Abstract
Photosynthetic bacteria respond to alterations in light conditions by migrating to locations that allows optimal use of light as an energy source. Studies have indicated that photosynthesis-driven electron transport functions as an attractant signal for motility among purple photosynthetic bacteria. However, it is unclear just how the motility-based signal transduction system monitors electron flow through photosynthesis-driven electron transport. Recently, we have demonstrated that the purple photosynthetic bacterium Rhodospirillum centenum is capable of rapidly moving swarm cell colonies toward infrared light as well as away from visible light. Light-driven colony motility of R. centenum has allowed us to perform genetic dissection of the signaling pathway that affects photosynthesis-driven motility. In this study, we have undertaken sequence and mutational analyses of one of the components of a signal transduction pathway, Ptr, which appears responsible for transmitting a signal from the photosynthesis-driven electron transport chain to the chemotaxis signal transduction cascade. Mutational analysis demonstrates that cells disrupted for ptr are defective in altering motility in response to light, as well as defective in light-dependent release of methanol. We present a model which proposes that Ptr senses the redox state of a component in the photosynthetic cyclic electron transport chain and that Ptr is responsible for transmitting a signal to the chemotaxis machinery to induce a photosynthesis-dependent motility response.
Quantity and quality of light are important environmental factors that all photosynthetic organisms respond to. Plant cells use light as a signal for controlling many photomorphogenic processes such as chloroplast and seedling maturation and development. Motile unicellular microorganisms such as algae, cyanobacteria, and anoxygenic (non-oxygen-evolving) photosynthetic bacteria are capable of altering swimming behavior in response to light. These cells utilize photosensory behavior as a tool to migrate to locations that are optimal for photosynthetic growth.
Photosensory behavioral responses displayed by microorganisms generally fall into two categories (reviewed in reference 28). One response is phototaxis, which is movement toward or away from a light source using a mechanism(s) that involves measuring the direction of propagation of a specific light beam. A second motility behavior is the scotophobic response, which involves a random alteration in the direction of movement when motile cells experience a decrease in light intensity or cross a light/dark border. Algae and cyanobacteria typically display both phototactic and scotophobic responses, while anoxygenic photosynthetic bacteria usually display only a scotophobic response. The process of phototaxis has been extensively studied in several different algal species where specific light-absorbing photoreceptors are used to mediate photosensory responses (28). Detailed genetic and biochemical analyses of the archaeon Halobacterium salinarium also indicate that this species uses specific light-absorbing photoreceptors to mediate a scotophobic photosensory response (reviewed in references 13 and 28).
No detailed molecular genetic or biochemical characterization of the phototactic or scotophobic photosensory apparatus from prokaryotic cells has been published. There is evidence that the scotophobic response exhibited by prokaryotic purple bacteria is tightly coupled with photosynthesis-driven electron transport. For example, studies with Rhodospirillum rubrum (12), Rhodobacter sphaeroides (1, 2), and Rhodospirillum centenum (18) have established that a functional photosynthetic electron transport chain is required for a scotophobic response. Studies with chemical inhibitors of the electron transport chain also indicate that R. sphaeroides responds to a change in the rate of cyclic electron transport of the photosystem rather than to alterations of membrane potential or proton motive force (9, 10). It is also known that the chemotaxis cascade in purple bacteria constitutes a downstream signaling pathway that transduces the photosynthesis-driven photoresponse to the flagellar motor (16, 17, 26). This indicates that the purple bacterial photosensory response involves integrating information of photosynthetic electron transport with that of chemosensory responses. It also suggests that one or more components of the purple bacterial photosensory machinery may have structural features in common with chemotaxis receptors.
Recently, the purple bacterium R. centenum was shown to exhibit a unique photosensory behavior that allows an efficient genetic screen for photosensing-defective mutants. In liquid growth medium, this species forms swim cells with a single polar flagella that exhibit a classic scotophobic response. However, when R. centenum is grown on an agar-solidified medium, the cells differentiate into multiflagellated swarm cells that are capable of exhibiting a visible phototactic response en masse as colonies (24, 25). By visually screening for swarm colonies that are unable to respond to light, we have been able to obtain a collection of mutants that are defective in photosensory behavior (18). In this study, we report the cloning, sequencing, and mutational analysis of a gene, which we have named ptr (for phototransducer), that appears responsible for transducing a signal from the photosynthesis-driven electron transport chain to the chemotactic signal transduction cascade. Sequence analysis reveals that Ptr has a high degree of sequence homology to chemoreceptors. A targeted chromosomal disruption of ptr results in a specific defect in the ability of both swim and swarm cells of R. centenum to perceive both visible and infrared light.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The wild-type R. centenum strain ATCC 51752 (8, 25) and a ptr disrupted strain, ZJF6-4, were used in this study. Cells were cultured photosynthetically in CENS medium or aerobically in CENS or PYVS medium (25). Escherichia coli strain XL1-Blue (Stratagene) was used for routine DNA cloning purposes and grown in Luria-Bertani medium at 37°C. When needed for plasmid selection, appropriate antibiotics were used in E. coli strains at concentrations of 150 μg/ml for ampicillin, 50 μg/ml for spectinomycin, and 10 μg/ml for gentamicin. For R. centenum strains, spectinomycin was used at 10 μg/ml and kanamycin was used at 40 μg/ml on agar plates. Light-driven phototactic colony migration assays were conducted on 0.8% PYVS agar plates as described previously (25).
DNA manipulation.
All restriction and DNA modification enzymes were purchased from New England Biolabs and used according to the manufacturer's instructions. Standard molecular methods were followed as described by Sambrook et al. (29). A clone of the mini-Tn5-Spr (spectinomycin resistance gene)-disrupted ptr gene was constructed by isolating genomic DNA from R. centenum strain ZJF6-4 as described by Jiang et al. (18). The chromosomal DNA was digested with NcoI, dephosphorylated with alkaline phosphatase, and ligated into plasmid vector pZJD7 that was also digested with NcoI. The ligation reaction product was then transformed into E. coli strain XL1-Blue with recombinant plasmid pPRCN, obtained by selecting for both ampicillin resistance and Spr. The DNA insertion in plasmid pPRCN is about 6.7 kb, which brings the total length of sequence flanking the interposon to about 4.7 kb.
One targeted mutant in the ptr locus (ZJCΔptr1) was constructed using a suicide vector to promote allelic replacement through recombination (Fig. 1A). A plasmid containing deletion of ptr (pPRCNΔ1) was constructed as follows. First, plasmid pPRCN (described above) was digested with SfiI, which removed the mini-Tn5 interposon Spr omega cassette as well as all of the ptr coding sequence between the interposon insertion site and the ptr stop codon. Second, a previously described nonpolar Spr gene cassette (17) was ligated with SfiI-digested pPRCN to give rise to plasmid pPRCNΔ1. An NcoI fragment from pPRCNΔ1 that contained the nonpolar Spr gene was then ligated to pGmLacZ (17) to give rise to pGm-PRCNΔ1, which was transformed in E. coli S17-1 (λ pir) for mating with wild-type R. centenum. Transconjugants were selected on CENS plates with kanamycin, spectinomycin, and 5-bromo-4-chloroindolyl-β-d-galactopyranoside (X-Gal) as an indicator dye. The putative deletion mutants LacZ− Spr Gms were confirmed by PCR analysis with relevant primers (data not shown).
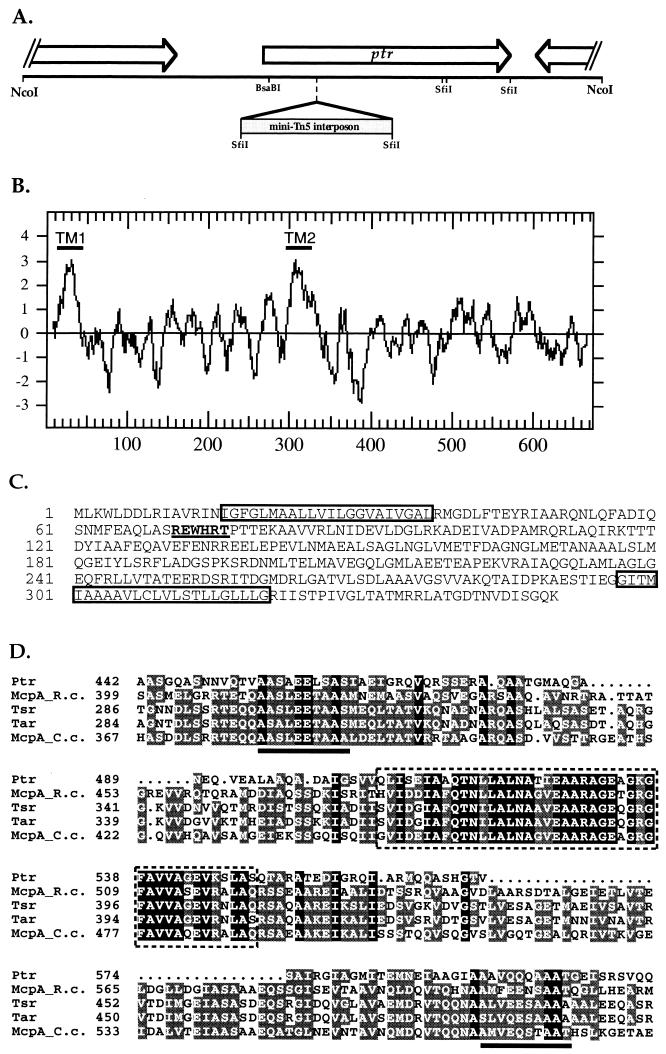
FIG. 1.
(A) Restriction and gene map of the ptr locus. ORFs are depicted as boxes with arrows. Two partial ORFs flank the ptr gene. The mini-Tn5 interposon insertion site in mutant ZJF6-4 is indicated. (B) Hydrophobicity plot map of Ptr. The two peaks of high hydrophobicity labeled TM1 and TM2 designate two putative transmembrane domains. (C) N-terminal sequence of Ptr. The two putative transmembrane domains are boxed, and the putative b-type heme-binding motif is underlined and in boldface. (D) Sequence alignment of carboxyl-terminal sequences of Ptr and known chemoreceptors. The putative methylation sites are underlined, and the highly conserved domain is boxed with dashed lines. Tsr and Tar, E. coli serine and aspartate chemoreceptors, respectively; McpA_R.c., McpA from Rhodospirillum centenum; McpA_C.c., McpA from Caulobacter crescentus.
DNA sequencing.
Double-stranded DNA sequencing was accomplished by primer walking using an ABI automated DNA sequencer (Applied Biosystems model 373; Perkin-Elmer). Sequence data were analyzed and assembled with the Sequencher program (Gene Codes Corporation, Ann Arbor, Mich.).
Methanol evolution assay.
Release of 3H-labeled methanol from R. centenum cells that undergo a decrease or increase in light intensity was measured using a modified volatile methanol evolution assay as described originally by Kehry et al. (19) and Kirby et al. (20). Cells were grown photosynthetically in minimal CENMED medium containing 0.2 mM l-methionine to approximately 200 Klett-Summerson photometer units (red filter no. 66). The cells were then washed three times with CENMED containing chloramphenicol (0.1 mg/ml) and suspended in the same medium to a final density of 500 Klett-Summerson units. Six milliliters of washed cells was then placed in a glass screw-cap tube containing 60 μCi of l-[methyl-3H]methionine (80 Ci/mmol, 5 μCi/μl; Amersham Life Science) and incubated for 2.5 h with a tungsten light source emitting a light intensity of 100 μW/cm2. After incubation, the cells were washed with 15 ml of CENMED medium containing 0.2 mM l-methionine and then loaded into a translucent 32-mm-diameter 0.45-μm-pore-size Acrodisc (Gelman Science) to form a cell filter paste. The cell filter paste was then attached to a Pharmacia high-pressure liquid chromatograph and washed continuously under low illumination (<5 μW/cm2) for 20 min with 40°C CENMED medium containing 0.2 mM l-methionine at a flow rate of 1.0 ml/min. A step-down light response was achieved by an increase in the infrared light on the cell filter paste to 100 μW/cm2 for 20 s followed by a shift-down to low-illumination conditions. Uncapped Eppendorf tubes containing 0.5-ml fractions of the cell filter paste flowthrough were then placed into 20-ml glass scintillation vials that contained 8 ml of nonaqueous scintillation fluid (Amersham Life Science). Volatile methanol released from the Eppendorf tube was then trapped in the scintillation fluid by incubating tightly capped scintillation vials at room temperature for 24 h. The Eppendorf tubes were then removed from the scintillation vials, and the scintillation fluid containing trapped methanol was quantified in a scintillation counter (model TRI-CARB2100TR; Packard Instrument Company, Downers Grove, Ill.).
Nucleotide sequence accession number.
The nucleotide sequence for ptr in R. centenum has been deposited in the GenBank database under accession number AF064528.
RESULTS
Isolation and characterization of the ptr gene.
In a previous study, we described the isolation of a collection of mini-Tn5-Spr mutants that exhibited an absence of scotophobic and phototactic behavior in response to changes in infrared and/or visible light intensity (18). One mutant strain, ZJF6-4, is particularly intriguing since both swim and swarm cells of this mutant are defective in responding to changes in both visible and infrared light. Unlike chemotactic mutants of R. centenum (17), this mutant strain exhibits normal chemotaxis to pyruvate and acetate, indicating that the chemotaxis machinery is intact. This finding, in combination with the fact that the strain exhibits normal photosynthetic growth, leads us to conclude that this mutant strain contains a specific defect in a gene that governs both visible and infrared photosensory responses. To analyze the gene disrupted in ZJF6-4, we constructed a genomic library from which we obtained a clone containing the mini-Tn5-Spr and flanking regions by selecting for resistance to spectinomycin. Sequence analysis of the cloned fragment revealed that the mini-Tn5-Spr transposon disrupted an open reading frame (ORF) containing a 671-amino-acid, 71-kDa protein (Fig. 1A). Since the mini-Tn5-Spr-disrupted gene appears to specifically affect the photosensory signaling pathway (see below), we have named the disrupted gene ptr for phototransducer.
A search of the GenBank database with the translated ptr sequence reveals a high degree of homology to a number of eubacterial and archaeal chemotaxis receptors (Fig. 1). Most conspicuous is a highly conserved carboxyl-terminal domain extending from amino acid residues 508 to 550 that in chemoreceptors is known to participate in binding of the chemotactic signaling molecule CheW (Fig. 1D) (23). Flanking this region are two putative CheR/CheB methylation/demethylation sites (Fig. 1D). One notable alteration is that the distance between the methylation/demethylation sites and the CheW/CheA docking site is approximately 45 amino acids shorter in Ptr than in other eubacterial methyl-accepting chemotaxis proteins (MCPs) (Fig. 1D). A hydrophobicity plot of Ptr (Fig. 1B) also indicated the presence of two putative transmembrane domains at the amino terminus, with one extending from amino acid residues 16 to 37 and the other extending from residues 297 to 320. Assuming that Ptr serves a receptor function similar to that of other known MCPs, the 260-amino-acid region between the two putative transmembrane domains could function as a sensory domain that is localized to the periplasm. In this context, it is interesting that the putative periplasmic loop contains a sequence motif, Glu-Trp-His-Arg (starting at residue 72), that is nearly identical to a b-type heme-binding sequence present in cytochrome b6 (Fig. 1C) (7).
In addition to ptr, the clone also contains a partial ORF of 400 amino acid residues that has sequence similarity to 3-methylcrotonyl coenzyme A carboxylase and propionyl coenzyme A carboxylase from various plant and bacterial sources. This ORF is located 600 bp upstream of, and transcribed in the same direction as, ptr (Fig. 1A). A second partial ORF (151 amino acid residues) with an unknown function is also present in the clone that is located downstream of ptr. This ORF is transcribed in the direction opposite ptr transcription (Fig. 1A).
Functional characterization of the ptr locus.
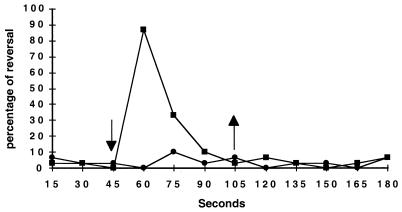
To confirm that the mini-Tn5-Spr disruption of ptr is responsible for the phenotype observed with strain ZJF6-4, we constructed a targeted chromosomal deletion of ptr using allelic exchange, creating the mutant ZYCΔptr1 (see Materials and Methods). As observed for the original ZJF6-4 mutant, ZYCΔptr1 displayed normal wild-type photosynthetic growth and a normal chemotactic response to pyruvate and serine (data not shown). To quantitatively evaluate the swim cell scotophobic photoresponse, we recorded the individual rotational movement of cells attached to a microscope slide and measured their response to a decrease in light intensity (17). The observed average percentage of 30 independently tracked cells of wild-type and ZJCΔptr1 mutant cells during a 3 min interval is shown in Fig. 2. After recording rotation direction during a 45-s interval, we subjected the cells to a 75% decrease in light intensity by inserting a neutral density filter into the microscope's light beam. After 105 s, the cells were returned to the original high light intensity. As indicated in Fig. 2, the proportion of wild-type cells responding to a reduction in light intensity increased from a background of ∼3% to more than 85%. In contrast, the percentage of ZYJΔptr1 cells that reversed remained at background levels, indicating that disruption of ptr results in a loss of the scotophobic reversal response. There was also no increased reversal frequency of either wild-type or ZYJΔptr1 cells to an increase in light intensity, which confirms previous observations that a scotophobic reversal response is observed only during a decrease in light intensity. However, an obvious increase of speed of rotation was observed for both strains upon the increase in light intensity (a process known as photokinesis), which is presumably caused by a light-driven increase in photosynthesis-generated membrane potential (11).
FIG. 2.
Reversal frequencies of swimming cells of wild-type (■) and ZJCΔptr1 mutant (●) R. centenum in response to a decrease in light intensity (↓) and an increase in light intensity to the original level (↑).
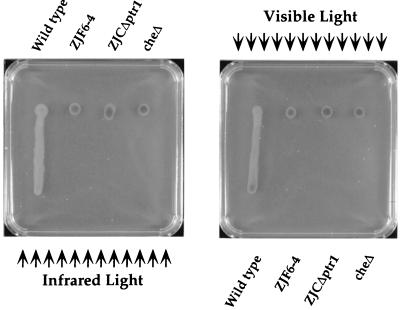
We also assayed the effect of deleting ptr on the phototactic colony motility exhibited by swarm cells. As shown in Fig. 3, deletion of ptr resulted in loss of both visible and infrared light-driven colony motility of strain ZYJΔptr1. Microscopic observation of individual cell movement in swarm colonies also indicates no effect of visible or infrared light on individual cell reversal in ZYJΔptr1 (data not shown). In contrast, individual cells in wild-type swarm colonies undergo a rapid loss of motility when subjected to a decrease in light intensity (17).
FIG. 3.
Phototactic colony migration assays of mutants ZJF6-4 and ZJCΔptr1 along with the wild type and a chemotaxis operon deletion mutant (17) as controls. Arrows indicate the direction of light that causes positive (colony as a unit moves toward the light source) and negative (colony moves away from the light source) phototactic responses.
We also performed trans complementation of ZYCΔptr1 with a plasmid-encoded copy of ptr. For this analysis, we constructed a replicating plasmid that contained the ptr gene along with 570 bp of intergenic DNA upstream of ptr. When this plasmid was mated into ZYCΔptr1, the strain regained normal scotophobic as well as phototactic responses to visible and infrared light during swim and swarm cell phases, respectively (data not shown). This confirmed that the observed lack of light-driven motility changes in ZYCΔptr1 is a consequence of disruption of ptr and not a result of polarity effects on neighboring gene expression.
Demethylation of Ptr during adaptation of the scotophobic response.
Interactions of chemical attractants with MCPs are known to cause a transient increase in the methylation state of the MCP receptors. The formation of a specific ligand-receptor interaction is thought to cause a conformational change in MCPs that promotes a subsequent interaction with CheR. CheR then methylates the MCP at three to four sites within two conserved methylation domains. The methyl groups are then released as methanol by phosphorylated CheB, with the rate of demethylation being stimulated by a decrease in attractant level (reviewed in reference 31). The presence of two putative methylation/demethylation sites in Ptr implies that the light-mediated scotophobic and phototactic responses that are mediated by Ptr may involve a methylation/demethylation process similar to that of chemoreceptors.
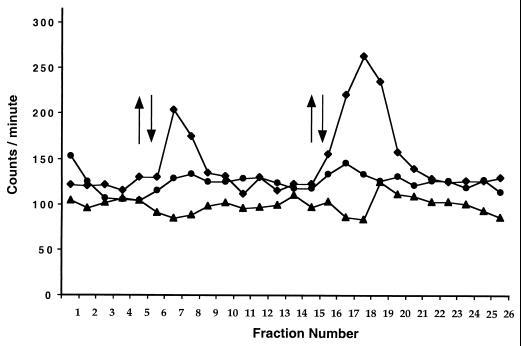
To assay for Ptr methylation, we conducted a light-dependent volatile methanol evolution assay based on previously developed methylation/demethylation assays of chemoreceptors. In this assay, we harvested [3H]methionine-grown cells onto a filter to form a cell paste, continually washed the cell filter paste with unlabeled growth medium to remove unincorporated [3H]methionine, and then subjected it to a step-up followed by a step-down in light intensity, with fractions of the cell filter paste wash assayed for elution of [3H]methanol as described in Materials and Methods. As shown in Fig. 4, wild-type cells subjected to a step-down in infrared light intensity released a burst of volatile methanol into the wash. In contrast, the ZYJΔptr1 mutant strain challenged with a similar step-down in light intensity did not produce methanol (Fig. 4). Indeed, the methanol elution profile of the ptr mutant was very similar to that observed with an R. centenum cheB-disrupted control strain that is incapable of stimulating removal of methyl groups from chemoreceptors (Fig. 4) (17). Also in congruence with motility assays is the absence of a release of [3H]methanol during a step-up in light intensity (Fig. 4). As discussed above, a step-up of light intensity increases the speed of swimming but does not affect the reversal frequency. These results indicate that a step-down in light intensity, which stimulates the scotophobic tumbling response, involves a Ptr-dependent release of methanol in a manner similar to that observed with chemoreceptors.
FIG. 4.
Infrared light-induced release of [3H]methanol with nonradioactive chase. The y axis denotes the measured scintillation counts from tritium-labeled methanol; the x axis denotes the collected fractions. A upward arrow indicates that the light source was turned on and the illumination of cell paste was started; a downward arrow indicates that the light source was switched off. Data for wild-type, cheB mutant, and ZJCΔptr1 mutant strains are represented by diamonds, triangles, and circles, respectively.
DISCUSSION
Even though it was observed over a century ago that photosynthetic bacteria exhibit light-induced alterations in motility (6), there has been little progress in determining how photosynthetic bacteria control photosensory behavior. Early studies by Manten (21) and Clayton (4) established that wavelengths of light that are attractants are also the same as those that are absorbed by the photosystem to promote photosynthesis-driven electron transport in purple photosynthetic bacteria (reviewed in reference 28). The scotophobic response has also been shown to require a functioning photosystem (reaction center), as well as components of the photosynthesis-driven electron transport system such as a functioning cytochrome bc1 complex (18). It has been proposed that the generation of electrons by photosynthesis is the signal that governs phototaxis in anoxygenic photosynthetic bacteria (10). This scenario is in contrast to what is thought to occur in the archaeon H. salinarium, in which separate retinal photoreceptors (sensory rhodopsins I and II) absorb light, resulting in conformational changes of the photoreceptors. Light excited sensory rhodopsin then interacts with MCP-like transducer proteins that interact with the Che proteins (3, 13, 27, 30) in a way not unlike that proposed for Ptr.
To obtain a better understanding of the nature of the purple bacterial photosensory transduction system, we previously performed a genetic screen for R. centenum mutants that are defective in light perception (18). One mutant strain isolated, ZJF6-4, exhibited characteristics of a disruption of a key component in the photosensory signal transduction system. Specifically, ZJF6-4 swim cells were observed to be defective in the scotophobic response, while swarm cells were defective in both positive and negative phototaxis. In this study, we cloned an ORF that was disrupted in ZJF6-4 and demonstrated by sequence analysis that it codes for a protein with a high degree of sequence similarity to bacterial and archaeal chemoreceptors. The observation that Ptr has structural features in common with MCPs clearly strengthens our earlier conclusion that the photosensory signaling pathway in purple photosynthetic bacteria involves the che gene products that are also responsible for mediating chemotaxis (17). Since only limited number of chemicals have been tested on R. centenum for chemotactic response, Ptr may serve an additional role as a bona fide chemoreceptor, similar to the transducer HtrII from H. salinarium, which transmits the blue light signal from sensory rhodopsin II and also serves as a chemoreceptor for serine (14). Presumably, a light-dependent signal perceived by Ptr is integrated into the chemosensory signal transduction cascade via interactions with CheW/CheAY (Fig. 5). Indeed, integration of a signal from the photosystem (i.e., photosynthesis electron flow) with signals derived from chemoreceptors should allow cells the ability to effectively migrate in response to either light or chemical gradients, depending on which signal is dominant. This should allow maximum flexibility for migration to positions conducive to optimal growth.
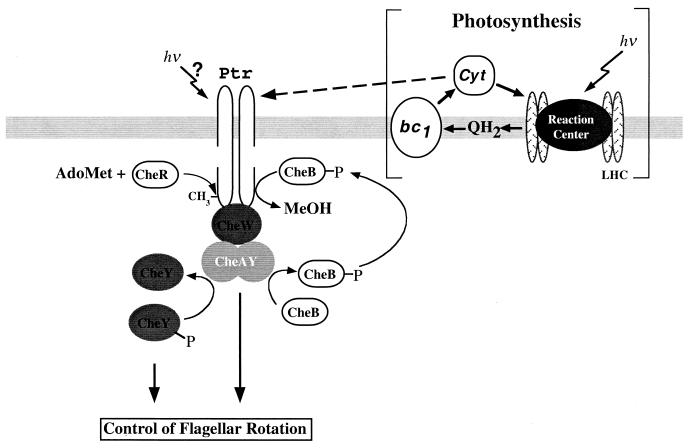
FIG. 5.
Proposed model of Ptr that senses the redox state of cytochrome (Cyt) c in the periplasmic space. LHC, light harvesting center; AdoMet, adenosylmethionine; MeOH, methanol.
Sequence analysis indicates that Ptr exhibits a high degree of identity with other MCPs in the putative cytoplasmic output domain, specifically in the region that interacts with CheW. One noticeable change in the output domain is closer positioning of the two putative methylation domains to the CheW docking domain. In comparison to E. coli Tsr (5), it appears that Ptr is missing the linker region that exists between α helix 7 and the methylated α helix 6; α9 helix, which contains the second methylation domain, is also significantly (approximately 10 helical turns) shorter. It remains to be seen whether these structural alterations are a unique feature of Ptr or are common to other MCPs from R. centenum. Like other chemoreceptors, Ptr does not show a high degree of sequence identity with other MCPs in the putative periplasmic sensory domain. If the periplasmic domain of Ptr monitors the redox state of a component of the photosynthesis-driven electron transport chain, then it may contain a redox-responsive center. In this vein, it is interesting that this domain has the sequence REWHRT, which is similar to the heme b-binding sites (underlined) found in cytochrome b561 from E. coli (KSWHET) and cytochrome b6 from Spinacium oleracea (RSVHRW) (7). Consequently, ptr may contain a heme in the periplasmic sensory domain. In some respects, this would be similar to the case for the heme-containing aerotaxis transducers, a notable difference being that characterized aerotaxis transducers have no periplasmic domains (15).
One puzzle is how disruption of Ptr leads to a loss of both positive (movement toward infrared light) and negative (movement away from visible light) phototactic colony motility of R. centenum. It is known that positive and negative phototaxes require functioning reaction center and cytochrome bc1 complexes, indicating that both processes involve a measure of photosynthesis electron transport (10, 18). It is also known that both infrared and visible light can be absorbed by photopigments and drive photosynthesis electron transport (22). Indeed, the observation that both light types drive photosynthesis and yet lead to such different motility responses indicates that Ptr responds not only to alterations in photosynthesis-driven electron transport but also to electron transport as well as to an additional component(s) that distinguishes between visible or infrared light. Another possibility may be that light directly modifies the activity of Ptr itself. Based on this study and previous studies of photosensory responses in R. centenum and other purple photosynthetic bacteria, a testable model can be proposed. As depicted in Fig. 5, a component of the periplasmic photosynthetic cyclic electron transport chain such as a cytochrome c may reduce a heme bound to the periplasmic loop of Ptr. This could cause a conformational change in Ptr that affects integration with the chemotaxis machinery. At this early stage of molecular analysis of bacterial photoperception, we cannot rule out that other components in the photosystem, such as the reaction center or cytochrome bc1 complex, may interact with Ptr to transmit signals generated by light perception. Clearly, additional characterization of Ptr and other upstream components is needed to determine how these two different types of light are capable of transmitting different signals through Ptr. Direct visualization of light-driven colony motility of R. centenum swarm colonies makes this a good model system to unravel additional details of bacterial photoperception.
ACKNOWLEDGMENTS
We thank members of the Indiana University Photosynthetic Bacteria Group for helpful comments.
This work was supported by funding from NIH GM58050.
REFERENCES
- 1.Armitage J P, Evans M C W. The reaction center in the phototactic and chemotactic responses of Rhodopseudomonas sphaeroides. FEMS Microbiol Lett. 1981;11:89–92. [Google Scholar]
- 2.Armitage J P, Ingham C, Evans M C W. Role of proton motive force in phototactic and aerotactic responses of Rhodopseudomonas sphaeroides. J Bacteriol. 1985;161:967–972. doi: 10.1128/jb.161.3.967-972.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanck A, Oesterhelt D, Ferrando E, Schegk E S, Lottspeich F. Primary structure of sensory rhodopsin I, a prokaryotic photoreceptor. EMBO J. 1989;8:3963–3971. doi: 10.1002/j.1460-2075.1989.tb08579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton R K. Studies in the phototaxis of Rhodospirillum rubrum. III. Quantitative relations between stimulus and response. Arch Microbiol. 1953;19:141–165. doi: 10.1007/BF00446397. [DOI] [PubMed] [Google Scholar]
- 5.Danielson M A, Bass R B, Falke J J. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. J Biol Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelmann T W. Bakterium photometricum. Ein Beitrag zur vergleichenden Physiologie des Licht- und farbensinnes. Pfluegers Arch Gesamte Physiol Menschen Tiere. 1883;30:95–124. [Google Scholar]
- 7.Esposti M D. Prediction and comparison of the haem-binding sites in membrane haemoproteins. Biochim Biophys Acta. 1989;977:249–265. doi: 10.1016/s0005-2728(89)80079-9. [DOI] [PubMed] [Google Scholar]
- 8.Favinger J, Stadtwald R, Gest H. Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Antonie Leeuwenhoek. 1989;55:291–296. doi: 10.1007/BF00393857. [DOI] [PubMed] [Google Scholar]
- 9.Gauden D E, Armitage J P. Electron transport-dependent taxis in Rhodobacter sphaeroides. J Bacteriol. 1995;177:5853–5859. doi: 10.1128/jb.177.20.5853-5859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grishanin R N, Gauden D E, Armitage J P. Photoresponses in Rhodobacter sphaeroides: role of photosynthetic electron transport. J Bacteriol. 1997;179:24–30. doi: 10.1128/jb.179.1.24-30.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häder D P. Photosensory behavior in procaryotes. Microbiol Rev. 1987;51:1–21. doi: 10.1128/mr.51.1.1-21.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harayama S, Iino T. Phototaxis and membrane potential in the photosynthetic bacterium Rhodospirillum rubrum. J Bacteriol. 1977;131:34–41. doi: 10.1128/jb.131.1.34-41.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoff W D, Jung K H, Spudich J L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 14.Hou S, Brooun A, Yu H S, Freitas T, Alam M. Sensory rhodopsin II transducer HtrII is also responsible for serine chemotaxis in the archaeon Halobacterium salinarum. J Bacteriol. 1998;180:1600–1602. doi: 10.1128/jb.180.6.1600-1602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou S, Larsen R W, Boudko D, Riley C W, Karatan E, Zimmer M, Ordal G W, Alam M. Myoglobin-like aerotaxis transducers in archaea and bacteria. Nature. 2000;403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Z-Y, Bauer C E. Analysis of a chemotaxis operon from Rhodospirillum centenum. J Bacteriol. 1997;179:5712–5719. doi: 10.1128/jb.179.18.5712-5719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Z-Y, Gest H, Bauer C E. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J Bacteriol. 1997;179:5720–5727. doi: 10.1128/jb.179.18.5720-5727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Z-Y, Rushing B G, Bai Y, Gest H, Bauer C E. Isolation of Rhodospirillum centenum mutants defective in phototactic colony motility by transposon mutagenesis. J Bacteriol. 1998;180:1248–1255. doi: 10.1128/jb.180.5.1248-1255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehry M R, Doak T G, Dahlquist F W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984;259:11828–11835. [PubMed] [Google Scholar]
- 20.Kirby J R, Kristich C J, Feinberg S L, Ordal G W. Methanol production during chemotaxis to amino acids in Bacillus subtilis. Mol Microbiol. 1997;24:869–878. doi: 10.1046/j.1365-2958.1997.3941759.x. [DOI] [PubMed] [Google Scholar]
- 21.Manten A. Phototaxis in the purple bacterium Rhodospirillum rubrum, and the relation between phototaxis and photosynthesis. Antonie Leeuwenhoek. 1948;14:65–86. doi: 10.1007/BF02272681. [DOI] [PubMed] [Google Scholar]
- 22.Noguchi T, Hayashi H, Tasumi M. Factors controlling the efficiency of energy transfer from carotenoids to bacteriochlorophyll in purple photosynthetic bacteria. Biochim Biophys Acta. 1990;1017:280–290. [Google Scholar]
- 23.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 24.Ragatz L, Jiang Z-Y, Bauer C E, Gest H. Phototactic purple bacteria. Nature. 1994;370:104. [Google Scholar]
- 25.Ragatz L, Jiang Z-Y, Bauer C E, Gest H. Macroscopic phototactic behavior of the purple photosynthetic bacterium Rhodospirillum centenum. Arch Microbiol. 1995;163:1–6. doi: 10.1007/BF00262196. [DOI] [PubMed] [Google Scholar]
- 26.Romagnoli S, Armitage J P. Roles of chemosensory pathways in transient changes in swimming speed of Rhodobacter sphaeroides induced by changes in photosynthetic electron transport. J Bacteriol. 1999;181:34–39. doi: 10.1128/jb.181.1.34-39.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudolph J, Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 1995;14:667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rushing B G, Jiang Z-Y, Gest H, Bauer C E. Phototactic behavior. In: Lederberg J, editor. Encyclopedia of Microbiology. 2nd ed. Vol. 3. San Diego, Calif: Academic Press; 2000. pp. 618–624. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Seidel R, Scharf B, Gautel M, Kleine K, Oesterhelt D, Engelhard M. The primary structure of sensory rhodopsin II: a member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc Natl Acad Sci USA. 1995;92:3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]