and 15.93 ± 6.08 h, respectively, which were significantly longer than that of the free PM administration group (3.04 ± 1.74 h and 8.22 ± 1.65 h for plasma and BM cells, respectively). Additionally, the free PM concentration in plasma peaked at 0.083 h after free PM administration, with a Cmax of 5264 ± 333 ng/mL, which was 7.36 folds than the Cmax of PMF administration, while their exposures (AUC0-∞) in plasma and BM cells have similar values. Furthermore, at 48 h after PMF administration, we found that the free PM concentrations in plasma and BM cells were 25.95 ± 1.73 ng/mL (44.24 ± 2.95 nM) and 423.31 ± 55.58 ng/107 cells (72.16 ± 9.47 nM), which were significantly higher than the IC50 values of CML cells, while no free PM was detected in plasma of the free PM administration mice. These results suggest that PMF has a sustained-release effect after intravenous administration, which could maintain a long-term anti-tumor activity. The rapid increase of excessive drug concentration in plasma may cause acute cardiotoxicity and more serious dose-limiting toxicity. We further performed a maximum tolerated dose (MTD) study of free PM and PMF by using a single dose of intravenous administration on Balb/c mice. The results showed that the MTD for PMF administration was 350 mg/kg, while the MTD for free PM was 150 mg/kg. Collectively, the injectable formulation of PM not only has a sustained-release effect but also reduces the dose-limiting toxicity.
Table 1.
Pharmacokinetic parameters of the free PM and PMF after a single intravenous administration of 10 mg/kg in T315I-CML mouse model (Mean ± SD; 3 mice/time point/group).
| Parameters | Free PM |
PMF |
||
|---|---|---|---|---|
| Plasma | Bone Marrow | Plasma | Bone Marrow | |
| AUC0-∞ (ng/L*h or ng/107 cell*h) | 6612 ± 788 | 65306 ± 9276 | 5105 ± 1119 | 65053 ± 3625 |
| MRT0-∞ (h) | 3.89 ± 0.17 | 13.78 ± 1.18 | 21.83 ± 6.12 | 22.61 ± 3.15 |
| t1/2z (h) | 3.04 ± 1.74 | 8.22 ± 1.65 | 18.99 ± 6.22 | 15.93 ± 6.08 |
| Tmax (h) | 0.08 ± 0 | 0.50 ± 0 | 0.08 ± 0 | 1.03 ± 0.96 |
| CLz (L/h/kg) | 1.53 ± 0.17 | 0.16 ± 0.12 | 2.02 ± 0.41 | 0.15 ± 0.01 |
| Cmax (ng/mL or ng/107 cell) | 5264 ± 333 | 10458 ± 3570 | 715 ± 103 | 5072 ± 594 |
3.4. PMF prolongs the survival of BCR-ABL(T315I) induced CML mice and impairs the LSCs
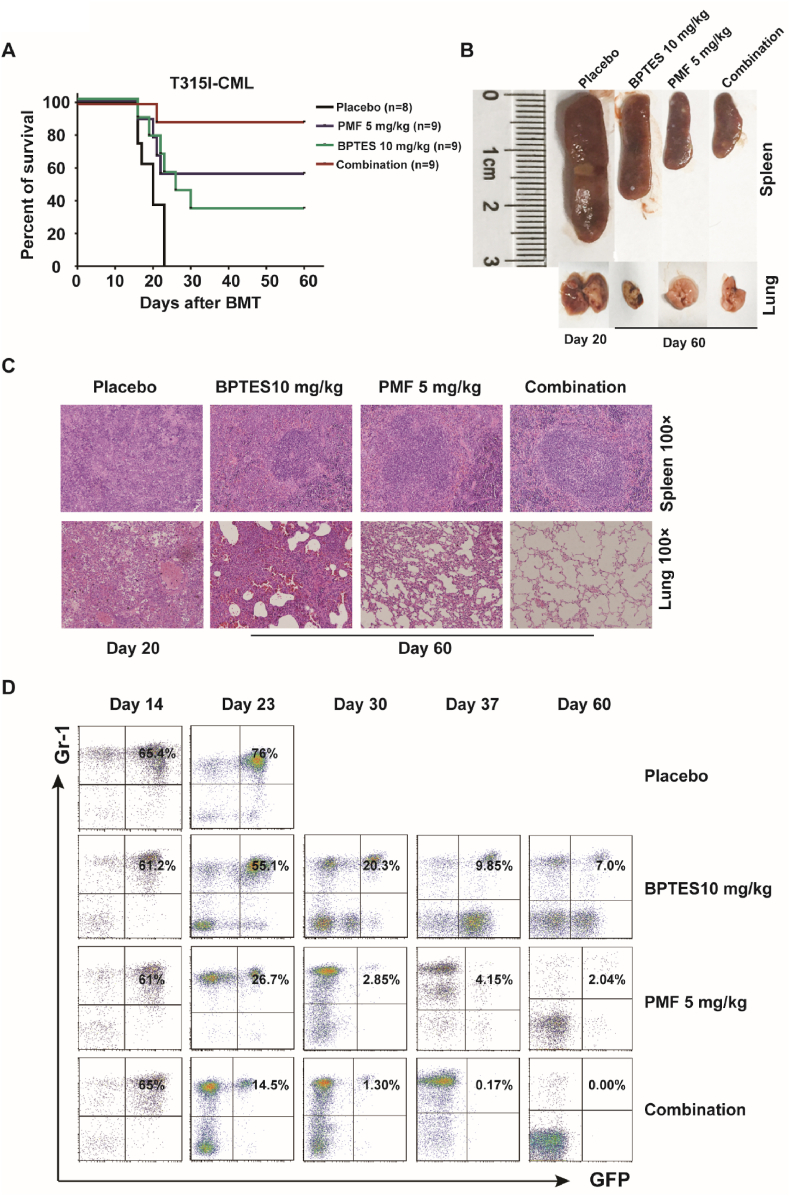
The greatest obstacle that needs to be overcome for CML cure is TKI resistance and the persistence of LSCs. To investigate the therapeutic effects of PMF on TKI resistant CML and LSCs, we established a BCR-ABL(T315I) induced CML mouse model and treated these mice with two doses of PMF. As expected, the diseased mice did not respond to IM, while PMF significantly prolonged the survival of the disease mice. On day 90 post bone marrow transplantation (BMT), the majority disease mice were still alive (10/14 and 7/8 respectively) in the PMF treated groups (Fig. 2A). Histopathological analysis demonstrated that the infiltration of myeloid leukemia cells was less severe in lungs and spleens for the mice received PMF treatment (Fig. 2B–C). During disease progression, leukemia cells in PB grew continually and maintained high tumor burden in Placebo and IM treatment groups. But in PMF treatment groups, leukemia cells decreased gradually and eventually almost disappeared after four-week treatment (Fig. 2D).
Fig. 2.
PMF prolongs the survival of BCR-ABL(T315I) induced CML mice and impairs the LSCs. (A) Kaplan-Meier survival curves for recipients of BCR-ABL(T315I) induced CML, which were treated with Placebo, IM (100 mg/kg), PMF (5 mg/kg or 10 mg/kg). (B) Overall appearance of the lungs and spleens of BCR-ABL (T315I) induced CML mice with treatment of Placebo and IM; but not PMF. (C) Photomicrographs of H&E staining lung and spleen sections from recipients of BCR-ABL (T315I) induced CML with treatment of Placebo, IM, and PMF (5 mg/kg). Original magnification × 100. (D) FACS analysis showed gradual disappearance of GFP+Gr-1+ cells in PB for recipients of BCR-ABL (T315I) induced CML with PMF treatment; but not Placebo and IM. (E–F) CML were induced with BCR-ABL(T315I), 14 days after BMT, mice treated with a single dose of PMF (n = 3, each group) for 48 h, and the number of LSCs in BM were counted and apoptosis levels were analyzed with Annexin-V/7-AAD staining. (G) On day 14, CML mice were sacrificed and 1 × 104 GFP + cells were seeded into 6 well plate containing 2 mL M3234 medium with indicated concentration of free PM. The total colonies were counted on day 7. (H–J) On day14, CML mice were sacrificed and 5 × 106 cells plated into 6 cm dish with stem cell condition. After 3 days, changing fresh medium and adding indicated concentrations of free PM for 48 h. LT-LSCs (CD34− LSC) and ST-LSCs + MPP (CD34+ LSC) were determined with FACS analysis. (K) On day14 post BMT, CML mice were treated with PMF at dose of 5 mg/kg and 10 mg/kg for 4 weeks, and leukemia cells in PB were monitored during the time. When the treatment was stopped for 2 weeks, leukemia cells reappeared. When these mice were treated with PMF again, leukemia cells were decreased. (L) The number of LSCs in the BM of PMF treated group mice was analyzed with FACS on day 14 and day 90 post BMT (n = 3, each group). Results were represented with the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 were considered as significant difference.
We further investigated the effects of PMF on LSCs and their progenitor cells, as shown in Fig. 2E–F and Fig. S5A, PMF significantly inhibited LSCs and progenitor cells, as well as induced LSCs apoptosis. In a stem cell culture condition, the proliferation and colony formation ability of LSCs were also inhibited by free PM (Fig. 2G–J). To examine whether PMF can completely eradicate LSCs in CML mice, PMF treatment was suspended for two weeks. During the time, leukemia cells reappeared in PB, which indicates that LSCs were not completely eradicated following four-week PMF treatment. When the disease relapsed mice were retreated with PMF, leukemia cells were reduced again (Fig. 2K). On day 90, we sacrificed the remaining mice and found that a few leukemia cells still remained in PB, spleen and BM (Fig. S5B). Further analysis showed that a residual LSC population was also detectable in BM (Fig. 2L). However, H&E staining showed that spleens and lungs physiological structure of recipients received long-term PMF treatment were comparable to that of healthy mouse (Fig. S5C). These results indicated that the continuous administration of PMF effectively suppressed leukemia development in BCR-ABL(T315I) induced CML mice and significantly extended the long-term survival. Unfortunately, PMF alone could not completely eradicate LSCs.
3.5. Multiple critical signaling pathways for LSCs survival are dysregulated by PMF
To further investigate the mechanism that prevented the complete eradication of LSCs by inhibiting of HDAC I/IIb, the LSCs from BCR-ABL(T315I) induced CML mice were flow-sorted after PMF treatment for RNA-seq. Gene profiling analysis revealed that 2672 genes were significantly increased and 773 genes were decreased (change fold≥2, P < 0.05) (Fig. 3A–B). GSEA analysis demonstrated that many important genes and signaling pathways for LSCs survival and proliferation were significantly changed (Supplementary Table 2a). As shown in Fig. 3C–D, the expression of genes in the “MYC_TARGETS” and “E2F_TARGETS” categories were significantly decreased. The MYC and E2F signaling pathways are essential for LSCs self-renewal and survival. Whereas, the expression of genes in the “KRAS_SIGNALING_DN” pathway was increased, which indicated a potent suppression of KRAS targets by PMF (Fig. 3G). Additionally, genes within the mTOR, Oxidative Phosphorylation and UPR categories were enriched with a decreased expression (Fig. 3H–J). Moreover, there was also a decreased expression of genes associated with “DNA_REPAIR” and the “G2M_checkpoint” (Fig. 3E–F). Interferon-γ, TNF response and apoptosis associated genes were enriched with an increased expression (Fig. 3K and Supplementary Table 2a).
Fig. 3.
Multiple critical signaling pathways for LSC survival were dysregulated by PMF.
(A) Heat map shows the expression pattern of 3415 genes and hierarchical clusters of the genes in GFP+ LSK population cells from two biological replicates, which were treated with PMF (10 mg/kg) for 24 h. Among these genes, the expression of 2672 genes were increased and 773 genes were decreased. (B) GSEA showed “MORF_HDAC2” gene set was enrichment in GFP+ LSK cells after PMF treatment. (C–K) GSEA was used to generate whole transcriptome comparisons, and enriched gene sets with increased or decreased gene expression were shown including “MYC TARGETS V1”, “E2F TARGETS”, “HALLMARK G2M checkpoint”, “HALLMARK DNA REPAIR”, “HALLMARK KRAS SIGNALING DN”, “HALLMARK MTORC1 SIGNALING”, “HALLMARK OXIDATIVE PHOSPHORYLATION”, “HALLMARK UNFOLDED PROTEIN RESPONSE” and “INTERFERON GAMMA RESPONSE”. (L) Left: the relative expression of c-Myc, E2f1, Ezh2, Alox5 and Alox15 in the RNA-seq data; right: On day 14 post BMT, CML mice were treat with Placebo or PMF (10 mg/kg) for 24 h and LSCs were sorted with FACS (n = 3, each group). The change of mRNA level of BCR-ABL, c-Myc, E2f1, Ezh2, Alox5 and Alox15 were examined with RT-PCR. (M) Left: the relative expression of P21 and P27 in RNA-seq data; Right: the change of P21 and P27 on LSCs were confirmed with RT-PCR. (N–O) β-Catenin and Alox15 expression in LSCs were analyzed after PMF or BPTES treatment with cellular immunofluorescence; the nucleus was stained with DAPI. Results were represented with the mean ± SEM. *P < 0.05, and **P < 0.01 were considered as significant difference.
To confirm the RNA-seq analysis results, genes within the aforementioned gene categories were selected and further validated with RT-PCR (Figs. S6A–D). Consistently, PMF significantly dysregulated the expression of c-Myc, β-Catenin, E2f1, Ezh2, Alox5 and Alox15 in LSCs, which had been previously identified as being essential for LSC survival and maintenance (Fig. 3L). Previously, we found that free PM upregulated Alox15 and β-Catenin expression in cell lines (Fig. 1H, Figs. S1D–F). While, in vivo assays showed that PMF caused Alox15 expression increase, but β-Catenin expression decrease (Fig. 3N–O). Furthermore, PMF also increased the expression of P27 and P21 in LSCs (Fig. 3M). Additionally, free PM showed the similar impacts of these targets on mature leukemia cells from the model mice in vitro (Figs. S6E–G). Cumulatively, these results suggested that PMF potently restrained LSCs by repressing multiple targets and signaling pathways, which are important for LSC maintenance and survival.
3.6. PMF potently disrupts stemness properties and skewed differentiation of LSCs
Epigenetic acetylation modifications can widely alter gene expression profiling. We analyzed the effect of PMF on LSC biological processes with C5BP in MSigDB. As shown in Fig. S7A, the top 20 activated and suppressed gene sets, following PMF treatment mostly suppressed genes associated with ribosome biogenesis, translation process, RNA metabolism, and mitochondria metabolism, while the activated gene sets associated with immune response, cell migration, and chemotaxis. GSEA demonstrated that PMF significantly altered gene sets related stemness and differentiation of LSCs. As shown in Figs. S7B–C, the expression of genes associated with “hematopoietic cell lineage” and the “B cell receptor signaling pathway” were increased, which indicated that the differentiation of LSCs was disrupted. Moreover, categories including genes related to LSC numbers (LIANG hematopoiesis stem cell number small vs huge DN, HUANG Gata2 targets UP), self-renewal (BYSTRYKH hematopoiesis stem cell and brain QTL CIS, TAKEDA targets of Nup98 Hoxa9 fusion 8D DN, JAATINEN hematopoietic stem cell DN), stemness and differentiation (RAMALHO stemness DN, DIAZ chronic myelogenous leukemia DN, TONKS targets of Runx1 Runx1t1 fusion erythrocyte UP, GAL leukemic stem cell DN) were enriched and increased (Supplementary Table 2b).
Furthermore, gene expression profiling revealed that PMF treatment increased the expression of 31/43 (72.09%) genes previously critically associated with the determination of lymphoid specialization and function and decreased the expression of 13/38 (34.2%) genes previously associated with myeloid specification and function [30] (Figs. S7F–G). These results suggested that PMF reversed the tendency of LSCs flowing into myeloid linage. Overall, targeting HDAC I/IIb effectively prevented CML and impaired the survival of LSCs by potently disrupting the stemness properties and skewed the differentiation of LSCs.
3.7. PMF increases glutaminolysis in LSCs
Interestingly, further gene profiling analysis showed that genes involved in the “GLUTAMATE_RECEPTOR_SIGNALING_PATHWAY” were enriched and their expression was increased (Fig. 4A). GLS1, the crucially metabolic enzyme of glutaminolysis, which is essential for survival of multiple cancers [31]. We found that free PM markedly caused the GLS1 expression increased in LAMA84 and K562 cell lines (Fig. 4B), as well as the increased of mRNA and protein levels of Gls1 in primary leukemia cells post free PM exposure in vitro (Fig. 4C). Furthermore, immunofluorescence staining revealed that PMF treatment also significantly increased Gls1 expression in LSCs (Fig. 4D). As shown in Fig. 4E, glutamine metabolic product α-KG enters TCA cycle then further generate Succinate-CoA and NADH by α-KDGH. To measure whether glutaminolysis was also affected in a manner corresponding to GLS1 level increased, the glutamate, α-KG, and Succinyl-CoA content were assessed both in vitro and in vivo. As shown in Fig. 4F, K562 cells treated with different concentration of free PM, the contents of glutamate, α-KG and Succinyl-CoA in cells were increased in dose-dependent manner. Additionally, leukemia cells isolated from BCR-ABL(T315I) induced CML mice post PMF treatment also showed higher contents of glutamate, α-KG and Succinyl-CoA compared to that in leukemia cells of Placebo treatment mice (Fig. 4G–I). Together, these results suggested that PMF increased LSC glutaminolysis by elevating Gls1 expression. Previously studies already showed that targeting the glutamine transporter ASCT2 prevented AML development and progression [32], BCR-ABL positive CD34+ cells utilize enhanced glutamine metabolism to maintain TCA cycle activity in glycolytic cells [33], and Imatinib treatment results in activated of the mitochondrial TCA cycle in human Ph + leukemia cells [34].
Fig. 4.
PMF increases glutamine metabolism by increasing GLS1 expression. (A) GSEA demonstrated that PMF significantly enriched and increased the expression of genes involved in “GO REGULATION GLUTAMATE RECEPTOR SIGNALING PATHWAY” gene set. (B) LAMA84 and K562 cells were treated with free PM at indicated concentration for 24 h, then cells were harvested for western blotting analysis with anti GLS1 antibody. GAPDH served as the total protein loading control. (C) Primary leukemia cells of spleen from CML mice were treated with free PM at indicated concentrations for 24 h, then cells were harvested for mRNA examination with RT-PCR and western blotting analysis with anti Gls1 antibody, Gapdh served as the total protein loading control. (D) On day 14 post BMT, CML mice were treated with Placebo or PMF (10 mg/kg, n = 3, each group) for 24 h. LSCs were sorted by FACS and Gls1 expression was examined with cellular immunofluorescence, and nucleus was stained with DAPI. (E) A schematic diagram of glutaminolysis and TCA cycle. (F) K562 cells were treated with free PM at different concentration for 24 h, the contents of glutamate, α-ketoglutarate and Succinyl-CoA contents in cells were measured with LC-MS/MS. (G–I) CML mice treated with PMF (10 mg/kg) for 24 h, the contents of glutamate, α-ketoglutarate and Succinyl-CoA in the spleen were measured with LC-MS/MS. The relative content was normalized to that in Placebo treated mice. Results were represented with the mean ± SEM. *P < 0.05, and **P < 0.01 were considered as significant difference.
3.8. Combination of PMF and glutaminolysis inhibition abolishes LSCs in vivo
Based on previous results, we speculate that treatment of PMF and simultaneous inhibition of glutaminolysis could obtain a better inhibitory effect on LSCs. Therefore, PMF was used to treat BCR-ABL(T315I) induced CML mice simultaneously with BPTES, a selective inhibitor of GLS1. As shown in Fig. 5A, PMF and BPTES monotherapy effectively prolonged recipient survival (5/9 and 3/9 mice were still alive on day 60 post BMT, respectively). However, in the combination treatment group, most mice (8/9) were still alive at experimental endpoint. Histological analysis of lungs and spleens showed that there were no obvious leukemia cells infiltration and displayed a normal physiological structure in PMF or combination treatment groups compared to that in Placebo and BPTES treatment groups (Fig. 5B and C). Additionally, FACS analysis revealed leukemia cells grew continually in the Placebo groups, whereas in the mono or co-therapy groups, leukemia cell percentage was gradually decreased. In the PMF and cotreatment groups, the leukemia cells burden was lower than that in the BPTES group mice. On day 60 post BMT, only a small percentage of leukemia cells remained in PMF treatment group, whereas no leukemia cells were detected in PB in co-therapy group (Fig. 5D). Additionally, the effects of free PM combined with inhibition of glutaminolysis on Ph + CML cell lines were analyzed in vitro. As shown in Figs. S8A–H, when absence of glutamine or presence of GLS1 inhibitor, free PM showed stronger anti-tumor activity and induced more cell apoptosis. Importantly, free PM treatment and simultaneous targeting of GLS1 also synergistically inhibited cell survival and induced apoptosis of human CD34+ CML cells isolated from newly diagnosed and TKI treatment relapsed patients (Fig. 6A–B).
Fig. 5.
Targeting glutaminolysis in combination with PMF synergistically prevents CML in mouse model. (A) Kaplan-Meier survival curves for BCR-ABL(T315I) induced CML mice, which were treated with Placebo, PMF (5 mg/kg), BPTES (10 mg/kg), and their combination. (B) The overall appearance of the lungs and spleens of BCR-ABL(T315I) induced CML mice treated with Placebo, BPTES, PMF alone, or the combination of PMF and BPTES. (C) Photomicrographs of H&E staining lung and spleen sections from BCR-ABL(T315I) induced CML treated with Placebo, BPTES, PMF or combination treatment. Original magnification × 100. (D) Leukemia cells (GFP+Gr-1+) in PB of CML mice treated with Placebo, BPTES, PMF alone or in combination of PMF and BPTES.
Fig. 6.
The combination of free PM and BPTES achieves synergistic effect in suppressing CD34+cells from CML patients. (A) CD34+ CML cells from five CML patients PB were cultured under human stem cell culture condition and treated with free PM and BPTES at indicated concentration for 48 h. Cells were stained with CD34 monoclonal antibody and then analyzed cell apoptosis by Annexin-V/PI. (B) Alive CD34+cells numbers were counted with trypan blue staining and FACS analysis. Relative viable cells were normalized with that in control treatment. Results were represented with the mean ± SEM. “NS” means no significance; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered as significant difference.
We further investigated the impact of combination therapy on leukemia cells and LSCs in vivo (Fig. 7A). As shown in Fig. 7B–D, PMF or PMF/BPTES cotreatment more effectively impeded leukemia cells growth in BM compared to that in BPTES or Placebo treatment groups, and BPTES displayed a weaker therapeutic effect. Notably, no leukemia cells were detected in BM of the cotreatment group mice after three-week treatment. The analysis of leukemia burden in PB showed similar results (Figs. S8I–K). Importantly, compared to PMF or BPTES treatment alone, cotreatment showed more effect in restraining LSCs. After three-week continuous treatment, no LSCs were found in the disease mouse BM (Fig. 7E–F). Moreover, PMF/BPTES cotreatment displayed stronger LSC growth inhibition and colony formation (Fig. 7G–J). Cumulatively, these results suggested that blocking glutaminolysis and HDAC I/IIb simultaneous could effectively eradicate LSCs.
Fig. 7.
Simultaneous inhibition of glutaminolysis and HDACI/IIb abolishes LSCs in vivo.
(A) Overview of the experimental design. Donor mice were treated with 5-Fu for 4 days and BM cells were transduced with BCR-ABL (T315I) virus, then were transplanted into lethally irradiated recipients. On 14 day post BMT, recipients were randomly divided into four groups and treated with Placebo, PMF, BPTES, or combination of PMF and BPTES. The treatment continued for 3 weeks, and GFP+Gr-1+cells and LSCs were analyzed weekly. (B–D) Total BM cell numbers, the percentages and numbers of GFP+Gr-1+ cell in the BM were analyzed after weekly treatment (n = 3, each group). (E–F) The percentages and numbers of LSCs in BM were analyzed after weekly treatment (n = 3, each group). (G) On day 14 post BMT, CML mice were sacrificed and 1 × 104 GFP + cells were seeded into 6 well plate containing 2 mL M3234 medium with free PM, BPTES, or the combination at the indicated concentration. The total colonies were counted on day 7 post seeding. (H–J) On day 14 post BMT, CML mice were sacrificed and 5 × 106 cells were plated into 6 cm dish under stem cell culture condition. After 3 days, the medium was changed and free PM and BPTES were added at the indicated concentration for 48 h. The numbers of LSCs, LT-LSCs (CD34− LSC) and ST-LSC + MPPs (CD34+ LSC) were determined with FACS analysis. Results were represented with the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 were considered as significant difference.
3.9. Inhibition of glutaminolysis alters multiple signaling pathways and synergizes with PMF in suppressing LSCs
To gain an insight into the underlying mechanism of LSC elimination following glutaminolysis and HDAC I/IIb inhibition, BCR-AB(T315I) induced CML mice were treated with BPTES and LSCs were flow-sorted for RNA-seq. Gene profiling analysis showed that 710 genes were significantly increased and 532 genes were decreased (change fold≥2, P < 0.05) (Fig. S9A). The effects of BPTES upon LSCs were analyzed by GSEA of C5BP in MSigDB. The top 20 gene sets with increased or decreased expression were shown in Fig. S9B. The gene sets with decreased expression mostly involved in ribosome biogenesis, translation process, RNA metabolism, mitochondria metabolism, amino acid metabolism, and protein maturation, whereas the gene sets with increased expression were mostly those involved in immune and inflammatory response and carbohydrate breakdown. After BPTES treatment, significantly altered gene sets were listed in Supplementary Tables 3a–b. Glutamine is predominately metabolized to enter the TCA cycle via GLS1 in mitochondria (Fig. 4E). It was shown that genes involved in “Oxidative phosphorylation” were decreased and enriched in LSCs when treated with BPTES (Fig. 8A). Additionally, multiple signaling pathways related LSCs survival and proliferation were repressed after BPTES treatment, including “MYC_TARGETS”, “DNA_REPAIR”, “E2F_TARGETS”, “NOTCH_SIGNALING” and “P53_PATHWAY” (Supplementary Table 3a). GSEA results also showed that BPTES significantly altered gene sets related to stemness maintenance of LSCs. Some critical gene sets, such as Notch, EWS/FLI1, and Hoxa9 were decreased after BPTES treatment (Fig. 8B–D and Supplementary Table 3b).
Fig. 8.
Inhibition of Gls1 alters multiple signaling pathways in LSCs and synergizes with PMF in suppressing LSCs. (A–C) GSEA showed enrichment of gene sets with decreased expression including “HALLMARK OXIDATIVE PHOSPHORYLATION”, “NOTCH SIGNALING PATHWAY”, “EWSR1 FLI1 FUSION UP” and (D) increased expression for “HOXA9 DN.V1 UP” in GFP+ LSK cells post BPTES treatment. (E–F) The expression of gene in KEGG pathways were enriched and significantly altered (change fold≥2, P < 0.05) post PMF and BPTES treatment. The Venn diagrams demonstrate that the expression of gene in the listed KEGG pathways were decreased post PMF treatment but were increased post BPTES treatment, as well as the expression of genes involved in the listed pathways were decreased after BPTES treatment, but increased after PMF treatment. (G) Left panel: the relative mRNA level of c-Myc, E2f1, Ezh2, Alox5 and Alox15 in RNA-seq data; Right panel: CML mice were induced by BCR-ABL(T315I), on day 14 post BMT, and recipients were treated with Placebo or BPTES (10 mg/kg) for 24 h, then LSCs were sorted with FACS (n = 3, each group). The mRNA levels of BCR-ABL, c-Myc, E2f1, Ezh2, Alox5 and Alox15 in LSCs were examined with RT-PCR. (H) LAMA84 cells were cultured in presence of the indicated concentration of free PM and BPTES for 24 h, then cells were harvested for western blotting analysis with anti BCR-ABL, c-MYC, EZH2, ALOX5, ALOX15, HOXA9, HSP90, P21 and P27 antibodies. GAPDH served as the total protein loading control. (I) A schematic diagram for the regulatory mechanisms of synergistic effects of PM and BPTES. Results were represented with the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 were considered as significant difference.
The subsequent analysis focused on identifying gene expression pathways with KEGG. In the PMF treatment group. We found that 70 KEGG pathways were enriched (among them, 36 associating with decreased expression and 34 associating with increased expression). In the BPTES group, 47 KEGG pathways were enriched (21 associating with increased expression and 26 associating with decreased expression). Interestingly, among the 34 increased KEGG pathways in PMF treated group, 23 of them were enriched with decreased expression in the BPTES treatment group. Conversely, among the 21 KEGG pathways with increased expression following BPTES treatment, the expression of 10 genes were decreased in PMF treated group (Fig. 8E–F, and Supplementary Table 4). These results potentially illustrated that the combination of PMF and BPTES had synergistic inhibitory effects on LSCs through compensating against the molecular cellular escape routes when treated individually. Finally, we found that inhibition of Gls1 did not change the expression of BCR-ABL, c-Myc, Ezh2, and Alox5, but significantly decreased Alox15 expression in LSCs (Fig. 8G). Moreover, western blotting results showed that the cotreatment synergistically decreased expression levels of BCR-ABL, c-MYC, EZH2, ALOX5, HOXA9 and HSP90; synergistically increased the expression of P21 but not P27 (Fig. 8H). Overall, these results reveal a possible molecular mechanism behind the synergistic effects on LSCs following PMF and BPTES combination treatment (Fig. 8I).
4. Discussion
The eradication of LSCs is essential to achieve long-term remission and cure of CML, which cannot be achieved using TKI treatments alone [35]. Besides, the high frequency of TKI resistant BCR-ABL mutations results in their treatment failure [36]. Previous studies have demonstrated that targeting multiple pathways is required for improving outcomes of CML treatment [37]. Epigenetic regulation widely affects gene expression and targeting epigenetic elements has been considered as a promising strategy to treat the multiple hematological malignancies [38]. HDACs inhibitor has been proven to be a successful case, however, low activity and lack of selectivity largely limit the application of existing HDACis [15,16,39].
The novel highly selective HDAC I/IIb inhibitor, PM, has better selectivity and inhibitory activity on HDAC I/IIb than all currently marketed HDACis [1,2]. However, the free PM has poor solubility in water and low bioavailability, thus hindering its further pharmacodynamic studies in vivo. HP-β-CD owns advantages of high aqueous solubility, non-toxicity, and excellent biocompatibility, and its unique truncated cone molecular structure, which can encapsulate hydrophobic drugs [[40], [41], [42]]. We found that PM molecules could combine with the ring center of HP-β-CD to form relatively stable host-guest (PM/HP-β-CD) non-covalent inclusion complexes through multiple supramolecular forces including hydrogen interaction, Pi-sigma interaction, and van der Waals' force (Fig. S3). There is a dynamic balance between the inclusion complex and free PM in serum. Once the free PM is absorbed by tumor cells, the inclusion complex will continuously release free PM from the cavity of the HP-β-CD. Thus, this bioactive material can not only improve the solubility and stability of free PM but also has a sustained release effect, which maintains a longer anti-tumor effect and reduces acute toxicity. Pharmacokinetic studies of free PM and PMF as well as acute toxicity studies further confirmed the advantage of this bioactive material (Table 1 and Fig. S4). Furthermore, we found that the specific molar ratios of PM/HP-β-CD, arginine, meglumine and mannitol could formation of alkaline in situ salts, and further improve the solubility and stability of the free PM in normal saline. Hydroxamic acids are generally less soluble and stable [43,44], our study demonstrated a suitable bioactive material for improving the solubility and stability of hydroxamic acids, as well as improving their pharmacokinetic properties and reducing toxicity.
Currently, PMF has been approved by the FDA and NMPA for treatment of relapsed and refractory B-cell malignancies in clinical trials. It displayed an outstanding anti-tumor activity on Ph+ B-ALL mouse model (including T315I mutation) without relapsed [1]. Based upon previous studies, we hypothesized that targeting HDAC I/IIb could be a way for CML stem cells elimination. Free PM showed excellent anti-leukemia activity on Ph + CML in vitro including human CD34+ CML cells from patients. PMF alone enabled T315I-CML mice achieving long-term remission, but still incompletely eliminated LSCs and cured disease. By analyzing transcriptome expression profiling of LSCs under different conditions, we found that targeting glutaminolysis and HDAC I/IIb could sufficiently abolish LSCs in the mouse model. Overall, our studies provided a potential strategy for CML cure.
PMF significantly disrupted several pivotal signaling pathways and biological processes for the survival and maintenance of LSCs in BCR-ABL(T315I) induced CML mice (Supplementary Table 2). It is well known that mTOR signaling is essential for HSC engraftment and hematopoiesis [45] and its high activity results in leukemia cell resistance to TKI [46]. Previous studies have demonstrated that primitive CML cells rely on upregulated oxidative metabolism for survival and targeting mitochondrial oxidative phosphorylation efficiently suppresses CML LSCs [47]. Rapid proliferating CML cells also rely on degradation of unfolded proteins, therefore, targeting ER stress core protein HSP90 effectively prevents TKI resistance CML and suppresses LSCs [24]. Hox9 and Meis1 are necessary for maintaining leukemia transformation and LSCs expansion [48]. CBFA2T3 has been identified to control hematopoietic cell fate decisions and inactivation of CBFA2T3 impairs the rapid expansion of short-term stem cells [49]. Interestingly, these two target sets were enriched and significantly decreased by PMF (Figs. S7D–E).
Although PMF treatment achieved promising therapeutic effects on BCR-ABL(T315I) induced CML mice, PMF alone could not incompletely eradicate LSCs and cure CML (Fig. 2). Epigenetic modification widely changes gene expression and promotes to generate new tumorigenesis events, which reprograms cancer cells to escape drug treatment with high-potential [50,51]. For example, Alox15, which was proved to be essential factors for LSCs survival had increased expression (Fig. 1H, S1B-D, 3L, and 3O) and leukemia cell glutaminolysis was also increased post PM treatment both in vivo and in vitro (Fig. 4). Previous studies have indicated that either targeting ALOX15 or glutaminolysis would be an efficient way for the suppression or abolition of LSCs [27,52]. However, due to the limitations of ALOX15 inhibitor, this study focused more upon targeting glutaminolysis.
Previously studies revealed that the glutamine importer SLC1A5 and glutamine uptake are enhanced in human cord blood CD34+ cells transduced with BCR-ABL, coinciding with an increased susceptibility to the glutaminase inhibitor [33]. Recently study suggested that upregulated GLS1 levels played an important role in aging, and glutaminolysis inhibition ameliorates various age-associated disorders [53]. All of these indicate that targeting glutaminolysis by inhibiting GLS1 in combination with PMF would achieve significantly therapeutic effects on CML. Further studies demonstrated that targeting glutaminolysis impeded the oxidative phosphorylation and amino acid metabolism, caused ER stress and restricted protein maturation, and destroyed the stemness of LCSs (Fig. S9B and Supplementary Table 2b). Most importantly, Alox15 was significantly repressed when inhibition of glutaminolysis (Figs. 8G and 3O). The combination of targeting GLS1 and inhibition HDAC I/IIb achieved an improved therapeutic effect compared to alone treatment both in vitro and in vivo. Ultimately, the combination achieved complete LSCs abolishment and CML cure in mouse model (Fig. 5, Fig. 6, Fig. 7). The therapeutic mechanism of this combination therapy on LSCs was also further investigated using the gene-profiling comparisons of LSCs post BPTES treatment. GO and KEGG analysis showed that the combination inhibition of GLS1 and HDAC I/IIb had complementary and synergistic effect for each other (Figs. S7A and S9B, and Fig. 8E–F and H). These findings explained the effects of PMF and BPTES in eradication of LSCs and CML cure (Fig. 8I).
Currently, a few studies of HDAC inhibitors in targeting LSCs are performed, which limits the application of HDACi in stem cell driven hematological malignance. Our studies firstly demonstrated the therapeutic effects on LSCs by collaborating targeting HDAC I/IIb and glutaminolysis with inhibitor, which greatly enhance the understanding of epigenetics and cancer metabolism in LSC biology. Moreover, this study will help guide for the clinical trial application of PMF in the treatment of relapsed and refractory CML in the future.
5. Conclusions
In summary, PM has higher selectivity and inhibitory activity for HDAC I/IIb than currently marketed HDACis. The lyophilized powder of PMF inclusion complex greatly increases the solubility of free PM in normal saline, improves PM stability and pharmacokinetic properties, and reduces toxicity. By using BCR-ABL(T315I) induced TKI resistance mouse model, it was demonstrated that PMF significantly prevented CML disease progression and diminished its LSCs through repressing many essential targets for LSC survival including c-Myc, β-Catenin, E2f, Ezh2, Alox5, and mTOR, and interrupting multiple critical biologic processes for LSCs maintenance. However, PMF treatment alone does not completely eliminate LSCs, but enhanced glutaminolysis by raising GLS1 expression. Importantly, PMF administration collaborating targeting glutaminolysis achieved an improved therapeutic effect and markedly abolished LSCs in mouse. Together, our studies provide a new therapeutic strategy for eliminating CML LSCs by simultaneous targeting HDAC I/IIb and glutaminolysis, which may guide PMF clinical trials in the future for CML patients hosting BCR-ABL TKI resistant mutations.
Ethics approval and consent to participate
All animal studies were performed in accordance with the guidelines approved by the Institutional Animal Care and Use Committees of Sichuan University. The patient samples were obtained from West China Hospital which were approved by the clinical ethics committee in West China Hospital, Sichuan University (Chengdu, China). All patients signed the informed consent form, and met all requirements of the Declaration of Helsinki.
CRediT authorship contribution statement
Qiang Qiu: Formal analysis, Writing – original draft, designed the experiments, performed the experiments, analyzed the data and wrote the original draft. Linyu yang: Formal analysis, Writing – original draft, designed the experiments, performed the experiments, analyzed the data and wrote the original draft. Yunyu Feng: Formal analysis, performed bioinformatic analysis, discussed the results. Zejiang Zhu: performed the experiments, discussed the results. Ning Li: performed the experiments, discussed the results. Li Zheng: performed the experiments, discussed the results. Yuanyuan Sun: performed the experiments, discussed the results. Cong Pan: gave assistance, discussed the results. Huandi Qiu: gave assistance, discussed the results. Xue Cui: gave assistance, discussed the results. Wei He: gave assistance, discussed the results. Fang Wang: performed the experiments, discussed the results. Yuyao Yi: performed the experiments, commented on the manuscript. Minghai Tang: performed the experiments, discussed the results. Zhuang Yang: discussed the results and commented on the manuscript. Yunfan Yang: gave assistance, discussed the results. Zhihui Li: discussed the results and commented on the manuscript. Lijuan Chen: Writing – review & editing, developed the projects, reviewed and edited the manuscript, discussed the results and commented on the manuscript. Yiguo Hu: Writing – review & editing, conceived the projects, designed the experiments, reviewed and edited the manuscript, discussed the results and commented on the manuscript. .
Declaration of competing interest
The authors declare no conflict interests.
Acknowledgements
The authors greatly appreciate the State Key Laboratory of Biotherapy & Collaborative Innovation Center for Biotherapy for their support; especially the staff of the flow cytometry and animal facility of the State Key Laboratory of Biotherapy and West China Hospital; Klarke M. Sample for editing the manuscript language.
This work was supported by grants from the Fundamental Research Funds for the Central Universities (2021SCU12022 to L. Yang), the 1.3.5 Project for Disciplines of Excellence (to Z. Li and L. Chen), West China Hospital, Sichuan University, the National Natural Science Foundation of China (82104211 to L. Yang), and the National Natural Science Foundation of China (81541092 and 81770103 to Y. Hu).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.08.006.
Contributor Information
Lijuan Chen, Email: chenlijuan125@163.com.
Yiguo Hu, Email: huyiguo@scu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Yang L., Qiu Q., Tang M., Wang F., Yi Y., Yi D., Yang Z., Zhu Z., Zheng S., Yang J., Pei H., Zheng L., Chen Y., Gou L., Luo L., Deng X., Ye H., Hu Y., Niu T., Chen L. Purinostat mesylate is a uniquely potent and selective inhibitor of HDACs for the treatment of BCR-ABL-induced B-cell acute lymphoblastic leukemia. Clin. Cancer Res. 2019;25(24):7527–7539. doi: 10.1158/1078-0432.CCR-19-0516. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y., Wang X., Xiang W., He L., Tang M., Wang F., Wang T., Yang Z., Yi Y., Wang H., Niu T., Zheng L., Lei L., Li X., Song H., Chen L. Development of purine-based hydroxamic acid derivatives: potent histone deacetylase inhibitors with marked in vitro and in vivo antitumor activities. J. Med. Chem. 2016;59(11):5488–5504. doi: 10.1021/acs.jmedchem.6b00579. [DOI] [PubMed] [Google Scholar]
- 3.Holyoake T., Jiang X., Eaves C., Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94(6):2056–2064. [PubMed] [Google Scholar]
- 4.Sattler M., Griffin J.D. Molecular mechanisms of transformation by the BCR-ABL oncogene. Semin. Hematol. 2003;40(2 Suppl 2):4–10. doi: 10.1053/shem.2003.50034. [DOI] [PubMed] [Google Scholar]
- 5.Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., Sawyers C.L. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 6.Khorashad J.S., Kelley T.W., Szankasi P., Mason C.C., Soverini S., Adrian L.T., Eide C.A., Zabriskie M.S., Lange T., Estrada J.C., Pomicter A.D., Eiring A.M., Kraft I.L., Anderson D.J., Gu Z., Alikian M., Reid A.G., Foroni L., Marin D., Druker B.J., O'hare T., Deininger M.W. BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: frequency and clonal relationships. Blood. 2013;121(3):489–498. doi: 10.1182/blood-2012-05-431379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holyoake T.L., Helgason G.V. Do we need more drugs for chronic myeloid leukemia? Immunol. Rev. 2015;263(1):106–123. doi: 10.1111/imr.12234. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt M., Rinke J., Schafer V., Schnittger S., Kohlmann A., Obstfelder E., Kunert C., Ziermann J., Winkelmann N., Eigendorff E., Haferlach T., Haferlach C., Hochhaus A., Ernst T. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28(12):2292–2299. doi: 10.1038/leu.2014.272. [DOI] [PubMed] [Google Scholar]
- 9.Vicente-Duenas C., Hauer J., Cobaleda C., Borkhardt A., Sanchez-Garcia I. Epigenetic priming in cancer initiation. Trends in cancer. 2018;4(6):408–417. doi: 10.1016/j.trecan.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Holtz M.S., Forman S.J., Bhatia R. Nonproliferating CML CD34+ progenitors are resistant to apoptosis induced by a wide range of proapoptotic stimuli. Leukemia. 2005;19(6):1034–1041. doi: 10.1038/sj.leu.2403724. [DOI] [PubMed] [Google Scholar]
- 11.Koschmieder S., Vetrie D. Epigenetic dysregulation in chronic myeloid leukaemia: a myriad of mechanisms and therapeutic options. Semin. Cancer Biol. 2018;51:180–197. doi: 10.1016/j.semcancer.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Fiskus W., Pranpat M., Bali P., Balasis M., Kumaraswamy S., Boyapalle S., Rocha K., Wu J., Giles F., Manley P.W., Atadja P., Bhalla K. Combined effects of novel tyrosine kinase inhibitor AMN107 and histone deacetylase inhibitor LBH589 against Bcr-Abl-expressing human leukemia cells. Blood. 2006;108(2):645–652. doi: 10.1182/blood-2005-11-4639. [DOI] [PubMed] [Google Scholar]
- 13.George P., Bali P., Annavarapu S., Scuto A., Fiskus W., Guo F., Sigua C., Sondarva G., Moscinski L., Atadja P., Bhalla K. Combination of the histone deacetylase inhibitor LBH589 and the hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;105(4):1768–1776. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B., Strauss A.C., Chu S., Li M., Ho Y., Shiang K.D., Snyder D.S., Huettner C.S., Shultz L., Holyoake T., Bhatia R. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17(5):427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nebbioso A., Manzo F., Miceli M., Conte M., Manente L., Baldi A., De Luca A., Rotili D., Valente S., Mai A., Usiello A., Gronemeyer H., Altucci L. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009;10(7):776–782. doi: 10.1038/embor.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanginlar C., Logie C. HDAC11 is a regulator of diverse immune functions. Biochimica et Biophysica Acta. Gene. Regular Mech. 2018;1861(1):54–59. doi: 10.1016/j.bbagrm.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Raedler L.A. Farydak (Panobinostat): first HDAC inhibitor approved for patients with relapsed multiple myeloma. Am Health Drug. Benefit. 2016;9:84–87. Spec Feature) [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman M.L., Yang N., Sharma K.K., Balys M., Corbett C.A., Jordan C.T., Becker M.W., Steidl U., Abdel-Wahab O., Levine R.L., Marcucci G., Roboz G.J., Hassane D.C. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Mol. Cancer Therapeut. 2014;13(8):1979–1990. doi: 10.1158/1535-7163.MCT-13-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi J., Singh S., Hua W.K., Cai Q., Chao S.W., Li L., Liu H., Ho Y., Mcdonald T., Lin A., Marcucci G., Bhatia R., Huang W.J., Chang C.I., Kuo Y.H. HDAC8 inhibition specifically targets inv(16) acute myeloid leukemic stem cells by restoring p53 acetylation. Cell Stem Cell. 2015;17(5):597–610. doi: 10.1016/j.stem.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16(10):619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leone R.D., Zhao L., Englert J.M., Sun I.M., Oh M.H., Sun I.H., Arwood M.L., Bettencourt I.A., Patel C.H., Wen J., Tam A., Blosser R.L., Prchalova E., Alt J., Rais R., Slusher B.S., Powell J.D. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366(6468):1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S., Ilaria R.L., Jr., Million R.P., Daley G.Q., Van Etten R.A. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 1999;189(9):1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y., Swerdlow S., Duffy T.M., Weinmann R., Lee F.Y., Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc. Natl. Acad. Sci. U.S.A. 2006;103(45):16870–16875. doi: 10.1073/pnas.0606509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng C., Brain J., Hu Y., Goodrich A., Kong L., Grayzel D., Pak R., Read M., Li S. Inhibition of heat shock protein 90 prolongs survival of mice with BCR-ABL-T315I-induced leukemia and suppresses leukemic stem cells. Blood. 2007;110(2):678–685. doi: 10.1182/blood-2006-10-054098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., Hu Y., Zhang H., Peng C., Li S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat. Genet. 2009;41(7):783–792. doi: 10.1038/ng.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie H., Peng C., Huang J., Li B.E., Kim W., Smith E.C., Fujiwara Y., Qi J., Cheloni G., Das P.P., Nguyen M., Li S., Bradner J.E., Orkin S.H. Chronic myelogenous leukemia- initiating cells require polycomb group protein EZH2. Cancer Discov. 2016;6(11):1237–1247. doi: 10.1158/2159-8290.CD-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Peng C., Abraham S.A., Shan Y., Guo Z., Desouza N., Cheloni G., Li D., Holyoake T.L., Li S. Arachidonate 15-lipoxygenase is required for chronic myeloid leukemia stem cell survival. J. Clin. Invest. 2014;124(9):3847–3862. doi: 10.1172/JCI66129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y., Chen Y., Douglas L., Li S. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. 2009;23(1):109–116. doi: 10.1038/leu.2008.262. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Z., Wen J., Xu Y., Pei H., Li D., Tang M., Bai P., He J., Yang Z., Chen L. Therapeutic efficacy of an injectable formulation of purinostat mesylate in SU-DHL-6 tumour model. Ann. Med. 2022;54(1):743–753. doi: 10.1080/07853890.2022.2045347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., Weissman I.L. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. U.S.A. 2005;102(26):9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L., Venneti S., Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu. Rev. Biomed. Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 32.Ni F., Yu W.-M., Li Z., Graham D.K., Jin L., Kang S., Rossi M.R., Li S., Broxmeyer H.E., Qu C.-K. Critical role of ASCT2-mediated amino acid metabolism in promoting leukaemia development and progression. Nature Metabolism. 2019;1(3):390–403. doi: 10.1038/s42255-019-0039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sontakke P., Koczula K.M., Jaques J., Wierenga A.T., Brouwers-Vos A.Z., Pruis M., Gunther U.L., Vellenga E., Schuringa J.J. Hypoxia-Like signatures induced by BCR-ABL potentially alter the glutamine uptake for maintaining oxidative phosphorylation. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0153226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottschalk S., Anderson N., Hainz C., Eckhardt S.G., Serkova N.J. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells, Clinical cancer research : an. official journal of the American Association for Cancer Research. 2004;10(19):6661–6668. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y., Li S. Survival regulation of leukemia stem cells, Cellular and molecular life sciences. CM. 2016;73(5):1039–1050. doi: 10.1007/s00018-015-2108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoklosa T., Poplawski T., Koptyra M., Nieborowska-Skorska M., Basak G., Slupianek A., Rayevskaya M., Seferynska I., Herrera L., Blasiak J., Skorski T. BCR/ABL inhibits mismatch repair to protect from apoptosis and induce point mutations. Cancer Res. 2008;68(8):2576–2580. doi: 10.1158/0008-5472.CAN-07-6858. [DOI] [PubMed] [Google Scholar]
- 37.Bixby D., Talpaz M. Seeking the causes and solutions to imatinib-resistance in chronic myeloid leukemia. Leukemia. 2011;25(1):7–22. doi: 10.1038/leu.2010.238. [DOI] [PubMed] [Google Scholar]
- 38.Fong C.Y., Morison J., Dawson M.A. Epigenetics in the hematologic malignancies. Haematologica. 2014;99(12):1772–1783. doi: 10.3324/haematol.2013.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guedes-Dias P., Oliveira J.M. Lysine deacetylases and mitochondrial dynamics in neurodegeneration. Biochim. Biophys. Acta. 2013;1832(8):1345–1359. doi: 10.1016/j.bbadis.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Brewster M.E., Loftsson T. Cyclodextrins as pharmaceutical solubilizers. Adv. Drug Deliv. Rev. 2007;59(7):645–666. doi: 10.1016/j.addr.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Qiu N., Cheng X., Wang G., Wang W., Wen J., Zhang Y., Song H., Ma L., Wei Y., Peng A., Chen L. Inclusion complex of barbigerone with hydroxypropyl-beta-cyclodextrin: preparation and in vitro evaluation. Carbohydr. Polym. 2014;101:623–630. doi: 10.1016/j.carbpol.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 42.Yan C., Liang N., Li Q., Yan P., Sun S. Biotin and arginine modified hydroxypropyl-beta-cyclodextrin nanoparticles as novel drug delivery systems for paclitaxel. Carbohydr. Polym. 2019;216:129–139. doi: 10.1016/j.carbpol.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 43.Yang J., Ma Y., Luo Q., Liang Z., Lu P., Song F., Zhang Z., Zhou T., Zhang J. Improving the solubility of vorinostat using cyclodextrin inclusion complexes: the physicochemical characteristics, corneal permeability and ocular pharmacokinetics of the drug after topical application. Eur. J. Pharmaceut. Sci. 2022;168 doi: 10.1016/j.ejps.2021.106078. [DOI] [PubMed] [Google Scholar]
- 44.Chaudhuri S., Fowler M.J., Baker C., Stopka S.A., Regan M.S., Sablatura L., Broughton C.W., Knight B.E., Stabenfeldt S.E., Agar N.Y.R., Sirianni R.W. Beta-cyclodextrin-poly (beta-Amino ester) nanoparticles are a generalizable strategy for high loading and sustained release of HDAC inhibitors. ACS Appl. Mater. Interfaces. 2021;13(18):20960–20973. doi: 10.1021/acsami.0c22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo F., Zhang S., Grogg M., Cancelas J.A., Varney M.E., Starczynowski D.T., Du W., Yang J.Q., Liu W., Thomas G., Kozma S., Pang Q., Zheng Y. Mouse gene targeting reveals an essential role of mTOR in hematopoietic stem cell engraftment and hematopoiesis. Haematologica. 2013;98(9):1353–1358. doi: 10.3324/haematol.2012.080424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Airiau K., Mahon F.X., Josselin M., Jeanneteau M., Belloc F. PI3K/mTOR pathway inhibitors sensitize chronic myeloid leukemia stem cells to nilotinib and restore the response of progenitors to nilotinib in the presence of stem cell factor. Cell Death Dis. 2013;4:e827. doi: 10.1038/cddis.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuntz E.M., Baquero P., Michie A.M., Dunn K., Tardito S., Holyoake T.L., Helgason G.V., Gottlieb E. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 2017;23(10):1234–1240. doi: 10.1038/nm.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins C.T., Hess J.L. Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets. Oncogene. 2016;35(9):1090–1098. doi: 10.1038/onc.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinauer N., Guo C., Zhang J. Emerging roles of MTG16 in cell-fate control of hematopoietic stem cells and cancer. Stem Cell. Int. 2017;2017 doi: 10.1155/2017/6301385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Barrios O., Galaras A., Trincado J.L., Azagra A., Collazo O., Meler A., Agraz-Doblas A., Bueno C., Ballerini P., Cazzaniga G., Stam R.W., Varela I., De Lorenzo P., Valsecchi M.G., Hatzis P., Menendez P., Parra M. HDAC7 is a major contributor in the pathogenesis of infant t(4;11) proB acute lymphoblastic leukemia. Leukemia. 2021;35(7):2086–2091. doi: 10.1038/s41375-020-01097-x. [DOI] [PubMed] [Google Scholar]
- 51.Crispo F., Condelli V., Lepore S., Notarangelo T., Sgambato A., Esposito F., Maddalena F., Landriscina M. Metabolic dysregulations and epigenetics: a bidirectional interplay that drives tumor progression. Cells. 2019;8(8) doi: 10.3390/cells8080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacque N., Ronchetti A.M., Larrue C., Meunier G., Birsen R., Willems L., Saland E., Decroocq J., Maciel T.T., Lambert M., Poulain L., Hospital M.A., Sujobert P., Joseph L., Chapuis N., Lacombe C., Moura I.C., Demo S., Sarry J.E., Recher C., Mayeux P., Tamburini J., Bouscary D. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126(11):1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johmura Y., Yamanaka T., Omori S., Wang T.W., Sugiura Y., Matsumoto M., Suzuki N., Kumamoto S., Yamaguchi K., Hatakeyama S., Takami T., Yamaguchi R., Shimizu E., Ikeda K., Okahashi N., Mikawa R., Suematsu M., Arita M., Sugimoto M., Nakayama K.I., Furukawa Y., Imoto S., Nakanishi M. Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science. 2021;371(6526):265–270. doi: 10.1126/science.abb5916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.