Abstract
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the central nervous system that usually affects young adults. The development of MS is closely related to the changes in the metabolome. Metabolomics studies have been performed using biofluids or tissue samples from rodent models and human patients to reveal metabolic alterations associated with MS progression. This review aims to provide an overview of the applications of metabolomics that for the investigations of the perturbed metabolic pathways in MS and to reveal the potential of metabolomics in personalizing treatments. In conclusion, informative variations of metabolites can be potential biomarkers in advancing our understanding of MS pathogenesis for MS diagnosis, predicting the progression of the disease, and estimating drug effects. Metabolomics will be a promising technique for improving clinical care in MS.
Keywords: Metabolomics, Multiple sclerosis, EAE
Multiple sclerosis (MS) is a chronic demyelinating disorder of the central nervous system (CNS) with inflammatory and degenerative components [[1], [2], [3]]. It is a significant cause of neurologic morbidity in young adults and has a significant economic impact on the healthcare system [4]. Although the exact reason for causing MS remains unknown to date itself, genetic and environmental factors appear to play a role in its etiopathogenesis [[5], [6], [7], [8]]. Multiple systems biology approaches such as genomics, epigenomics, and proteomics have been applied to the study of MS and have yielded valuable insights into the pathogenesis of the disease. Despite these advances, there remains a need for additional tools to understand the precise etiopathogenesis of MS. There is also a significant unmet need for diagnostic and prognostic biomarkers in MS, especially in progressive forms of the disease [[9], [10], [11], [12], [13]].
Metabolomics is a promising technique that explores small molecules (<1500 Da) in various biological matrices including cells, biofluids such as serum, plasma, cerebrospinal fluid (CSF), urine, excrement, tissue, and exhaled gas [14]. Untargeted and targeted approaches have been developed on various analytical platforms for metabolomic studies [5,15,16]. Data analysis methods for metabolomics are growing, but no consensus currently exists on the optimal data analysis and interpretation approach. Since metabolites are the end products of the different physiological and pathological processes ongoing in the body, this technique can provide information that cannot be gleaned from other technologies, such as genomics, transcriptomics, or proteomics [17]. Metabolomics has gained prominence in recent years for its utility in identifying potential biomarkers of disease and providing insight into the pathogenesis of the disease. It targets small molecules and can provide information not readily obtained from genomics, transcriptomics, or proteomics [18]. It can also offer new insights into disease mechanisms by identifying metabolic pathways that are perturbed. Additionally, this technique may help personalize treatments by identifying new markers of treatment response and disease progression [[19], [20], [21], [22], [23]]. Increasing evidence shows that metabolomics can provide putative biomarkers, insights into the pathophysiology of the disease, and aid in precision medicine for patients with MS [11,16,24,25]. Hereafter, we review the processes of metabolomic study and the applications of metabolomics for the care of MS.
Procedures of metabolomic study
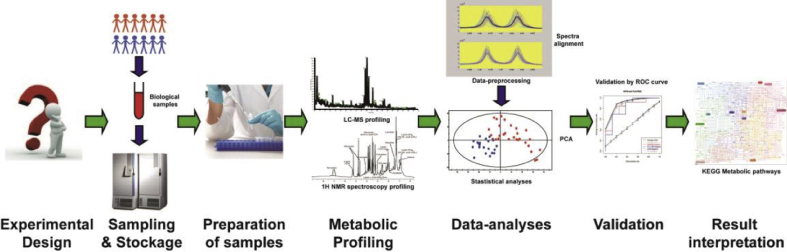
The workflow of metabolomics experiments consists of study design, sample collection and preparation, the manifestation of metabolome based on selected analytical platforms (mass spectrometry coupled with separative systems, and nuclear magnetic resonance (NMR)), processing of data generated, and data analysis and interpretation [6,24,26,27], as shown Fig. 1.
Fig. 1.
Schematic flow-chart emerging the general process of metabolomic study. Abbreviations: LC-MS, liquid chromatography-mass spectrometry; NMR, nuclear magnetic resonance; ROC, receiver operating characteristic.
Experimental design
Before the experimental operations, a proper experimental design is essential to ensure that the data obtained can be appropriately interpreted. This involves the inclusion of a sufficient number of subjects in each group to maximize power to detect inter-group differences, matching for appropriate covariates that are known to affect metabolites of interest, consideration of proper sample collection procedures, and relevant data analysis techniques.
Sample collection and preparation for analysis
Almost all biological materials, including biofluids, cells, tissue, and feces, can be analyzed via metabolomics. However, regarding existing studies concerning MS, biofluids are more popular than other types of sampling, which will be discussed later.
Standardized and optimized procedures for sample collection and storage improve sample quality and reproducibility of metabolomics experiments [28]. Several metabolites may be affected by factors such as fed vs. fasting state, medications, time of day when blood is collected, and processing time [29]. Failure to account for such factors could introduce unwanted, leading to either false-positive or false-negative results [30].
The sample preparation step depends on the analytical platform utilized. For example, NMR approaches do not require special preparation of the samples and hence, are less likely to alter metabolites. On the other hand, mass spectrometry-based approaches generally require sample preparation involving extraction with various solvents [6,31,32].
Most studies examine the metabolites present in the biological matrix at a particular time point, thus providing a snapshot of the metabolome (Table 1: provides a compendium of previous metabolomic studies and the various sample types and platforms used as their findings). It is now possible to introduce a metabolite with an isotope (13C) into a biological system and then measure the concentrations of metabolites containing the isotope, thus enabling a more dynamic assessment of specific metabolic pathways [33]. This can help determine the source of an intracellular molecule, the fate of a particular metabolite, the flux at one metabolic reaction, or the cellular redox balance, thus providing information that cannot be obtained from steady-state metabolomics experiments [[34], [35], [36]].
Table 1.
Summary of prior studies and findings involving MS or MS rodent model (EAE).
| Population | Sample; Platform | Disease vs. Control | Involved Pathways | Study |
|---|---|---|---|---|
| EAE (C57BL mice): 5 Control: 5 |
Plasma; mass spectrometry | ↓: α-linolenic acid, glutamine, glutamate, tryptophan, and arachidonic acid | tryptophan metabolism, fatty acid biosynthesis, pentose pathway, linoleic acid metabolism, arachidonic acid metabolism, and polyunsaturated fatty acid metabolism | Poisson et al. [97] |
| EAE (C57BL mice): 10 Control: 10 |
Plasma/urine; mass spectrometry | ↓: Phenylalanine, tyrosine, tryptophan taurine, arginine, proline, and hypotaurine | Phenylalanine, tyrosine, and tryptophan biosynthesis; arginine and proline metabolism; tyrosine metabolism; and taurine and hypotaurine metabolism | Singh et al. [95] |
| Group 1: RRMS 50, SPMS 20, PPMS 17, and control 49 | Serum; mass spectrometry | ↑: quinolinic acid; in progressive MS; ↑ quinolinic acid kynurenic acid, and kynurenine/tryptophan ratios (which were related to higher EDSS); ↓: Tryptophan and NAD | tryptophan metabolism-endogenous | Lim et al. [72] |
| Group 2: RRMS 44, SPMS 15, MS 518, and control 167 | Serum; HPLC |
↑: hypoxanthine, xanthine, uric acid, inosine, uracil, β-pseudouridine, uridine creatinine, and lactate. Levels correlated with disease severity EDSS | Energy metabolism, nucleotide metabolism, and xanthine metabolism | Lazzarino et al. [101] |
| RRMS: 30 SPMS: 16 Control: 10 |
CSF; mass spectrometry | Alterations in expression of 5-hydroxytryptophan, kynurenate, and N-acetylserotonin 5-hydroxyindoleactate in SPMS; Uridine, deoxyuridine, thymine, and glutamine altered in SPMS | Amino acid metabolism and nucleotide metabolism | Herman et al. [65] |
| MS:27 Control: 27 |
Plasma; mass spectrometry | ↑: Gamma-glutamyl amino acids, glutathione metabolites; ↓: Caffeine/xanthine metabolites, benzoate metabolites | Amino acid metabolism, redox metabolism, benzoate metabolism, and xenobiotic metabolism | Bhargava et al. [99] |
| MS: 73 Controls: 28 |
Plasma; NMR |
↑: 3-OH-butyrate, acetoacetate, acetone, alanine, and choline; ↓: glucose, 5-OH-tryptophan, and tryptophan | Energy metabolism and tryptophan metabolism | Cocco et al. [5] |
| MS: 15 Non-MS: 17 |
CSF; NMR |
↑: Threonate, myo-inositol, and choline; ↓: 3-hydroxybutyrate, citrate, phenylalanine, 2-hydroxyisovalerate, and mannose | Energy metabolism and phospholipid metabolism | Reinke et al. [61] |
| MS:23 Controls:28 |
Serum; NMR |
↑: Glucose; ↓: Valine | Glucose metabolism and amino acid metabolism | Mehrpour et al. [102] |
| Pediatric MS: 66 Controls: 66 |
Serum; mass spectrometry | ↓ Levels of tryptophan and indole lactate in pediatric MS; Kynurenine level predicted the relapse rate, indole acetate related to SDMT, and indole propionate related to EDSS and SDMT | tryptophan metabolism-endogenous and gut microbiota related | Nourbakhsh et al. [41] |
| MS 32 Non-MS neural-inflammatory: 32 Non-MS neural-disorders: 18 |
CSF; NMR |
↑: (RRMS) lactate; ↓: Formate | Energy metabolism | Simone et al. [103] |
| MS 30 (RRMS and PPMS) | CSF; NMR |
↑: Acetate; ↓: Formate | Energy metabolism | Bernardes et al. [93] |
| MS: 50 NMOSD: 57 Control: 17 |
CSF; NMR |
↑: Pyroglutamate, acetone, formate, and 2-hydroxybutyrate; ↓: Citrate, glucose, and acetate |
Energy metabolism and fatty acid biosynthesis | Kim et al. [104] |
| Group 1: MS 238, controls 74 Group 2: MS 61, controls 41 |
Serum; mass spectrometry | MS vs. Control: lysophosphatidylethanolamine and sphingomyelin; Predictors of MS progression: hydrocortisone, glutamic acid, tryptophan, eicosapentaenoic acid, and 13S-hydroxyoctadecadienoic acid | Lipid metabolism, steroid metabolism, and amino acid metabolism | Villoslada et al. [71] |
| MS: 18 Controls 18 |
Plasma; mass spectrometry | ↑, 5-oxoproline; lactate, N1-methylinosine, N2, N2 dimethylguanosine, pseudouridine, sphingosine-1-phosphate, and sphinganine-1-phosphate | Sphingolipid metabolism, redox metabolism, and nucleotide metabolism | Bhargava et al. [105] |
| RRMS: 17 Control 17 |
Plasma; mass spectrometry | ↑: glutamate; ↓:C10:1 carnitine, leucine, valine, isoleucine |
Amino acid catabolism, glutamate metabolism, and energy metabolism | Kasakin et al. [106] |
| MS cohorts: PPMS 33 and RRMS 10 Control: 33 PD cohort: PD: 40 Control: 20 |
Plasma; mass spectrometry | ↓: PPMS vs (RRMS, PD, HC): LysoPE (18.2) and LysoPC (20:0) ↓: PPMS vs HC: tiglylcarnitine ↑: PPMS vs RRMS: tiglylcarnitine ↓: PPMS vs HC: gamma-linolenic acid, L-Tryptophan, LysoPC (20:0) ↑: (RRMS and PD vs HC:) gamma-linolenic acid, L-Tryptophan, LysoPC (20:0) ↓: PPMS citrulline vs HC ↓: PPMS vs HC: L-Tryptophan |
PPMS vs. HC: glycerophospholipid metabolism, linoleic acid metabolism, arginine biosynthesis, and kynurenine pathway | Stoessel et al. [40] |
| MS: 514 RRMS: 72.71% Progressive MS: 27.29% Controls: 214 |
Plasma/serum; mass spectroscopy | ↓:phenyllactate, 3-(-4-hydroxyphenyl)-lactate, indolelactate, imidazole lactate, kynurenine, kynurenate, tryptophan and phenylalanine ↑: p-cresol glucuronide, p-cresol sulfate, and phenylacetylglutamine. altered ratios of phenylacetylglutamine/phenyllactate, indole acetate/indolelactate, p-cresol glucuronide/3-(4-hydroxyphenyl)lactate. |
lactate-related metabolites in AAA pathways (tryptophan, phenylalanine metabolism), BCAA-related metabolites, other AAA metabolites, bile acid metabolism, xanthine metabolites, acetylated amino acids |
Fitzgerald et al. [79] |
| MS: 90 Controls:90 |
Serum; mass spectroscopy | ↓: indoleproprionate, indoleactate ↑:p-cresol sulfate |
Tryptophan degradation pathway | Levi et al. [78] |
| RRMS: 13 Controls: 13 |
Plasma, cerebral cortex; mass spectroscopy | ↓: methionine (plasma) | DNA and histone H3 methylation in cerebral cortex, methionine metabolism, regulation of mitochondrial electron transport complex proteins(brain). | Singhal et al. [80] |
CSF: cerebrospinal fluid; EAE: experimental autoimmune encephalomyelitis, EDSS: expanded disability status scale; HC: healthy control, HPLC: high-performance liquid chromatography; MS: multiple sclerosis; NAD: nicotinamide adenine dinucleotide, NMOSD: neuromyelitis optica spectrum disorder, PD: Parkinson's disease, PPMS: primary progressive multiple sclerosis; RRMS: relapsing-remitting multiple sclerosis; SDMT: symbol digit modalities test, SPMS: secondary progressive multiple sclerosis; ↑: increase; ↓: decrease.
Data analysis
This step involves the identification of metabolites and the determination of the relative abundance. Metabolite identification varies depending on the platform and the approach (targeted vs. untargeted) applied [37]. In targeted experiments, standards are generally run, and hence there is less uncertainty regarding metabolite identification [38]. In the untargeted method, the spectra are processed using proprietary or freely available software and compared to the public libraries for metabolite identification [39]. The data should also be inspected for outliers and samples or metabolites with high amounts of missing values. At this step, samples or metabolites that do not meet quality control criteria should be removed. Following these steps, a set of metabolite features that are robustly quantified is obtained [40,41].
There is a large amount of correlation between various metabolites that may arise from the fact that metabolites may belong to the same metabolic pathway or in a specific experiment may be subject to factors leading to them being altered together. Correlation network analysis is a method that aims to identify networks within the metabolomics data without taking into account prior knowledge regarding known metabolic pathways. Individual metabolites that act as nodes and connections are based on calculated correlation coefficients. A partial correlation is used to construct such networks. The coefficient is calculated based on the correlation between two metabolites while considering the correlation with the remaining metabolite set. Similarly, weighted correlation network analysis can also be applied to metabolomics data to identify the clusters of highly correlated metabolites and their relationship with other clusters of metabolites or with phenotypic variables of interest [42,43].
The neutral network analysis is also applied in the combinational analysis among multiple “omics” data. Massive information provided by integral omics enables an overall insight into intercellular and intracellular mechanisms, including the mutation processes, deregulated signaling pathways, and the interactions between proteins and small molecules in the duration of disease development [17]. Since short-gun profiling approaches are generally used in most omics analyses, the functions of resulted features derived from various pathways are complex. Thus, network analysis provides an understanding of synergy functions for matched genes, mRNAs, proteins, and metabolites. This promising approach has been applied in MS [44]. In general, the multiple omics network analysis can be realized via two different strategies, which have been both utilized in MS research. One method is based on data fusion, where the correlation between variables obtained from various omics analyses is parallelly calculated. Then, the correlations are ranked and a linkage between variables with high correlation is constructed. Consequently, a network illustrating the connections between defined variables is established.
As an example, Blanchet et al. combined metabolomics and proteomics on innovative dimension reduction and variable selection [45]. Their study tests the correlation between a selective protein and a metabolite. The contribution of feature metabolite for a discriminant model is calculated in the group comparison and the in-line variations between a candidate metabolite and at least three peptides of the candidate protein. This non-database-dependent method appears to be a high throughput screening of valuable variables. It provides evidence to explore unknown interactions between metabolites and proteins. However, the candidate metabolites and proteins were determined with a small number of MS model animals and controls, prone to false-positive errors.
Another method revealing the correlation between proteins and metabolites is based on network analysis with the help of the database covering comprehensive known signaling and metabolic pathways. Until now, there are various open-source repositories, such as KEGG (www.kegg.jp) [46] and HMDB (www.hmdb.ca) [47]. With these datasets, one would expect that the investigation of metabolite function or relationship between candidate metabolite and gene/protein can be simply performed with a search in the pathway library. However, this is not the case because one metabolite is usually involved in a wealth of metabolic and signaling pathways. Thus, pathway enrichment is needed before the analysis of function.
Several software tools exist for performing metabolite set enrichment analysis and visualizing these results. Metaboanalyst (www.metaboanalyst.ca) [48] is an online platform for metabolomics data analysis that has several tools for pathway enrichment analysis. KEGG and Metaboanalyst and Metlin (https://metlin.scripps.edu) [49], a vast database of metabolites with their MS-derived ions, serve for ingenuity pathway analyses is an online software allowing comprehensive analyses and interpretation of data of omics. With the application of ingenuity pathway analyses, regulations of metabolites are intuitively related to the regulations of upstream genes, mRNAs, and proteins, helping to understand underlying mechanisms associated with metabolomic findings [50,51].
Applications
This section illustrates the applications of metabolomics in MS and its preclinical animal model, experimental autoimmune encephalomyelitis (EAE).
Metabolic characteristics and biomarkers determined in human MS
MS is empirically categorized into four subtypes: relapsing-remitting multiple sclerosis (RRMS), secondary progressive multiple sclerosis (SPMS), primary progressive multiple sclerosis (PPMS), and progressive relapsing multiple sclerosis (PRMS). According to the latest McDonald criteria 2017, the diagnosis of MS primarily relies on the imaging examination and test of IgG oligoclonal bands in CSF. However, the limitations of these tests are also well known [52]. In addition, the prognosis of MS in patients varies substantially across individual patients [53]. Thus, the diagnosis and prognosis of MS are challenging with existing clinical methods. Increasing studies have demonstrated that metabolomics is a promising tool for the diagnosis and prognosis of MS. One important reason is that metabolite variations are sensible to both genetic and environmental factors contributing to the occurrence of MS. Besides, metabolomics is also known to provide evidence for exhibiting individual differences in patients, which makes the precise treatment possible [54]. Here, metabolomic studies facing to human patients of MS are first reviewed, in the light of samplings analyzed.
CSF
CSF is the secretion product of the CNS that fills the ventricles and the subarachnoid space of the brain and spinal column. It protects the brain from physical shocks, stores nutrition, and washes away waste from brain tissue [55,56]. CSF findings are an essential part of the diagnosis and prognosis of CNS diseases, especially for progressive MS [57]. Apart from numerous pediatric studies executed previously [58], people realize that analysis of small molecules in CSF is also of great significance for clinical applications. Metabolomics is a promising tool that provides an overview of metabolic variations in the CSF [59]. Because of direct interchanges between CSF and brain tissue, metabolic alterations in CSF are informative that reflect existing physiological shifts [59,60]. Therefore, CSF analysis has been generally applied in metabolomics-based MS studies.
In Reinke et al. [61], authors performed 800 MHz 1H-NMR spectroscopic analyses of CSF specimens to identify biomarkers of MS, yielding reproducible detection of 15 metabolites from MS (n = 15) and non-MS (n = 17) patients. Mean levels of choline, myoinositol, and threonate were increased, whereas 3-hydroxybutyrate, citrate, phenylalanine, 2-hydroxyisovalerate, and mannose were decreased in MS-derived CSF (p < 0.05), suggesting alterations to energy and phospholipid metabolism. Multivariate hierarchal cluster analysis indicated a high correlation within the metabolite profiles, significantly clustering samples into the two clinical groups. CSF metabolomics can yield quantitative biomarkers and insights into the pathogenesis of MS. An earlier work based on NMR-based metabolic profiling aimed to determine characteristic features to predict inflammation plaques in MS patients [62]. Accordingly, a higher level of lactate and reduced phenylalanine in CSF were revealed to be associated with MS-induced inflammation.
By using high-resolution mass spectrometry, the metabolome of CSF was investigated by Herman et al. [63]. Tryptophan (Trp) metabolism alteration was underlined. Pathophysiological differences between SPMS and relapsing-remitting multiple sclerosis (RRMS) were shown. The authors further identified potential biomarkers of disease progression. Their results supported the hypothesis that the CSF metabolome might explore changes that occur in the transition between the RRMS and SPMS pathologies. Another study based on a targeted analysis of patients' CSF was executed by Markianos et al., which concerned the relevance between levels of discriminant metabolites and the severity and disability in MS. In their work, high-performance liquid chromatography without mass spectrometry was used to determine levels of targeted metabolites such as noradrenaline, homovanillic acid, and 5-hydroxyindoleacetate in the CSF of RRMS patients. They proved that the deregulation of 5-hydroxyindoleacetate was predictable for MS severity and for the rate of disability, which was negatively correlated with MS-related clinical scores [64].
A novel analytical approach is proposed to scrutinize and combine information from biomarkers originating from multiple sources to discover a condensed set of biomarkers that, in combination, could distinguish SPMS from RRMS [65]. CSF samples collected from sixteen SPMS patients were compared to 30 RRMS patients and ten healthy participants with a five-year follow-up. Metabolomics data was integrated with magnetic resonance imaging and protein variables. The combination of two metabolites (20β-dihydrocortisol and indolepyruvate) with three magnetic resonance imaging and six proteic variables has the potential to distinguish SPMS from RRMS. The work is of interest for providing a method to estimate an aggravated progression of MS during the long-term follow-up. Without further variable screening, the resulted marker was relatively complex but logical due to a critical consideration of clinical scores and molecular changes.
Plasma and serum
Blood-derived samples are most employed in metabolomics studies due to their accessibility and comprehensive contents that represent a whole situation for the human body. Except for massive electrolytes, proteins, and peptides, various small molecules, both hydrophile and hydrophobic, are carried in blood [66]. Even though it has been claimed that whole blood is more informative for metabolomic investigation [67], the applications of plasma and serum are largely more common due to the exclusion of complexities caused by red blood cell metabolites. In comparison with CSF, findings in plasma or serum are less intuitive due to the filtration of the blood-brain barrier; however, they are less invasive for patients. Besides, CSF analysis is not indispensable for diagnosing early cases of MS according to the MS diagnostic guideline [68]. Thus, plasma and serum metabolomics analysis is still a potential method, especially for diagnosis and prognosis in the early stages of MS [61].
In a recent study by Stoessel et al. [40], the researchers utilized untargeted high-resolution metabolomics to plasma samples to identify primary progressive MS-specific signatures, validated the findings in independent sex- and age-matched primary progressive MS and healthy control cohorts and built discriminatory models by partial least square discriminant analysis. This signature was compared to sex- and age-matched RRMS patients, to patients with Parkinson's disease and healthy controls. They also investigated these metabolites in a longitudinal cohort of primary progressive MS patients over 24 months. The partial least square discriminant analysis yielded predictive models for classification along with a set of 20 primary progressive MS-specific informative metabolite markers. These metabolites suggest disease-specific alterations in glycerophospholipid and linoleic acid pathways.
Without regrouping, according to the disease stage among MS patients, people could differentiate MS from healthy controls using gas chromatography-mass spectrometry-based metabolome analysis in plasma [69]. Their findings coincided with one NMR-based study discussed hereinbefore [61]. Abnormal reductions in fructose, myo-inositol, pyroglutamate, threonate, and leucine were confirmed in the MS patients, whose levels of glutamine, glutamic acids, and ornithine were enhanced. Based on receiver operating characteristic curve analysis, the integration set of discriminant metabolites showed good predictability for the MS cohort. A slightly earlier work, which was fairly similar to this one, was derived with 1H-NMR analysis discovered consentaneous results. An excellent receiver operating characteristic curve prediction of MS was performed with the pattern model [5].
Plasma samples were also collected to characterize the overlapping clinical features and provide a molecular signature of RRMS, aquaporin-4 antibody neuromyelitis optica spectrum disorder, and myelin oligodendrocyte glycoprotein (MOG) antibody disease [70]. The metabolites identified could have potential use for disease monitoring and diagnosis.
In the study of Villoslada et al. [71] metabolomic profiling (lipids and amino acids) was performed by ultra-high-performance liquid chromatography coupled to mass spectrometry in serum samples. Data analysis was performed using parametric methods, principal component analysis, and partial least square discriminant analysis to address the differences between cases and controls and subgroups based on disease severity. They identified metabolomics signatures with high accuracy for classifying patients from healthy controls and classifying patients with a medium to high disability. Among them, sphingomyelin and lysophosphatidylethanolamine were the metabolites that showed a more robust pattern in the time series analysis for discriminating between patients and controls. Moreover, levels of hydrocortisone, glutamic acid, Trp, eicosapentaenoic acid, 13S-hydroxyoctadecadienoic acid, lysophosphatidylcholines, and lysophosphatidyletha-nolamines were associated with more severe disease.
In work by Lim et al. [72] the authors showed that serum kynurenine pathway (KP) parameters are strongly associated with the MS subtype, correlating with disease severity scores. The changing levels of KP metabolites observed also provide a mechanistic insight that may explain the transition from the milder RRMS form to the more debilitating SPMS disease form. Furthermore, they infer that KP profiling is likely to be relevant to the pathogenesis of other diseases characterized by inflammation and neurodegeneration, like Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis, where aberrant KP metabolism has been observed. Their results also suggest that strategies aimed at rebalancing the KP could be helpful therapeutic approaches in slowing neurodegeneration in MS. A relevant study conducted by Nourbakhsh et al. [41] showed a higher relative abundance of Trp and indole lactate, known gut microbiota-derived Trp metabolites, were associated with a lower risk of MS. Consistent findings were elsewhere attained in a recent metabolomics study focusing on the efficacy of treatment [73]. The researchers observed metabolic variations between the patients who positively and negatively responded to interferon beta-1a. Consequently, Trp and its derivative, kynurenine, were considered to account for outcomes. Of note, although the upregulation of Trp from a lower level for MS patients was associated with an optimal signal for the outcome, even a slightly higher level of Trp was revealed in therapeutic non-responders. These paradoxical findings were partly due to the deregulation of kynurenine derivatives and the accumulation of Trp in the blood. Given that an upregulated kynurenic acid pathway is an essential indicator of inflammation of the CNS [74,75], this pathway is, therefore, regarded as a double-edged sword [76,77].
Other than feature variations in the KP metabolism, various metabolic features in the host serum metabolome can be viewed due to the interplays between the host and the gut microbiome. By combining the profiling of microbiome and serum metabolites from 129 MS patients, Levi and colleagues revealed that depleted indolepropionate, which is a neuroprotective anti-oxidant associated with the reduced butyrate-producing bacteria, was generally found in the MS patients [78]. The study confirmed the contribution of microbiome to the pathology and etiology of MS and emphasized the functions of gut microbiome-producing metabolites such as short-chain fatty acids and indoles in MS. A similar study focused on the microbiome-induced aromatic amino acids (AAAs) in serum was also carried out in a large cohort of MS patients. The researchers found that the imbalance of AAAs in the MS patients was responsible for the dysregulation of downstream inflammation-related metabolites such as indoleacetate, phenylacetylglutamine, p-cresol sulfate, and p-cresol glucuronide [79]. Another recent work witnessed the disorders of bile acid metabolism in the serum of MS patients by untargeted and targeted metabolomics. The human gut flora converts primary bile acids into secondary bile acids. In this work, the authors found it possible to differentiate RRMS from PMS or healthy controls in adults and healthy controls from MS in pediatric-onset patients, based on primary and secondary bile acids profiling.
The deregulations in the methionine pathway, which are associated with CNS methylation, were investigated in another study [80]. People have examined the relationship between alterations in circulating methionine metabolites and markers of methylation linked to MS pathology. They found a significant reduction in plasma methionine levels in early-stage RRMS patients compared to controls.
Urine
Like blood, urine is also a metabolite-rich biofluid that is easy to obtain without invasive sampling in the clinic. Previously, abnormal proteic alterations in urine were demonstrated to be earlier than the onset of MS [81]. Hence, one would expect that metabolome profiling for urine provides warning signatures for MS diagnosis and prognosis. Several studies focusing on the feature of urinary metabolome have been performed to better understand the pathology of MS. For example, Gebregiworgis et al. showed that a set of 8 metabolites in urine were potential markers for MS [82]. In a study from Bert et al., MS metabolic profiling was otherwise compared with other neurological diseases and robust controls [83]. The investigators further performed a similar urinary metabolomic analysis in animal models. Their results showed a consensus of variations in the aspartate pathway in humans and rodent urine.
Brain tissue
Despite surgical treatments that have been reported for patients who do not respond to drug therapy [84], a surgical operation is not predominant among MS therapeutic strategies, which seems to close the door on brain tissue analysis for MS diagnosis. However, with the help of advanced computational technologies associated with magnetic resonance imaging, a noninvasive metabolite scan on the white matter was realized by Vingara et al. [85]. By comparing RRMS patients to healthy controls, glutamic acid, creatine, and N-acetyl-aspartate and choline were the key variables decreased in RRMS. Even though no novel metabolite markers were revealed for RRMS diagnosis, this study inspires future metabolome imaging for brain diseases.
Experimental autoimmune encephalomyelitis (EAE)
The EAE model that induces autoimmune inflammatory disorders of the CNS has long been used to mimic MS in rodents and monkeys [86]. Disease in these models is initiated from an inflammatory response to antigens in which immunization against myelin autoantigens triggers T-cell responses and eventual myelin destruction [87]. It is a series of comprehensive conditions caused by the cross-talk of immunopathology and neurological lesions, leading to MS features, such as demyelination, axon loss, and glial cell proliferation. In practice, three primary myelin proteins, or characteristic peptides contained in these proteins, are used within Freund's adjuvant to induce EAE, including MOG, myelin basic protein, and proteolipid protein. Among these three proteins, MOG and proteolipid protein are commonly used in MS-like models, which lead to the chronic EAE model and relapse remitting EAE model, respectively [88]. It should be emphasized that none of the existing modeling methods can reproduce precisely the same inflammatory and demyelination mechanism of MS [89]. Even so, EAE models are still widely used to research the process of inflammation and injury in the CNS and develop novel medicines [90]. More than the studies in humans that have been mentioned before, EAE model animals are valuable subjects that have been analyzed in substantial metabolomic studies. Like the human studies, researches concerning metabolomic analyses in EAE rodents are also discussed below according to the samples collected.
CSF
As discussed herein, integral metabolomics-proteomics analysis is promising for biomarker exploration and personalized treatment [51], which is well proved by a typical study upon the fusion between metabolomics and proteomics [45]. After a network analysis, discriminant metabolites refined from the comparison between EAE rats and healthy controls involved in the deregulations of energy supply and amino acid metabolism in the CSF of EAE rats. Interestingly, these turnovers were linked with the deregulations of T kininogen 1, complement C3, and ceruloplasmin. The first two proteins are well-known inflammation-related proteins. Ceruloplasmin was reported as a biomarker for hypothalamic–pituitary–adrenal axis deregulation. Such findings proved that the combination of metabolomics and proteomics helps to improve the understanding of physiological alterations and the relevant functions of refined features.
A study in myelin essential protein-induced EAE rats also analyzed the CSF samples of the rats [91]. The study followed the metabolome changes along with the progression from the EAE onset to the peak of disease. Their findings revealed that downregulated arginine, alanine, and branched amino acids (BCAA) were correlated with the early stage of EAE modeling. At the same time, the increases of glutamine, O-phosphoethanolamine, BCAAs, and putrescine were the signatures in the EAE rats for the peak of disease, relative to the controls. Altogether, interestingly, accordant turnovers in amino acid (AA) or AA derivates and xenobiotics are widely found in the comparisons between the EAE model and healthy controls. Variations in the energy-related pathways such as glycolysis, tricarboxylic acid (TCA) cycle, and fatty acids are other general signs of the development of MS. It is worth noting that these metabolic alterations are in accordance with those obtained in human MS patients.
Serum and plasma
A comprehensive coverage of metabolites in the plasma of EAE model mice was shown in the study of Mangalam et al. with integral data acquired from UPLC-MS and GC-MS. According to the pathway analysis of 44 potential biomarkers, metabolisms of tryptophan, histidine, linoleic acid, and d-arginine/d-ornithine were summarized to be varied in EAE mice [88]. It should be underlined that these results were similar to the findings in human patients. Typically, the dysregulations in the tryptophan and lipid pathways support that the pathological alterations in the CNS are related to aberrant regulations against inflammation during the development of MS. Based on the MOG-induced EAE model, Lee and co-workers showed a 30-day course of metabolite variations in BCAAs, long-chain phospholipids, and fatty acids in the model rats [92]. With pathway enrichment and correlative analyses between these metabolites and two inflammatory markers, pathways involved in pro-inflammation, anti-inflammation and neuroprotection were revealed as predominant shifts for the mice.
It was demonstrated by Bernardes et al. that metabolite variations in EAE model rodents could be otherwise quantified with biochemical methods using specific enzyme-linked immunosorbent assay kits, other than NMR and MS platforms [93]. As relative to the instrumental analysis, this approach requires neither complicated sample preparation nor a high volume of sample. However, the cost of the kit and the throughput are the drawbacks. According to their results, physical exercises are salutary to prevent mass loss and reduce the clinical score. Unexpectedly, except for cholesterol, metabolites such as serum glucose and triglyceride were neither dependent on the sport of mice nor predictable for the peak of inflammation. Compared to other studies, the shift of metabolite patterns should be attributed to the experiment duration. As the study lasted for more than six weeks, metabolites might be stabilized due to a chronic mechanism of sensitization and inflammation induced by MOG [89].
Urine
To have an overview of metabolic variations of MS-mimicked model animals. Some studies investigated both the urine and the plasma of EAE rats. One of the studies by Dickens et al. was able to distinguish chronic relapsing EAE mice from controls with the silent disease (10 and 28 days after modeling) and active disease (14 and 38 days after modeling) [94]. The results showed that disturbances in glucose, citrate, taurine, trimethylamine oxide (TMAO), creatine, and fatty acids were responsible for separating chronic relapsing EAE and naïve mice. Another similar study revealed general disorders in amino acids, xenobiotics, and nucleotides in the urine and plasma of EAE animals [95]. Except for glucose, taurine, and creatinine, most of the other refined metabolic markers from the two studies were barely overlapped due to the different analytical instruments utilized. The study based on NMR determined the feature variations of metabolites over time, while the study using the UPLC-MS platform focused on the expected changes of metabolites detected in both urine and serum. These two works remarkably exhibited that the metabolic variations in plasma were not associated with those in urine.
Tissue
A study by Battini et al. collected whole CNS tissue and optic nerves [96]. They compared CNS metabolome between myelin primary protein-induced EAE mice and controls using a high-resolution magic angle spinning based 1H-NMR approach. The study indicated that energy-associated metabolites such as glucose and lactate and various amino acids were elevated in EAE mice. A significant decrease of N-acetyl-aspartate was another typical feature in the whole CNS and optic nerves in the model animals.
Applications of metabolomics for identifying a therapeutic target in MS
Metabolomics studies were also applied to investigate the therapeutic effects of drug-like metabolites in EAE animals and human patients. In one study, the workers found multiple pathways related to fatty acid metabolisms that vary between MOG-induced EAE mice [97]. The decreased levels of ω-3 and ω-6 polyunsaturated fatty acids were typical signs of EAE mice. In their following work, treatment of resolvin D1, a downstream metabolite of ω-3, to the EAE mice hinders the progression of MS-like disease. Their further investigation of the resolvin D1 effect revealed underlying mechanisms involving the induction of regulatory T cells, the polarization of monocytes/macrophages and microglia into the M2 phenotype, and the induction of inflammatory responses.
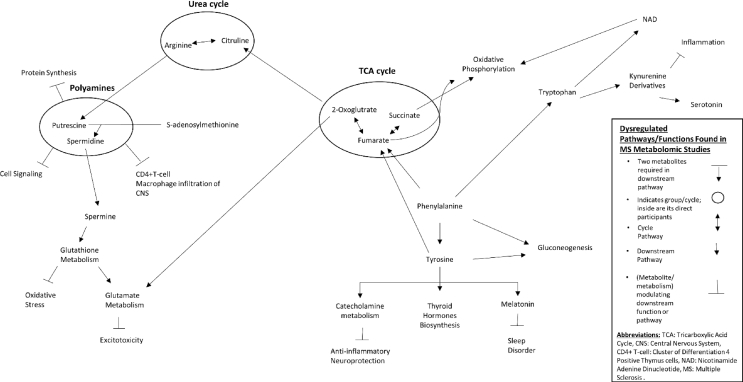
Notably, other pharmaceutical research on metabolomic techniques has been performed with model animals and with cells and human patients. Fig. 2 provides a map of known pathways that are perturbed in MS. For example, one study tested the neuron protective effect of methionine enkephalin using a capillary electrophoresis-tandem mass spectrometry-based approach [98]. Significantly increased AAs and glycylglycine were witnessed in the extracted cell metabolome after supplementation of methionine enkephalin to glioma cells. Another study using ultra-high-performance liquid chromatography-mass spectroscopy focused on determining the effect of vitamin D in healthy people and MS patients [99]. Alterations in metabolites associated with oxidative stress and xenobiotic metabolism were shown in the plasma of healthy controls, while such variations were not present in the plasma of MS patients after the vitamin D treatment.
Fig. 2.
Pathway networks found to be perturbed in Multiple Sclerosis via metabolomic studies.
Additionally, supplementation of acetate and methionine was determined to be salutary for improving MS outcomes [80,100]. In turn, these two metabolites' deficit is considered alarm of MS aggravation. As discussed above, turnover in energy supply, especially in the TCA cycle, and methylation within CNS is responsible for these features. As these experiments were conducted in EAE model animals, further confirmation is required for clinical applications in human patients.
Concluding remarks
According to an overview of previous metabolomic studies in MS patients and EAE model animals, variations in comprehensive metabolites and metabolic pathways have been refined to be associated with the occurrence and progression of MS. Accordance findings are concentrated with several of the following pathways. First, a downregulated aspartate-N-acetyl-aspartate pathway accompanied with increased glutamate was defined within CSF is specific for RRMS. Indeed, these indicators of neuron conditions have been discovered for a long time. Second, the enhanced conversion of tryptophan into kynurenine is associated with MS occurrence and worse outcomes. In particular, implications for Trp-KP turnovers may be more than an occurrence of inflammation in the CNS and a disturbance in the gut microbe. Finally, decreased levels of BCAAs in the blood are negative signs linked with abnormal energetic metabolism for MS prognosis. Decreased BCAAs and increased TCA intermediates and lactate are common indicators of a broad spectrum of inflammation. These findings are suggested to be proofs rather than biomarkers for the diagnosis or prognosis of MS. To avoid bias, integral analysis that combines metabolomics with other data such as other omics and clinical variables is recommended.
To conclude, increasing data have provided evidence that metabolomics is a valuable tool in various aspects of MS clinical care. Informative variations of metabolites can be potential biomarkers for MS diagnosis, predict the progression of the disease, and estimate drug effects. We, therefore, have reason to believe that metabolomics will be a promising technique for improving clinical care for MS.
Funding
The study was supported by the Grants for Scientific Research of BSKY(XJ201729) of Anhui Medical University, the Research Fund of Anhui Medical University, and Anhui Natural Science Foundation (2008085QH364).
Author contributions
The design of the study was directed by Dr. Rui. Dr. Liu worked on the redaction of the manuscript. Jeffrey Waters helps to process the figures and table. All the authors contributed to correcting the manuscript.
Informed consent statement
The informed consent statement is not applicable to the study.
Data availability
The authors declare that there is no data to display.
Conflicts of interest
The authors claim that there are no conflicts of interest.
Acknowledgments
The authors thank Dr. Zeng Li and Stevens Stephanie for their correction suggestions.
Footnotes
Peer review under responsibility of Chang Gung University.
Contributor Information
Zhicheng Liu, Email: liuzhicheng@ahmu.edu.cn.
Bin Rui, Email: happyruibin12@gmail.com.
References
- 1.Giovannoni G. Multiple sclerosis cerebrospinal fluid biomarkers. Dis Markers. 2006;22:187–196. doi: 10.1155/2006/509476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murgia F., Lorefice L., Poddighe S., Fenu G., Secci M.A., Marrosu M.G., et al. Multi-platform characterization of cerebrospinal fluid and serum metabolome of patients affected by relapsing-remitting and primary progressive multiple sclerosis. J Clin Med. 2020;9:863. doi: 10.3390/jcm9030863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filippatou A.G., Lambe J., Sotirchos E.S., Fitzgerald K.C., Aston A., Murphy O.C., et al. Association of body mass index with longitudinal rates of retinal atrophy in multiple sclerosis. Mult Scler. 2020;26:843–854. doi: 10.1177/1352458519900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pieragostino D., D'Alessandro M., di Ioia M., Rossi C., Zucchelli M., Urbani A., et al. An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis. Mol Biosyst. 2015;11:1563–1572. doi: 10.1039/c4mb00700j. [DOI] [PubMed] [Google Scholar]
- 5.Cocco E., Murgia F., Lorefice L., Barberini L., Poddighe S., Frau J., et al. (1)H-NMR analysis provides a metabolomic profile of patients with multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2016;3:e185. doi: 10.1212/NXI.0000000000000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin R., Charlesworth J., van der Mei I., Taylor B.V. The genetics of multiple sclerosis. Pract Neurol. 2012;12:279–288. doi: 10.1136/practneurol-2012-000276. [DOI] [PubMed] [Google Scholar]
- 7.Graber J.J., Dhib-Jalbut S. Biomarkers of disease activity in multiple sclerosis. J Neurol Sci. 2011;305:1–10. doi: 10.1016/j.jns.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Kang J., Zhu L., Lu J., Zhang X. Application of metabolomics in autoimmune diseases: insight into biomarkers and pathology. J Neuroimmunol. 2015;279:25–32. doi: 10.1016/j.jneuroim.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Botas A., Campbell H.M., Han X., Maletic-Savatic M. Metabolomics of neurodegenerative diseases. Int Rev Neurobiol. 2015;122:53–80. doi: 10.1016/bs.irn.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Tumani H., Hartung H.P., Hemmer B., Teunissen C., Deisenhammer F., Giovannoni G., et al. Cerebrospinal fluid biomarkers in multiple sclerosis. Neurobiol Dis. 2009;35:117–127. doi: 10.1016/j.nbd.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Ebers G.C. Environmental factors and multiple sclerosis. Lancet Neurol. 2008;7:268–277. doi: 10.1016/S1474-4422(08)70042-5. [DOI] [PubMed] [Google Scholar]
- 12.Comabella M., Montalban X. Body fluid biomarkers in multiple sclerosis. Lancet Neurol. 2014;13:113–126. doi: 10.1016/S1474-4422(13)70233-3. [DOI] [PubMed] [Google Scholar]
- 13.Gandhi R., Healy B., Gholipour T., Egorova S., Musallam A., Hussain M.S., et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013:73729–73740. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 14.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 15.Dendrou C.A., Fugger L., Friese M.A. Immunopathology of multiple sclerosis. Nat Rev Immunol. 2015;15:545–558. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- 16.Park S.J., Kim J.K., Kim H.H., Yoon B.A., Ji D.Y., Lee C.W., et al. Integrative metabolomics reveals unique metabolic traits in Guillain-Barre Syndrome and its variants. Sci Rep. 2019;9:1077. doi: 10.1038/s41598-018-37572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohart F., Gautier B., Singh A., Le Cao K.A. mixOmics: an R package for 'omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Housley W.J., Pitt D., Hafler D.A. Biomarkers in multiple sclerosis. Clin Immunol. 2015;161:51–58. doi: 10.1016/j.clim.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Ottervald J., Franzen B., Nilsson K., Andersson L.I., Khademi M., Eriksson B., et al. Multiple sclerosis: identification and clinical evaluation of novel CSF biomarkers. J Proteomics. 2010;73:1117–1132. doi: 10.1016/j.jprot.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Andersen S.L., Briggs F.B.S., Winnike J.H., Natanzon Y., Maichle S., Knagge K.J., et al. Metabolome-based signature of disease pathology in MS. Mult Scler Relat Disord. 2019;31:12–21. doi: 10.1016/j.msard.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtzke J.F. Geography in multiple sclerosis. J Neurol. 1977;215:1–26. doi: 10.1007/BF00312546. [DOI] [PubMed] [Google Scholar]
- 23.Kahana E., Zilber N., Abramson J.H., Biton V., Leibowitz Y., Abramsky O. Multiple sclerosis: genetic versus environmental aetiology: epidemiology in Israel updated. J Neurol. 1994;241:341–346. doi: 10.1007/BF00868444. [DOI] [PubMed] [Google Scholar]
- 24.Olsson T., Barcellos L.F., Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13:25–36. doi: 10.1038/nrneurol.2016.187. [DOI] [PubMed] [Google Scholar]
- 25.Teunissen C.E., Khalil M. Neurofilaments as biomarkers in multiple sclerosis. Mult Scler. 2012;18:552–556. doi: 10.1177/1352458512443092. [DOI] [PubMed] [Google Scholar]
- 26.Bielekova B., Martin R. Development of biomarkers in multiple sclerosis. Brain. 2004;127:1463–1478. doi: 10.1093/brain/awh176. [DOI] [PubMed] [Google Scholar]
- 27.Bhargava P., Calabresi P.A. Metabolomics in multiple sclerosis. Mult Scler. 2016;22:451–460. doi: 10.1177/1352458515622827. [DOI] [PubMed] [Google Scholar]
- 28.Yin P., Peter A., Franken H., Zhao X., Neukamm S.S., Rosenbaum L., et al. Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clin Chem. 2013;59:833–845. doi: 10.1373/clinchem.2012.199257. [DOI] [PubMed] [Google Scholar]
- 29.Pinto J., Domingues M.R., Galhano E., Pita C., Almeida Mdo C., Carreira I.M., et al. Human plasma stability during handling and storage: impact on NMR metabolomics. Analyst. 2014;139:1168–1177. doi: 10.1039/c3an02188b. [DOI] [PubMed] [Google Scholar]
- 30.Gertsman I., Barshop B.A. Promises and pitfalls of untargeted metabolomics. J Inherit Metab Dis. 2018;41:355–366. doi: 10.1007/s10545-017-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Shen T., Rui B., Zhou W., Zhou X., Shang C., et al. CeCaFDB: a curated database for the documentation, visualization and comparative analysis of central carbon metabolic flux distributions explored by 13C-fluxomics. Nucleic Acids Res. 2015;43:D549–D557. doi: 10.1093/nar/gku1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim S.M., Gold R. Genomics, proteomics, metabolomics: what is in a word for multiple sclerosis? Curr Opin Neurol. 2005;18:231–235. doi: 10.1097/01.wco.0000169738.06664.3b. [DOI] [PubMed] [Google Scholar]
- 33.Chen J., Zheng H., Liu H., Niu J., Liu J., Shen T., et al. Improving metabolic flux estimation via evolutionary optimization for convex solution space. Bioinformatics. 2007;23:1115–1123. doi: 10.1093/bioinformatics/btm050. [DOI] [PubMed] [Google Scholar]
- 34.Niedenfuhr S., Wiechert W., Noh K. How to measure metabolic fluxes: a taxonomic guide for (13)C fluxomics. Curr Opin Biotechnol. 2015;34:82–90. doi: 10.1016/j.copbio.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Klein S., Heinzle E. Isotope labeling experiments in metabolomics and fluxomics. Wiley Interdiscip Rev Syst Biol Med. 2012;4:261–272. doi: 10.1002/wsbm.1167. [DOI] [PubMed] [Google Scholar]
- 36.Cortassa S., Caceres V., Bell L.N., O'Rourke B., Paolocci N., Aon M.A. From metabolomics to fluxomics: a computational procedure to translate metabolite profiles into metabolic fluxes. Biophys J. 2015;108:163–172. doi: 10.1016/j.bpj.2014.11.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cajka T., Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem. 2016;88:524–545. doi: 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths W.J., Koal T., Wang Y., Kohl M., Enot D.P., Deigner H.P. Targeted metabolomics for biomarker discovery. Angew Chem Int Ed Engl. 2010;49:5426–5445. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- 39.Schrimpe-Rutledge A.C., Codreanu S.G., Sherrod S.D., McLean J.A. Untargeted metabolomics strategies-challenges and emerging directions. J Am Soc Mass Spectrom. 2016;27:1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoessel D., Stellmann J.P., Willing A., Behrens B., Rosenkranz S.C., Hodecker S.C., et al. Metabolomic profiles for primary progressive multiple sclerosis stratification and disease course monitoring. Front Hum Neurosci. 2018;12:226. doi: 10.3389/fnhum.2018.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nourbakhsh B., Bhargava P., Tremlett H., Hart J., Graves J., Waubant E. Altered tryptophan metabolism is associated with pediatric multiple sclerosis risk and course. Ann Clin Transl Neurol. 2018;5:1211–1221. doi: 10.1002/acn3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buck D., Hemmer B. Biomarkers of treatment response in multiple sclerosis. Expert Rev Neurother. 2014;14:165–172. doi: 10.1586/14737175.2014.874289. [DOI] [PubMed] [Google Scholar]
- 43.Lawton K.A., Brown M.V., Alexander D., Li Z., Wulff J.E., Lawson R., et al. Plasma metabolomic biomarker panel to distinguish patients with amyotrophic lateral sclerosis from disease mimics. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15:362–370. doi: 10.3109/21678421.2014.908311. [DOI] [PubMed] [Google Scholar]
- 44.Villoslada P., Baranzini S. Data integration and systems biology approaches for biomarker discovery: challenges and opportunities for multiple sclerosis. J Neuroimmunol. 2012;248:58–65. doi: 10.1016/j.jneuroim.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Blanchet L., Smolinska A., Attali A., Stoop M.P., Ampt K.A., van Aken H., et al. Fusion of metabolomics and proteomics data for biomarkers discovery: case study on the experimental autoimmune encephalomyelitis. BMC Bioinformatics. 2011;12:254. doi: 10.1186/1471-2105-12-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wishart D.S., Tzur D., Knox C., Eisner R., Guo A.C., Young N., et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia J., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith C.A., O'Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R., et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 50.Lin Z., Sun X., Xie H., Zhang T., Zu X., Qiao L., et al. Plasma metabolomics coupled with MetaboAnalyst and Ingenuity Pathway Analysis characterizes linoleic acid metabolism disorder in patients with spleen-yang-deficiency syndrome. European Journal of Integrative Medicine. 2018;19:72–79. [Google Scholar]
- 51.Del Boccio P., Rossi C., di Ioia M., Cicalini I., Sacchetta P., Pieragostino D. Integration of metabolomics and proteomics in multiple sclerosis: from biomarkers discovery to personalized medicine. Proteomics Clin Appl. 2016;10:470–484. doi: 10.1002/prca.201500083. [DOI] [PubMed] [Google Scholar]
- 52.Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G., et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17:162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- 53.Oh J., Vidal-Jordana A., Montalban X. Multiple sclerosis: clinical aspects. Curr Opin Neurol. 2018;31:752–759. doi: 10.1097/WCO.0000000000000622. [DOI] [PubMed] [Google Scholar]
- 54.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Nature Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 55.Segal M.B. Extracellular and cerebrospinal fluids. J Inherit Metab Dis. 1993;16:617–638. doi: 10.1007/BF00711896. [DOI] [PubMed] [Google Scholar]
- 56.Andersson M., Alvarez-Cermeno J., Bernardi G., Cogato I., Fredman P., Frederiksen J., et al. Cerebrospinal fluid in the diagnosis of multiple sclerosis: a consensus report. J Neurol Neurosurg Psychiatry. 1994;57:897–902. doi: 10.1136/jnnp.57.8.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stangel M., Fredrikson S., Meinl E., Petzold A., Stuve O., Tumani H. The utility of cerebrospinal fluid analysis in patients with multiple sclerosis. Nat Rev Neurol. 2013;9:267–276. doi: 10.1038/nrneurol.2013.41. [DOI] [PubMed] [Google Scholar]
- 58.Maurer M.H. Proteomics of brain extracellular fluid (ECF) and cerebrospinal fluid (CSF) Mass Spectrom Rev. 2010;29:17–28. doi: 10.1002/mas.20213. [DOI] [PubMed] [Google Scholar]
- 59.Quinones M.P., Kaddurah-Daouk R. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis. 2009;35:165–176. doi: 10.1016/j.nbd.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 60.Wishart D.S., Lewis M.J., Morrissey J.A., Flegel M.D., Jeroncic K., Xiong Y., et al. The human cerebrospinal fluid metabolome. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:164–173. doi: 10.1016/j.jchromb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Reinke S.N., Broadhurst D.L., Sykes B.D., Baker G.B., Catz I., Warren K.G., et al. Metabolomic profiling in multiple sclerosis: insights into biomarkers and pathogenesis. Mult Scler. 2014;20:1396–1400. doi: 10.1177/1352458513516528. [DOI] [PubMed] [Google Scholar]
- 62.Lutz N.W., Viola A., Malikova I., Confort-Gouny S., Audoin B., Ranjeva J.P., et al. Inflammatory multiple-sclerosis plaques generate characteristic metabolic profiles in cerebrospinal fluid. PLoS One. 2007;2:e595. doi: 10.1371/journal.pone.0000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herman S., Åkerfeldt T., Spjuth O., Burman J., Kultima K. Biochemical differences in cerebrospinal fluid between secondary progressive and relapsing–remitting multiple sclerosis. Cells. 2019;8:84. doi: 10.3390/cells8020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Markianos M., Koutsis G., Evangelopoulos M.E., Mandellos D., Karahalios G., Sfagos C. Relationship of CSF neurotransmitter metabolite levels to disease severity and disability in multiple sclerosis. J Neurochem. 2009;108:158–164. doi: 10.1111/j.1471-4159.2008.05750.x. [DOI] [PubMed] [Google Scholar]
- 65.Herman S., Khoonsari P.E., Tolf A., Steinmetz J., Zetterberg H., Akerfeldt T., et al. Integration of magnetic resonance imaging and protein and metabolite CSF measurements to enable early diagnosis of secondary progressive multiple sclerosis. Theranostics. 2018;8:4477–4490. doi: 10.7150/thno.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Psychogios N., Hau D.D., Peng J., Guo A.C., Mandal R., Bouatra S., et al. The human serum metabolome. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stringer K.A., Younger J.G., McHugh C., Yeomans L., Finkel M.A., Puskarich M.A., et al. Whole blood reveals more metabolic detail of the human metabolome than serum as measured by 1H-NMR spectroscopy: implications for sepsis metabolomics. Shock. 2015;44:200–208. doi: 10.1097/SHK.0000000000000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon J.H., Li D., Traboulsee A., Coyle P.K., Arnold D.L., Barkhof F., et al. Standardized MR imaging protocol for multiple sclerosis: consortium of MS Centers consensus guidelines. AJNR Am J Neuroradiol. 2006;27:455–461. [PMC free article] [PubMed] [Google Scholar]
- 69.Poddighe S., Murgia F., Lorefice L., Liggi S., Cocco E., Marrosu M.G., et al. Metabolomic analysis identifies altered metabolic pathways in Multiple Sclerosis. Int J Biochem Cell Biol. 2017;93:148–155. doi: 10.1016/j.biocel.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 70.Jurynczyk M., Probert F., Yeo T., Tackley G., Claridge T.D.W., Cavey A., et al. Metabolomics reveals distinct, antibody-independent, molecular signatures of MS, AQP4-antibody and MOG-antibody disease. Acta Neuropathol Commun. 2017;5:95. doi: 10.1186/s40478-017-0495-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Villoslada P., Alonso C., Agirrezabal I., Kotelnikova E., Zubizarreta I., Pulido-Valdeolivas I., et al. Metabolomic signatures associated with disease severity in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2017;4:e321. doi: 10.1212/NXI.0000000000000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim C.K., Bilgin A., Lovejoy D.B., Tan V., Bustamante S., Taylor B.V., et al. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci Rep. 2017;7:41473. doi: 10.1038/srep41473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lorefice L., Murgia F., Fenu G., Frau J., Coghe G., Murru M.R., et al. Assessing the metabolomic profile of multiple sclerosis patients treated with interferon beta 1a by (1)H-NMR spectroscopy. Neurotherapeutics. 2019;16:797–807. doi: 10.1007/s13311-019-00721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campbell B.M., Charych E., Lee A.W., Moller T. Kynurenines in CNS disease: regulation by inflammatory cytokines. Front Neurosci. 2014;8:12. doi: 10.3389/fnins.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedraz-Petrozzi B., Elyamany O., Rummel C., Mulert C. Effects of inflammation on the kynurenine pathway in schizophrenia - a systematic review. J Neuroinflammation. 2020;17:56. doi: 10.1186/s12974-020-1721-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lovelace M.D., Varney B., Sundaram G., Franco N.F., Ng M.L., Pai S., et al. Current evidence for a role of the kynurenine pathway of tryptophan metabolism in multiple sclerosis. Front Immunol. 2016;7:246. doi: 10.3389/fimmu.2016.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwidzinski E., Bechmann I. Ido expression in the brain: a double-edged sword. J Mol Med. 2007;85:1351–1359. doi: 10.1007/s00109-007-0229-7. [DOI] [PubMed] [Google Scholar]
- 78.Levi I., Gurevich M., Perlman G., Magalashvili D., Menascu S., Bar N., et al. Potential role of indolelactate and butyrate in multiple sclerosis revealed by integrated microbiome-metabolome analysis. Cell Rep Med. 2021;2:100246. doi: 10.1016/j.xcrm.2021.100246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fitzgerald K.C., Smith M.D., Sotirchos E.S., Kornberg M.D., Douglas M., Nourbakhsh B., et al. Multi-omic evaluation of metabolic alterations in multiple sclerosis identifies shifts in aromatic amino acid metabolism. Cell Rep Med. 2021;2:100424. doi: 10.1016/j.xcrm.2021.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singhal N.K., Freeman E., Arning E., Wasek B., Clements R., Sheppard C., et al. Dysregulation of methionine metabolism in multiple sclerosis. Neurochem Int. 2018;112:1–4. doi: 10.1016/j.neuint.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 81.Zhao M., Wu J., Li X., Gao Y. Early urinary candidate biomarkers in a rat model of experimental autoimmune encephalomyelitis. BioRxiv. 2017 doi: 10.1098/rsos.230118. 205294 [Preprint][cited 2019 Feb 2]:[26p] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gebregiworgis T., Nielsen H.H., Massilamany C., Gangaplara A., Reddy J., Illes Z., et al. A urinary metabolic signature for multiple sclerosis and neuromyelitis optica. J Proteome Res. 2016;15:659–666. doi: 10.1021/acs.jproteome.5b01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.’t Hart B.A., Vogels J.T., Spijksma G., Brok H.P., Polman C., van der Greef J. 1H-NMR spectroscopy combined with pattern recognition analysis reveals characteristic chemical patterns in urines of MS patients and non-human primates with MS-like disease. J Neurol Sci. 2003;212:21–30. doi: 10.1016/s0022-510x(03)00080-7. [DOI] [PubMed] [Google Scholar]
- 84.Montano N., Papacci F., Cioni B., Di Bonaventura R., Meglio M. What is the best treatment of drug-resistant trigeminal neuralgia in patients affected by multiple sclerosis? A literature analysis of surgical procedures. Clin Neurol Neurosurg. 2013;115:567–572. doi: 10.1016/j.clineuro.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 85.Vingara L.K., Yu H.J., Wagshul M.E., Serafin D., Christodoulou C., Pelczer I., et al. Metabolomic approach to human brain spectroscopy identifies associations between clinical features and the frontal lobe metabolome in multiple sclerosis. Neuroimage. 2013;82:586–594. doi: 10.1016/j.neuroimage.2013.05.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Constantinescu C.S., Farooqi N., O'Brien K., Gran B. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS) Br J Pharmacol. 2011;164:1079–1106. doi: 10.1111/j.1476-5381.2011.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mix E., Meyer-Rienecker H., Hartung H.P., Zettl U.K. Animal models of multiple sclerosis--potentials and limitations. Prog Neurobiol. 2010;92:386–404. doi: 10.1016/j.pneurobio.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mangalam A., Poisson L., Nemutlu E., Datta I., Denic A., Dzeja P., et al. Profile of circulatory metabolites in a relapsing-remitting animal model of multiple sclerosis using global metabolomics. J Clin Cell Immunol. 2013;4 doi: 10.4172/2155-9899.1000150. 10.4172/2155-9899.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lassmann H., Bradl M. Multiple sclerosis: experimental models and reality. Acta Neuropathol. 2017;133:223–244. doi: 10.1007/s00401-016-1631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lassmann H. Pathogenic mechanisms associated with different clinical courses of multiple sclerosis. Front Immunol. 2019;9:3116. doi: 10.3389/fimmu.2018.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noga M.J., Dane A., Shi S., Attali A., van Aken H., Suidgeest E., et al. Metabolomics of cerebrospinal fluid reveals changes in the central nervous system metabolism in a rat model of multiple sclerosis. Metabolomics. 2012;8:253–263. doi: 10.1007/s11306-011-0306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee G., Hasan M., Kwon O.S., Jung B.H. Identification of altered metabolic pathways during disease progression in EAE mice via metabolomics and lipidomics. Neuroscience. 2019;416:74–87. doi: 10.1016/j.neuroscience.2019.07.029. [DOI] [PubMed] [Google Scholar]
- 93.Bernardes D., Oliveira-Lima O.C., da Silva T.V., Juliano M.A., dos Santos D.M., Carvalho-Tavares J. Metabolic alterations in experimental autoimmune encephalomyelitis in mice: effects of prior physical exercise. Neurophysiology. 2016;48:117–121. [Google Scholar]
- 94.Dickens A.M., Larkin J.R., Davis B.G., Griffin J.L., Claridge T.D., Sibson N.R., et al. NMR-based metabolomics separates the distinct stages of disease in a chronic relapsing model of multiple sclerosis. J Neuroimmune Pharmacol. 2015;10:435–444. doi: 10.1007/s11481-015-9622-0. [DOI] [PubMed] [Google Scholar]
- 95.Singh J., Cerghet M., Poisson L.M., Datta I., Labuzek K., Suhail H., et al. Urinary and plasma metabolomics identify the distinct metabolic profile of disease state in chronic mouse model of multiple sclerosis. J Neuroimmune Pharmacol. 2019;14:241–250. doi: 10.1007/s11481-018-9815-4. [DOI] [PubMed] [Google Scholar]
- 96.Battini S., Bund C., Moussallieh F.M., Cicek A.E., De Seze J., Namer I.J. Metabolomics approaches in experimental allergic encephalomyelitis. J Neuroimmunol. 2018;314:94–100. doi: 10.1016/j.jneuroim.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 97.Poisson L.M., Suhail H., Singh J., Datta I., Denic A., Labuzek K., et al. Untargeted plasma metabolomics identifies endogenous metabolite with drug-like properties in chronic animal model of multiple sclerosis. J Biol Chem. 2015;290:30697–30712. doi: 10.1074/jbc.M115.679068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao C., Du H., Xu L., Wang J., Tang L., Cao Y., et al. Metabolomic analysis revealed glycylglycine accumulation in astrocytes after methionine enkephalin administration exhibiting neuron protective effects. J Pharm Biomed Anal. 2015;115:48–54. doi: 10.1016/j.jpba.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 99.Bhargava P., Fitzgerald K.C., Calabresi P.A., Mowry E.M. Metabolic alterations in multiple sclerosis and the impact of vitamin D supplementation. JCI Insight. 2017;2 doi: 10.1172/jci.insight.95302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chevalier A.C., Rosenberger T.A. Increasing acetyl-CoA metabolism attenuates injury and alters spinal cord lipid content in mice subjected to experimental autoimmune encephalomyelitis. J Neurochem. 2017;141:721–737. doi: 10.1111/jnc.14032. [DOI] [PubMed] [Google Scholar]
- 101.Lazzarino G., Amorini A.M., Petzold A., Gasperini C., Ruggieri S., Quartuccio M.E., et al. Serum compounds of energy metabolism impairment are related to disability, disease course and neuroimaging in multiple sclerosis. Mol Neurobiol. 2017;54:7520–7533. doi: 10.1007/s12035-016-0257-9. [DOI] [PubMed] [Google Scholar]
- 102.Mehrpour M., Kyani A., Tafazzoli M., Fathi F., Joghataie M.T. A metabonomics investigation of multiple sclerosis by nuclear magnetic resonance. Magn Reson Chem. 2013;51:102–109. doi: 10.1002/mrc.3915. [DOI] [PubMed] [Google Scholar]
- 103.Simone I.L., Federico F., Trojano M., Tortorella C., Liguori M., Giannini P., et al. High resolution proton MR spectroscopy of cerebrospinal fluid in MS patients. Comparison with biochemical changes in demyelinating plaques. J Neurol Sci. 1996;144:182–190. doi: 10.1016/s0022-510x(96)00224-9. [DOI] [PubMed] [Google Scholar]
- 104.Kim H.H., Jeong I.H., Hyun J.S., Kong B.S., Kim H.J., Park S.J. Metabolomic profiling of CSF in multiple sclerosis and neuromyelitis optica spectrum disorder by nuclear magnetic resonance. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhargava P., Fitzgerald K.C., Venkata S.L.V., Smith M.D., Kornberg M.D., Mowry E.M., et al. Dimethyl fumarate treatment induces lipid metabolism alterations that are linked to immunological changes. Ann Clin Transl Neurol. 2019;6:33–45. doi: 10.1002/acn3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kasakin M.F., Rogachev A.D., Predtechenskaya E.V., Zaigraev V.J., Koval V.V., Pokrovsky A.G. Targeted metabolomics approach for identification of relapsing-remitting multiple sclerosis markers and evaluation of diagnostic models. Medchemcomm. 2019;10:1803–1809. doi: 10.1039/c9md00253g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that there is no data to display.