Abstract
ToxT, a member of the AraC family of transcriptional regulators, controls the expression of several virulence factors in Vibrio cholerae. In the classical biotype of V. cholerae, expression of toxT is regulated by the same environmental conditions that control expression of the virulence determinants cholera toxin and the toxin coregulated pilus. Several genes that activate toxT expression have been identified. To identify genes that repress toxT expression in nonpermissive environmental conditions, a genetic screen was used to isolate mutations which alter the expression of a toxT-gusA transcriptional fusion. Several mutants were isolated, and the mutants could be divided into two classes. One class of mutants exhibited higher expression levels of toxT-gusA at both the nonpermissive pH and temperature, while the second class showed elevated toxT-gusA expression only at the nonpermissive pH. One mutant from the second class was chosen for further characterization. This mutant was found to carry a TnphoA insertion in a homolog of the Escherichia coli pepA gene. Disruption of pepA in V. cholerae resulted in elevated levels of expression of cholera toxin, tcpA, toxT, and tcpP at the noninducing pH but not at the noninducing temperature. Elevated levels of expression of toxT and tcpP at the nonpermissive pH in the pepA mutant were abolished in tcpP toxR mutant and aphB mutant backgrounds, respectively. A putative binding site for PepA was identified in the tcpPH-tcpI intergenic region, suggesting that PepA may act at the level of tcpPH transcription. Disruption of pepA caused only partial deregulation at the noninducing pH, suggesting the involvement of additional factors in the pH regulation of virulence genes in V. cholerae.
Vibrio cholerae is a gram-negative bacillus that is the etiologic agent for the diarrheal illness cholera. Most epidemics of cholera are caused by strains belonging to serotype O1, which can be divided into classical and El Tor biotypes (28). Cholera toxin and the toxin-coregulated pilus (TCP) are among the important V. cholerae virulence factors. Cholera toxin is a heterodimeric secreted protein which consist of two subunits, A and B. The A subunit is enzymatically active and causes elevation of intracellular cyclic AMP (cAMP). A pentamer of B subunits, associated with a single A subunit, binds the holotoxin to the ganglioside GM1 receptor on eukaryotic cells (17, 18, 35). The genes that encode cholera toxin, ctxA and ctxB, are arranged as an operon and are contained within the genome of a filamentous bacteriophage, CTXφ, that lysogenizes V. cholerae (36, 56). The second major V. cholerae virulence factor, TCP, is a type IV pilus. A cluster of 12 genes are involved in the processing and assembly of the pilus on the surface of the bacterium. The major subunit of TCP is encoded by tcpA and the tcp gene cluster is expressed as an operon from a promoter upstream of tcpA (3, 50). The genes of the tcp operon, as well as adjacent genes, have recently been shown to be encoded by another filamentous bacteriophage, VPIφ (29).
ToxT, a member of the AraC family of transcriptional regulators, controls the transcription of the ctxAB and the tcp operons (15, 24). The toxT gene is contained within the tcp gene cluster, and toxT transcription occurs as part of the tcp operon, as well as from a promoter immediately upstream of toxT itself (3). Activation of toxT transcription depends on two pairs of proteins, ToxR-ToxS and TcpP-TcpH, which act synergistically at the toxT promoter (7, 22). ToxR is a transmembrane protein with an amino-terminal, cytoplasmic DNA-binding domain that acts as a transcriptional activator, while ToxS encodes a periplasmic protein that facilitates dimerization and activation of ToxR (14, 41, 42, 44). TcpP is a transmembrane protein, which, like ToxR, has homology with members of the bacterial two-component family of response regulators (22). TcpH, which is encoded by a gene that forms an operon with tcpP, enhances the activity of TcpP as a transcriptional activator of toxT (7). Recently, expression of the tcpPH operon has been shown to be positively regulated by the proteins AphA and AphB. AphA has no known homolog in the database, while AphB is homologous to members of the LysR family of transcriptional regulators (30, 47).
Due to the important role of ToxR in activating virulence genes and because it was the first regulator identified, the regulatory circuit controlling virulence gene expression in V. cholerae is called the ToxR regulon. The current model for controlling virulence gene expression in V. cholerae is that of a regulatory cascade (13). According to this model, AphA and AphB activate expression of TcpP and TcpH which, in turn, act synergistically with ToxR and ToxS to positively regulate toxT expression. Finally, ToxT activates expression of its dependent genes, which are collectively called the ToxT-dependent branch of the ToxR regulon, leading to production of cholera toxin and TCP. A different branch of the ToxR regulon involves the direct control of expression of the outer membrane proteins OmpU and OmpT by ToxR, and this branch is called the ToxT-independent branch (9, 11, 15, 32).
Expression of cholera toxin and TCP, and of their activator ToxT, in classical strains of V. cholerae is strongly regulated by environmental signals, such as pH, temperature, amino acid concentration, and osmolarity (43, 44). The environmental conditions that activate expression of V. cholerae virulence genes are called ToxR-inducing conditions (30°C and pH 6.5), whereas conditions of pH 8.4 and 37°C, termed ToxR-noninducing conditions, result in repression of the regulon (43). Expression of the tcpPH operon in the classical cholera strain is regulated by the same environmental conditions that regulate expression of the ToxT-dependent branch of the ToxR regulon, whereas expression of toxR has been shown to be constitutive in different environmental conditions. These observations have led to the proposal that the regulated expression of tcpPH couples environmental signals to the expression of toxT and ToxT-dependent genes (7).
As described above, significant knowledge exists about the role of transcriptional activators of virulence genes in V. cholerae. However, surprisingly little information is available about the mechanisms used by the bacterium to sense its environment and the regulatory pathways that transduce these signals to affect regulation of the ToxR regulon in different environmental conditions, particularly the regulatory signals that repress expression in nonpermissive environmental conditions. Recently, it was reported that disruption of the genes, cya and crp, which encode adenylate cyclase (cAMP) and the cAMP receptor protein (CRP), respectively, in a classical V. cholerae strain derepresses the expression of cholera toxin and TCP at the nonpermissive pH of 8.4 (46). A putative consensus binding site for the cAMP-CRP complex overlaps the −35 sequence of the tcpA promoter, and in the crp mutant, derepression of ctx expression also occurs in the toxR background. Therefore, the authors of that study hypothesized that the cAMP-CRP complex acts as a repressor at the level of toxT expression, perhaps by modulating activity at the tcpA promoter. According to this model, disruption of cya or crp would derepress tcpA from pH regulation, causing increased expression of toxT, and consequently cholera toxin and TCP, as a result of increased readthrough transcription from the upstream tcpA promoter (46).
The present study was undertaken to identify additional negative regulators of the ToxR regulon. To isolate mutants showing altered expression of the toxT promoter in the noninducing environmental conditions, we performed transposon mutagenesis on a classical strain of V. cholerae carrying a toxT-gusA transcriptional fusion. By inserting the toxT-gusA fusion within lacZ, separate from the rest of the tcpA operon, we hoped to identify novel genes exerting direct transcriptional control over either toxT expression or one of its regulators upstream in the regulatory cascade, and not genes such as cya or crp, that act at the tcpA promoter. Here, we report the identification of several mutants causing deregulated toxT-gusA expression at either the noninducing pH or the noninducing temperature, and we present a detailed characterization of one of them, a homolog of pepA from Escherichia coli that mediates negative regulation of the ToxR regulon at the noninducing pH in V. cholerae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are shown in Table 1. All strains were maintained at −70°C in Luria-Bertaini (LB) medium containing 15% glycerol. LB medium contained 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter. Ampicillin (25 or 100 μg/ml), streptomycin (100 μg/ml), tetracycline (15 μg/ml), or kanamycin (45 μg/ml) were added as appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotypea | Source or reference |
|---|---|---|
| V. cholerae | ||
| O395 | Wild type (classical Ogawa); Smr | 36 |
| C6709 | Wild type (El Tor Inaba); Smr | 5 |
| JB29 | O395 lacZ::toxT-gusA; Smr | This study |
| JB62 | JB29 eno::TnphoA; Smr Kmr | This study |
| JB65 | JB29 rhlB::TnphoA; Smr Kmr | This study |
| JB66 | JB29 cadB::TnphoA; Smr Kmr | This study |
| JB69 | JB29 pepA::TnphoA; Smr Kmr | This study |
| JB70 | JB29 yhdA::TnphoA; Smr Kmr | This study |
| JB73 | JB29 argH::TnphoA; Smr Kmr | This study |
| JB98 | JB29 pepA::pJB10; Smr Apr | This study |
| JB104 | YM2-34 pepA::pJB10; Smr Apr | This study |
| JB115 | JB29 lap::pJB12; Smr Apr | This study |
| JB123 | JB29 ΔpepA; Smr | This study |
| JB129 | JB123 toxR::pVM55; Smr Apr | This study |
| JB131 | JB123 tcpP::pYM2-16; Smr Apr | This study |
| JB132 | YM2-34 ΔpepA; Smr | This study |
| JB134 | JB29 toxR::pVM55; Smr Apr | This study |
| JB135 | JB29 tcpP::pYM2-16; Smr Apr | This study |
| JB148 | YM2-34 aphB::pJB15; Smr Apr | This study |
| JB150 | JB132 aphB::pJB15; Smr Apr | This study |
| JB152 | JB123 ΔpepA::pJB14; Smr Apr | This study |
| YM2-34 | O395 lacZ::tcpP-gusA; Smr | 45 |
| E. coli | ||
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 (φ80dlacZΔM15) | 20 |
| DH5α λpir | λpir lysogen of DH5α | Laboratory strain |
| SM10 λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir R6K | 43 |
| Plasmids | ||
| pUJ10 | Plasmid for the construction of divergent fusions to lacZ/phoA reporters; Apr | 12 |
| pGP704 | Suicide vector; pBR322 derivative with oriR6K mobRP4; Apr | 19 |
| pT689 | A p6891MCS derivative which contains the rrnB transcriptional terminator from plasmid pKK223-3; Apr | 45 |
| pCVD442 | Positive selection suicide vector; Apr | 16 |
| pYM2-16 | pGP704 derivative carrying a 230-bp internal tcpP fragment; Apr | Y. M. Murley and S. B. Calderwood, unpublished data |
| pVM55 | Suicide plasmid pJM703.1, containing an internal fragment of toxR; Apr | 43 |
| pJB4 | pUJ10 derivative carrying the toxT-gusA fusion; Apr | This study |
| pJB6 | pT689 derivative carrying the toxT-gusA fusion; Apr | This study |
| pJB10 | pGP704 derivative carrying an internal pepA fragment; Apr | This study |
| pJB12 | pGP704 derivative carrying an 800-bp internal fragment of lap; Apr | This study |
| pJB13 | pCVD442 derivative carrying a pepA allele containing an in-frame deletion; Apr | This study |
| pJB14 | pGP704 derivative carrying a wild-type copy of pepA; Apr | This study |
| pJB15 | pGP704 derivative carrying a 584-bp internal fragment of aphB; Apr | This study |
Sm, streptomycin; Ap, ampicillin; Km, kanamycin; Tc, tetracycline.
Bacteria were grown in either LB medium at pH 6.5 and 30°C to activate expression of ToxR-dependent virulence gene expression (inducing conditions) (43) or in LB medium at pH 6.5 and 37°C (noninducing temperature), LB medium at pH 8.4 and 30°C (noninducing pH), or LB medium at pH 8.4 and 37°C (noninducing temperature and pH) to repress virulence gene expression. Cultures were examined for the presence of autoagglutination, which reflects expression of TCP, after overnight growth with moderate aeration as described elsewhere (52).
In vitro manipulation of DNA.
Restriction analyses and cloning were done by standard techniques as described earlier (2). Enzymatic reagents were purchased from New England Biolabs (Beverley, Mass.) or Boehringer Mannheim (Indianapolis, Ind.) and used as specified by the manufacturer. Amplification of DNA by PCR was carried out as previously described (26).
Construction of a chromosomal toxT-gusA fusion.
A 761-bp toxT promoter-containing fragment that extends from within the tcpF gene to the start codon of toxT was PCR amplified from O395 using the primer pair RT19B (5′-AAAGGATCCTATGATATTGTGAATGTTGGTGGTG-3′, BamHI site underlined) and PAC59 (5′-AAACAGGATTTCTATATACATTAGTTTGAAAAG-3′; PstI site underlined) and cloned into the BglII and PstI sites of plasmid pUJ10. The phoA gene in pUJ10 was replaced with the promoterless gusA gene, encoding β-glucuronidase, from plasmid pWM2 (37), to create plasmid pJB4. The DNA fragment containing the toxT-gusA fusion was excised by digestion with XbaI and NotI and cloned into pT689, to yield plasmid pJB6. Plasmid pJB6 was introduced into O395 by electroporation, and the toxT::gusA fusion was exchanged by double homologous recombination into the lacZ gene as previously described (6), resulting in strain JB29. Insertion of the fusion into the chromosome at the lacZ locus was confirmed by Southern blot analysis.
TnphoA mutagenesis and determination of sequences adjacent to transposon insertions.
A library of random transposon insertion mutants derived from strain JB29 was constructed by TnphoA mutagenesis as previously described (51). Appropriate dilutions of the mutant library were plated onto LB agar plates, at pH 8.4, containing 40 μg of X-glucuronidase (Sigma Chemical Co., St. Louis, Mo.) per ml and incubated for 48 h at 30°C. Colonies that appeared blue on plates, reflecting elevated toxT-gusA expression under these noninducing conditions, were isolated for further characterization by quantitative β-glucuronidase assays.
To identify the DNA region that harbored the transposon in a given mutant, the TnphoA-chromosomal junction was amplified by three rounds of PCR amplification with a set of nested TnphoA-specific primers (PAC35, 5′-TATCGCCCTGAGCAGCCCGG; JB15L, 5′-AGCGGCAGTCTGATCACCCG; and JB16L, 5′-CTTCGGCATAATTACGTGCG) and random primers containing restriction site-specific sequences for 4- or 6-bp cutters at their 3′ end (JB11R, 5′-NNNNNNNNNNGATC; JB12R, 5′-NNNNNNNNNNTCGA; JB13R, 5′-NNNNNNNNNNGGATCC; and JB14R, 5′-NNNNNNNNNNGAATTC), as previously described (21). The amplicons were gel purified, recovered with a Compass kit (American Bioanalytical, Natick, Mass.), and sequenced with a TnphoA-specific primer (JB28L, 5′-TTCCAGAACAGGGCAAAACG) that lies 5′ of the primer used in the third round of amplification.
DNA sequencing was performed at the Massachusetts General Hospital Department of Molecular Biology in the DNA Sequencing Core Facility by using ABI Prism DiTerminator Cycle sequencing with AmpliTaq DNA polymerase FS and an ABI377 DNA sequencer (Perkin-Elmer/Applied Biosystems, Foster City, Calif.). The sequences obtained were analyzed by using the University of Wisconsin Genetics Computer Group package, version 8.0 (Madison, Wis.) and at the National Center for Biotechnology Information via the BLAST program (1).
Strain constructions.
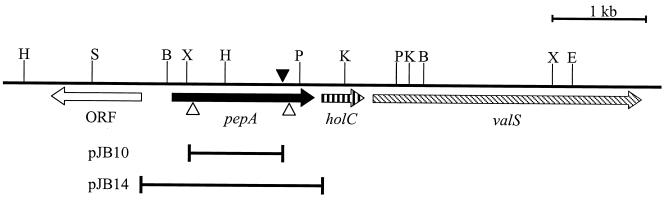
To construct a polar mutation in the pepA gene, a 938-bp XbaI-EcoRI fragment was amplified by PCR from the O395 chromosome with the primer pair JB29R (5′-GCGGTGATCTAGAGGGTAAAC-3′; XbaI site underlined) and JB30L (5′-CAACGGAATTCAGTACGTCACACA-3′; EcoRI site underlined), ligated to XbaI-EcoRI-digested suicide vector pGP704, and transformed into E. coli DH5α λpir to create plasmid pJB10. The recombinant plasmid was mobilized into JB29 and YM2-34 by conjugation, using the E. coli donor strain SM10λpir, to create strains JB98 and JB104, respectively. Disruption of the pepA locus was verified by Southern blot analysis.
To construct a nonpolar deletion in pepA, we first created pJB13, a derivative of the positive selection suicide vector pCVD442 carrying an in-frame deletion in pepA. To engineer the in-frame deletion in pepA, we performed overlap extension PCR using O395 chromosomal DNA as the template. The first round of PCR was performed with the primer pairs JB33R (5′-TTGCAGCAAGTCGACACTGTTTTCTGC-3′; SalI site is underlined) and JB34L (5′-CACCGCGCTGATGTGATGGCCCACACCGGGTACTTGATGCAG-3′; bases complementary to the 3′ fragment are underlined) to generate the 852-bp 5′ fragment; and with the primer pair JB35R (5′-CTGCATCAAGTACCCGGTGTGGGCCATCACATCAGCGCGGTG-3′; bases complementary to the 5′ fragment are underlined) and JB36L (5′-AGTCTACGAGCTCGGTAAAGGTACGC-3′; SacI site is underlined) to amplify the 800-bp 3′ fragment. The amplicons of the first round were purified and used as templates in a second PCR with the outer primer pairs JB33R and JB36L to generate a 1,610-bp fragment encompassing the pepA gene but containing a 957 bp in-frame deletion in its coding sequence. The 1.6-kbp PCR product was digested with SalI and SacI and cloned into similarly digested pCVD442 to generate pJB13. Plasmid pJB13 was mobilized into JB29 or YM2-34 by conjugation, and allelic replacement of the chromosomal copy of pepA was achieved as previously described (16). The deletion and the loss of vector sequences from the chromosome were confirmed by PCR amplification of the pepA region with flanking primers and sequencing across the deletion junction.
To complement the pepA mutation in JB123, a wild-type copy of pepA was amplified from O395 using the primers JB31R (5′-TGGTAGGAATTCCACCGTCAG-3′; EcoRI site underlined) and JB32L (5′-TGCGGTAGAGATCTTGGTCTC-3′; BglII site underlined). The 1,935-bp amplicon, which also included the upstream ORF-pepA and pepA-holC intergenic regions (see Fig. 2), was digested with EcoRI and BglII and cloned into pGP704. The recombinant plasmid, called pJB14, was conjugated into JB123 to force its integration at the pepA locus based on the homology between the wild-type and mutant copies of pepA, creating the merodiploid strain, JB152.
FIG. 2.
Schematic representation of the V. cholerae pepA locus and its surrounding region. ORFs are shown as arrows pointing in the direction of transcription. The site of insertion of TnphoA in JB69 is marked by a solid arrowhead. The open arrowheads indicate the fragment of pepA deleted in JB123. The 938-bp insert in plasmid pJB10 and the 1,935-bp insert in pJB14 used to complement the mutation in pepA are shown below the pepA ORF as black bars. H, HindIII; B, BamHI; X, XbaI; P, PstI; K, KpnI; E, EcoRI; S, SalI.
Disruptions in tcpP, aphB, and lap were made as described above for pepA with the pGP704 derivative vectors pYM2-16, pJB15, and pJB12, respectively, which carry internal fragments of these genes. Plasmid pVM55 was used for creating toxR disruptions as previously described (43).
Assays.
β-Glucuronidase assays were performed as described previously, with some minor modifications (27). For toxT-gusA fusion-containing strains, overnight cultures (approximately 16 h of incubation) were used for β-glucuronidase assays. Assays were performed by harvesting 500 μl of bacterial culture, pelleting the cells by centrifugation, washing them once with 50 mM phosphate buffer (pH 7.0), and resuspending the cells in 500 μl of phosphate buffer. Cell were lysed (20 to 250 μl, depending on the expected β-glucuronidase activity) by vortexing them for 20 s after the addition of 10 μl of toluene. The volume of the reaction tubes was brought up to 900 μl with 50 mM phosphate buffer, and samples incubated at 37°C for 10 min. p-Nitrophenyl-β-d-glucuronide (100 μl; Sigma) was added to start the reactions, and the samples were incubated at 37°C for 20 to 60 min. Reactions were stopped by the addition of 400 μl of 3 M 2-amino-2-methylpropanediol (Sigma), vortexed for 20 s, and centrifuged for 10 min, and the absorbance of the supernatants at 420 nm was measured. Similar assays were performed with the tcpP-gusA fusion-containing strains, except that overnight cultures in LB medium were subcultured the next day and samples harvested after 6 h of incubation at the inducing or noninducing conditions (expression of the fusion in inducing conditions was highest at this time point). Cholera toxin was assayed in culture supernatants after overnight growth in inducing or noninducing conditions, using GM-1 enzyme-linked immunosorbent assay (ELISA) as previously described (25).
Protein analysis.
Total cell lysates prepared from V. cholerae cells grown to stationary phase overnight were diluted 1:100 in distilled water, and the protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, Calif.). For immunoblot analysis, 40 μg of each protein sample was subjected to electrophoresis on a 15% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. The membrane was probed with a polyclonal rabbit anti-TcpA antibody (a gift of John J. Mekalanos) overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit antibody (Amersham Pharmacia Biotech, Piscataway, N.J.) for 1 h. The blot was developed using the ECL kit (Amersham Pharmacia) exposed to film for up to 1 min.
Nucleotide sequence accession number.
The sequence of the V. cholerae O395 pepA gene has been deposited in the GenBank database under accession no. AF282267.
RESULTS
Isolation of mutants showing deregulated expression of toxT in noninducing environmental conditions.
To isolate mutants of V. cholerae exhibiting enhanced toxT expression in noninducing growth conditions, we constructed a pool of approximately 50,000 independent TnphoA insertion mutants derived from the parental strain JB29. Subsequently, the mutant pool was plated onto LB agar plates, with the pH adjusted to 8.4 and containing 40 μg of the chromogenic substrate X-glucuronidase per ml, and incubated at 30°C for 48 h. Normally, growth of the parental strain JB29 at pH 8.4 and 30°C represses expression of toxT-gusA and colonies are white on the indicator medium for the first 24 to 48 h. We were interested in isolating colonies that appeared blue, since they might represent mutants exhibiting elevated toxT expression in noninducing conditions. Of the several dozen mutants initially isolated from plates, 16 reproducibly showed increased β-glucuronidase activity in noninducing conditions in quantitative liquid assays and were chosen for further analysis.
To determine the insertion sites of the transposon in the various mutants, we amplified the DNA region upstream of the junction with the transposon by PCR by using a series of nested primers specific to TnphoA and a set of random primers containing restriction site-specific sequences at the 3′ end as described in Materials and Methods. After three rounds of PCR amplification, the amplicons were gel purified and sequenced with a TnphoA-specific primer that read outward from the transposon toward the adjacent unknown chromosomal sequence. Six mutants—JB62, JB65, JB66, JB69, JB70, and JB73—yielded sufficient sequence information to enable us to identify the site of transposon insertion by similarity searches.
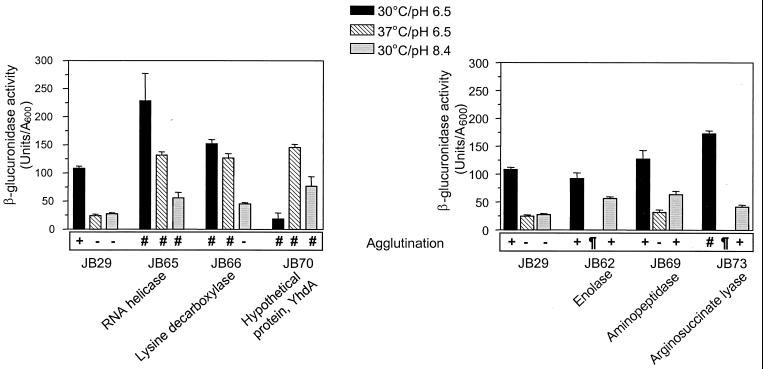
These mutants were characterized further with regard to toxT-gusA expression in different environmental conditions (Fig. 1). Based on the β-glucuronidase activities at either the noninducing pH or temperature, the six mutants could be divided into two groups. The first group, which included JB65, JB66, and JB70, showed slightly higher β-glucuronidase activities than the control at the noninducing pH but significantly higher activities at the noninducing temperature. For reasons that are unclear, JB70 reproducibly expressed very low levels of toxT-gusA in inducing conditions. All three strains in this group showed significant growth defects and, with the exception of the growth of JB66 at pH 8.4, grew too poorly in all conditions tested for us to assess their agglutination phenotypes.
FIG. 1.
β-Glucuronidase activity in three different growth conditions of V. cholerae O395 carrying a toxT-gusA fusion and six transposon-generated mutant strains. The left panel shows the control strain compared with three mutants exhibiting defective regulation of toxT-gusA expression in response to both temperature and pH. The right panel shows the control strain and three mutants exhibiting defective regulation in response to pH. The agglutination phenotypes are shown at the bottom of each graph. The symbols used for agglutination in the figure represent the following: +, agglutination; −, no agglutination; ¶, strain failed to grow; and #, strain grew too poorly to allow analysis of agglutination. Also shown in the figure are the proteins in the database showing the highest degree of homology to the disrupted sequences in each of the mutants. The data represent means plus the standard deviations (SDs) of two independent experiments, each done in duplicate.
The second group, including JB62, JB69, and JB73, exhibited higher toxT-gusA expression at the noninducing pH compared to the control. At the noninducing temperature, JB69 expressed the fusion at levels comparable to that of the parent, while JB62 and JB73 failed to grow reproducibly; JB73 also had nearly twofold-higher toxT-gusA expression in inducing conditions than the parental strain. Elevated toxT-gusA expression at pH 8.4 in JB62, JB69, and JB73 was accompanied by agglutination of cultures after overnight growth, suggesting that these mutants were also expressing elevated levels of the TCP in these normally nonpermissive conditions. The mutant JB69, which showed defective pH regulation of toxT-gusA expression and had no obvious growth defect, was selected for further detailed characterization.
Sequence analysis of the pepA gene.
We utilized PCR amplification and sequencing of the junctional fragment with TnphoA in JB69 and demonstrated that the transposon had inserted within pepA (Fig. 2). We searched the V. cholerae genome in the TIGR database and located the complete sequence of pepA and the adjacent chromosomal region. Analysis of an approximately 6-kb fragment in the vicinity of pepA revealed four open reading frames (ORFs) (Fig. 2). We identified the 447-bp coding region of a putative protein 82 bp downstream of pepA and transcribed in the same orientation, which was 41% identical (49% similar) to the χ subunit of DNA polymerase III holoenzyme from E. coli encoded by holC (accession no. P28905) (8). A third ORF, located 92 bp downstream of the holC homolog and oriented similarly, encoded a 953-amino-acid putative protein with 75% amino acid identity to valyl-tRNA-synthetase from E. coli (accession no. P07118). A fourth ORF, located upstream of pepA and transcribed divergently, encoded a putative protein with 42% identity to a 40.4-kDa hypothetical transmembrane protein from E. coli (accession no. P39340). The organization of these four ORFs in V. cholerae is strikingly similar to that of E. coli. Based on the significant similarity to E. coli genes and on their conserved arrangement on the chromosome, we have named these ORFs pepA, holC, and valS.
We designed oligonucleotide primers to amplify overlapping segments of the pepA gene from the classical biotype strain O395 and the El Tor strain C6709 by PCR. Direct sequencing of both strands of PCR amplified products revealed 30 nucleotide differences between the pepA coding sequences from the classical and the El Tor strains. However, all of these base changes were silent, resulting in identical deduced amino acid sequences of PepA from the two strains, which were also identical to that of El Tor strain N16961 in the TIGR database. The mole percent G+C content of pepA from O395 was 50%, which is consistent with that previously observed in V. cholerae (33). A putative ribosome binding site, AGGAG, is centered 8 bp upstream of the apparent pepA initiation codon. The pepA ORF is predicted to encode a protein of 503 amino acids with a calculated Mr of 54,617 and pI of 6.37. The insertion of the transposon in JB69 was found to have occurred immediately before a guanine nucleotide at position 1085 of the pepA ORF, resulting in deletion of the terminal 141 residues of the pepA gene product. The deduced amino acid sequence of PepA had a very high degree of similarity to PepA from E. coli (81% identity, 87% similarity; accession no. X86443), PhpA from Pseudomonas aeruginosa (55% identity; accession no. AF054622), PepA from P. putida (53% identity; accession no. AJ010261), and PepA from Haemophilus influenzae (61% identity; accession no. U32843), including perfect conservation of the residues believed to be involved in the active site of the enzyme based on crystallographic studies of bovine lens leucine aminopeptidase (4) (Fig. 3).
FIG. 3.
Alignment of the amino acid sequences of the PepA homologs from V. cholerae (V. chol), E. coli, P. aeruginosa (P. aeru), H. influenzae (H. infl), and P. putida (P. puti). An asterisk indicates that the residue at that position is conserved among all the sequences shown. The residues shown in boldface are in the active site of the enzyme based on the crystal structure of bovine lens leucine aminopeptidase (4).
Effect of pepA disruption on toxT-gusA expression.
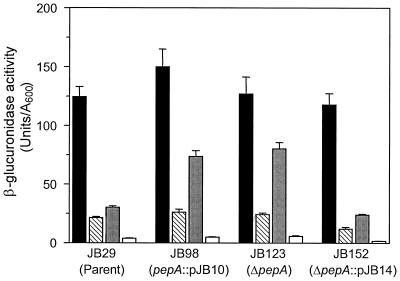
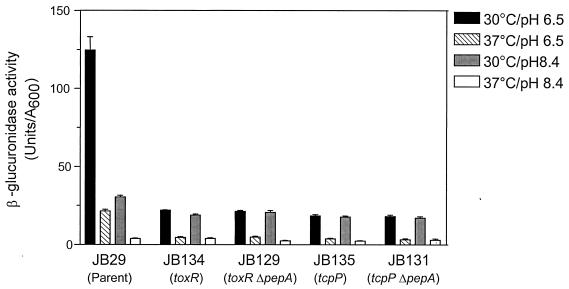
To determine whether the phenotype of JB69 was actually due to the insertion of TnphoA into the pepA gene or to an unlinked second-site mutation or polar effect on a downstream gene, we constructed additional pepA mutations in the toxT-gusA fusion strain JB29. Strain JB98 was created by forcing integration of a pepA internal fragment cloned in suicide vector pGP704, into the chromosome of JB29 at the pepA locus, thereby disrupting the ORF after codon 362. Analysis of toxT-gusA expression in JB98 revealed the same phenotype as the transposon mutant JB69, with approximately threefold-higher expression of the fusion in cultures grown at the nonpermissive pH of 8.4 (Fig. 4). It is worth noting that JB69 and JB98 still exhibited some residual regulation at pH 8.4, with toxT-gusA expression levels at pH 8.4 approximately 50% of those at pH 6.5, suggesting that classical V. cholerae may have regulatory pathways mediating pH regulation of toxT expression independently of pepA. Importantly, the disruption of pepA resulted in neither any difference in the maximal levels of toxT-gusA expression in inducing conditions (pH 6.5, 30°C) compared to the parent strain, nor did it influence the negative regulation of the fusion by the nonpermissive temperature. Similarly, when cultures were incubated at the nonpermissive conditions of 37°C and pH 8.4, expression was downregulated to the same extent as with the wild-type strain. These results suggest that pepA mediates negative regulation of toxT expression at the noninducing pH but not at the noninducing temperature.
FIG. 4.
β-Glucuronidase activity in different growth conditions of various V. cholerae strains carrying a transcriptional toxT-gusA fusion in the lacZ locus. The data represent the means + the SDs of two independent experiments, each done in triplicate. Columns: ■, 30°C, pH 6.5; ▧, 37°C, pH 6.5; , 30°C, pH 8.4; □, 37°C, pH 8.4.
As mentioned above, the pepA gene lies immediately upstream of the V. cholerae holC and valS homologs, therefore raising the possibility that the three genes might be expressed as a polycistronic message from the pepA promoter. The disruption of pepA by insertional mutagenesis in JB98 could result in polar effects on the two downstream genes, confounding the results obtained above. To address this possibility, we engineered an in-frame pepA deletion in JB29, extending from codon 69 to 387, creating strain JB123. Similar to the results obtained with JB98, toxT-gusA expression in JB123 was comparable to that of JB98 and was partially derepressed at the noninducing pH. We therefore concluded that the phenotype observed with strains JB69, JB98, and JB123 was the result of pepA disruption and not due to polar effects on downstream genes.
We also asked whether it was possible to rescue the pepA phenotype by introducing a single copy of the wild-type allele into strain JB123. We inserted a complete copy of pepA, including its upstream region, into the suicide vector pGP704 and introduced it into JB123 by conjugation. Integration of the plasmid at the pepA locus as a result of sequence homology between the introduced copy of pepA and the partially deleted pepA allele on the chromosome resulted in the merodiploid strain, JB152. This strain was found to have normal regulation of toxT-gusA expression when grown in medium at pH 8.4 (Fig. 4), confirming the role of pepA in pH regulation of toxT.
Deregulated expression of cholera toxin and TcpA at pH 8.4 in pepA mutants.
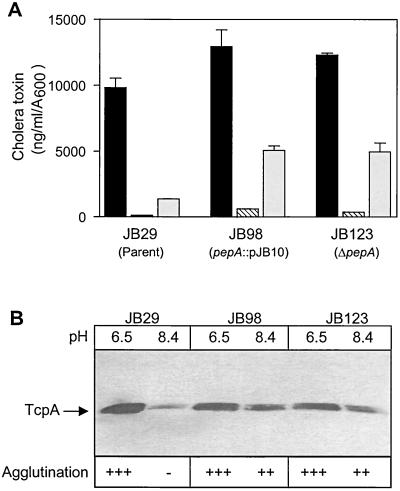
ToxT controls the expression of the two major virulence factors of V. cholerae, cholera toxin and the TCP (15). Since disruption of pepA caused elevated toxT expression at the noninducing pH, we hypothesized that deregulated expression of ToxT at pH 8.4 should also result in elevated expression of the genes controlled by it. To test whether the deregulated toxT expression in pepA mutants was similarly reflected in regulation of cholera toxin production, we performed cholera toxin ELISAs on culture supernatants of the pepA mutants grown in various conditions (Fig. 5A). Cholera toxin production in strains JB98 and JB123, containing the polar and in-frame pepA deletions, respectively, was only slightly elevated in inducing conditions or at the noninducing temperature. However, when the strains were grown at the noninducing pH, cholera toxin expression was three- to fourfold higher in the pepA mutants than in the control. Interestingly, the expression of cholera toxin was not completely deregulated in the pepA mutants, a finding that is consistent with our results from toxT-gusA expression assays.
FIG. 5.
Effect of pepA disruption on expression of virulence factors. (A) Cholera toxin production in various V. cholerae strains. Supernatants were assayed after overnight growth in LB medium in the conditions indicated. The data represent the means + the SDs of two separate experiments, each done in duplicate. Columns: ■, 30°C, pH 6.5; ▧, 37°C, pH 6.5; □, 30°C, pH 8.4. (B) Immunoblot of TcpA production and agglutination phenotype in various V. cholerae. Strains were grown overnight at 30°C in LB medium adjusted either to pH 6.5 or 8.4, as indicated above each lane. A total of 40 μg of total protein was loaded into each lane. The position of the TcpA band is indicated on the left. The agglutination phenotype of each culture is shown at the bottom of the gel. The symbols used in the figure indicate the following: +++, agglutination with the formation of a large cell pellet, with almost complete clearing of the supernatant; ++, agglutination with a moderately sized pellet, with some residual turbidity of the supernatant; and −, no agglutination.
We also measured cholera toxin production in strain JB152, in which an in-frame pepA deletion was complemented with a wild-type pepA allele. Similarly to the parent strain, cholera toxin production in JB152 showed an approximately 10-fold downregulation at the nonpermissive pH. Also, the level of cholera toxin production at pH 8.4 was similar to that of JB29, demonstrating that the wild-type copy of pepA had successfully restored pH regulation in this mutant (data not shown).
Like cholera toxin, expression of TCP in the classical biotype is also downregulated dramatically when V. cholerae is grown either at 37°C, at pH 8.4, or both. To test whether the pepA mutants showed elevated levels of TcpA at a noninducing pH, we performed immunoblot analysis with cell extracts prepared from cultures grown at pH 6.5 or at pH 8.4 (Fig. 5B). The parent strain showed approximately 10-fold lower levels of TcpA protein at pH 8.4 than at pH 6.5, while the pepA mutants JB98 and JB123 showed only twofold repression of TcpA at pH 8.4.
Expression of TCP in liquid cultures can be inferred by monitoring the agglutination of cells (52), which results in the formation of a cell pellet at the bottom of the test tube accompanied by almost complete clearing of the supernatant. Consistent with the results obtained from the immunoblot analysis, JB98 and JB123 cultures agglutinated at the noninducing pH, whereas the control strain failed to do so. Perhaps as a function of slightly lower levels of TcpA expression, the pellets formed by the pepA mutants JB98 and JB123 were smaller, and the supernatant was more turbid, at pH 8.4 compared to those formed at pH 6.5. None of the strains agglutinated at the noninducing temperature (data not shown), again suggesting that pepA mediates pH but not temperature regulation of toxT expression and the genes regulated by it.
Mutation in pepA does not change the requirement for ToxR and TcpP for toxT expression.
As mentioned above, the expression of toxT is dependent on the transcriptional activators ToxR and TcpP. The results suggesting that the pepA gene product functions as a negative regulator of virulence gene expression led us to question whether disruption of pepA could cause elevated expression of toxT at pH 8.4 in the absence of ToxR or TcpP. We tested this possibility directly by constructing disruptions in either toxR or tcpP in the wild-type and pepA backgrounds and then assaying toxT-gusA expression in different conditions of pH and temperature. Disruption of either toxR or tcpP in the wild-type background resulted in dramatically lower levels of expression of the fusion in all four growth conditions (Fig. 6). Expression levels in the double mutants were also very low and comparable to those of the single mutants. These results suggest that loss of PepA function does not change the requirement for the transcriptional activators ToxR and TcpP for toxT expression in either inducing or noninducing conditions and raise the possibility that PepA may act indirectly on toxT, through either toxR or tcpP. However, our results do not exclude the possibility that PepA may act at more than one level in the virulence gene regulatory cascade.
FIG. 6.
Comparison of toxT-gusA expression in parent and single and double toxR, tcpP, and pepA disruption mutants. β-Glucuronidase activity was assayed after overnight growth of cultures in different growth conditions. The data represent the means + the SDs of two independent experiments, each done in triplicate.
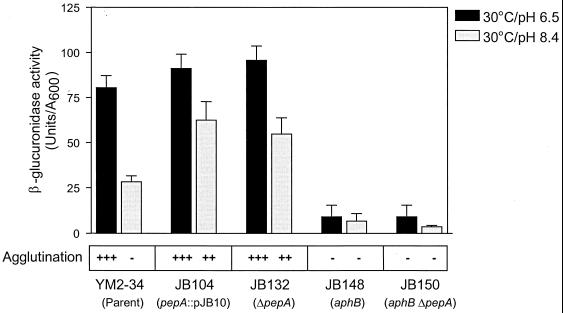
Disruption of pepA partially relieves negative regulation of tcpP expression by noninducing pH.
Recent reports from our own and other laboratories have established the important role of TcpP and TcpH in transcription of toxT in classical V. cholerae (7, 22). Transcription of tcpPH in classical V. cholerae is regulated by pH and temperature, while the expression of toxR has been shown to be constitutive in different environmental conditions. These observations have led to the hypothesis that regulated expression of TcpP and TcpH may couple environmental growth conditions to transcription of toxT and ToxT-dependent virulence genes in V. cholerae (7).
Our observations that pepA mutants showed partially defective pH regulation of toxT and its dependent genes raised the possibility that pepA may be necessary for negative regulation of tcpPH expression at pH 8.4. To test this hypothesis, we constructed a polar pepA disruption and an in-frame pepA deletion in the strain YM2-34 (45), which contains a tcpP-gusA fusion cloned in the lacZ locus in a classical V. cholerae background, resulting in the strains JB104 and JB132, respectively. When these pepA mutant strains were grown in LB medium, pH 6.5, no significant difference in the levels of β-glucuronidase activity were found compared to the parental control (Fig. 7). However, when cultures were grown at the noninducing pH of 8.4, JB104 and JB132 expressed β-glucuronidase activity at levels approximately twofold higher than did YM2-34, suggesting that pepA disruption partially relieves the negative regulation of tcpPH expression at the noninducing pH. These same two mutants also agglutinate in the nonpermissive pH, similar to the results described above.
FIG. 7.
β-Glucuronidase activity of V. cholerae parent and pepA and aphB single and double disruption mutants. All strain carry a transcriptional tcpP-gusA fusion in the lacZ locus. Overnight cultures in LB broth were subcultured into LB medium adjusted to either pH 6.5 or pH 8.4 and incubated at 30°C for 6 h before the samples were harvested for assays. The data represent the means + the SDs of two independent experiments, each done in duplicate. The agglutination phenotypes were assessed after overnight growth of cultures (see Fig. 5 for symbols).
Recently, two proteins, AphB and AphA, have been shown to function synergistically to activate tcpPH expression in V. cholerae, although the expression of neither aphA nor aphB is strongly regulated by environmental conditions (30, 47). To test whether the elevated tcpP expression at pH 8.4 in the pepA mutant strain could be observed in the absence of the tcpP transcriptional activator, AphB, we engineered an aphB disruption by insertional mutagenesis in JB123 (ΔpepA) and in the parental strain, JB29. As shown in Fig. 7, disruption of aphB dramatically reduced expression of the tcpP-gusA fusion at both the inducing and the noninducing pH in the aphB single mutant, as well as in the pepA aphB double mutant, suggesting that AphB is required for transcriptional activation of tcpPH even in the absence of the negative regulator, PepA. These results, along with those presented above, lead us to conclude that pepA mediates pH regulation of the ToxT-dependent branch of the ToxR regulon by acting as a negative regulator at a level upstream of tcpPH transcription in the virulence gene regulatory cascade in classical V. cholerae.
The V. cholerae lap gene does not mediate pH regulation of the ToxR regulon.
Recently, Toma and Honma reported the identification of a V. cholerae gene, lap, encoding a 501-amino-acid protein with homology to the Vibrio proteolyticus aminopeptidase and showed that this was an active leucine aminopeptidase by the ability of the recombinant lap gene product to cleave the substrate leucyl-p-nitroanilide (53). The lap gene was found by PCR analysis to be widely distributed among V. cholerae strains but was absent in other bacterial species examined. While the pepA gene identified in our screen encodes a protein with homology to leucine aminopeptidases from E. coli and other species, there is no sequence similarity between it and the V. cholerae lap gene. To test whether the lap gene, like pepA, also mediates environmental regulation of virulence gene expression in V. cholerae, we engineered a disruption in lap by insertional mutagenesis with the suicide plasmid pJB12, creating the strain JB115. Analysis of toxT-gusA expression and agglutination of JB115 in cultures grown at either the nonpermissive pH or temperature revealed no difference between JB115 and the parental strain JB29 (data not shown). Thus, the lap gene product, unlike that of pepA, does not appear to have a role in pH regulation of the ToxR regulon.
DISCUSSION
The expression of virulence determinants in classical strains of V. cholerae is coordinately regulated by several regulatory proteins and is strongly influenced by signals derived from the host and the environment. We report here the identification of two classes of transposon-generated mutants showing altered regulation of toxT and its dependent genes when grown at a noninducing pH or noninducing temperature. The successful isolation of several different mutants exhibiting deregulated pH and temperature control suggests that environmental regulation of virulence genes in V. cholerae is a complex process involving multiple genes and regulatory pathways. While several positive regulators of the ToxR regulon in V. cholerae have been identified, we know little about negative regulation of this regulon. We demonstrate here that pepA acts as a negative regulator of virulence determinants at the noninducing pH, suggesting that downregulation of gene expression in nonpermissive conditions may also play an important role in controlling the ToxR regulon. The fact that disruption of pepA deregulates pH but not temperature regulation of toxT also suggests that these two environmental signals are perceived differently by the organism and have separable signaling pathways in the bacterium.
It is now well established that many pathogenic bacteria have evolved from related nonpathogenic species by acquiring virulence genes that remain clustered together on the bacterial chromosome, forming pathogenicity islands (31). Since these horizontally acquired genes are often regulated by the same environmental signals that regulate non-virulence-associated genes, it is possible that horizontally acquired genes may utilize or adapt to existing regulatory circuits modulating gene expression in response to environmental cues. Therefore, it is not surprising that the horizontally acquired, phage-encoded ctxAB and tcp operons in V. cholerae have acquired regulation by genes such as toxR that are located elsewhere on the primordial bacterial chromosome. Evidence for the regulation of accessory genetic elements by ancestral chromosomal components, such as cAMP and CRP (46), AphA and AphB (30, 47), and the NQR complex (23), points toward a significant interplay between the metabolic and virulence functions of this bacterium. PepA appears to be another example of an ancestral chromosomal gene that is involved in the regulation of phage-encoded virulence genes and presumably has other cellular metabolic functions besides regulation of virulence genes, as has been hypothesized for ToxR and cAMP-CRP.
The pepA gene product in E. coli is a multifunctional protein. It has been shown to be an active leucine aminopeptidase (55), a member of the multiprotein complex that resolves plasmid multimers into monomers to result in heritable stability of plasmid ColE1 (48), and a repressor in the pyrimidine-mediated negative regulation of the carAB operon that encodes carbamoylphosphate synthetase (10). Recently, a role for PhpA, the P. aeruginosa homolog of PepA, was demonstrated in the regulation of virulence determinants. Disruption of phpA in an algB genetic background resulted in increased expression of the alginate biosynthetic operon, suggesting that phpA may act as a negative regulator of virulence-associated genes in this pathogenic bacterium as well (57).
Considering the fact that PepA homologs serve a remarkably wide range of functions, there are several possible ways in which PepA could exert a negative regulatory effect on the ToxR regulon in V. cholerae. The regulatory role of PepA could be dependent on its enzymatic activity and could depend on an exopeptidase activity to activate a target repressor protein, or inactivate a transcriptional activator of tcpPH, at the noninducing pH. For example, the removal of an N-terminal methionine residue results in the activation of glutamine phosphoribosylpyrophosphate aminotransferase that requires an N-terminal cysteine residue for its activity (54). AphA and AphB have been shown to activate tcpPH expression but do not appear to be strongly regulated themselves by the environmental signals that modulate tcpPH transcription (30, 47). It is conceivable that PepA could act directly or indirectly to modify AphA or AphB activity. However, by analogy with E. coli, it appears less likely that the role of aminopeptidase as a pH regulator in V. cholerae is dependent on its enzymatic activity. Aminopeptidases have broad substrate specificity and can cleave N-terminal amino acids from peptides of various sizes and sequences (38, 39), making it less likely that there are specific targets for PepA activity. Moreover, it has been demonstrated that the aminopeptidase activity of PepA in E. coli is separable from its regulatory function and that the aminopeptidase activity is not required for either plasmid ColE1 Xer site-specific recombination or for the repression of the carAB operon (10, 34). However, it is possible that the regulatory role of PepA may be dependent on the degradation or modification of a peptide that functions as an inducer or repressor of the ToxR regulon or as a signaling molecule in specific environmental conditions.
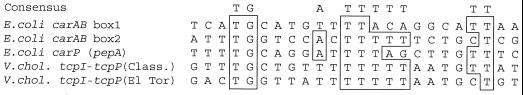
Another intriguing possibility is that PepA may mediate pH regulation by functioning as a DNA-binding protein and directly affecting transcription. In E. coli, PepA has been conclusively shown to be a sequence-specific DNA-binding protein involved both in the regulation of carAB, the carbamoylphosphate synthetase operon, and as an autorepressor at the pepA promoter (10). Sequence alignment of five experimentally identified DNA-binding sites of PepA, three of which are shown in Fig. 8, has revealed that a number of positions that are AT-rich are strongly conserved (10). There are two PepA target sites, 25 to 30 bp in length and 65 nucleotides apart, in the carAB operon in E. coli and Salmonella enterica serovar Typhimurium and in the ColEI cer site but only one site in the promoter region of the pepA gene itself. Based on the consensus sequence proposed by Charlier et al. (10), we have identified a putative PepA target site in the tcpPH-tcpI intergenic region (Fig. 8). This site is centered 133 bp upstream of the tcpPH transcriptional start site and 49 bp upstream of the tcpI transcriptional start (45). In the classical biotype, the putative target sequence matches the consensus in 9 of 10 positions, while in the El Tor biotype, it matches the consensus in 8 of 10 positions (Fig. 8).
FIG. 8.
Sequence alignment of the DNA-binding sites for PepA in the control regions of the E. coli carAB operon and the E. coli pepA gene was used to generate an AT-rich consensus shown above the sequences (10). Bases conserved in the classical and El Tor V. cholerae tcpI-tcpP intergenic region with the consensus sequence are boxed.
We speculate that PepA could act by binding to this putative target site in the tcpPH control region and interfering with transcriptional regulation. It is also possible that binding of PepA to the target site could interfere with the expression of tcpI, which in turn may be involved in regulating other genes in the ToxR regulon. Whether, in fact, PepA can directly bind to the tcpPH promoter and repress its activity either by inducing changes in DNA topology or by potentially competing with AphA or AphB for target sites in the tcpI-tcpPH intergenic region is currently under investigation. We are also investigating whether expression of pepA itself is regulated by environmental cues in V. cholerae.
Our results showing that disruption of the lap gene, encoding another V. cholerae leucine aminopeptidase with no sequence homology to PepA, does not alter regulation of toxT are consistent with the conclusion that PepA has distinct regulatory roles not shared by other cellular aminopeptidases. E. coli and serovar Typhimurium have several different aminopeptidases (39, 40). In serovar Typhimurium, metabolic analysis of strains carrying mutations in the genes encoding aminopeptidases A, B, D, and N suggests that the enzymes function to break down exogenously supplied peptides for use as nutrients. In addition, they degrade endogenous peptides generated from cleavage of cellular protein. However, only PepA appears to have additional regulatory functions in Xer recombination and regulation of pyrimidine biosynthesis (10, 38, 39).
We have isolated several mutants that show deregulated toxT expression at the noninducing pH alone or at both the noninducing pH and the noninducing temperature. As yet, we do not know the precise role of the genes disrupted in these deregulated mutants. It is possible that one or more of these genes may participate in the same control pathway as PepA. In E. coli, cer site-specific recombination involves a multiprotein complex in which PepA appears to serve an accessory role (49). Therefore, it is possible that PepA may play a role as a member of a protein complex that controls gene regulation at the noninducing pH, and disruption of pepA would interfere with the regulation mediated by this complex. This model would also explain why only partial deregulation at the noninducing pH is observed in pepA mutants, since the loss of PepA would result in defective pH sensing but perhaps not the total loss of function of the pH-sensing complex.
ACKNOWLEDGMENTS
We thank Barry Wanner, William Metcalf, and Patrick Piggot for the gift of plasmids and John Mekalanos for the anti-TcpA antibody. We also thank Joan Butterton, Yvette Murley, Camille Kotton, and members of the Calderwood laboratory for the gift of reagents and helpful advice and Costi Sifri for comments on the manuscript.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases, RO1 AI44487, to S.B.C.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Brown R C, Taylor R K. Organization of tcp, acf, and toxTgenes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 4.Burley S K, David P R, Sweet R M, Taylor A, Lipscomb W N. Structure determination and refinement of bovine lens leucine aminopeptidase and its complex with bestatin. J Mol Biol. 1992;224:113–140. doi: 10.1016/0022-2836(92)90580-d. [DOI] [PubMed] [Google Scholar]
- 5.Butterton J R, Beattie D T, Gardel C L, Carroll P A, Hyman T, Killeen K P, Mekalanos J J, Calderwood S B. Heterologous antigen expression in Vibrio choleraevector strains. Infect Immun. 1995;63:2689–2696. doi: 10.1128/iai.63.7.2689-2696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterton J R, Ryan E T, Acheson D W, Calderwood S B. Coexpression of the B subunit of Shiga toxin 1 and EaeA from enterohemorrhagic Escherichia coli in Vibrio choleraevaccine strains. Infect Immun. 1997;65:2127–2135. doi: 10.1128/iai.65.6.2127-2135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 8.Carter J R, Franden M A, Lippincott J A, McHenry C S. Identification, molecular cloning and characterization of the gene encoding the chi subunit of DNA polymerase III holoenzyme of Escherichia coli. Mol Gen Genet. 1993;241:399–408. doi: 10.1007/BF00284693. [DOI] [PubMed] [Google Scholar]
- 9.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxTmutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 10.Charlier D, Hassanzadeh G, Kholti A, Gigot D, Pierard A, Glansdorff N. carP, involved in pyrimidine regulation of the Escherichia colicarbamoylphosphate synthetase operon encodes a sequence-specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColEI multimers. J Mol Biol. 1995;250:392–406. doi: 10.1006/jmbi.1995.0385. [DOI] [PubMed] [Google Scholar]
- 11.Crawford J A, Kaper J B, DiRita V J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiRita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 14.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 15.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coliby using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill D M. The arrangement of subunits in cholera toxin. Biochemistry. 1976;15:1242–1248. doi: 10.1021/bi00651a011. [DOI] [PubMed] [Google Scholar]
- 18.Gill D M, Meren R. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: basis of the activation of adenylate cyclase. Proc Natl Acad Sci USA. 1978;75:3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg M B, Boyko S A, Calderwood S B. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Harrison R W, Miller J C, D'Souza M J, Kampo G. Easy gene walking. BioTechniques. 1997;22:650–653. doi: 10.2144/97224bm17. [DOI] [PubMed] [Google Scholar]
- 22.Hase C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hase C C, Mekalanos J J. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio choleraeis a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmgren J. Comparison of the tissue receptors for Vibrio cholerae and Escherichia colienterotoxins by means of gangliosides and natural cholera toxoid. Infect Immun. 1973;8:851–859. doi: 10.1128/iai.8.6.851-859.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. [Google Scholar]
- 27.Jefferson R A, Burgess S M, Hirsh D. β-Glucuronidase from Escherichia colias a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. . (Erratum, 8:316.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaolis D K, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 30.Kovacikova G, Skorupski K. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPHpromoter to activate expression of the ToxR virulence cascade. J Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C A. Pathogenicity islands and the evolution of bacterial pathogens. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 32.Li C C, Crawford J A, DiRita V J, Kaper J B. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol Microbiol. 2000;35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 33.Mazel D, Dychinco B, Webb V A, Davies J. A distinctive class of integron in the Vibrio choleraegenome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 34.McCulloch R, Burke M E, Sherratt D J. Peptidase activity of Escherichia coliaminopeptidase A is not required for its role in Xer site-specific recombination. Mol Microbiol. 1994;12:241–251. doi: 10.1111/j.1365-2958.1994.tb01013.x. [DOI] [PubMed] [Google Scholar]
- 35.Mekalanos J J, Collier R J, Romig W R. Enzymic activity of cholera toxin. I. New method of assay and the mechanism of ADP-ribosyl transfer. J Biol Chem. 1979;254:5849–5854. [PubMed] [Google Scholar]
- 36.Mekalanos J J, Swartz D J, Pearson G D, Harford N, Groyne F, de Wilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 37.Metcalf W W, Wanner B L. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene. 1993;129:17–25. doi: 10.1016/0378-1119(93)90691-u. [DOI] [PubMed] [Google Scholar]
- 38.Miller C G. Genetic mapping of Salmonella typhimuriumpeptidase mutations. J Bacteriol. 1975;122:171–176. doi: 10.1128/jb.122.1.171-176.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller C G, Mackinnon K. Peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974;120:355–363. doi: 10.1128/jb.120.1.355-363.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller C G, Schwartz G. Peptidase-deficient mutants of Escherichia coli. J Bacteriol. 1978;135:603–611. doi: 10.1128/jb.135.2.603-611.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller V L, DiRita V J, Mekalanos J J. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 45.Murley Y M, Behari J, Griffin R, Calderwood S B. Classical and El Tor biotypes of Vibrio cholerae differ in timing of transcription of tcpPHduring growth in inducing conditions. Infect Immun. 2000;68:3010–3014. doi: 10.1128/iai.68.5.3010-3014.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skorupski K, Taylor R K. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPHoperon. Mol Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 48.Stirling C J, Colloms S D, Collins J F, Szatmari G, Sherratt D J. xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J. 1989;8:1623–1627. doi: 10.1002/j.1460-2075.1989.tb03547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Summers D K. Derivatives of ColE1 cershow altered topological specificity in site-specific recombination. EMBO J. 1989;8:309–315. doi: 10.1002/j.1460-2075.1989.tb03378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor R, Shaw C, Peterson K, Spears P, Mekalanos J. Safe, live Vibrio cholerae vaccines? Vaccine. 1988;6:151–154. doi: 10.1016/s0264-410x(88)80019-7. [DOI] [PubMed] [Google Scholar]
- 51.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoAgene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toma C, Honma Y. Cloning and genetic analysis of the Vibrio choleraeaminopeptidase gene. Infect Immun. 1996;64:4495–4500. doi: 10.1128/iai.64.11.4495-4500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tso J Y, Hermodson M A, Zalkin H. Glutamine phosphoribosylpyrophosphate amidotransferase from cloned Escherichia coli purF. NH2-terminal amino acid sequence, identification of the glutamine site, and trace metal analysis. J Biol Chem. 1982;257:3532–3536. [PubMed] [Google Scholar]
- 55.Vogt V M. Purification and properties of an aminopeptidase from Escherichia coli. J Biol Chem. 1970;245:4760–4769. [PubMed] [Google Scholar]
- 56.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 57.Woolwine S C, Wozniak D J. Identification of an Escherichia coli pepA homolog and its involvement in suppression of the algB phenotype in mucoid Pseudomonas aeruginosa. J Bacteriol. 1999;181:107–116. doi: 10.1128/jb.181.1.107-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]