Abstract
Age-related macular degeneration (AMD), the most prevalent visual disorder among the elderly, is confirmed as a multifactorial disease. Studies demonstrated that genetic factors play an essential role in its pathogenesis. Our study aimed to make a relatively comprehensive study about biological functions of AMD related genes and crosstalk of their enriched pathways. 1691 AMD genetic studies were reviewed, GO enrichment and pathway crosstalk analyses were conducted to elucidate the biological features of these genes and to demonstrate the pathways that these genes participate. Moreover, we identified novel AMD-specific genes using shortest path algorithm in the context of human interactome. We retrieved 176 significantly AMD-related genes. GO results showed that the most significant term in each of these three GO categories was: signaling receptor binding (PBH = 4.835 × 10−7), response to oxygen-containing compound (PBH = 2.764 × 10−21), and extracellular space (PBH = 2.081 × 10−19). The pathway enrichment analysis showed that complement pathway is the most enriched. The pathway crosstalk study showed that the pathways could be divided into two main modules. These two modules were connected by cytokine-cytokine receptor interaction pathway. 42 unique genes potentially participating AMD development were obtained. The aberrant expression of the mRNA of FASN and LRP1 were validated in AMD cell and mouse models. Collectively, our study carried out a comprehensive analysis based on genetic association study of AMD and put forward several evidence-based genes for future study of AMD.
Keywords: GO analyses, pathway crosstalk, gene identification, AMD, lipid metabolism
Introduction
Age-related macular degeneration (AMD) is a major cause of irreversible blindness and visual impairment in the elderly of industrialized countries (Gehrs et al., 2006; Klein et al., 2011). AMD leads to progressive central vision loss because of macular atrophy and choroidal neovascularization (Lambert et al., 2016). Currently, no efficient medical or surgical treatment is available for geographic atrophy (GA), also known as the “dry” form of AMD, while anti-vascular endothelial growth factor (VEGF) therapies have been used for treating neovascular AMD, also known as the “wet” form (Campa and Harding, 2011). As one of the most severe eye diseases, the mechanisms of AMD pathogenesis remain elusive.
In the past several decades, researches have demonstrated that AMD is a multi-factorial disease. Both genetic and environmental factors influence the development of AMD. Many risk factors have been confirmed to contribute to AMD progression, including aging, smoking, oxidative stress, sunlight exposure, and genetic factors (Lambert et al., 2016). Identification of risk factors has become one of the main aspects of AMD research in recent years due to their strong correlation with prevalence of AMD. One study showed that the risk of developing late AMD was increased approximately 4-fold for those with a family history of AMD (Smith and Mitchell, 1998). Also, numerous studies about gene polymorphism have been carried out. They have elucidated a lot different genetically susceptive factors for AMD, such as complement factor H (CFH) (Klein et al., 2013), Apolipoprotein E (APOE) (McKay et al., 2011), vascular endothelial growth factor (VEGF) (Miller et al., 2013), and hepatic lipase (LIPC) (Neale et al., 2010). Despite considerable success in deciphering AMD genetic risk factors, the intact mechanism is still veiled. Recently, a meta-analysis of genome-wide association studies (GWAS) for advanced AMD estimated that currently identified loci account for nearly 55% of the heritability of advanced AMD (Yu et al., 2011). On the one hand, a complicated disease tends to be influenced by lots of genes with small or mild effects rather than one or two major genes with large effects. A comprehensive analysis of potentially causal genes within a pathway and/or a network framework might provide some important insights beyond the conventional single-gene analyses (Goeman and Buhlmann, 2007; Glazko and Emmert-Streib, 2009; Jia et al., 2011b; Hu et al., 2017). On the other hand, the disease proteins always tend to interact with each other instead of scattering randomly in the human interactome and form one or several connected subgraphs (Xu and Li, 2006; Goh et al., 2007; Feldman et al., 2008). So, identification of existing AMD-related genes and delineation of the AMD subnetwork may enable us to predict the potential AMD-associated genes, which provide us a more thorough understanding of AMD pathogenesis.
In this study, we firstly established a relatively ample collection of genes genetically associated with AMD. Then, we performed functional enrichment analyses to identify the significant gene ontology (GO) terms and pathways within these retrieved genes. To further explore the pathogenesis of AMD in a more specific manner, we analyzed the crosstalk of AMD-related pathways. Moreover, AMD-associated subnetwork was extracted using shortest path algorithm in the context of the human protein-protein interactome. Subsequently, we made a prediction of candidate genes based on the betweenness in the AMD-specific network. This study provides insights in pathogenesis of AMD and contributes to identify novel genes related with AMD.
Materials and methods
Identification of AMD-Related genes
Candidate genes associated with AMD were collected by retrieving the human genetic association studies deposited in PUBMED (http://www.ncbi.nlm.nih.gov/pubmed/). Similar with references (Sullivan et al., 2004; Hu et al., 2017), we searched for studies about AMD with the term (age-related macular degeneration [MeSH]) and (polymorphism [MeSH] or genotype [MeSH] or alleles [MeSH]) not (neoplasms [MeSH]). By 4 January 2020, a total of 1,691 publications were retrieved for the disorder. We reviewed the abstract of all 1,691 publications to select genetic association studies of AMD. Among the selected publications, we only focused on the genes that are statistically significantly related to the incidence of AMD. Moreover, we reviewed the full report of publications that contain significant association to ensure the conclusion was supported by the research. After reviewing, we incorporated those genes into our study and set up a gene collection named AMDgset.
Functional enrichment analysis of AMD-Related genes
The functional feature of the AMD-related genes were analyzed by ToppGene (Chen et al., 2009). ToppGene is a web-based system that contains information from different resources and is able to be used in detecting the biological themes out of the candidate gene lists, including evaluating the enrichment significance of GO terms. Here, we employed the criterion that only the GO terms of biological processes with both p value and false discovery rate (FDR) value smaller than 0.05 were accepted as the significantly enriched GO term. p values were calculated with Fisher’s exact test and FDR values were performed by Benjamini and Hochberg (BH) method (P BH ). Due to the advantages of combining multi-databases, ToppGene was also selected to analyze the pathways enriched in the candidate genes. Basically, we uploaded the genes with their symbols and/or corresponding NCBI Entrez Gene IDs into the server and compared with the genes included in each canonical pathway based on the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg) and Biocarta (www.biocarta.com) pathway databases. All the pathways contained two or more candidate genes were extracted, with each of them assigned a p value to denote overlap significance between the pathway and the input genes via Fisher’s exact test. Thereafter, we only considered the pathways with FDR value less than 0.05 as significantly enriched pathways. FDR values were also performed by BH method (P BH ).
Pathway crosstalk analysis
Crosstalk analysis between pathways was evaluated by the Jaccard Coefficient (JC) = and the Overlap Coefficient (OC) = , where A and B is the list of genes included in the two tested pathways. Here we administrate the following procedure to establish the pathway crosstalk:
1. Select a set of pathways for crosstalk analysis. Only the pathways with P BH value less than 0.05 were used. Meanwhile, pathways containing less than two candidate genes were removed because pathways with too few genes might have insufficient biological information.
2. Count the number of shared candidate genes between any pair of pathways. Pathway pair with less than two overlapped genes was removed.
3. Calculate the overlap of all pathway pairs and rank them. All the pathway pairs were ranked according to their JC and OC value.
4. Visualize the selected pathway crosstalk with the software Cytoscape [35].
Identification of AMD-specific genes based on human interactome
The disease proteins (the products of disease genes) are not dispersed randomly in the interactome, but tend to interact with each other, forming one or several connected subinteractome that we call the disease module. A total of 176 genes were already included in AMD disease module in our study. To identify novel AMD-related genes, we firstly adopted a relatively complete human interactome from a recent study which contained 138,427 physical interactions between 13,460 proteins, including protein-protein and regulatory interactions, metabolic pathway interactions, and kinase-substrate interactions (Menche et al., 2015). Secondly, Subnet, a Java-based stand-alone program for extracting subnetworks using the pairwise K-shortest path algorithms, was employed to extract AMD-specific genes (Lemetre et al., 2013). Here, we used the concept of betweenness (the number of shortest paths connect all pairs of genes in AMDgset and the path should contain a given gene as an inner gene) to evaluate novel AMD associated genes. It is possible that genes with high betweenness may participate more pathological processes of AMD than those with low betweenness. As a gene in a given network, its betweenness may be influenced by the primary structure of the network. For instance, the cut-vertex of the network may always have high betweenness regardless of the distribution of known genes, therefore, a permutation test was conducted to eliminate this phenomenon. We randomly selected the same number of genes as the number of AMDgset from human interactome 100 times and recalculated the shortest paths between these randomly selected genes. The permutation FDR of the shortest path genes was defined as.
FDR i = , where betweennessactual and betweennessrandom was the number of shortest paths that across gene i among AMDgset and randomly selected genes respectively. Count (betweennessactual > betweennessrandom) denoted the count of times when betweennessrandom was greater than betweennessactual. According to Jiang et al.’s work, only genes with betweennessactual > 1,000 and FDR <0.05 were included. Besides, significant AMD specific genes should meet the criteria that count (betweennessrandom) < 50 so that we could furtherly exclude hub genes in the background network (Jiang et al., 2013).
Cell culture
Adult human RPE cell line ARPE-19 cell was purchased from MEISENCTCC company (Hangzhou, China). DMEM/F12 culture media (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, United States), 100 U/mL penicillin and 100 mg/ml streptomycin was used in cell culture. All cells were incubated at 37°C under an atmosphere of 5% CO2. For further analysis, cells were seeded in 6- or 96-well plate as needed.
Cell viability assay
After the Sodium iodate (SI, Sigma-aldrich, San Francisco, CA, United States) treatment, the cell viability was measured with CCK-8 kit (Yeasen, Shanghai, China) according to the manufacturer’s protocol then was detected with a microplate reader (BioTek, VT, United States). Propidium Iodide (PI) staining assay was also used to evaluate the cell viability. Briefly, after treatment, cells were incubated with PI (10 μg/ml) and Hoechst for 10 min before imaging at 550 nm.
Mice
C57BL/6J male mice (6–8 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology (Beijing, China). The animal experiments were all performed according to the ARRIVE guidelines and the ARVO Statement for the Use of Animals in Ophthalmic and Vision. All animal experiments were authorized by the ethical committee of Shanghai 10th People’s Hospital. All animals were given free access to food and drinking water. Mice were housed in a pathogen-free room with constant temperature (22°C) under a 12 h light-dark cycle. SI was dissolved in sterile saline at the concentrations of 4 mg/ml. The solution was given as a single dose at the concentration of 40 mg/kg intraperitoneally. The mice were sacrificed after 2 days.
Hematoxylin and eosin staining
The mice were sacrificed after 2 days and eyes were fixed in 4% paraformaldehyde for 24 h. After fixation, paraffin-embedded serially sections of 3 μm were cut carefully and then stained with hematoxylin-eosin (H&E). Photos of the sections were taken using an upright light microscope (Leica Microsystems).
Quantitative PCR
After treatment, total RNA was extracted by EZ-press RNA purification Kit (Roseville, MN, United States) and RNA concentration was determined with NanoDrop 3,300 (Thermo Fisher Scientific). cDNA was synthesized from 1 μg of total RNA using HiScript III first Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The qPCR analysis was performed using ChamQ universal SYBR qPCR Master Mix (Vazyme). The contents of different mRNA targets in different groups were calculated by ΔΔCt method. Primers were synthesized by Sangon Biotech (Sangon Biotech, Shanghai, China). Primers used in the experiments were as follows: human APOA1 (F: 5′- CCCTGGGATCGAGTGAAGGA-3′; R: 5′- CTGGGACACATAGTCTCTGCC-3′), human FASN (F: 5′- AAGGACCTGTCTAGGTTTGATGC-3′; R: 5′- TGGCTTCATAGGTGACTTCCA-3′), human ABCG5 (F: 5′- TGGACCAGGCAGATCCTCAAA-3′; R: 5′- CCGTTCACATACACCTCCCC-3′), human LRP1 (F: 5′- CTATCGACGCCCCTAAGACTT-3′; R: 5′- CATCGCTGGGCCTTACTCT-3′), mouse APOA1 (F: 5′- CTTGGCACGTATGGCAGCA-3′; R: 5′- CCAGAAGTCCCGAGTCAATGG-3′), mouse FASN (F: 5′- GGAGGTGGTGATAGCCGGTAT-3′; R: 5′- TGGGTAATCCATAGAGCCCAG-3′), mouse ABCG5 (F: 5′- AGAGGGCCTCACATCAACAGA-3′; R: 5′- CTGACGCTGTAGGACACATGC-3′), mouse LRP1 (F: 5′- CCACTATGGATGCCCCTAAAAC-3′; R: 5′- GCAATCTCTTTCACCGTCACA-3′), human NCK1 (F: 5′- CAACATGCCCGCTTATGTGAA-3′; R: 5′- CATGACGATCACCTTTGTCCC-3′), human PTPN11 (F: 5′- GAACTGTGCAGATCCTACCTCT-3′; R: 5′- TCTGGCTCTCTCGTACAAGAAA-3′), human PNN (F: 5′- GTCGCCGTGAGAACTTTGC-3′; R: 5′- GGTCCTCCTCCACTATCTGAGA-3′), human CNGB1 (F: 5′- GGACCCCTCGGAAGACCAA-3′; R: 5′- CTCAGGATTCGGTTCTGGTTC-3′).
Statistical analysis
Each experiment was repeated at least thrice. Graphpad Prism 9 was used to perform statistical analyses. All data was expressed as the mean ± SEM, statistical differences were determined by Student’s t-test for comparison between two groups. p < 0.05 was considered to be statistically significant.
Results
Retrieve of genes reported to Be associated with AMD
With the criteria described above, publications showing significant association of gene(s) with the disease were collected; those insignificant results were excluded. A detailed list of genes that have been reported to be significantly associated with AMD is provided in Table 1. We constructed a gene set (referred to as AMD-related genes gene set (AMDgset)) which contains 176 genes significantly associated with AMD. Among them, the complement family (C2, C3, C9, CFH, CFHR1, CFHR2) contained the maximum members and was considered to play a pivotal role in AMD pathogenesis. AMDgset also contained cytochrome proteins (CYP1A2, CYP46A1, CYP2R1), vascular endothelial growth factor A (VEGFA), and anti-oxidative proteins (SOD2, SOD3), which are highly associated with intraretinal environment. At the meantime, some other proteins such as collagen family (COL4A3, COL8A1, COL10A1, COL15A1), matrix metallopeptidase (MMP2, MMP9, MMP20), and toll like receptor (TLR2, TLR3, TLR4) were also reported to be associated with AMD. Our results showed the diversity of AMD related genes and indicated the multifactorial characteristic of AMD in terms of genetics.
TABLE 1.
Genes retrieved from human genetic association studies.
| Gene Symbol | Gene ID | Full Name |
|---|---|---|
| ABCG1 | 9619 | ATP binding cassette subfamily G member 1 |
| ABCG8 | 64241 | ATP binding cassette subfamily G member 8 |
| ABHD2 | 11057 | abhydrolase domain containing 2 |
| ACAD10 | 80724 | acyl-CoA dehydrogenase family member 10 |
| ACE | 1636 | angiotensin I converting enzyme |
| ADAMTS9 | 56999 | ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| ALDH3A2 | 224 | aldehyde dehydrogenase 3 family member A2 |
| ANGPT2 | 285 | angiopoietin 2 |
| APOE | 348 | apolipoprotein E |
| ARHGAP21 | 57584 | Rho GTPase activating protein 21 |
| ARMS2 | 387715 | age-related maculopathy susceptibility 2 |
| ASPM | 259266 | abnormal spindle microtubule assembly |
| B3GLCT | 145173 | beta 3-glucosyltransferase |
| BCO1 | 53630 | beta-carotene oxygenase 1 |
| BCO2 | 83875 | beta-carotene oxygenase 2 |
| C2 | 717 | complement C2 |
| C20orf85 | 128602 | chromosome 20 open reading frame 85 |
| C3 | 718 | complement C3 |
| C4A | 720 | complement C4A (Rodgers blood group) |
| C6orf223 | 221416 | chromosome 6 open reading frame 223 |
| C9 | 735 | complement C9 |
| CACNG3 | 10368 | calcium voltage-gated channel auxiliary subunit gamma 3 |
| CAPN5 | 726 | calpain 5 |
| CATSPER2 | 117155 | cation channel sperm associated 2 |
| CCL2 | 6347 | C-C motif chemokine ligand 2 |
| CCR2 | 729230 | C-C motif chemokine receptor 2 |
| CCR3 | 1232 | C-C motif chemokine receptor 3 |
| CD36 | 948 | CD36 molecule |
| CD63 | 967 | CD63 molecule |
| CETP | 1071 | cholesteryl ester transfer protein |
| CFB | 629 | complement factor B |
| CFD | 1675 | complement factor D |
| CFH | 3075 | complement factor H |
| CFHR1 | 3078 | complement factor H related 1 |
| CFHR2 | 3080 | complement factor H related 2 |
| CFHR3 | 10878 | complement factor H related 3 |
| CFHR4 | 10877 | complement factor H related 4 |
| CFHR5 | 81494 | complement factor H related 5 |
| CFI | 3426 | complement factor I |
| CLUL1 | 27098 | clusterin like 1 |
| CNN2 | 1256 | calponin 2 |
| COL10A1 | 1300 | collagen type X alpha 1 chain |
| COL15A1 | 1306 | collagen type XV alpha 1 chain |
| COL4A3 | 1285 | collagen type IV alpha 3 chain |
| COL8A1 | 1295 | collagen type VIII alpha 1 chain |
| CRP | 1401 | C-reactive protein [Homo sapiens |
| CST3 | 1471 | cystatin C |
| CTRB1 | 1504 | chymotrypsinogen B1 |
| CTRB2 | 440387 | chymotrypsinogen B2 |
| CX3CR1 | 13051 | chemokine (C-X3-C motif) receptor 1 |
| CXCL8 | 3576 | C-X-C motif chemokine ligand 8 |
| CYP1A2 | 1544 | cytochrome P450 family 1 subfamily A member 2 |
| CYP2R1 | 120227 | cytochrome P450 family 2 subfamily R member 1 |
| CYP46A1 | 10858 | cytochrome P450 family 46 subfamily A member 1 |
| DAPL1 | 92196 | death associated protein like 1 |
| DDR1 | 780 | discoidin domain receptor tyrosine kinase 1 |
| ELN | 2006 | elastin |
| ELOVL4 | 6785 | ELOVL fatty acid elongase 4 |
| ERCC2 | 2068 | ERCC excision repair 2, TFIIH core complex helicase subunit |
| ERCC6 | 2074 | ERCC excision repair 6, chromatin remodeling factor |
| ESR1 | 2099 | estrogen receptor 1 |
| F13B | 2165 | coagulation factor XIII B chain |
| FADS1 | 3992 | fatty acid desaturase 1 |
| FADS2 | 9415 | fatty acid desaturase 2 |
| FBLN5 | 10516 | fibulin 5 |
| FCGR2A | 2212 | Fc fragment of IgG receptor IIa |
| FGD6 | 55785 | FYVE, RhoGEF and PH domain containing 6 |
| FGL1 | 2267 | fibrinogen like 1 |
| FILIP1L | 11259 | filamin A interacting protein 1 like |
| FKBPL | 63943 | FK506 binding protein like |
| FLT1 | 2321 | fms related tyrosine kinase 1 |
| FPR1 | 2357 | formyl peptide receptor 1 |
| FRK | 2444 | fyn related Src family tyrosine kinase |
| GAS6 | 2621 | growth arrest specific 6 |
| GPX1 | 2876 | glutathione peroxidase 1 |
| GPX3 | 2878 | glutathione peroxidase 3 |
| GRK5 | 2869 | G protein-coupled receptor kinase 5 |
| GSTM1 | 2944 | glutathione S-transferase mu 1 |
| HLA-B | 3106 | major histocompatibility complex, class I, B |
| HLA-C | 3017 | major histocompatibility complex, class I, C |
| HLA-DQB1 | 3119 | major histocompatibility complex, class II, DQ beta 1 |
| HMCN1 | 83872 | hemicentin 1 |
| HMOX1 | 3162 | heme oxygenase 1 |
| HMOX2 | 3163 | heme oxygenase 2 |
| HTRA1 | 5654 | HtrA serine peptidase 1 |
| IER3 | 8870 | immediate early response 3 |
| IGF1R | 3480 | insulin like growth factor 1 receptor |
| IL17A | 3605 | interleukin 17A |
| IL17RC | 84818 | interleukin 17 receptor C |
| IL1B | 3553 | interleukin 1 beta |
| KCTD10 | 83892 | potassium channel tetramerization domain containing 10 |
| KDR | 3791 | kinase insert domain receptor |
| KMT2E | 55904 | lysine methyltransferase 2E |
| LIPC | 3990 | lipase C, hepatic type |
| LOXL1 | 4016 | lysyl oxidase like 1 |
| LRP6 | 4040 | LDL receptor related protein 6 |
| MALL | 7851 | mal, T cell differentiation protein like |
| MMP2 | 4313 | matrix metallopeptidase 2 |
| MMP20 | 9313 | matrix metallopeptidase 20 |
| MMP9 | 4318 | matrix metallopeptidase 9 |
| MRPL10 | 124995 | mitochondrial ribosomal protein L10 |
| MT2A | 4502 | metallothionein 2A |
| MTHFR | 4524 | methylenetetrahydrofolate reductase |
| MTR | 4548 | 5-methyltetrahydrofolate-homocysteine methyltransferase |
| MYRIP | 25924 | myosin VIIA and Rab interacting protein |
| NFE2L2 | 4780 | nuclear factor, erythroid 2 like 2 |
| NOS2 | 4843 | nitric oxide synthase 2 |
| NOS3 | 4846 | nitric oxide synthase 3 |
| NPC1L1 | 29881 | NPC1 like intracellular cholesterol transporter 1 |
| NPHP1 | 4867 | nephrocystin 1 |
| NPLOC4 | 55666 | NPL4 homolog, ubiquitin recognition factor |
| NQO1 | 1728 | NAD(P)H quinone dehydrogenase 1 |
| OSBP2 | 23762 | oxysterol binding protein 2 |
| P2RX4 | 5025 | purinergic receptor P2X 4 |
| P2RX7 | 5027 | purinergic receptor P2X 7 |
| PGF | 5228 | placental growth factor |
| PILRA | 29992 | paired immunoglobin like type 2 receptor alpha |
| PILRB | 29990 | paired immunoglobin like type 2 receptor beta |
| PLEKHA1 | 59338 | pleckstrin homology domain containing A1 |
| PON1 | 5444 | paraoxonase 1 |
| PPARG | 5468 | peroxisome proliferator activated receptor gamma |
| PPARGC1A | 10891 | PPARG coactivator 1 alpha |
| PRKDC | 5591 | protein kinase, DNA-activated, catalytic polypeptide |
| PRKN | 5071 | parkin RBR E3 ubiquitin protein ligase |
| PRLR | 5618 | prolactin receptor |
| PTCHD3 | 374308 | patched domain containing 3 |
| RAD51 | 5888 | RAD51recombinase |
| RAD51B | 5890 | RAD51 paralog B |
| RDH5 | 5959 | retinol dehydrogenase 5 |
| RGS10 | 6001 | regulator of G protein signaling 10 |
| RHO | 6010 | rhodopsin [Homo sapiens |
| RLBP1 | 6017 | retinaldehyde binding protein 1 |
| ROBO1 | 6091 | roundabout guidance receptor 1 |
| RORA | 6095 | RAR related orphan receptor A |
| RORB | 6096 | RAR related orphan receptor B |
| RXRA | 6256 | retinoid X receptor alpha |
| SCARB1 | 949 | scavenger receptor class B member 1 |
| SELP | 6403 | selectin P |
| SERPINF1 | 5176 | serpin family F member 1 |
| SERPING1 | 710 | serpin family G member 1 |
| SIRT1 | 23411 | sirtuin 1 |
| SKIV2L | 6499 | Ski2 like RNA helicase |
| SLC16A8 | 23539 | solute carrier family 16 member 8 |
| SLC44A4 | 80736 | solute carrier family 44 member 4 |
| SMUG1 | 23583 | single-strand-selective monofunctional uracil-DNA glycosylase |
| SOD2 | 6648 | superoxide dismutase 2 |
| SOD3 | 6649 | superoxide dismutase 3 |
| SPEF2 | 79925 | sperm flagellar 2 |
| SRPK2 | 6733 | SRSF protein kinase 2 |
| STRC | 161497 | stereocilin |
| SYN3 | 8224 | synapsin III |
| TF | 7018 | transferrin |
| TFR2 | 7036 | transferrin receptor 2 |
| TFRC | 7037 | transferrin receptor |
| TGFBR1 | 7046 | transforming growth factor beta receptor 1 |
| TIMP3 | 7078 | TIMP metallopeptidase inhibitor 3 |
| TLR2 | 7097 | toll like receptor 2 |
| TLR3 | 7098 | toll like receptor 3 |
| TLR4 | 7099 | toll like receptor 4 |
| TMEM97 | 27346 | transmembrane protein 97 |
| TNF | 7124 | tumor necrosis factor |
| TNFRSF10A | 8797 | TNF receptor superfamily member 10a |
| TNMD | 64102 | tenomodulin |
| TNXB | 7148 | tenascin XB |
| TRPM1 | 4308 | transient receptor potential cation channel subfamily M member 1 |
| TRPM3 | 80036 | transient receptor potential cation channel subfamily M member 3 |
| TSPAN10 | 83882 | tetraspanin 10 |
| UBE3D | 90025 | ubiquitin protein ligase E3D |
| UNG | 7374 | uracil DNA glycosylase |
| VDR | 7421 | vitamin D receptor |
| VEGFA | 7422 | vascular endothelial growth factor A |
| VLDLR | 7436 | very low density lipoprotein receptor |
| VTN | 7448 | vitronectin |
| ZBTB41 | 226470 | zinc finger and BTB domain containing 41 |
Gene ontology enrichment analysis
To reveal a more specifically functional feature of these genes, we performed GO enrichment analysis with ToppGene and incorporated the top 10 GO terms of each category (Table 2). Results showed that the most significant term in each of these three GO categories was: signaling receptor binding (PBH = 4.835 × 10−7), response to oxygen-containing compound (PBH = 2.764 × 10−21), and extracellular space (PBH = 2.081 × 10−19), respectively (Figure 1). It has long been presumed that aberration of cytokine-cytokine receptor activation is the main early AMD manifestation as mononuclear phagocytes (MPs) are observed on large drusen (Combadiere et al., 2007). Moreover, immunostaining of central retinal pigment epithelium (RPE) flatmounts reveal that IBA-1+ MPs and CCR2+ monocytes (Mos), can be detected within geographic zone and on drusen, are seldom present in healthy age-matched central donor RPE (Sennlaub et al., 2013; Eandi et al., 2016). These atypical appearances of monocytes can be explained by a combination of abnormal signaling receptor binding, including age-related increase of CCL2, deficiency of CX3CL1 as well as pro-inflammatory pattern of interleukins (Guillonneau et al., 2017). We also noticed that lipid (e.g., protein-lipid complex binding, lipoprotein particle binding, lipid binding), oxidative (e.g., response to oxygen-containing compound, reactive oxygen species metabolic process) and extracellular matrix (ECM) (e.g., ECM, ECM component, proteinaceous ECM) related GO terms were enriched in the genes of AMDgset. These results were in accordance with previous researches which demonstrated lipid deposition, oxidative stress, and ECM alteration played prominent roles in AMD pathogenesis (Nita et al., 2014; Jun et al., 2019). Our GO results indicated the AMDgset is relatively reliable for subsequent analysis.
TABLE 2.
Gene Ontology (GO) terms enriched with AMDgset (Top 10 terms).
| Go terms | P a | P BH b | Observed |
|---|---|---|---|
| Molecular Function | |||
| GO:0005102: signaling receptor binding | 5.783×10-10 | 4.835×10-7 | 41 |
| GO:0071814: protein-lipid complex binding | 2.408×10-9 | 5.919×10-8 | 7 |
| GO:0071813: lipoprotein particle binding | 2.408×10-9 | 6.711×10-7 | 7 |
| GO:0008289: lipid binding | 1.89×10-8 | 6.711×10-7 | 24 |
| GO:1901681: sulfur compound binding | 1.019×10-7 | 3.949×10-6 | 14 |
| GO:0017127: cholesterol transporter activity | 1.045×10-7 | 1.455×10-5 | 5 |
| GO:0060089: molecular transducer activity | 1.247×10-7 | 1.455×10-5 | 38 |
| GO:0038023: signaling receptor activity | 1.571×10-7 | 1.49×10-5 | 34 |
| GO:0034185: apolipoprotein binding | 2.246×10-7 | 1.642×10-5 | 5 |
| GO:0032934: sterol binding | 2.282×10-7 | 1.823×10-5 | 7 |
| Biological Process | |||
| GO:1901700: response to oxygen-containing compound | 5.695×10-25 | 2.764×10-21 | 64 |
| GO:0009611: response to wounding | 3.818×10-24 | 9.267×10-21 | 50 |
| GO:1903034: regulation of response to wounding | 8.509×10-21 | 1.377×10-17 | 34 |
| GO:0050727: regulation of inflammatory response | 8.855×10-20 | 1.075×10-16 | 29 |
| GO:0006954: inflammatory response | 1.297×10-19 | 1.259×10-16 | 39 |
| GO:0032101: regulation of response to external stimulus | 5.007×10-19 | 4.051×10-16 | 45 |
| GO:0033993: response to lipid | 4.416×10-18 | 2.824×10-15 | 44 |
| GO:0001525: angiogenesis | 4.654×10-18 | 2.824×10-15 | 31 |
| GO:0010035: response to inorganic substance | 1.413×10-17 | 7.622×10-15 | 33 |
| GO:0072593: reactive oxygen species metabolic process | 3.054×10-17 | 1.482×10-14 | 24 |
| Cellular Component | |||
| GO:0005615: extracellular space | 5.038×10-22 | 2.081×10-19 | 57 |
| GO:0009986: cell surface | 6.597×10-13 | 1.362×10-10 | 34 |
| GO:0031012: extracellular matrix | 2.024×10-11 | 2.786×10-9 | 23 |
| GO:0044420: extracellular matrix component | 4.704×10-11 | 4.857×10-9 | 14 |
| GO:0005578: proteinaceous extracellular matrix | 3.289×10-10 | 2.717×10-8 | 20 |
| GO:0009897: external side of plasma membrane | 5.808×10-10 | 3.998×10-8 | 18 |
| GO:0072562: blood microparticle | 7.148×10-10 | 4.217×10-8 | 13 |
| GO:0005604: basement membrane | 5.02×10-9 | 2.592×10-7 | 11 |
| GO:0098552: side of membrane | 5.07×10-8 | 2.327×10-6 | 20 |
| GO:0044433: cytoplasmic vesicle part | 5.038×10-22 | 2.444×10-6 | 22 |
FIGURE 1.
The top 10 GO terms of each category. The GO terms were divided into 3 parts according to cellular component, biological process and molecular function.
Pathway enrichment analysis in AMDgset
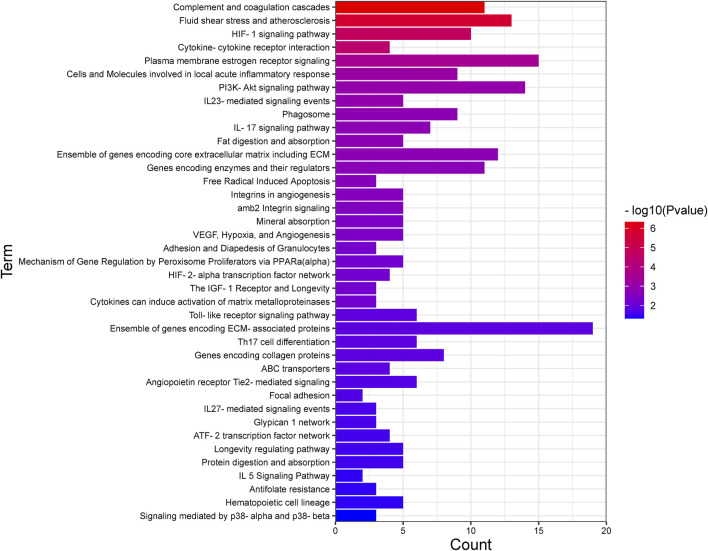
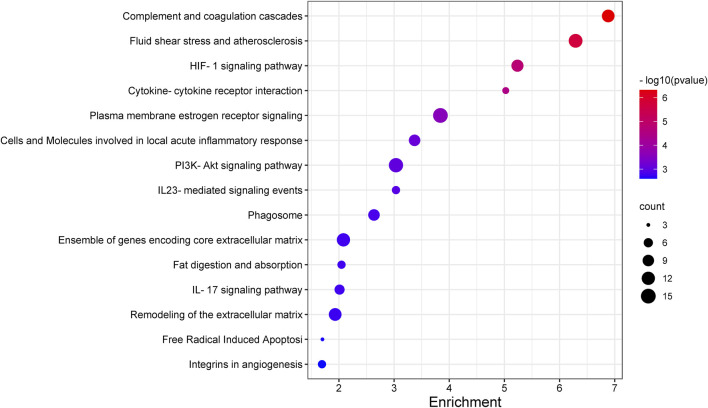
Recognizing the biochemical pathways enriched in the candidate genes will help us to make a better understanding about the specific intracellular signaling related to AMD. We used ToppGene and found 39 significant enrichment pathways for AMD (Figure 2; Table 3). The top 15 pathways were showed in Figure 3. Since numerous complement related genes were included in AMDgset, complement and coagulation cascades pathway was the most significantly enriched pathway in AMDgset. The result suggested the importance of complement system in the pathogenesis of AMD (Despriet et al., 2009; Baas et al., 2010). Also, results showed that IL-23, IL-17, IL-27 and IL-5 mediated signaling pathways were significantly enriched. IL-17 was confirmed to be elevated in the serum of AMD patients. Coughlin et al. demonstrated that IL-17 could mediate the local inflammation augmenting which is triggered by choroidal neovascularization (CNV) lesions (Coughlin et al., 2016). Moreover, consist with GO analysis, the Fat digestion related pathway was testified as enriched pathway, indicating a prominent role of lipid metabolism in the development of AMD. Furthermore, several canonical pathways such as Free Radical Induced Apoptosis pathway (Jarrett and Boulton, 2012) and VEGF, Hypoxia, and Angiogenesis pathway (Bressler, 2009) were verified in our study as well.
FIGURE 2.
All the pathways enriched in AMDgset ranked by significance.
TABLE 3.
Pathways enriched in AMDgset.
| Pathways | P a | P BH b | Genes included in Pathways |
|---|---|---|---|
| Complement and coagulation cascades | 2.404×10–9 | 4.712×10–7 | CFH, VTN, CFI, F13B, CFB, CFD, SERPING1, C2, C3, C4A, C9 |
| Fluid shear stress and atherosclerosis | 1.532×10–8 | 2.002×10–6 | HMOX1, HMOX2, GSTM1, NFE2L2, NQO1, CCL2, KDR, TNF, MMP2, MMP9, IL1B, NOS3, VEGFA |
| HIF–1 signaling pathway | 3.59×10–7 | 1.716×10–5 | FLT1, ANGPT2, HMOX1, TF, TFRC, IGF1R, TLR4, NOS2, NOS3, VEGFA |
| Cytokine–cytokine receptor interaction | 8.867×10–7 | 3.476×10–5 | FLT1, IL17A, IL17RC, TNFRSF10A, CCR2, TGFBR1, CCL2, KDR, CCR3, TNF, IL1B, PRLR, CX3CR1, CXCL8, VEGFA |
| Plasma membrane estrogen receptor signaling | 1.274×10–5 | 2.628×10–4 | ESR1, IGF1R, MMP2, MMP9, NOS3 |
| Cells and Molecules involved in local acute inflammatory response | 4.264×10–5 | 7.268×10–4 | SELP, C3, TNF, CXCL8 |
| PI3K–Akt signaling pathway | 6.555×10–5 | 1.028×10–3 | COL4A3, FLT1, VTN, ANGPT2, PGF, RXRA, IGF1R, TLR2, TLR4, KDR, TNXB, NOS3, PRLR, VEGFA |
| IL23–mediated signaling events | 7.585×10–5 | 1.144×10–3 | IL17A, CCL2, TNF, IL1B, NOS2 |
| Phagosome | 1.009×10–4 | 1.465×10–3 | HLA–B, HLA–DQB1, TFRC, FCGR2A, CD36, SCARB1, TLR2, TLR4, C3 |
| Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans | 1.221×10–4 | 1.668×10–3 | COL4A3, COL8A1, COL10A1, FBLN5, VTN, COL15A1, GAS6, KERA, HMCN1, ELN, FGL1, TNXB |
| Fat digestion and absorption | 1.254×10–4 | 1.668×10–3 | ABCA1, CD36, SCARB1, NPC1L1, ABCG8 |
| IL–17 signaling pathway | 1.277×10–4 | 1.668×10–3 | IL17A, IL17RC, CCL2, TNF, MMP9, IL1B, CXCL8 |
| Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix | 1.426×10–4 | 1.803×10–3 | HTRA1, SERPINF1, MMP20, F13B, TIMP3, ADAMTS9, LOXL1, CST3, SERPING1, MMP2, MMP9 |
| Free Radical Induced Apoptosis | 1.933×10–4 | 2.368×10–3 | GPX1, TNF, CXCL8 |
| Integrins in angiogenesis amb2 Integrin signaling | 2.186×10–4 | 2.521×10–3 | COL4A3, VTN, IGF1R, KDR, VEGFA |
| Mineral absorption | 2.679×10–4 | 3×10–3 | SELP, VTN, TNF, MMP2, MMP9 |
| VEGF, Hypoxia, and Angiogenesis | 3.57×10–4 | 3.782×10–3 | HMOX1, HMOX2, TF, MT2A, VDR |
| Adhesion and Diapedesis of Granulocytes | 3.803×10–4 | 3.822×10–3 | FLT1, KDR, NOS3, VEGFA |
| Mechanism of Gene Regulation by Peroxisome Proliferators via PPARa(alpha) | 5.66×10–4 | 5.283×10–3 | SELP, TNF, CXCL8 |
| Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix | 6.509×10–4 | 5.934×10–3 | RXRA, PPARGC1A, CD36, TNF, NOS2 |
| The IGF–1 Receptor and Longevity | 7.013×10–4 | 6.039×10–3 | ACE, TNF, IL1B |
| HIF–2–alpha transcription factor network | 7.013×10–4 | 6.039×10–3 | IGF1R, SOD2, SOD3 |
| Toll–like receptor signaling pathway | 7.087×10–4 | 6.039×10–3 | FLT1, SIRT1, KDR, VEGFA |
| Ensemble of genes encoding ECM–associated proteins including ECM–affilaited proteins, ECM regulators and secreted factors | 1.749×10–3 | 1.224×10–2 | TLR2, TLR3, TLR4, TNF, IL1B, CXCL8 |
| Pathways | P a | P BH b | Genes included in Pathways |
|---|---|---|---|
| Th17 cell differentiation | 1.842×10–3 | 1.313×10–2 | IL17A, HLA–DQB1, RXRA, TGFBR1, RORA, IL1B |
| Genes encoding collagen proteins | 1.89×10–3 | 1.323×10–2 | COL4A3, COL8A1, COL10A1, COL15A1 |
| ABC transporters | 2.055×10–3 | 1.389×10–2 | ABCA1, ABCA4, ABCG1, ABCG8 |
| Angiopoietin receptor Tie2–mediated signaling | 2.61×10–3 | 1.734×10–2 | ANGPT2, TNF, MMP2, NOS3 |
| Focal adhesion | 2.815×10–3 | 1.839×10–2 | COL4A3, FLT1, VTN, PGF, IGF1R, KDR, TNXB, VEGFA |
| Glypican 1 network | 3.637×10–3 | 2.263×10–2 | FLT1, TGFBR1, VEGFA |
| IL27–mediated signaling events | 3.637×10–3 | 2.263×10–2 | IL17A, TNF, IL1B |
| ATF–2 transcription factor network | 4.287×10–3 | 2.626×10–2 | PPARGC1A, MMP2, NOS2, CXCL8 |
| Longevity regulating pathway | 4.378×10–3 | 2.64×10–2 | PPARG, SIRT1, PPARGC1A, IGF1R, SOD2 |
| Protein digestion and absorption | 4.592×10–3 | 2.727×10–2 | COL4A3, COL10A1, COL15A1, ELN, CTRB1 |
| Antifolate resistance | 6.018×10–3 | 3.511×10–2 | MTHFR, TNF, IL1B |
| IL 5 Signaling Pathway | 6.09×10–3 | 3.511×10–2 | CCR3, IL1B |
| Hematopoietic cell lineage | 6.298×10–3 | 3.578×10–2 | HLA–DQB1, TFRC, CD36, TNF, IL1B |
| Signaling mediated by p38–alpha and p38–beta | 8.462×10–3 | 4.672×10–2 | ESR1, PPARGC1A, NOS2 |
FIGURE 3.
The top 15 pathways enriched in AMDgset.
Crosstalk among significantly enriched pathways
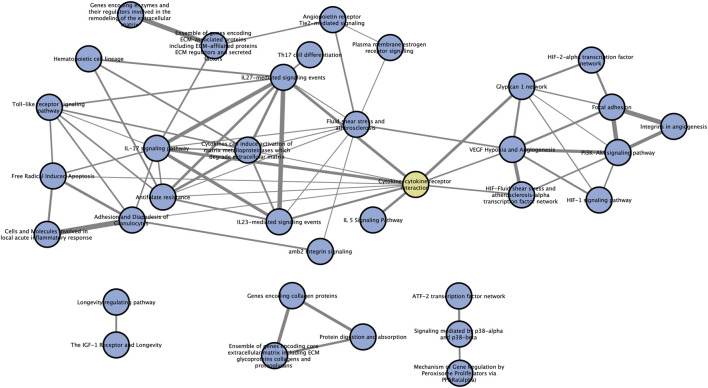
Pathways always exert their functions interactively instead of independently. So, we performed a pathway crosstalk analysis among 39 significantly enriched pathways to elaborate their relationships in this disorder. According to the assumption that two pathways were considered to crosstalk if they shared two or more genes of AMDgset (Jia et al., 2011a), we extracted 142 pathway interactions which met the criterion for crosstalk analysis (Table 4). Then we calculated their overlapping level according to the average score of coefficients JC and OC. Furthermore, to make a brief view of the complicate network of pathway crosstalk, we only chose the top 50% overlapped interactions (edges) and their related pathways (nodes) to build the pathway crosstalk (Figure 4). As it was reflected in our results, the pathways could be grouped into two major modules. Each module contained a relatively centralized crosstalk. This phenomenon indicated that the pathways in the same module might take part in a common biological process. The smaller one mainly contained pathways associated with hypoxia, antioxidation and angiogenesis. The bigger module was consisted of pathways related to immune system, inflammation response and ECM. Moreover, results also clearly showed that the two modules were jointed by cytokine-cytokine receptor interaction pathway instead of operating independently.
TABLE 4.
Pathway crosstalk information.
| Pathway A | Pathway B | Score |
| Cells and Molecules involved in local acute inflammatory response | Adhesion and Diapedesis of Granulocytes Focal adhesion | 0.87500 |
| lntegrins in angiogenesis | Focal adhesion | 0.81250 |
| Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.80556 |
| IL23-mediated signaling events | IL27-mediated signaling events | 0.80000 |
| PI3K-Akt signaling pathway | Focal adhesion | 0.78571 |
| IL-17 signaling pathway | IL27-mediated signaling events | 0.71429 |
| PI3K-Akt signaling pathway | lntegrins in angiogenesis | 0.67857 |
| VEGF Hypoxia and Angiogenesis | HIF-Fluid shear stress and atherosclerosis-alpha transcription factor network | 0.67500 |
| Ensemble of genes encoding core extracellular matrix including ECM glycoproteins collagens and proteoglycans | Genes encoding collagen proteins | 0.66667 |
| IL23-mediated signaling events | ||
| PI3K-Akt signaling pathway | IL-17 signaling pathway | 0.65000 |
| Genes encoding collagen proteins | VEGF Hypoxia and Angiogenesis | 0.64286 |
| Cytokine-cytokine receptor interaction | Protein digestion and absorption | 0.62500 |
| Cytokine-cytokine receptor interaction | IL-17 signaling pathway | 0.61607 |
| Cytokine-cytokine receptor interaction | IL27-mediated signaling events | 0.60000 |
| Free Radical Induced Apoptosis | Glypican 1 network | 0.60000 |
| Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | Adhesion and Diapedesis of Granulocytes | 0.58333 |
| Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | IL27-mediated signaling events | 0.58333 |
| IL27-mediated signaling events | Antifolate resistance | 0.58333 |
| Cytokine-cytokine receptor interaction | ||
| Ensemble of genes encoding core extracellular matrix including ECM glycoproteins collagens and proteoglycans | Antifolate resistance | 0.58333 |
| VEGF Hypoxia and Angiogenesis | IL 5 Signaling Pathway | 0.56667 |
| HIF-Fluid shear stress and atherosclerosis alpha transcription factor network | Protein digestion and absorption | 0.55385 |
| Cells and Molecules involved in local acute inflammatory response | Focal adhesion | 0.54167 |
| VEGF Hypoxia and Angiogenesis | Focal adhesion | 0.54167 |
| HIF-Fluid shear stress and atherosclerosis alpha transcription factor network | Free Radical Induced Apoptosis | 0.53333 |
| ATF-Fluid shear stress and atherosclerosis transcription factor network | Glypican 1 network | 0.53333 |
| Cytokine-cytokine receptor interaction | Glypican 1 network | 0.53333 |
| HIF-1 signaling pathway | Signaling mediated by p38-alpha and p38-beta | 0.53333 |
| IL23-mediated signaling events | IL23-mediated signaling events | 0.52500 |
| IL23-mediated signaling events | VEGF Hypoxia and Angiogenesis | 0.51136 |
| amb2 Integrin signaling | Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | 0.50000 |
| Mechanism of Gene Regulation by Peroxisome Proliferators via PPARa(alpha) | ||
| The IGF-1 Receptor and Longevity | Antifolate resistance | 0.50000 |
| Adhesion and Diapedesis of Granulocytes | 0.50000 | |
| Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | Signaling mediated by p38-alpha and p38-beta | 0.50000 |
| IL27-mediated signaling events | Longevity regulating pathway | 0.50000 |
| Antifolate resistance | Hematopoietic cell lineage | 0.50000 |
| Fluid shear stress and atherosclerosis | ||
| Fluid shear stress and atherosclerosis | Hematopoietic cell lineage | 0.50000 |
| IL-17 signaling pathway | Hematopoietic cell lineage | 0.50000 |
| VEGF Hypoxia and Angiogenesis | 0.48214 | |
| Free Radical Induced Apoptosis | Angiopoietin receptor Tie2-mediated signaling | 0.48214 |
| Adhesion and Diapedesis of Granulocytes | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.48214 |
| Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | Toll-like receptor signaling pathway | 0.47619 |
| Toll-like receptor signaling pathway | Toll-like receptor signaling pathway | 0.47619 |
| Toll-like receptor signaling pathway | Toll-like receptor signaling pathway | |
| Th17 cell differentiation | 0.47619 | |
| PI3K-Akt signaling pathway | IL27-mediated signaling events | 0.47619 |
| Cytokine-cytokine receptor interaction | Antifolate resistance | 0.47619 |
| Cytokine-cytokine receptor interaction | IL27-mediated signaling events | 0.47619 |
| HIF-1 signaling pathway | HIF-Fluid shear stress and atherosclerosis-alpha transcription factor network | 0.47500 |
| IL-17 signaling pathway | VEGF Hypoxia and Angiogenesis | 0.46875 |
| IL-17 signaling pathway | HIF-Fluid shear stress and atherosclerosis-alpha transcription factor network | 0.46875 |
| IL-17 signaling pathway | PI3K-Akt signaling pathway | 0.46667 |
| IL-17 signaling pathway | Free Radical Induced Apoptosis | 0.45833 |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | Adhesion and Diapedesis of Granulocytes | 0.45833 |
| Focal adhesion | Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | 0.45833 |
| HIF-1 signaling pathway | Antifolate resistance | 0.45833 |
| Angiopoietin receptor Tie2-mediated signaling | 0.45395 | |
| Fluid shear stress and atherosclerosis | ||
| Fluid shear stress and atherosclerosis | Glypican 1 network | 0.44444 |
| Fluid shear stress and atherosclerosis | Glypican 1 network | 0.42424 |
| Fluid shear stress and atherosclerosis | IL-17 signaling pathway | 0.41071 |
| Fluid shear stress and atherosclerosis | Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | 0.40476 |
| Fluid shear stress and atherosclerosis | IL27-mediated signaling events | 0.40476 |
| Fluid shear stress and atherosclerosis | Antifolate resistance | 0.40476 |
| PI3K-Akt signaling pathway | Plasma membrane estrogen receptor signaling | 0.40000 |
| IL23-mediated signaling events | 0.40000 | |
| IL-17 signaling pathway | amb2 Integrin signaling | 0.40000 |
| Cytokine-cytokine receptor interaction | Glypican 1 network | 0.40000 |
| Cytokine-cytokine receptor interaction | Toll-like receptor signaling pathway | 0.40000 |
| Cytokine-cytokine receptor interaction | Free Radical Induced Apoptosis | 0.39583 |
| Plasma membrane estrogen receptor signaling | Adhesion and Diapedesis of Granulocytes | 0.39583 |
| Cells and Molecules involved in local acute inflammatory response | Cytokines can induce activation of matrix metalloproteinases which degrade extracellular matrix | 0.39583 |
| Fat digestion and absorption | Antifolate resistance | 0.39583 |
| lntegrins in angiogenesis | Angiopoietin receptor Tie2-mediated signaling | 0.38596 |
| lntegrins in angiogenesis | amb2 Integrin signaling | 0.38596 |
| amb2 Integrin signaling | ABC transporters | 0.38596 |
| Mechanism of Gene Regulation by Peroxisome Proliferators via PPARa (alpha) | VEGF Hypoxia and Angiogenesis | 0.38596 |
| Free Radical Induced Apoptosis | HIF-Fluid shear stress and atherosclerosis-alpha transcription factor network | 0.38596 |
| Angiopoietin receptor Tie2-mediated signaling | 0.38596 | |
| Adhesion and Diapedesis of Granulocytes | ATF-Fluid shear stress and atherosclerosis transcription factor network | 0.38596 |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.38596 | |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.38596 |
| 0.38596 | ||
| Cells and Molecules involved in local acute inflammatory response | IL27-mediated signaling events | |
| IL23-mediated signaling events | 0.38596 | |
| amb2 Integrin signaling | Toll-like receptor signaling pathway | |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.32500 | |
| Cells and Molecules involved in local acute inflammatory response | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.37500 |
| 0.31111 | ||
| HIF-1 signaling pathway | IL-17 signaling pathway | 0.37500 |
| HIF-1 signaling pathway | HIF-Fluid shear stress and atherosclerosis-alpha transcription factor network | |
| Cytokine-cytokine receptor interaction | Angiopoietin receptor Tie2-mediated signaling | 0.36111 |
| Cytokine-cytokine receptor interaction | Toll-like receptor signaling pathway | 0.33333 |
| Plasma membrane estrogen receptor signaling | ||
| IL23-mediated signaling events | Th17 cell differentiation | 0.33333 |
| IL23-mediated signaling events | amb2 Integrin signaling | 0.33333 |
| Mechanism of Gene Regulation by Peroxisome Proliferators via PPARa(alpha) | Mechanism of Gene Regulation by Peroxisome Proliferators via PPARa(alpha) | 0.33333 |
| Hematopoietic cell lineage | 0.32500 | |
| Fluid shear stress and atherosclerosis | Hematopoietic cell lineage | 0.32500 |
| PI3K-Akt signaling pathway | HIF-Fluid shear stress and atherosclerosis-alpha transcription factor network | 0.32500 |
| IL23-mediated signaling events | Angiopoietin receptor Tie2-mediated signaling | 0.32500 |
| IL23-mediated signaling events | Toll-like receptor signaling pathway | 0.31667 |
| Toll-like receptor signaling pathway | Th17 cell differentiation | 0.31250 |
| Cytokine-cytokine receptor interaction | Hematopoietic cell lineage | 0.31111 |
| Fluid shear stress and atherosclerosis | Cells and Molecules involved in local acute inflammatory response | 0.31111 |
| Cells and Molecules involved in local acute inflammatory response | Cytokine-cytokine receptor interaction | 0.31111 |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.30882 | |
| 0.27143 | ||
| IL-17 signaling pathway | amb2 Integrin signaling | 0.30000 |
| IL-17 signaling pathway | Hematopoietic cell lineage | |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | ATF-Fluid shear stress and atherosclerosis transcription factor network | 0.30000 |
| Fluid shear stress and atherosclerosis | 0.30000 | |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.30000 | |
| Fluid shear stress and atherosclerosis | ||
| Phagosome | Focal adhesion | 0.25758 |
| Fat digestion and absorption | 0.28846 | |
| Phagosome | Hematopoietic cell lineage | |
| HIF-1 signaling pathway | Plasma membrane estrogen receptor signaling | 0.28750 |
| HIF-1 signaling pathway | lntegrins in angiogenesis | 0.28333 |
| HIF-1 signaling pathway | ||
| Ensemble of genes encoding core extracellular matrix including ECM glycoproteins collagens and proteoglycans | Mineral absorption | 0.28333 |
| Focal adhesion | 0.27692 | |
| Plasma membrane estrogen receptor signaling | ||
| 0.27692 | ||
| Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix | 0.27692 |
| amb2 Integrin signaling | 0.27574 | |
| Ensemble of genes encoding core extracellular matrix including ECM glycoproteins collagens and proteoglycans | ||
| Fluid shear stress and atherosclerosis | lntegrins in angiogenesis | 0.27143 |
| 0.27143 | ||
| Fluid shear stress and atherosclerosis | lntegrins in angiogenesis | |
| Cytokine-cytokine receptor interaction | Mineral absorption | 0.26667 |
| Plasma membrane estrogen receptor signaling | Hematopoietic cell lineage | |
| IL-17 signaling pathway | Focal adhesion | 0.26250 |
| Cytokine-cytokine receptor interaction | PI3K-Akt signaling pathway | 0.26250 |
| Th17 cell differentiation | 0.26250 | |
| Cytokine-cytokine receptor interaction | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.26250 |
| Cytokine-cytokine receptor interaction | 0.25882 | |
| lntegrins in angiogenesis | 0.12795 | |
| 0.25758 | ||
| Plasma membrane estrogen receptor signaling | Hematopoietic cell lineage | .25595 |
| Phagosome | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | |
| Fluid shear stress and atherosclerosis | Toll-like receptor signaling pathway | 0.25556 |
| Fluid shear stress and atherosclerosis | Toll-like receptor signaling pathway | 0.25556 |
| Cytokine-cytokine receptor interaction | HIF-1 signaling pathway | 0.24762 |
| PI3K-Akt signaling pathway | PI3K-Akt signaling pathway | |
| Toll-like receptor signaling pathway | Toll-like receptor signaling pathway | 0.24359 |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.22549 | |
| PI3K-Akt signaling pathway | 0.22500 | |
| Fluid shear stress and atherosclerosis | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins collagens and proteoglycans | 0.22286 |
| Fluid shear stress and atherosclerosis | PI3K-Akt signaling pathway | 0.22222 |
| HIF-1 signaling pathway | Focal adhesion | 0.21212 |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | Phagosome | 0.58333 |
| PI3K-Akt signaling pathway | Focal adhesion | 0.19022 |
| 0.17788 | ||
| PI3K-Akt signaling pathway | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.17763 |
| 0.16993 | ||
| HIF-1 signaling pathway | Phagosome | 0.16667 |
| Complement and coagulation cascades | Cytokine-cytokine receptor interaction | |
| HIF-1 signaling pathway | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix | 0.15887 |
| HIF-1 signaling pathway | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.15873 |
| Fluid shear stress and atherosclerosis | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix | |
| Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins ECM regulators and secreted factors | 0.14348 | |
| Complement and coagulation cascades | 0.51136 | |
| 0.14091 | ||
| 0.13846 | ||
| 0.13636 | ||
| 0.12795 |
FIGURE 4.
Pathway crosstalk among AMDgset-enriched pathways. Nodes denote pathways while edges represent crosstalk between pathways. The yellow node represents “cytokine-cytokine receptor interaction” pathway which acts as the joint of two main modules. The width of edges is depended on the score of specific pathway pair, wider edge indicates stronger correlation.
Identification of genes related to AMD
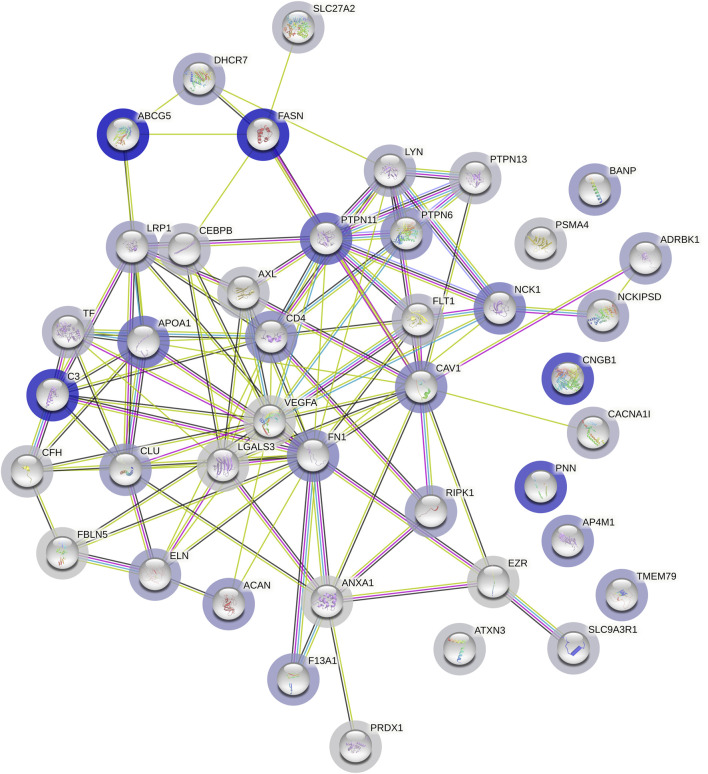
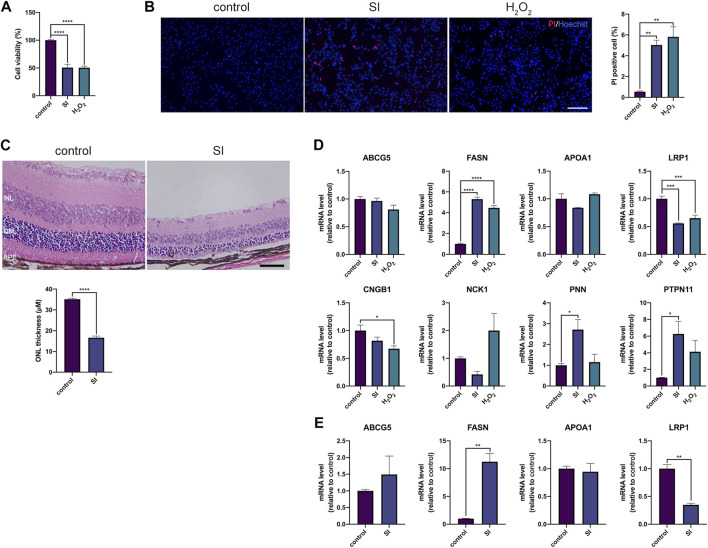
To make a more comprehensive list of AMD related genes, we used shortest path algorithm based on the background human interactome which contained 13,460 nodes and 138,427 edges and provided by a recent study (Menche et al., 2015). The primary analysis extracted 4,587 genes participated in AMD protein-protein interaction (PPI) network. We discarded genes of which the betweenness was below 1,000 and conducted permutation test. Finally, in our collection, we obtained 42 genes highly associated with AMD (Table 5). The PPI network among the 42 genes were showed in Figure 5. There were 7 genes belonged to AMDgset, including C3, ELN, TF, FLT1, CFH, VEGFA and FBLN5 (Stone et al., 2004; Fang et al., 2009; Anderson et al., 2010; Yamashiro et al., 2011; Wysokinski et al., 2013; Owen et al., 2014), indicating our results identified many novel genes that are potentially associated with AMD. The genes associated with lipid metabolism had high betweenness, such as ABCG5, FASN, APOA1, and LRP1. Han et al., reported that higher APOA1 level increased the risk of AMD (Han et al., 2021). Since these genes were not included in the AMDgset, we intended to make a brief validation on their potential in further investigation of AMD. We used sodium iodate (SI) and H2O2 to treat RPE cells and establish an AMD cell model (Elliot et al., 2006; Tao et al., 2013). Moreover, we used SI to induce an AMD mouse model (Carido et al., 2014)Hanus, 2016 #2412}. The results of CCK-8 and PI staining confirmed RPE cell death and indicated that the AMD cell model was successfully established (Figures 6A,B). The results of H&E staining showed the AMD-like phenotype in the retina of the mouse under SI treatment (Figure 6C). Then we evaluated the mRNA levels of several genes with high betweenness including ABCG5, FASN, APOA1, LRP1, CNGB1, NCK1, PNN1, and PTPN11. The qRT-PCR results showed that FASN was up-regulated while LRP1 was downregulated in AMD cell and mouse model (Figures 6D,E). Storck et al., reported that selective deletion of LRP1 in the brain endothelium of C57BL/6 mice strongly reduced brain efflux of injected Aβ (1–42) (Storck et al., 2016). Since Aβ is also a crucial component of drusen, our results suggest that the downregulation of LRP1 might promote drusen formation in AMD. The function of FASN is to promote saturated fatty acid (SFA) synthesis. Previous study confirmed that SFA was associated significantly with increased risk of AMD (Agron et al., 2021). Therefore, the upregulation of FASN might exert a pro-AMD effect through promoting SFA synthesis. The mRNA levels of ABCG5 and APOA1 were relatively low in RPE cells and were not significantly altered (Figures 6D,E). We speculated that these genes might participate in AMD pathogenesis by acting in other tissues such as liver or intestine where they modulate fat digestion and absorption. Moreover, besides genes associated with lipid metabolism, some other genes in our collection were reported to participate in AMD progression or therapy e.g. NCK1 and EZR (Murad et al., 2014; Dubrac et al., 2016). The mRNA level of NCK1 was upregulated in the H2O2 AMD cell model (Figure 6D). Previous study showed that NCK1 knockdown was associated with neovascular inhibition (Dubrac et al., 2016). However, the mRNA level of NCK1 was slightly decreased in the SI AMD cell model, the reason might be different damage mode between SI and H2O2 PTPN11 was reported to be a diagnostic marker of AMD (Li et al., 2022). We also detected a significant upregulation of PTPN11 in the SI AMD cell model, indicating a potential role of PTPN11 in RPE degeneration. The exact role of NCK1 and PTPN11 in AMD progression needs further investigation in more AMD models. These results confirmed that our novel AMD gene collection have significant importance in guiding further investigation on AMD.
TABLE 5.
Shortest path genes with betweenness greater than 1,000.
| Gene ID | Official Symbol | Official Full Name | Betweenness |
|---|---|---|---|
| 64240 | ABCG5 | ATP binding cassette subfamily G member 5 | 5123 |
| 2194 | FASN | fatty acid synthase | 4885 |
| 718 | C3 a | complement C3 | 4533 |
| 1258 | CNGB1 | cyclic nucleotide gated channel beta 1 | 3931 |
| 5411 | PNN | pinin, desmosome associated protein | 3892 |
| 5781 | PTPN11 | protein tyrosine phosphatase, non-receptor type 11 | 3207 |
| 335 | APOA1 | apolipoprotein A1 | 2980 |
| 4690 | NCK1 | NCK adaptor protein 1 | 2640 |
| 857 | CAV1 | caveolin 1 | 2468 |
| 2335 | FN1 | fibronectin 1 | 2421 |
| 9179 | AP4M1 | adaptor related protein complex 4 subunit mu 1 | 2330 |
| 920 | CD4 | CD4 molecule | 2310 |
| 5777 | PTPN6 | protein tyrosine phosphatase, non-receptor type 6 | 2248 |
| 176 | ACAN | aggrecan | 2218 |
| 54971 | BANP | BTG3 associated nuclear protein) | 2118 |
| 84283 | TMEM79 | transmembrane protein 79 | 2100 |
| 2162 | F13A1 | coagulation factor XIII A chain | 2073 |
| 1191 | CLU | clusterin | 2002 |
| 2006 | ELN a | elastin | 1926 |
| 156 | GRK2 | G protein-coupled receptor kinase 2 | 1911 |
| 8737 | RIPK1 | receptor interacting serine/threonine kinase 1 | 1899 |
| 4035 | LRP1 | LDL receptor related protein 1 | 1885 |
| 1717 | DHCR7 | 7-dehydrocholesterol reductase | 1862 |
| 51517 | NCKIPSD | NCK interacting protein with SH3 domain | 1843 |
| 4067 | LYN | LYN proto-oncogene, Src family tyrosine kinase | 1698 |
| 7018 | TF a | transferrin | 1591 |
| 8911 | CACNA1I | calcium voltage-gated channel subunit alpha1 I | 1580 |
| 2321 | FLT1a | fms related tyrosine kinase 1 | 1501 |
| 1051 | CEBPB | CCAAT/enhancer binding protein beta | 1458 |
| 5783 | PTPN13 | protein tyrosine phosphatase, non-receptor type 13 | 1426 |
| 9368 | SLC9A3R1 | SLC9A3 regulator 1 | 1413 |
| 11001 | SLC27A2 | solute carrier family 27 member 2 | 1411 |
| 5685 | PSMA4 | proteasome subunit alpha 4 | 1342 |
| 3075 | CFH a | complement factor H | 1323 |
| 558 | AXL | AXL receptor tyrosine kinase | 1289 |
| 4287 | ATXN3 | ataxin 3 | 1261 |
| 3958 | LGALS3 | galectin 3 | 1143 |
| 5052 | PRDX1 | peroxiredoxin 1 | 1112 |
| 7430 | EZR | ezrin | 1083 |
| 7422 | VEGFA a | vascular endothelial growth factor A | 1056 |
| 10516 | FBLN5 a | fibulin 5 | 1045 |
| 301 | ANXA1 | annexin A1 | 1044 |
Genes included in AMDgset.
FIGURE 5.
Protein-protein interaction network of the 42 genes in AMD gene collection. The blue halo around the gene indicates high betweenness while the gray halo indicates low betweenness.
FIGURE 6.
(A) CCk-8 results of RPE cells that were under SI (40 mM) or H2O2 (200 μM) treatment for 4 h (B) Representative images and the corresponding statistical result of PI staining. The cells were under SI (40 mM) or H2O2 (200 μM) treatment for 4 h; scale bar = 200 μm (C) H&E staining of retinal sections from mice at 2 days after 40 mg/kg SI injection; scale bar = 50 μm (D) Quantification of mRNA expression of indicated genes in RPE cells. The cells were under SI (40 mM) or H2O2 (200 μM) treatment for 4 h (E) Quantification of mRNA expression of indicated genes in RPE-choroid complex in mouse that were treated with SI for 2 days **p < 0.01, ***p < 0.001, ****p < 0.0001, compared versus control. INL: inner nuclear layer, ONL: outer nuclear layer, RPE: retinal pigmented epithelium.
Discussion
Studies have confirmed that there is a strong correlation between a family history of AMD and the subsequent development of both dry and wet form of the disease. Genetic factors play a potential role in the etiology of AMD, explaining 46%–71% of the variation in the overall severity of the disease, while environmental factors take charge of the rest (Seddon et al., 2005). According to Yu et al., we only have recognized half of genetic risk factors of AMD (Yu et al., 2011). Therefore, making predictions based on the identified genetic risk factors and a comprehensive human interactome could be valuable to take a glimpse into the unknown half. A previous study about AMD related GO analysis showed a variant result with ours as they found the most significant terms are plasma membrane, cell surface receptor linked signal transduction and intracellular signaling cascade (Zhang et al., 2013). The inconformity between our results may ascribe to the method we chose genes and the quantity of genes we retrieved. In our study, we firstly established a relatively comprehensive collection of the genes genetically associated with AMD. Then, we proceeded GO enrichment and pathway enrichment analyses to demonstrate the most significant biological functions and cellular signaling related to AMD. Moreover, the results of crosstalk study showed a visualized interaction of pathways that we have identified. At last, we made a predictive list of potential AMD related genes by using shortest path algorithm and confirmed that FASN and LRP1 were potentially associated with AMD. By retrieving AMDgset from PUBMED, we obtained 176 genes which were reported significantly genetically related to AMD. Both dry and wet forms of AMD were included in our research. According to the clinical character of AMD, new vessels may invade the outer retina, subretinal space or subRPE space, resulting in macular neovascularization (MNV) at any stage of dry AMD (Fleckenstein et al., 2021). The natural course of AMD indicates that the pathogeneses of dry and wet AMD are common to a great extent. Therefore, it is of great significance to study the genetic risk factors and the pathway crosstalk in the combination of dry and wet AMD.
Our pathway analysis revealed that complement related pathway was enriched in AMDgset. This finding further consolidates the link between AMD and complement system. Precedent identification of several molecular components of the complement cascade in drusen suggests that complement activation is an important element in drusen biogenesis (Johnson et al., 2001). CFH binds to glycoaminoglycans (GAG) on host cells and apoptotic bodies and acts as a cofactor of Complement factor I (CFI) that cleaves C3b into iC3b and prevents membrane attack complex (MAC) formation (Atkinson and Goodship, 2007). Hageman et al. demonstrated that risk alleles decreased the function of CFH, which may lead to high MAC aggregation at the RPE-choroid interface and jeopardize the integrity of Bruch’s membrane (Hageman et al., 2005). However, Hageman et al. claimed that CFH immunoreactivity in the eye is stronger, not weaker, in AMD donor tissues. Calippe et al. recently showed that the AMD-associated CFH variant CFH(H402) contributes to AMD etiology by increasing subretinal macrophage accumulation through binding CD11b. Together with their results, there is a discrepancy with the function of CFH in AMD progression that need to be well studied in the future. Cipriani et al. recently revealed that AMD was associated with genetically driven elevated circulating levels of complement factor H related 4 (CFHR4). The role of complement factor H related 1 (CFHR1) is protecting intercapillary septa ECM from complement activation (Clark et al., 2014; McHarg et al., 2015), but this protective function may be diminished by elevation of CFHR4. Strong evidences indicate that these abnormities result in dysregulation of the complement cascade and aberrant activation of the immune system. Besides, we noticed that pathways associated with hypoxia and angiogenesis were also enriched in AMDgset. The mechanism may due to the limited blood supply which is caused by choroidal capillary atrophy and high oxygen demand in macula. This imbalance situation causes relative hypoxia, which furtherly up-regulates the expression of growth factors, such as VEGF family (Penfold et al., 2001).
In our pathway crosstalk analysis, we demonstrated two main components interacted with each other. One component was mainly predominated by inflammation related pathways while another was hypoxia-angiogenesis related pathways. The two modules were connected by cytokine-cytokine receptor interaction pathway (genes: TLR4, NOS2, NOS3, VEGFA) instead of operating separately. We attach much importance to the mediating role of cytokines-cytokine receptors signaling and speculate that the cytokines and chemokines related to macrophages, RPE cells and vessel endothelial cells play a central role in mediating two main modules of AMD associated pathways. TLR2/TLR4 plays a prominent role in recognizing pathogen-associated molecular pattern (PAMP) or damage-associated molecular patterns (DAMP) and activates NLRP3 inflammasome or NF-κB related pathways to modulate inflammation state (Schmitz and Orso, 2002; Allan et al., 2005; Schroder and Tschopp, 2010). The pro-inflammation, anti-angiogenic, potentially neurotoxic state is characterized by IL-1β, TNF-α, IL-6, CCL2 and iNOS, while the anti-inflammation, wound healing, fibrosis state is defined by VEGF, IL-10 and IL-1RA among others (Sica and Mantovani, 2012; Wynn and Vannella, 2016). It is interesting that our pathway crosstalk analysis also reflected this phenomenon. The larger module contained the acute inflammatory response and ECM degradation pathways, which indicated the pro-inflammatory state. Those potentially neurotoxic cytokines may contribute to RPE and photoreceptor degeneration and result in the geographic atrophy. The smaller module contained angiogenesis pathways, which indicated the anti-inflammatory state and CNV formation. Our pathway crosstalk study is of great significance as it reflects the pivotal role of cytokines and cytokine receptors in prompting early AMD to the two distinct types. It also indicated that there might be a possibility to modulate the specific type of cytokines in early AMD to control its progression. There are limited researches focused on the role of TLR4 and NOS family in AMD. Chen et al. demonstrated that TLR4 mediated subretinally-deposited amyloid-β induced angiogenic and inflammation (Chen et al., 2016). Imran A. Bhutto et al. showed that the decrease in retinal NOS1 in AMD eyes was probably related to neuronal degeneration. The decrease in NOS1 and NOS3 in AMD choroid could be associated with vasoconstriction and hemodynamic changes (Bhutto et al., 2010). We strongly propose that future studies should focus on these cytokines and cytokine receptors.
In our novel gene collection, besides the genes we have verified, CNGB1 is also a candidate gene that might participate AMD. CNGB1 is a gene encoding cyclic nucleotide-gated (CNG) channels proteins which are key components for signal transduction in rod outer segment and olfactory sensory neurons (OSNs) (Charbel Issa et al., 2018). It has been verified that AMD patients suffer from impaired dark adaptation, which indicates a rod deficiency (Flamendorf et al., 2015). Zhang et al. found that the amplitude of dark adaptive b-wave was significantly diminished in CNGB1 knockout mice, more importantly, these mice showed a rod-cone degeneration. These results strongly implicate that CNGB1 may account for the deteriorated dark adaptation in AMD especially in the dry form. Although the mRNA level of CNGB1 is decreased only in H2O2 AMD cell model, considering the fact that the cell model was established by RPE cells, further study should investigate the dysregulation of CNGB1 in photoreceptor cells in AMD model.
Although we have provided a new perspective on AMD associated genes, there are several limitations of our study. First, most of our results are based on literatures, so the partialness of some studies can affect our analysis. Second, the identification of AMD risk genes is a gradual process, as well as the background human interactome. The incomplete human interactome may bring some false-positive or false-negative results to our study. More importantly, the genes in our novel collection should be verified in more cell models and animal models of AMD.
Conclusion
Our study filled the gap in the integrated study in genetic field of AMD, and we revealed the potential relationships between these pathways as well as their operation pattern. Moreover, we demonstrated a relatively comprehensive AMD associated genes list and validated that the mRNA levels of FASN and LRP1 are dysregulated in both cell and mouse models of AMD, indicating they might regulate AMD progression directly.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by ethical committee of Shanghai 10th People’s Hospital.
Author contributions
CR: Conceptualization, Methodology, Validation, Investigation, Formal analysis, Data curation, Writing—Original Draft, Visulization; JY: Conceptualization, Writing—Review and Editing, Funding acquisition, Supervision.
Funding
This work was supported by NSFC (82000903) and NSFC (82101130) and Fundamental Research Funds for the Central Universities (22120180509).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.992328/full#supplementary-material
References
- Agron E., Mares J., Clemons T. E., Swaroop A., Chew E. Y., Keenan T. D. L., et al. (2021). Dietary nutrient intake and progression to late age-related macular degeneration in the age-related eye disease studies 1 and 2. Ophthalmology 128 (3), 425–442. 10.1016/j.ophtha.2020.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan S. M., Tyrrell P. J., Rothwell N. J. (2005). Interleukin-1 and neuronal injury. Nat. Rev. Immunol. 5 (8), 629–640. 10.1038/nri1664 [DOI] [PubMed] [Google Scholar]
- Anderson D. H., Radeke M. J., Gallo N. B., Chapin E. A., Johnson P. T., Curletti C. R., et al. (2010). The pivotal role of the complement system in aging and age-related macular degeneration: Hypothesis re-visited. Prog. Retin. Eye Res. 29 (2), 95–112. 10.1016/j.preteyeres.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J. P., Goodship T. H. (2007). Complement factor H and the hemolytic uremic syndrome. J. Exp. Med. 204 (6), 1245–1248. 10.1084/jem.20070664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas D. C., Ho L., Ennis S., Merriam J. E., Tanck M. W., Uitterlinden A. G., et al. (2010). The complement component 5 gene and age-related macular degeneration. Ophthalmology 117 (3), 500–511. 10.1016/j.ophtha.2009.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutto I. A., Baba T., Merges C., McLeod D. S., Lutty G. A. (2010). Low nitric oxide synthases (NOSs) in eyes with age-related macular degeneration (AMD). Exp. Eye Res. 90 (1), 155–167. 10.1016/j.exer.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler S. B. (2009). Introduction: Understanding the role of angiogenesis and antiangiogenic agents in age-related macular degeneration. Ophthalmology 116 (10), S1–S7. 10.1016/j.ophtha.2009.06.045 [DOI] [PubMed] [Google Scholar]
- Campa C., Harding S. P. (2011). Anti-VEGF compounds in the treatment of neovascular age related macular degeneration. Curr. Drug Targets 12 (2), 173–181. 10.2174/138945011794182674 [DOI] [PubMed] [Google Scholar]
- Carido M., Zhu Y., Postel K., Benkner B., Cimalla P., Karl M. O., et al. (2014). Characterization of a mouse model with complete RPE loss and its use for RPE cell transplantation. Invest. Ophthalmol. Vis. Sci. 55 (8), 5431–5444. 10.1167/iovs.14-14325 [DOI] [PubMed] [Google Scholar]
- Charbel Issa P., Reuter P., Kuhlewein L., Birtel J., Gliem M., Tropitzsch A., et al. (2018). Olfactory dysfunction in patients with CNGB1-associated retinitis pigmentosa. JAMA Ophthalmol. 136 (7), 761–769. 10.1001/jamaophthalmol.2018.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bardes E. E., Aronow B. J., Jegga A. G. (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311. Web Server issue. 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Bai Y., Zhao M., Jiang Y. (2016). TLR4 inhibitor attenuates amyloid-beta-induced angiogenic and inflammatory factors in ARPE-19 cells: Implications for age-related macular degeneration. Mol. Med. Rep. 13 (4), 3249–3256. 10.3892/mmr.2016.4890 [DOI] [PubMed] [Google Scholar]
- Clark S. J., Schmidt C. Q., White A. M., Hakobyan S., Morgan B. P., Bishop P. N. (2014). Identification of factor H-like protein 1 as the predominant complement regulator in bruch's membrane: implications for age-related macular degeneration. J. Immunol. 193 (10), 4962–4970. 10.4049/jimmunol.1401613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combadiere C., Feumi C., Raoul W., Keller N., Rodero M., Pezard A., et al. (2007). CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J. Clin. Invest. 117 (10), 2920–2928. 10.1172/JCI31692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin B., Schnabolk G., Joseph K., Raikwar H., Kunchithapautham K., Johnson K., et al. (2016). Connecting the innate and adaptive immune responses in mouse choroidal neovascularization via the anaphylatoxin C5a and γδT-cells. Sci. Rep. 6, 23794. 10.1038/srep23794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despriet D. D., van Duijn C. M., Oostra B. A., Uitterlinden A. G., Hofman A., Wright A. F., et al. (2009). Complement component C3 and risk of age-related macular degeneration. Ophthalmology 116 (3), 474–480.e2. 10.1016/j.ophtha.2008.09.055 [DOI] [PubMed] [Google Scholar]
- Dubrac A., Genet G., Ola R., Zhang F., Pibouin-Fragner L., Han J., et al. (2016). Targeting NCK-mediated endothelial cell front-rear polarity inhibits neovascularization. Circulation 133 (4), 409–421. 10.1161/CIRCULATIONAHA.115.017537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eandi C. M., Charles Messance H., Augustin S., Dominguez E., Lavalette S., Forster V., et al. (2016). Subretinal mononuclear phagocytes induce cone segment loss via IL-1β. Elife 5, e16490. 10.7554/eLife.16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot S., Catanuto P., Stetler-Stevenson W., Cousins S. W. (2006). Retinal pigment epithelium protection from oxidant-mediated loss of MMP-2 activation requires both MMP-14 and TIMP-2. Invest. Ophthalmol. Vis. Sci. 47 (4), 1696–1702. 10.1167/iovs.05-1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang A. M., Lee A. Y., Kulkarni M., Osborn M. P., Brantley M. A., Jr. (2009). Polymorphisms in the VEGFA and VEGFR-2 genes and neovascular age-related macular degeneration. Mol. Vis. 15, 2710–2719. [PMC free article] [PubMed] [Google Scholar]
- Feldman I., Rzhetsky A., Vitkup D. (2008). Network properties of genes harboring inherited disease mutations. Proc. Natl. Acad. Sci. U. S. A. 105 (11), 4323–4328. 10.1073/pnas.0701722105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamendorf J., Agron E., Wong W. T., Thompson D., Wiley H. E., Doss E. L., et al. (2015). Impairments in dark adaptation are associated with age-related macular degeneration severity and reticular pseudodrusen. Ophthalmology 122 (10), 2053–2062. 10.1016/j.ophtha.2015.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein M., Keenan T. D. L., Guymer R. H., Chakravarthy U., Schmitz-Valckenberg S., Klaver C. C., et al. (2021). Age-related macular degeneration. Nat. Rev. Dis. Prim. 7 (1), 31. 10.1038/s41572-021-00265-2 [DOI] [PubMed] [Google Scholar]
- Gehrs K. M., Anderson D. H., Johnson L. V., Hageman G. S. (2006). Age-related macular degeneration--emerging pathogenetic and therapeutic concepts. Ann. Med. 38 (7), 450–471. 10.1080/07853890600946724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G. V., Emmert-Streib F. (2009). Unite and conquer: univariate and multivariate approaches for finding differentially expressed gene sets. Bioinformatics 25 (18), 2348–2354. 10.1093/bioinformatics/btp406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman J. J., Buhlmann P. (2007). Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics 23 (8), 980–987. 10.1093/bioinformatics/btm051 [DOI] [PubMed] [Google Scholar]
- Goh K. I., Cusick M. E., Valle D., Childs B., Vidal M., Barabasi A. L. (2007). The human disease network. Proc. Natl. Acad. Sci. U. S. A. 104 (21), 8685–8690. 10.1073/pnas.0701361104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillonneau X., Eandi C. M., Paques M., Sahel J. A., Sapieha P., Sennlaub F. (2017). On phagocytes and macular degeneration. Prog. Retin. Eye Res. 61, 98–128. 10.1016/j.preteyeres.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Hageman G. S., Anderson D. H., Johnson L. V., Hancox L. S., Taiber A. J., Hardisty L. I., et al. (2005). A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 102 (20), 7227–7232. 10.1073/pnas.0501536102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Ong J. S., Hewitt A. W., Gharahkhani P., MacGregor S. (2021). The effects of eight serum lipid biomarkers on age-related macular degeneration risk: a mendelian randomization study. Int. J. Epidemiol. 50 (1), 325–336. 10.1093/ije/dyaa178 [DOI] [PubMed] [Google Scholar]
- Hu Y., Pan Z., Hu Y., Zhang L., Wang J. (2017). Network and pathway-based analyses of genes associated with Parkinson's disease. Mol. Neurobiol. 54 (6), 4452–4465. 10.1007/s12035-016-9998-8 [DOI] [PubMed] [Google Scholar]
- Jarrett S. G., Boulton M. E. (2012). Consequences of oxidative stress in age-related macular degeneration. Mol. Asp. Med. 33 (4), 399–417. 10.1016/j.mam.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P., Kao C. F., Kuo P. H., Zhao Z. (2011a). A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC Syst. Biol. 5, S12. Suppl 3. 10.1186/1752-0509-5-S3-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P., Zheng S., Long J., Zheng W., Zhao Z. (2011b). dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics 27 (1), 95–102. 10.1093/bioinformatics/btq615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Chen Y., Zhang Y., Chen L., Zhang N., Huang T., et al. (2013). Identification of hepatocellular carcinoma related genes with k-th shortest paths in a protein-protein interaction network. Mol. Biosyst. 9 (11), 2720–2728. 10.1039/c3mb70089e [DOI] [PubMed] [Google Scholar]
- Johnson L. V., Leitner W. P., Staples M. K., Anderson D. H. (2001). Complement activation and inflammatory processes in Drusen formation and age related macular degeneration. Exp. Eye Res. 73 (6), 887–896. 10.1006/exer.2001.1094 [DOI] [PubMed] [Google Scholar]
- Jun S., Datta S., Wang L., Pegany R., Cano M., Handa J. T. (2019). The impact of lipids, lipid oxidation, and inflammation on AMD, and the potential role of miRNAs on lipid metabolism in the RPE. Exp. Eye Res. 181, 346–355. 10.1016/j.exer.2018.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R., Chou C. F., Klein B. E., Zhang X., Meuer S. M., Saaddine J. B. (2011). Prevalence of age-related macular degeneration in the US population. Arch. Ophthalmol. 129 (1), 75–80. 10.1001/archophthalmol.2010.318 [DOI] [PubMed] [Google Scholar]
- Klein R., Myers C. E., Meuer S. M., Gangnon R. E., Sivakumaran T. A., Iyengar S. K., et al. (2013). Risk alleles in CFH and ARMS2 and the long-term natural history of age-related macular degeneration: the beaver dam eye study. JAMA Ophthalmol. 131 (3), 383–392. 10.1001/jamaophthalmol.2013.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N. G., ElShelmani H., Singh M. K., Mansergh F. C., Wride M. A., Padilla M., et al. (2016). Risk factors and biomarkers of age-related macular degeneration. Prog. Retin. Eye Res. 54, 64–102. 10.1016/j.preteyeres.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemetre C., Zhang Q., Zhang Z. D. (2013). SubNet: a Java application for subnetwork extraction. Bioinformatics 29 (19), 2509–2511. 10.1093/bioinformatics/btt430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fu J., Liu J., Feng H., Chen X. (2022). Diagnostic markers and molecular dysregulation mechanisms in the retinal pigmented epithelium and retina of age-related macular degeneration. J. Healthc. Eng. 2022, 3787567. 10.1155/2022/3787567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHarg S., Clark S. J., Day A. J., Bishop P. N. (2015). Age-related macular degeneration and the role of the complement system. Mol. Immunol. 67 (1), 43–50. 10.1016/j.molimm.2015.02.032 [DOI] [PubMed] [Google Scholar]
- McKay G. J., Patterson C. C., Chakravarthy U., Dasari S., Klaver C. C., Vingerling J. R., et al. (2011). Evidence of association of APOE with age-related macular degeneration: a pooled analysis of 15 studies. Hum. Mutat. 32 (12), 1407–1416. 10.1002/humu.21577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menche J., Sharma A., Kitsak M., Ghiassian S. D., Vidal M., Loscalzo J., et al. (2015). Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 347 (6224), 1257601. 10.1126/science.1257601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. W., Le Couter J., Strauss E. C., Ferrara N. (2013). Vascular endothelial growth factor a in intraocular vascular disease. Ophthalmology 120 (1), 106–114. 10.1016/j.ophtha.2012.07.038 [DOI] [PubMed] [Google Scholar]
- Murad N., Kokkinaki M., Gunawardena N., Gunawan M. S., Hathout Y., Janczura K. J., et al. (2014). miR-184 regulates ezrin, LAMP-1 expression, affects phagocytosis in human retinal pigment epithelium and is downregulated in age-related macular degeneration. FEBS J. 281 (23), 5251–5264. 10.1111/febs.13066 [DOI] [PubMed] [Google Scholar]
- Neale B. M., Fagerness J., Reynolds R., Sobrin L., Parker M., Raychaudhuri S., et al. (2010). Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc. Natl. Acad. Sci. U. S. A. 107 (16), 7395–7400. 10.1073/pnas.0912019107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita M., Strzalka-Mrozik B., Grzybowski A., Mazurek U., Romaniuk W. (2014). Age-related macular degeneration and changes in the extracellular matrix. Med. Sci. Monit. 20, 1003–1016. 10.12659/MSM.889887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen L. A., Morrison M. A., Ahn J., Woo S. J., Sato H., Robinson R., et al. (2014). FLT1 genetic variation predisposes to neovascular AMD in ethnically diverse populations and alters systemic FLT1 expression. Invest. Ophthalmol. Vis. Sci. 55 (6), 3543–3554. 10.1167/iovs.14-14047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold P. L., Madigan M. C., Gillies M. C., Provis J. M. (2001). Immunological and aetiological aspects of macular degeneration. Prog. Retin. Eye Res. 20 (3), 385–414. 10.1016/s1350-9462(00)00025-2 [DOI] [PubMed] [Google Scholar]
- Schmitz G., Orso E. (2002). CD14 signalling in lipid rafts: new ligands and co-receptors. Curr. Opin. Lipidol. 13 (5), 513–521. 10.1097/00041433-200210000-00007 [DOI] [PubMed] [Google Scholar]
- Schroder K., Tschopp J. (2010). The inflammasomes. Cell 140 (6), 821–832. 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Seddon J. M., Cote J., Page W. F., Aggen S. H., Neale M. C. (2005). The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch. Ophthalmol. 123 (3), 321–327. 10.1001/archopht.123.3.321 [DOI] [PubMed] [Google Scholar]
- Sennlaub F., Auvynet C., Calippe B., Lavalette S., Poupel L., Hu S. J., et al. (2013). CCR2(+) monocytes infiltrate atrophic lesions in age-related macular disease and mediate photoreceptor degeneration in experimental subretinal inflammation in Cx3cr1 deficient mice. EMBO Mol. Med. 5 (11), 1775–1793. 10.1002/emmm.201302692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A., Mantovani A. (2012). Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122 (3), 787–795. 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W., Mitchell P. (1998). Family history and age-related maculopathy: the blue mountains eye study. Aust. N. Z. J. Ophthalmol. 26 (3), 203–206. 10.1111/j.1442-9071.1998.tb01311.x [DOI] [PubMed] [Google Scholar]
- Stone E. M., Braun T. A., Russell S. R., Kuehn M. H., Lotery A. J., Moore P. A., et al. (2004). Missense variations in the fibulin 5 gene and age-related macular degeneration. N. Engl. J. Med. 351 (4), 346–353. 10.1056/NEJMoa040833 [DOI] [PubMed] [Google Scholar]
- Storck S. E., Meister S., Nahrath J., Meissner J. N., Schubert N., Di Spiezio A., et al. (2016). Endothelial LRP1 transports amyloid-β(1-42) across the blood-brain barrier. J. Clin. Invest. 126 (1), 123–136. 10.1172/JCI81108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P. F., Neale B. M., van den Oord E., Miles M. F., Neale M. C., Bulik C. M., et al. (2004). Candidate genes for nicotine dependence via linkage, epistasis, and bioinformatics. Am. J. Med. Genet. B Neuropsychiatr. Genet. 126B (1), 23–36. 10.1002/ajmg.b.20138 [DOI] [PubMed] [Google Scholar]
- Tao Z., Dai J., He J., Li C., Li Y., Yin Z. Q. (2013). The influence of NaIO(3)-induced retinal degeneration on intra-retinal layer and the changes of expression profile/morphology of DA-ACs and mRGCS. Mol. Neurobiol. 47 (1), 241–260. 10.1007/s12035-012-8366-6 [DOI] [PubMed] [Google Scholar]
- Wynn T. A., Vannella K. M. (2016). Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44 (3), 450–462. 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokinski D., Danisz K., Blasiak J., Dorecka M., Romaniuk D., Szaflik J., et al. (2013). An association of transferrin gene polymorphism and serum transferrin levels with age-related macular degeneration. Exp. Eye Res. 106, 14–23. 10.1016/j.exer.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y. (2006). Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics 22 (22), 2800–2805. 10.1093/bioinformatics/btl467 [DOI] [PubMed] [Google Scholar]
- Yamashiro K., Mori K., Nakata I., Tsuchihashi T., Horie-Inoue K., Nakanishi H., et al. (2011). Association of elastin gene polymorphism to age-related macular degeneration and polypoidal choroidal vasculopathy. Invest. Ophthalmol. Vis. Sci. 52 (12), 8780–8784. 10.1167/iovs.11-8205 [DOI] [PubMed] [Google Scholar]
- Yu Y., Bhangale T. R., Fagerness J., Ripke S., Thorleifsson G., Tan P. L., et al. (2011). Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum. Mol. Genet. 20 (18), 3699–3709. 10.1093/hmg/ddr270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Jiang M., Yuan F., Feng K. Y., Cai Y. D., Xu X., et al. (2013). Identification of age-related macular degeneration related genes by applying shortest path algorithm in protein-protein interaction network. Biomed. Res. Int. 2013, 523415. 10.1155/2013/523415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.