Abstract
Pikas (Lagomorpha: Ochotonidae) are small mouse-like lagomorphs. To investigate their adaptation to different ecological environments during their dispersal from the Qinghai-Xizang (Tibet) Plateau (QTP), we collected 226 pikas and measured 20 morphological characteristics and recorded habitat information. We also sequenced the genome of 81 specimens, representing 27 putative pika species. The genome-wide tree based on 4 090 coding genes identified five subgenera, i.e., Alienauroa, Conothoa, Lagotona, Ochotona, and Pika, consistent with morphometric data. Morphologically, Alienauroa and Ochotona had similar traits, including smaller size and earlier divergence time compared to other pikas. Consistently, the habitats of Alienauroa and Ochotona differed from those of the remaining subgenera. Phylogenetic signal analysis detected 83 genes significantly related to morphological characteristics, including several visual and hearing-related genes. Analysis of shared amino acid substitutions and positively selected genes (PSGs) in Alienauroa and Ochotona identified two genes, i.e., mitochondrial function-related TSFM (p.Q155E) and low-light visual sensitivity-related PROM1 (p.H419Y). Functional experiments demonstrated that TSFM-155E significantly enhanced mitochondrial function compared to TSFM-155Q in other pikas, and PROM1-419Y decreased the modeling of dynamic intracellular chloride efflux upon calcium uptake. Alienauroa and Ochotona individuals mostly inhabit different environments (e.g., subtropical forests) than other pikas, suggesting that a shift from the larger ancestral type and changes in sensory acuity and energy enhancement may have been required in their new environments. This study increases our understanding of the evolutionary history of pikas.
Keywords: Pikas, Genomics, Morphometrics, Adaptive evolution, Sensory and energy functions
INTRODUCTION
Pikas (genus Ochotona; Lagomorpha: Ochotonidae) are small mouse-like lagomorphs (Wang et al., 2020). Once widely distributed across Eurasia, Africa, and North America, most extant pikas are found in China, especially in the Hengduan Mountains, with a few species inhabiting North America, Central Asia, and the Russian Far-East (Lissovsky et al., 2019; Liu et al., 2017; Wang et al., 2020). Pikas are larger than mice but smaller than rabbits, and exhibit size differences among species. Extant pikas inhabit alpine areas, tundra, steppes, and other cold environments (Wang et al., 2020; Wilson and Smith, 2015) and play important ecological roles in stabilizing plant communities and promoting plant growth due to grazing (Smith At, 2018; Wang et al., 2020; Wilson and Smith, 2015).
The taxonomy and phylogenetic relationships within the genus Ochotona have been explored using different mitochondrial genes, nuclear genes, and exome data (Ge et al., 2013; Wilson DE, 2005; Koju et al., 2017; Lanier and Olson, 2009; Lissovsky, 2014; Liu et al., 2017; Melo-Ferreira et al., 2015; Niu Yd, 2004; Yu et al., 2020). Liu et al. (2017) proposed five subgenera (Conothoa, Ochotona, Pika, Alienauroa, and Lagotona) based on cytochrome b (cyt b) sequences, while Wang et al. (2020) grouped Lagotona into Pika and supported only four subgenera (Conothoa, Ochotona, Pika, and Alienauroa) based on whole-genome coding sequences. The genetic basis of high-altitude adaptations in pikas has also been investigated (Henry and Russello, 2013; Rankin et al., 2017; Wang et al., 2020), but with an emphasis on adaptations to hypoxia, ultraviolet radiation, and cold tolerance. However, pikas have occupied various ecological environments during their dispersal from the Qinghai-Xizang (Tibet) Plateau (QTP) to other parts of Eurasia and North America (Wang et al., 2020). Indeed, the major clades/subgenera of extant pikas are classified into four ecotypes according to their distribution and ecological environment. The Conothoa mountain group is mainly distributed in the Himalayas and surrounding habitats dominated by talus; the Alienauroa forest group is primarily found along the eastern edge of the QTP in humid forest areas dominated by moss and leaf litter; the Pika northern group is distributed at high latitudes with rock and steppe habitats; and the Ochotona shrub-steppe group is mainly distributed in the northern areas of the QTP dominated by shrub-steppe (Wang et al., 2020).
As pika species inhabit different environments, and environmental stress was a major driving force during pika evolution (Parsons, 2005; Kristensen et al., 2020), we hypothesized that certain functions and pathways may be under different selection in different pikas. To study the evolutionary history of pikas, we measured 20 external, cranial, and dental characteristics, and recorded habitat information of 226 adult pikas collected from 107 sampling sites in China. We also sequenced the genomes of 81 specimens representing 27 putative species of pika and reconstructed a genome-wide tree based on 4 090 coding genes. Based on extensive genomic, morphological, and habitat datasets, we: (1) assessed the morphological variation and evolution of different pikas; (2) explored the adaptations of different pikas and the relationship between molecular genetics and phenotypic characteristics; and (3) performed functional experiments based on cellular mutations.
MATERIALS AND METHODS
Ethics statement
All samples were obtained following the Guidelines of the American Society of Mammalogists (ASM guidelines) (Sikes et al., 2011) and the laws and regulations of China for the protection of terrestrial wild animals (State Council Decree 1992). The BGI Institute of Review Board of Bioethics and Biosafety (BGI-IRB) (No. FT17005) approved all collection and research protocols. Voucher specimens were deposited in the Sichuan Academy of Forestry, Chengdu, China.
Sample and data collection
In this study, we selected 226 pika specimens collected from 107 sampling sites in China over the past 30 years (33 samples were provided by the Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, China). Specimens were captured by randomly placing cage-type small-animal traps at the sampling sites. After capture, the pikas were anesthetized with ether on the spot and measured for external features. Of the captured individuals, 226 adult specimens were transported to the laboratory for processing, and the rest were released in situ. Upon return to the laboratory, the 226 adult specimens were anesthetized and euthanized by cervical dislocation. The pikas were then dissected, and tissue samples were extracted and stored in 95% analytical alcohol. All specimens were adults with intact skulls for morphometric analyses. Ecological conditions were recorded at the time of field sampling (e.g., elevation, habitat, landscape type). Species identification using morphological characteristics was based on the Taxonomic Index of the Ochotona in China (Feng, 1985), Taxonomic list and distribution of mammal species and subspecies in China (Wang, 2003), and previous articles (Liu et al., 2017; Wang, 1988; Gong et al., 2000), as well as drawings and morphological descriptions in the literature (Feng, 1985; Hoffmann Rs, 2005; Smith, 2009; Sokolov et al., 2009; Wilson et al., 1993). We also sequenced the whole genomes of 81 individuals, although some samples were sequenced at low coverage.
High-throughput sequencing
Total genomic DNA was extracted from muscle and liver tissue from the 81 samples using a Gentra Puregene Tissue Kit (Qiagen, USA) according to the manufacturer’s protocols. We constructed genomic libraries (paired-end libraries with insert sizes of ~300 bp) following BGISEQ 500 (Huang et al., 2017) or Illumina Hiseq 2500 (USA) (Zhang, 2021) protocols (details in Supplementary Table S1). We then sequenced each library and generated >45 Gb of data (10–30X) for one representative specimen of each morphologically distinct species, and ~5 Gb of data (1–3X) were generated for the remaining specimens of each species. In the high-coverage datasets, low-quality reads were excluded if one or more of the following criteria were met: (1) N-content >10%; (2) adapter-contaminated reads with adapter sequences overlapping reads by >50%; (3) read length below Q10 >20%. For the low-coverage datasets, the low-quality read filtering criteria and sequencing platform information are listed in Supplementary Table S1. In total, almost 2 Tb of 150-bp paired-end data were obtained.
Measurement of morphological characteristics
We measured 20 phenotypic characteristics in the 226 adults with intact skulls to construct the morphological dataset. Individuals were identified as adults by inspection of their teeth (see detailed criteria in Supplementary Methods). We measured external, cranial, and dental characteristics for all specimens. External measurements were recorded upon capture in the field to an accuracy of 0.5 mm. Cranial and dental characteristics were measured in the laboratory using an electronic vernier caliper to an accuracy of 0.01 mm. External measurements included head and body length (HBL), ear length (from notch to top of ear, EL), and hind foot length (HFL) excluding claws. Cranial measurements included skull greatest length (SGL), skull basal length (SBL), occipital condyle to nasal bone length (OCNL), zygomatic breadth (ZB), minimal interorbital width (minimum distance across frontal bone between orbits, IOW), skull height (vertical distance from ventral surface of bullae to top of cranium, SH), auditory bulla length (ABL), eyepit (eye socket) length (maximum inner diameter of long axis of eyepit, EPL), eyepit breadth (maximum inner diameter of short axis of eyepit, EPB), nasal bone length (NBL), and nasal bone breadth (NBB). Dental measurements included upper molar occlusal surface length (UOSL) and lower molar occlusal surface length (LOSL). We also recorded four discrete morphological features, i.e., presence of congenital tragus (CT); presence of oval foramen (OF); number of incisive and palatal foramen (IF and PF); and shape of skull (Skull). In addition, we calculated ratios between features, including ratio of ear length to head and body length (EHR) and ratios of auditory bulla length, eyepit length, and eyepit breadth to skull greatest length (ASR, ESLR, ESBR).
We analyzed morphometric variation among the 226 adult specimens using principal component analysis (PCA) in R v.4.0.2 (Null et al., 2011). We applied the Kaiser-Meyer-Olkin (KMO) and Bartlett’s tests in the R Psych package v2.1.9 (Revelle, 2020) to check PCA fitness. We performed Permutational Multivariate Analysis of Variance (PERMANOVA) in the R Vegan package v2.5.7 (Oksanen et al., 2019) to examine statistical differences at the species and subgenus levels. Heatmaps were drawn using the R package ComplexHeatmap v2.10.0 (Gu et al., 2016). Box plots were drawn using the R package ggplot2 v3.3.5, with the significance threshold determined using the t-test.
Molecular data pre-processing and construction of ortholog datasets
We downloaded the reference genome and full gene dataset of the North American Pika (Ochotona princeps, vOchPri2.0) from Ensemble (Hubbard et al., 2002) to obtain the orthologous genes of our sequenced samples. We then obtained the lineage-specific single-copy ortholog list of the Euarchontoglires group from the BUSCO database (v10.1) (Simão et al., 2015) and extracted the corresponding orthologous genes from the full gene dataset of O. princeps. To avoid mapping uncertainty that may be caused by paralogs, genes with high similarity within the gene set were removed using BLASTn (v2.6.0+, e value<1e-5) (Altschul et al., 1990). Subsequently, we obtained a O. princeps single-copy orthologous gene dataset with 5 684 complete genes. We aligned the whole-genome sequencing (WGS) data of each sequenced individual to the ortholog dataset using BWA-MEM v0.7.17 (Li, 2013) with default parameters and obtained the corresponding genes using the consensus calling function in BCFtools v.1.10.2 (Danecek et al., 2021). High-quality coding sequences (CDS) were extracted from the mapping results and subjected to pipeline analysis, as detailed in the Supplementary Methods.
We aligned the corresponding protein sequences using MAFFT v.7.453 (Katoh and Standley, 2013) and conducted CDS alignment using PAL2NAL v14 (Suyama et al., 2006) based on the protein alignments to obtain an individual-level nuclear gene CDS dataset. Representative sample sequences of each morphologically distinct species with the highest sequencing depth were selected to construct a species-level nuclear gene CDS dataset. In addition, four species with published genomes and protein gene sequences (Oryctolagus cuniculus, Peromyscus maniculatus bairdii, Mus musculus, and Rattus norvegicus) were designated as the outgroups. Their sequence data were downloaded from the NCBI database and combined with the corresponding CDS dataset of pikas. Species-level CDS was aligned using PRANK v.140603 (Loytynoja, 2014) based on protein alignments.
Phylogenetic analyses
For the combined individual-level nuclear gene dataset, we used ModelTest-NG v0.1.6 (Darriba et al., 2020) to identify the best model for each gene based on corrected Akaike Information Criterion (AICc) and the recommended parameters for tree construction. We then inferred the phylogenetic tree using RAxML v8.2.12 (Stamatakis, 2014) with the parameter -m GTRGAMMAI and 100 bootstrap replicates. The best-scoring maximum-likelihood (ML) trees for individual genes and their bootstrap replicate trees were used as input files for ASTRAL-III v.5.15.1 (Zhang et al., 2018) to generate a nuclear gene phylogenetic tree under the multi-locus bootstrapping model. We also ran ASTRAL-III without a multi-locus bootstrapping model. The stability of the tree topology was confirmed using default parameters with the dataset containing only the top-scoring ML trees and the dataset containing the processed top-scoring ML trees (with branches showing bootstrap support <10% removed), as recommended by ASTRAL. The final species tree was achieved using ASTRAL-III based on the multispecies coalescent model. Due to the limitations of ASTRAL when estimating branch length (i.e., branch lengths are only estimated for internal not terminal branches, are in coalescent units, and are prone to underestimation due to statistical noise in gene tree estimation), direct use of ASTRAL results was not conducive for subsequent analysis. Consequently, branch lengths of the final phylogenetic tree were re-estimated using substitutions per site with ExaML v.3.0.22 (Kozlov et al., 2015). We used the ASTRAL bootstrap replicate trees and determined bootstrap support in the phylogenetic tree using RAxML with the “-f b” parameter.
Divergence time inference
Divergence times of the species tree were estimated based on nuclear genes using MCMCTree in the PAML v.4.9h package (Yang, 2007) with approximate likelihood determined using the “REV” (GTR, model=7) model. All ambiguous codon sites were removed from the “species-level nuclear gene set” and only the third codon sites of them were extracted to generate a new supergene. TrimAl v1.4.15 (Capella-Gutierrez et al., 2009) was used to refine the alignment. The final species tree only retained topological information and was extracted as the input tree for subsequent dating analysis. Fossil calibration points were sourced from previous research (Meredith et al., 2011) and the Paleobiology Database (https://paleobiodb.org/), setting priors of divergence time for the tree nodes. The root was set to 79.5 million years ago (Ma). The Rodentia-Lagomorpha split, Leporidae-Ochotonidae split, and Mus-Rattus split were constrained at 71.5–94.1, 47.4–56.9, and 11–13 Ma, respectively. In addition, the oldest extant pika fossils (0.116–0.160 Ma) were used to calibrate the occurrence of Ochotonidae. The calibration constraints were specified with soft boundaries using 2.5% tail probabilities above and below their limits using the built-in function in MCMCTree. The prior for the mean substitution rate was estimated using BaseML. The independent rate model (clock=2 in MCMCTree) was used to specify the rate priors for internal nodes. The overall substitution rate (rgene_gamma) was set to G (1, 6.3085) and the rate drift parameter (sigma2_gamma) was set to G (1, 4.5). Each run was executed with 5 000 000 iterations as burn-in and sampling every 200 iterations until 100 000 samples were collected. Finally, MCMC analyses were run thrice with different random seed numbers to check convergence, with similar results found.
Phylogenetic comparative analyses
To investigate the relationship between molecular genetics and phenotypic characteristics in extant pikas, we measured phylogenetic signals, i.e., degree to which related species are phenotypically similar, using the Blomberg’s K statistic (Blomberg et al., 2003) implemented in RASP v4 (Yu et al., 2020). We calculated the average values of the continuous traits for each species, then extracted single-gene phylogenetic trees with complete representative species from the previously obtained best-scoring ML trees. We estimated the phylogenetic signal for each trait using different gene phylogenetic trees and the final species tree.
We selected the best-fit model by evaluating the AICc values of different models using the R phytools v.0.7-90 package (Revell, 2012). Among the models, the morphological characteristics with continuous data were more suitable for the Brownian motion model, while the discrete data were more suitable for an equal-rates (ER) model. According to the fitness test results, we performed fast estimation of the ML ancestral states for the continuous traits following the Brownian motion model and computed 95% confidence intervals (CIs) for these estimates using the fastAnc function in R phytools v.0.7-90 package (Revell, 2012). We evaluated the evolutionary traits of the discrete characteristics using the ace function with an ER model in the R ape v.5.4-1 package (Paradis and Schliep, 2019).
Evolutionary analyses
Based on morphological clustering, species in Alienauroa and Ochotona exhibited more similar morphological characteristics than other species, but O. curzoniae and O. dauurica showed significant deviations in morphological PCA and inhabited different environments compared to other species within Ochotona (Supplementary Table S2). A separate tree was generated by pruning these two species from the final species tree to avoid outlier effects on evolutionary analysis. Each species node in these two subgenera and their ancestral branches were then labeled as foreground clades. Subsequently, all gene sequences of the remaining 25 species were extracted from the species-level nuclear gene CDS dataset.
Selection analyses were performed using the CODEML program in the PAML package (Yang, 2007). We used the branch-site model to identify positively selected genes (PSGs) in the foreground clades. For each gene, we compared model A (“model 2, NSsites 2, fix_omega 0, omega 1; some sites in the foreground branch are under positive selection) against model A null (“model 2, NSsites 2, fix_omega 1, omega 1”; all sites evolved neutrally) and used the Chi-square test to evaluate the significance of the compared likelihood ratio tests (LRTs). Genes showing significant differences (P≤0.05) were considered as PSGs. In accordance with Yu et al. (2016), we searched for common amino acid substitutions in the foreground and background branches of the PSGs. We expanded the sample size to incorporate all individuals to exclude variation in individual species.
Functional gene annotations were downloaded from OrthoDB v10.1 (https://www.orthodb.org/) including Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and InterPro domains. Gene symbols were converted from the reference genome of O. princeps. Enrichment analyses were performed using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost) (Raudvere et al., 2019), with a Benjamini-Hochberg false discovery rate (FDR≤0.05) significance threshold and the background gene set derived from the Ensembl annotation of O. princeps (version: e104_eg51_p15_3922dba). Protein-protein interaction networks of PSGs were constructed using the STRING (Szklarczyk et al., 2017) protein interaction database. Cytoscape v3.8.2 (Shannon et al., 2003) was used to visualize the protein-protein interaction network.
Intracellular chloride ion measurement
We tested the functional effects of the substitution PROM1-H419Y following previous research (Hori et al., 2019). Mouse embryonic fibroblasts (MEFs) were maintained in 1640 medium (Gibco, Life Technologies, USA) with 10% fetal bovine serum (Gibco, Life Technologies, USA) supplemented with penicillin-streptomycin (Gibco, Life Technologies, USA). The chloride-sensitive fluorescent indicator MQAE (MCE, USA) was used according to the manufacturer’s instructions. Briefly, the MEFs were incubated with 5 mmol/L MQAE in Krebs-HEPES buffer (20 mmol/L HEPES-NaOH, 128 mmol/L NaCl, 2.5 mmol/L KCl, 2.7 mmol/L CaCl2, 1 mmol/L MgSO4, 16 mmol/L glucose) for 1 h at 37 °C. Calcium ionophore was added at a final concentration of 15 μmol/L (A23187, MCE) and temporal changes in chloride ions were detected using a FV3000 confocal microscope (Olympus, Japan) at 1 min intervals.
Cell Mito Stress Test
We performed cell mitochondrial functional analysis to investigate the functional changes in the substitution TSFM-Q155E. A Seahorse XF Cell Mito Stress Test Kit (Agilent) was used to evaluate the oxygen consumption rate (OCR). Briefly, 10 000 human umbilical vein endothelial cells (HUVECs) were seeded into an 8-well Agilent Seahorse XF Cell Culture Microplate, separately. Then, 1.5 µmol/L oligomycin, 2.0 µmol/L FCCP, and 0.5 µmol/L rotenone-antimycin A were dissolved in an assay medium and loaded into the sensor cartridge. Details on plasmid construction are provided in the Supplementary Methods.
RESULTS
Taxon sampling and datasets
We measured 20 morphological characteristics, including external, cranial, and dental characteristics, of 226 pika specimens collected over the past 30 years (see Methods, Supplementary Table S2). We also obtained environmental information on the collected specimens, including elevation, habitat, landscape type, and microhabitat (Supplementary Table S2). The specimens were assigned to five subgenera and 27 species (nominal or tentatively named) collected from 12 provinces and autonomous regions of China. Many specimens were topotypes (i.e., specimens collected at locality of original type) (Figure 1A; Supplementary Table S1). The complete analysis pipeline is shown in Supplementary Figure S1.
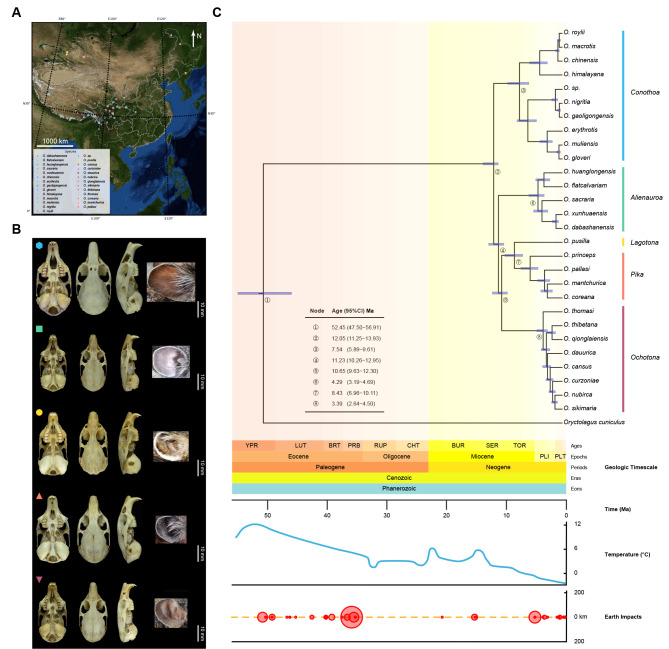
Figure 1.
Geographical distribution, morphological characteristics, and species tree with divergence times of pikas
A: Geographical distribution of 81 pika specimens used for phylogenetic analysis in this study. Sampling sites covered the major distribution areas of pikas in China. Marked longitudes of individuals sampled at the same latitude and longitude on map deviate 0.1 to the east. Different colored dots represent five different subgenera: Conothoa blue, Alienauroa green, Lagotona yellow, Pika orange, and Ochotona purple. Colors of subgenera in Figure 1B and 1C are consistent with that in Figure 1A. B: One species from each subgenus (O. gloveri, O. huanglongensis, O. pusilla, O. mantchurica, and O. thibetana from top to bottom) showing skull in ventral view, dorsal view, lateral view, and external ear characteristics from left to right. Scale bar: 10 mm (for skull pictures only). C: Divergence times in species tree were estimated based on nuclear gene using MCMCTree in the PAML package. Nodes with numbered circles show detailed time estimates from left table. Light purple bar on nodes represents 95% CI. Temperature data were obtained from previous study (Zachos et al., 2001). Geological timescales and Earth impacts were obtained from TIMETREE (http://www.timetree.org/).
We sequenced the genome of 81 individuals, resulting in 2.09 Tb of clean bases. Four samples were removed due to low library quality and thus were not used for subsequent analyses (Supplementary Table S1). We obtained 5 684 full-length single-copy orthologous genes spanning 872 931 477 nucleotides (coding sequence) of the 77 specimens from O. princeps, which were used for nuclear gene dataset construction. After removal of low confidence genes and potential paralogous genes, we retained 4 090 coding genes (“nuclear gene CDS dataset”), with average, maximum, and minimum coverages of 83.94%, 93.67%, and 63.40% for all samples, respectively (Supplementary Table S3).
Molecular phylogeny and morphological data
We reconstructed the phylogeny of extant pikas using nuclear genes. All pika samples were clearly separated from the Oryctolagus cuniculus (Leporidae) outgroup and constituted a monophyletic group in the nuclear gene phylogenetic tree. Furthermore, based on the phylogenetic tree (Supplementary Figure S2), the genus Ochotona was recovered as five independent evolutionary branches, representing the five subgenera Alienauroa, Conothoa, Lagotona, Ochotona, and Pika, consistent with the morphological results (Figure 1A, B). The inter-relationship within the genus was (Conothoa, (Alienauroa, ((Lagotona, Pika), Ochotona))), with Conothoa being the most basal clade.
We used the same method as Wang et al. (2020), but with a larger dataset and more loci for estimating the divergence time. We computed divergence times using the species tree based on nuclear genes and a Bayesian relaxed clock (MCMCTree) (Yang, 2007). From our estimation, extant pikas originated 12.05 Ma in the middle Miocene (Figure 1C), consistent with estimates from previous studies (11.62–14.65 Ma) (Ge et al., 2012; Koju et al., 2017; Lanier and Olson, 2009; Wang et al., 2020). Extant pikas rapidly diverged in their early speciation from 12.05 Ma to 10.65 Ma. Conothoa was the earliest clade to diverge (12.05 Ma), and the last common ancestor of Conothoa existed in the late Miocene (7.54 Ma). Alienauroa diverged 11.23 Ma in the Miocene, and the last common ancestor appeared in the late Neogene (4.29 Ma). Finally, Ochotona diverged 10.65 Ma, with the last common ancestor at 3.39 Ma, and Lagotona and Pika diverged 8.43 Ma. Compared with previous studies, the larger dataset and loci used for estimation here may result in later divergence times at both the subgenus and species level.
After obtaining the phylogenetic relationship, we next analyzed the morphological data. Initial observations of external, cranial, and dental characteristics revealed 27 distinct patterns (Supplementary Table S2), each representing a putative species of pika. This included 26 described species (Liu et al., 2017; Wang et al., 2020) and one previously evaluated but unnamed new species (Wang et al., 2020). We selected one species from each subgenus and noted its skull and external ear characteristics to distinguish unique features (Figure 1B). For example, O. huanglongensis (Alienauroa) had a flatter skull shape than the other subgenera and a distinctive triangular protrusion (congenital tragus) on its ear not found in any other subgenus. The incisive and palatal foramina were separate in O. mantchurica (Pika) but connected in the other subgenera. Additionally, O. gloveri (Conothoa) had an oval foramen, which had disappeared in the other subgenera (Figure 1B).
Taxonomic identifications were corroborated by PCA (Figure 2A), which indicated that the five subgenera were distinct from each other, although some samples were highly similar in several morphological characteristics. At the subgenus level, Conothoa (blue) and Pika (orange) were most similar and Alienauroa (green) and Ochotona (purple) were most similar, with their centers closer than to any other subgenera, despite not being evolutionarily close (Figure 1C). The PERMANOVA test results showed that the morphological characteristics of the five subgenera differed significantly (P<0.001), with significant differences between each subgenus pair (P<0.014) (Supplementary Table S4). Approximately two-thirds of the studied species differed significantly in the 20 morphological characteristics, indicating that a large proportion of species can be identified using morphometrics alone (see details in Supplementary Results).
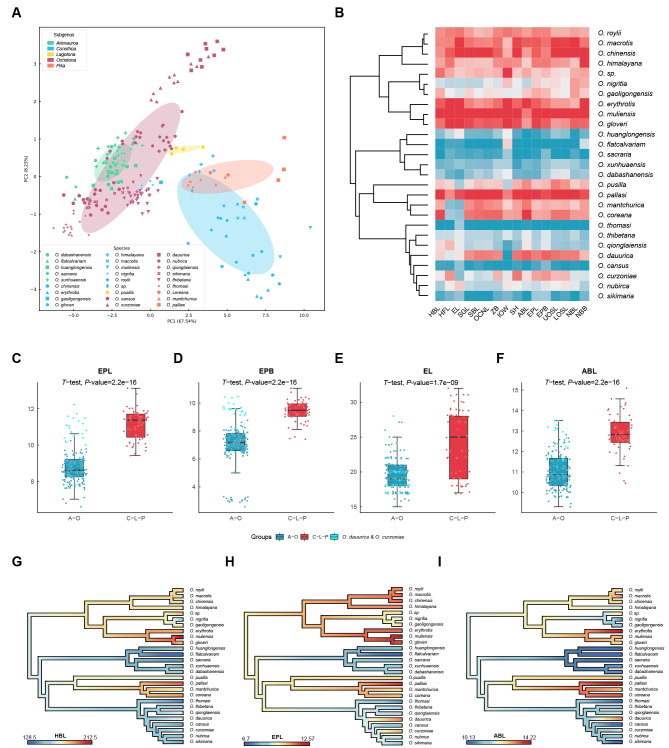
Figure 2.
Morphological characteristics of pikas
A: PCA of morphological characteristics showing five well-distinguished subgenera. B: Heatmap of 20 morphological characteristics. Values of each morphological characteristic are scaled. Redder colors indicate larger values, bluer colors indicate smaller values. C–F: Box plots of morphological characteristics EPL (eyepit length), EPB (eyepit breadth), EL (ear length), and ABL (auditory bulla length) between (Alienauroa and Ochotona) and (Conothoa, Lagotona, and Pika) groups. Both O. dauurica and O. curzoniae are highlighted in bright blue. G–I: Reconstructed ancestral states of morphological characteristics, including head and body length (HBL), EPL, and ABL along phylogenetic lineages of extant pikas.
Consistency in habitat changes and morphological variations
We also recorded collection site information for the 226 individuals, covering 107 sites ranging from 537–4 690 m in elevation (Supplementary Table S2). Through the combination of our results and other records (Wang et al., 2020), we identified that Alienauroa and Ochotona mainly inhabited low-latitude subtropical forest, with a five-layer vertical structure (megaphanerophyte, dunga-runga, high shrub, low shrub, and herbaceous). Beyond that, O. thomasi and O. nubrica (both Ochotona) inhabited high-altitude dense scrub with an abundant herbaceous layer, whereas O. dauurica and O. curzoniae (both Ochotona) inhabited prairie or shrub habitat with less vegetation. The other three subgenera (Pika, Lagotona, and Conothoa) resided in environments with simple vegetation structures, such as scree, arid valley, prairie, and desert scrub. Several species (e.g., O. roylii, O. gaoligongensis, O. nigritia, O. mantchurica, and O. coreana) lived along the edge of forest, with a simple two-layer vertical structure (megaphanerophyte and shrub (or herbaceous)). Overall, the habitats of Alienauroa and Ochotona were markedly different from those of the subgenera Pika, Lagotona, and Conothoa (Supplementary Table S2).
To test whether the different ecological environments had different effects on the evolution of morphological characteristics in pikas, we plotted a heatmap using the 20 morphological characteristics. Overall, the morphological characteristics of Alienauroa and Ochotona were smaller than those of the other three subgenera (Conothoa, Lagotona, and Pika), while O. dauurica and O. curzoniae were larger in size than other species in the subgenus Ochotona (Figure 2B). Box plots showed that most differences in size were significant between the (Alienauroa and Ochotona) and (Conothoa, Lagotona, and Pika) groups (Supplementary Figure S3), including EPL, EPB, EL, and ABL (Figure 2C–F). In total, 18 of the 20 morphological characteristics showed significant differences between the above two groups, except for EHR and ASR (Supplementary Figure S3).
We subsequently reconstructed the ancestral states of 24 morphological characteristics along phylogenetic lineages of extant pikas to further investigate their phenotypic evolution (Supplementary Figure S4 and Table S5). Notably, only Alienauroa species evolved a congenital tragus (CT). Furthermore, consistent with heatmap analysis, after the early divergence of Alienauroa and Ochotona, both evolved similar and smaller morphological characteristics compared with the other pikas and their ancestors (Supplementary and Table S2; fossil evidence in Supplementary Results). For example, Alienauroa and Ochotona individuals had shorter HBL (Figure 2G), EPL (Figure 2H), and ABL (Figure 2I). In contrast, Conothoa, Lagotona, and Pika traits remained as large as the ancestral types.
Phylogenetic signals and positive selection
To determine the effect of size trends in different phenotypic traits on genetics, we first measured phylogenetic signals. In total, 83 genes significantly related to at least one of the 16 continuous morphological characteristics were identified (Supplementary Table S6; Figure 3A). Among them, EPL had the greatest number of significantly related genes (40), including several genes related to visual ability, such as RPE65, NEIL3, and CROCC. In addition, EL had the second greatest number of significantly related genes (29), including several hearing-related genes, such as JAG1, and LARS2. We also detected other hearing-related genes that were significantly related to non-EL characteristics, such as LRP10 in ABL.
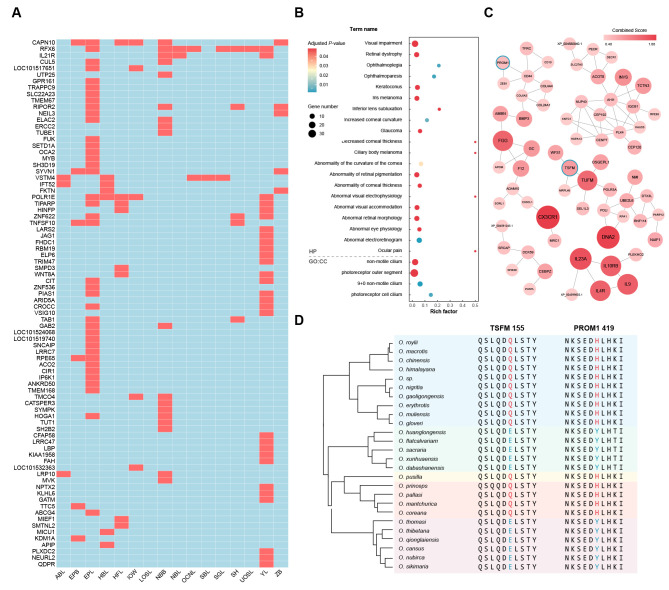
Figure 3.
Phylogenetic signal analysis and shared amino acid substitutions
A: Heatmap of phylogenetic signals, showing 83 genes significantly related (K>1, red) to at least one of the 16 continuous morphological characteristics, red indicates Blomberg’s K>1, blue indicates K<1. B: Enrichment analysis of PSGs, including GO:CC and HP categories. C: Protein‐protein interaction network of PSGs. Networks with less than three nodes were hidden. Size and color of nodes are proportional to combined score in STRING. TSFM and PROM1 are highlighted with a blue circle. D: Alignment of pika TSFM and PROM1 amino acid sequences. Amino acids unique to the foreground branch (Alienauroa and Ochotona, pruning O. curzoniae and O. dauurica) are shown in blue, and background branch (other pikas) are shown in red. Reference genome is O. princeps.
Based on phylogenetic signal, morphological variation, and habitat data, we hypothesized that different environments enabled certain genes, especially sensory-related genes, to undergo the same amino acid substitutions, resulting in similar morphological and functional changes in Alienauroa and Ochotona and divergence from other pika subgenera. Therefore, we identified shared amino acid substitutions between (Alienauroa and Ochotona) and (Conothoa, Lagotona, and Pika). To limit species variation, we included all 73 individuals, after excluding four O. curzoniae and O. dauurica individuals because they showed significant deviation in morphological PCA (Figure 2A), smaller morphological characteristics (Figure 2B), and simpler environments (Supplementary Table S2) compared to other Alienauroa and Ochotona species. Overall, we identified 408 common amino acid substitutions in 345 genes in all Alienauroa and Ochotona pikas.
We identified a total of 181 PSGs in the Alienauroa and Ochotona lineages, excluding O. curzoniae and O. dauurica (Supplementary Table S7). These PSGs were enriched in functional categories related to visual ability, such as photoreceptor cell cilium (GO:0097733), non-motile cilium (GO:0097730), and photoreceptor outer segment (GO:0001750). In addition, abundant human phenotype ontologies (HP) related to visual ability, such as abnormal visual electrophysiology (HP:0030453), abnormal eye physiology (HP:0012373), and retinal dystrophy (HP:0000556), were significantly enriched in the PSGs (Figure 3B; Supplementary Table S8). We performed protein-protein interaction analysis using all 181 PSGs and only illustrated networks with more than two genes (Figure 3C), which showed that the PSGs were highly correlated to each other. Among the above PSGs, RPE65 and NEIL3 were also detected in the phylogenetic signal. Other PSGs, such as PROM1, IQCB1, GUCA1B, and RP1, were also related to low-light visual sensitivity. In addition to vision-related genes, mitochondrial function-related genes were also identified as PSGs, including TSFM and TUFM.
Functional validation of substitutions in TSFM and PROM1
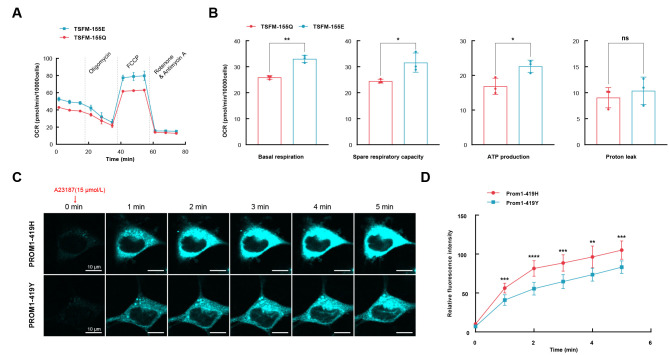
Of the common substitutions identified, 65 occurred in 53 genes also identified as PSGs (Supplementary Table S7). Combining amino acid mutations, positively selected genes, and the above-mentioned genes related to vision and mitochondrial function, we screened two loci, i.e., low-light visual sensitivity gene PROM1 (p.H419Y) and mitochondrial function-related gene TSFM (p.Q155E), for subsequent functional verification (Figure 3D). All sampled Alienauroa and Ochotona specimens had the Prom1-419Y and TSFM-155E substitutions, whereas all other pikas had Prom1-419H and TSFM-155Q (Figure 3D). TSFM is mainly located in the mitochondria and is associated with mitochondrial products and function (Scala et al., 2019). The effects of TSFM-155Q and TSFM-155E on mitochondrial function in HUVECs showed that TSFM-155E significantly improved basal respiration but did not change proton leak (Figure 4A, B; Supplementary Tables S9, S10). Additionally, the maximal respiratory rate was markedly higher in HUVECs that transfected TSFM-155E plasmids, indicating enhancement in maximal electron transport chain activity.
Figure 4.
Functional validation of identified variants on TSFM and PROM1
A: Effects of TSFM-155Q and TSFM-155E on mitochondrial function in HUVECs were assessed using a microfluidic extracellular flux analyzer (Seahorse XFp) to measure oxygen consumption rate (OCR). OCR was normalized per 10 000 HUVECs. B: Mitochondrial functional parameters for TSFM-155Q and TSFM-155. Data are mean±standard deviation (SD). n=3 per group. Unpaired t-test was applied. ns: No significance; *: P<0.0007; **: P<0.0003; ***: P<0.0002; ****: P<0.0001. C: We overexpressed Prom1-419Y and Prom1-419H plasmids in MEFs and detected temporal changes in intracellular chloride ion levels upon calcium uptake using chloride-sensitive fluorescent indicator MQAE. Fluorescence intensity of MQAE decreased proportionally with the increase in chloride ion concentration. MEFs were incubated with 5 mmol/L MQAE in Krebs-HEPES buffer (see Materials and Methods), and intracellular calcium uptake was provoked by adding 15 µmol/L of calcium ionophore A23187 to the medium (time 0). Temporal changes in fluorescence intensity were imaged every 1 min (0 to 5 min) after ionophore treatment under a FV3000 confocal microscope. Representative images are shown. Scale bar: 10 μm. D: Quantitative data for Figure 4C. Ten cells were selected from the Prom1-419Y and Prom1-419H cells, and fluorescence intensities at each time point were quantified. Data are mean±standard error of the mean (SEM).
PROM1 drives chloride ion efflux upon intracellular calcium ion uptake (Hori et al., 2019). Here, we investigated whether PROM1-H419Y affects PROM1 function. First, we overexpressed Prom1-419Y or Prom1-419H plasmids in MEFs, then used the chloride-sensitive fluorescent indicator MQAE to detect temporal changes in intracellular chloride ion levels upon calcium uptake. Significant chloride efflux was observed in the MEFs that exogenously expressed Prom1-419H when intracellular calcium uptake was provoked by the calcium ionophore A23187. Taken together, Prom1-419H promoted the modeling of dynamic intracellular chloride efflux upon calcium uptake compared to Prom1-419Y (Figure 4C, D; Supplementary Table S11).
DISCUSSION
We collected 226 pika specimens, covering most of their distribution in China (Lissovsky et al., 2019; Liu et al., 2017; Wang et al., 2020), and generated a comprehensive dataset to study their environmental adaptation. Results showed that morphological characteristics can be used to identify pika species, but species with similar morphology need to be further identified using genomic sequences. Furthermore, genomic and morphological analyses supported the classification of five subgenera (Alienauroa, Conothoa, Lagotona, Ochotona, and Pika) (Liu et al., 2017). The Alienauroa and Ochotona species were morphologically smaller than the other pika species and their ancestors for nearly all 20 morphological characteristics examined (Figure 2B–I; Supplementary Figures S3, S4). This is consistent with the fossil records for extinct pikas, such as Ochotonoides complicidens and Ochotona lagreli, which indicate that they were morphologically larger than extant pikas (Institute of Vertebrate Paleontology, 1960) (see Supplementary Results). The smaller body size of extant Alienauroa and Ochotona species may be associated with the dispersal of pikas into different habitats with different environmental conditions. Recent study suggested that extant pikas may have originated from high-elevation areas of the QTP, with later dispersal to other parts of Eurasia and North America (Wang et al., 2020). During dispersal, different species of pika would have occupied different ecological environments, which may have influenced certain morphological characteristics. For Alienauroa species, which inhabit mossy forest areas (Wang et al., 2020) (Supplementary Table S2), we identified microhabitats such as talus, moss, shrubs, grass, stone gaps, and bamboo forest. For Ochotona species, which are typical forest and scrub-grassland-steppe dwellers (Wang et al., 2020) (Supplementary Table S2), we identified several microhabitats, including fallen logs, bushes, bamboo forest, talus, and scrub-grassland (Supplementary Table S2). Pika, Lagotona, and Conothoa species, which belong to the northern and mountain groups (Wang et al., 2020), mainly inhabit scree, arid valley, shrub, prairie, and desert habitats, with much less vegetation than forests. Consistently, according to our results, Alienauroa and Ochotona species mostly inhabited subtropical forest habitats, whereas the subgenera Pika, Lagotona, and Conothoa mainly inhabited scree, arid valley, prairie, and desert scrub habitats (Supplementary Table S2). Dispersal into different environments may require different sensory and energy functions to avoid predators and find food, thus implying natural selection pressure for sensory and energy functions in Alienauroa and Ochotona species to adapt to their different environments.
Indeed, the phylogenetic signals suggested that the morphological evolution of different pikas focused on sensory functions, such as vision and hearing (Figure 3A). The identified PSGs in Alienauroa and Ochotona species (Supplementary Table S7) were mainly enriched in GO categories and HP ontologies related to visual ability (Supplementary Table S8). Several PSGs were closely related to low-light visual sensitivity, such as PROM1, RPE65, NEIL3, IQCB1, GUCA1B, and RP1. The PROM1 (prominin 1) gene, also known as CD133 and AC133, encodes a pentaspan transmembrane glycoprotein, which is concentrated at the base of the photoreceptor outer segment and acts as a key regulator of disk morphogenesis during early retinal development (Fujinami et al., 2020; Maw et al., 2000). Variants of PROM1 can cause diseases such as retinal degeneration, cone-rod dystrophies, and retinitis pigmentosa (Carss et al., 2017; Cehajic-Kapetanovic et al., 2019; Song et al., 2011). RPE65 is a protein-coding gene, and its protein is a component of the vitamin A visual cycle of the retina, which provides the 11-cis retinal chromophore of the photoreceptor opsin visual pigments (Jin et al., 2005). Mutations in RPE65 can cause various diseases, including Leber congenital amaurosis and early-onset retinal dystrophy, which can lead to poor vision in dim light (Jo et al., 2019; Kumaran et al., 2020; Motta et al., 2020). The NEIL3 (Nei-like DNA glycosylase 3) gene, which belongs to a class of DNA glycosylases homologous to the bacterial Fpg/Nei family, is necessary for normal retinal lamination and retinal neuronal differentiation by rescuing the retinal homeobox knockdown phenotype (Pan et al., 2018). Other identified PSGs, such as IQCB1, GUCA1B, and RP1, are also related to low-light visual sensitivity (Otto et al., 2005; Sato et al., 2005; Silva et al., 2020).
Among the identified PSGs, only PROM1 had a shared amino acid substitution (p.H419Y). All samples from Alienauroa and Ochotona contained Prom1-419Y, whereas the other pikas contained Prom1-419H. Thus, we overexpressed the Prom1-419Y and Prom1-419H plasmids in MEFs. Results showed that PROM1-419Y in Alienauroa and Ochotona inhibited the modeling of dynamic intracellular chloride efflux upon calcium uptake. Similarly, Prom1-knockout has a weakening effect on mice, resulting in light-dependent photoreceptor cell degeneration, which can be almost completely inhibited when Prom1-knockout mice are maintained in the dark (Dellett et al., 2015; Hori et al., 2019). As observed in the field, these pikas are diurnal animals (Smith At, 2018; Smith and Smith, 1986). This suggests that individuals with Prom1-knockout or reduced function, such as PROM1-419Y in Alienauroa and Ochotona species, may be better suited to living in subtropical forests, where additional layers of vegetation result in lower-light environments than found in the scree, arid valley, prairie, and desert scrub habitats of Pika, Lagotona, and Conothoa species. We must note that vision is determined by many genes and loci, and even the PROM1 gene has several mutations associated with retinal functions (Jespersgaard et al., 2019; Maw et al., 2000; Michaelides et al., 2010; Zhang et al., 2007).
In addition to vision-related genes, several mitochondrial function-related genes were also identified as PSGs, such as TSFM and TUFM. Mutations in the TSFM gene, which encodes a mitochondrial translation elongation factor enzyme, are associated with oxidative phosphorylation enzyme deficiency (Smeitink et al., 2006) and sensorineural hearing loss (Scala et al., 2019). TUFM encodes the protein Tu translation elongation factor involved in mitochondrial protein translation (Di Nottia et al., 2017). In addition, TSFM catalyzes the exchange of guanine nucleotides on the TUFM protein during elongation in mitochondrial protein translation (Wieden et al., 2002). Here, TSFM contained one amino acid substitution (p.Q155E), i.e., TSFM-155E in the Alienauroa and Ochotona samples and TSFM-155Q in all remaining pika species. Functional analysis indicated that TSFM-155E in Alienauroa and Ochotona individuals significantly enhanced mitochondrial function compared to TSFM-155Q in the other pikas (Figure 4A, B). Enhanced mitochondrial function may be related to the smaller body size of Alienauroa and Ochotona individuals. The mass-specific metabolic rate declines with increasing body size (Sukhotin et al., 2020). Larger organisms have lower metabolic turnover and energy demand per unit mass than smaller-bodied organisms across different animal taxa, different-sized organisms within a single species (Konarzewski and Ksiazek, 2013), and even within a single individual during ontogenesis (Maino and Kearney, 2014; Moses et al., 2008). We found that Alienauroa and Ochotona individuals were morphologically smaller than other pikas and their ancestors (Supplementary Figure S4 and Table S2). In addition, enhanced mitochondrial function can produce more adenosine triphosphate (ATP) to increase energy supply for activity (Gonzalez-Freire et al., 2015; Grevendonk et al., 2021; Hargreaves and Spriet, 2018), which may assist in the adaptation of Alienauroa and Ochotona species to subtropical forest with a five-layer vertical structure through increased athletic ability (Supplementary Table S2). However, this mutation may also contribute to hypoxia and cold tolerance, as previous studies have found that different pikas exhibit adaptive evolution to hypoxia and cold (Rankin et al., 2017; Wang et al., 2020). Thus, further functional tests should be performed to confirm the effects of this mutation.
Our study utilized years of field collections to perform a comprehensive morphological and phenotypic study of 27 extant pika species in China. Analysis supported five subgenera within the Ochotona genus, i.e., Alienauroa, Conothoa, Ochotona, Lagotona, and Pika. Morphometrics were sufficiently robust to identify most pika species and should be used in conjunction with genomics to expand and strengthen pika taxonomy in the future. Our morphological and phenotypic data identified two distinctive subgenera, i.e., Alienauroa and Ochotona, which were smaller in size and retained more PSGs related to sensory acuity and energy enhancement compared to the other studied and ancestral pikas. The Alienauroa and Ochotona individuals also inhabited different environments (e.g., subtropical forest), suggesting that the shift from a larger ancestral type and changes in sensory acuity and energy may have been caused by dispersal into new environments or led to dispersal into new environments. Thus, this study provides a comprehensive analysis of pika taxonomy and increases our understanding of the evolution of environmental adaptation in pikas during dispersal. However, potentially important environmental factors should be investigated in future studies to better explore how the ecological environment has impacted the morphological evolution and adaptations of different pikas.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
ACKNOWLEDGMENTS
We thank Rui Liao for assistance in collecting specimens in the field. We thank the Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, China for supporting our specimen inspection.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31470110, 31970399) and China National GeneBank (CNGB)
Contributor Information
Zhen-Xin Fan, Email: zxfan@scu.edu.cn.
Shao-Ying Liu, Email: shaoyliu@163.com.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Blomberg SP, Garland T Jr, Ives AR Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57(4):717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 3.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, et al Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. The American Journal of Human Genetics. 2017;100(1):75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cehajic-Kapetanovic J, Birtel J, McClements ME, Shanks ME, Clouston P, Downes SM, et al Clinical and molecular characterization of PROM1-related retinal degeneration. JAMA Network Open. 2019;2(6):e195752. doi: 10.1001/jamanetworkopen.2019.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al Twelve years of SAMtools and BCFtools. GigaScience. 2021;10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution. 2020;37(1):291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dellett M, Sasai N, Nishide K, Becker S, Papadaki V, Limb GA, et al Genetic background and light-dependent progression of photoreceptor cell degeneration in Prominin-1 knockout mice. Investigative Ophthalmology & Visual Science. 2015;56(1):164–176. doi: 10.1167/iovs.14-15479. [DOI] [PubMed] [Google Scholar]
- 9.Di Nottia M, Montanari A, Verrigni D, Oliva R, Torraco A, Fernandez-Vizarra E, et al Novel mutation in mitochondrial Elongation Factor EF-Tu associated to dysplastic leukoencephalopathy and defective mitochondrial DNA translation. Biochimica et Biophysica Acta-Molecular Basis of Disease. 2017;1863(4):961–967. doi: 10.1016/j.bbadis.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng ZJ, Zheng CL Studies on the pikas (genus: Ochotona) of China—Taxonomic notes and distribution. Acta Theriologica Sinica. 1985;5(4):269–289. [Google Scholar]
- 11.Fujinami K, Oishi A, Yang LZ, Arno G, Pontikos N, Yoshitake K, et al Clinical and genetic characteristics of 10 Japanese patients with PROM1-associated retinal disorder: a report of the phenotype spectrum and a literature review in the Japanese population. American Journal of Medical Genetics Part C:Seminars in Medical Genetics. 2020;184(3):656–674. doi: 10.1002/ajmg.c.31826. [DOI] [PubMed] [Google Scholar]
- 12.Ge DY, Wen ZX, Xia L, Zhang ZQ, Erbajeva M, Huang CM, et al Evolutionary history of lagomorphs in response to global environmental change. PLos One. 2013;8(4):e59668. doi: 10.1371/journal.pone.0059668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge DY, Zhang ZQ, Xia L, Zhang Q, Ma Y, Yang QS. 2012. Did the expansion of C4 plants drive extinction and massive range contraction of micromammals? Inferences from food preference and historical biogeography of pikas. Palaeogeography, Palaeoclimatology, Palaeoecology, 326–328: 160–171.
- 14.Gong ZD, Wang YX, Li ZH, Li SQ A new species of pika: pianma blacked pika, Ochotona nigritia (Lagomorpha: Ochotonidae) from Yunnan, China. Zoological Research. 2000;21(3):204–209. [Google Scholar]
- 15.Gonzalez-Freire M, de Cabo R, Bernier M, Sollott SJ, Fabbri E, Navas P, et al Reconsidering the role of mitochondria in aging. The Journals of Gerontology:Series A. 2015;70(11):1334–1342. doi: 10.1093/gerona/glv070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grevendonk L, Connell NJ, McCrum C, Fealy CE, Bilet L, Bruls YMH, et al Impact of aging and exercise on skeletal muscle mitochondrial capacity, energy metabolism, and physical function. Nature Communications. 2021;12(1):4773. doi: 10.1038/s41467-021-24956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu ZG, Eils R, Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 18.Hargreaves M, Spriet LL Exercise metabolism: fuels for the fire. Cold Spring Harbor Perspectives in Medicine. 2018;8(8):a029744. doi: 10.1101/cshperspect.a029744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry P, Russello MA Adaptive divergence along environmental gradients in a climate-change-sensitive mammal. Ecology and Evolution. 2013;3(11):3906–3917. doi: 10.1002/ece3.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hori A, Nishide K, Yasukuni Y, Haga K, Kakuta W, Ishikawa Y, et al Prominin-1 modulates Rho/ROCK-mediated membrane morphology and calcium-dependent intracellular chloride flux. Scientific Reports. 2019;9(1):15911. doi: 10.1038/s41598-019-52040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Liang XM, Xuan YK, Geng CY, Li YX, Lu HR, et al A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience. 2017;6(5):gix024. doi: 10.1093/gigascience/gix024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, et al The Ensembl genome database project. Nucleic Acids Research. 2002;30(1):38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Institute of Vertebrate Paleontology CaOS. 1960. Handbook of Vertebrate Fossils of China - Part of Mammals. Beijing: Science Press, 232. (in Chinese)
- 24.Jespersgaard C, Fang MY, Bertelsen M, Dang X, Jensen H, Chen YL, et al Molecular genetic analysis using targeted NGS analysis of 677 individuals with retinal dystrophy. Scientific Reports. 2019;9(1):1219. doi: 10.1038/s41598-018-38007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin M, Li S, Moghrabi WN, Sun H, Travis GH Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122(3):449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo DH, Song DW, Cho CS, Kim UG, Lee KJ, Lee K, et al CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Science Advances. 2019;5(10):eaax1210. doi: 10.1126/sciadv.aax1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K, Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koju NP, He K, Chalise MK, Ray C, Chen ZZ, Zhang B, et al Multilocus approaches reveal underestimated species diversity and inter-specific gene flow in pikas (Ochotona) from southwestern China. Molecular Phylogenetics and Evolution. 2017;107:239–245. doi: 10.1016/j.ympev.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Konarzewski M, Książek A Determinants of intra-specific variation in basal metabolic rate. Journal of Comparative Physiology B. 2013;183(1):27–41. doi: 10.1007/s00360-012-0698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlov AM, Aberer AJ, Stamatakis A ExaML version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics. 2015;31(15):2577–2579. doi: 10.1093/bioinformatics/btv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristensen TN, Ketola T, Kronholm I Adaptation to environmental stress at different timescales. Annals of the New York Academy of Sciences. 2020;1476(1):5–22. doi: 10.1111/nyas.13974. [DOI] [PubMed] [Google Scholar]
- 32.Kumaran N, Ali RR, Tyler NA, Bainbridge JWB, Michaelides M, Rubin GS Validation of a vision-guided mobility assessment for RPE65-associated retinal dystrophy. Translational Vision Science & Technology. 2020;9(10):5. doi: 10.1167/tvst.9.10.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanier HC, Olson LE Inferring divergence times within pikas (Ochotona spp. ) using mtDNA and relaxed molecular dating techniques. Molecular Phylogenetics and Evolution. 2009;53(1):1–12. doi: 10.1016/j.ympev.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 34.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997.
- 35.Lissovsky AA Taxonomic revision of pikas Ochotona (Lagomorpha, Mammalia) at the species level. Mammalia. 2013;78(2):199–216. [Google Scholar]
- 36.Lissovsky AA, Yatsentyuk SP, Koju NP Multilocus phylogeny and taxonomy of pikas of the subgenus Ochotona (Lagomorpha, Ochotonidae) Zoologica Scripta. 2019;48(1):1–16. doi: 10.1111/zsc.12325. [DOI] [Google Scholar]
- 37.Liu SY, Jin W, Liao R, Sun ZY, Zeng T, Fu JR, et al Phylogenetic study of Ochotona based on mitochondrial Cyt b and morphology with a description of one new subgenus and five new species. Acta Theriologica Sinica. 2017;37(1):1–43. doi: 10.1016/j.chnaes.2016.09.003. [DOI] [Google Scholar]
- 38.Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. In: Russell DJ. Multiple Sequence Alignment Methods. Totowa: Humana Press, 155–170.
- 39.Maino JL, Kearney MR Ontogenetic and interspecific metabolic scaling in insects. The American Naturalist. 2014;184(6):695–701. doi: 10.1086/678401. [DOI] [PubMed] [Google Scholar]
- 40.Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, et al A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Human Molecular Genetics. 2000;9(1):27–34. doi: 10.1093/hmg/9.1.27. [DOI] [PubMed] [Google Scholar]
- 41.Melo-Ferreira J, de Matos AL, Areal H, Lissovsky AA, Carneiro M, Esteves PJ The phylogeny of pikas (Ochotona) inferred from a multilocus coalescent approach. Molecular Phylogenetics and Evolution. 2015;84:240–244. doi: 10.1016/j.ympev.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, et al Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 43.Michaelides M, Gaillard MC, Escher P, Tiab L, Bedell M, Borruat FX, et al The PROM1 mutation p. R373C causes an autosomal dominant bull's eye maculopathy associated with rod, rod-cone, and macular dystrophy. Investigative Ophthalmology & Visual Science. 2010;51(9):4771–4780. doi: 10.1167/iovs.09-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moses ME, Hou C, Woodruff WH, West GB, Nekola JC, Zuo WY, et al Revisiting a model of ontogenetic growth: estimating model parameters from theory and data. The American Naturalist. 2008;171(5):632–645. doi: 10.1086/587073. [DOI] [PubMed] [Google Scholar]
- 45.Motta FL, Martin RP, Porto FBO, Wohler ES, Resende RG, Gomes CP, et al Pathogenicity reclasssification of RPE65 missense variants related to leber congenital amaurosis and early-onset retinal dystrophy. Genes. 2020;11(1):24. doi: 10.3390/genes11010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niu Yd, Wei FW, Li M, Liu Xm, Feng ZJ Phylogeny of pikas (Lagomorpha, Ochotona) inferred from mitochondrial cytochrome b sequences. Folia Zoologica -Praha. 2004;53(2):141–155. [Google Scholar]
- 47.Null R, Team R, Null R, Writing TC, Null R, Team R, et al. 2011. R: a language and environment for statistical computing. 1: 12–21.
- 48.Oksanen J, Blanchet FG, Kindt R, Legendre P, Wagner H. 2019. Vegan: community ecology package, v. 2.5--6.
- 49.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan SL, et al Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nature Genetics. 2005;37(3):282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 50.Pan Y, Kelly LE, El-Hodiri HM Identification of retinal homeobox (rax) gene-dependent genes by a microarray approach: the DNA endoglycosylase neil3 is a major downstream component of the rax genetic pathway. Developmental Dynamics. 2018;247(11):1199–1210. doi: 10.1002/dvdy.24679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paradis E, Schliep K Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35(3):526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 52.Parsons PA Environments and evolution: interactions between stress, resource inadequacy and energetic efficiency. Biological Reviews. 2005;80(4):589–610. doi: 10.1017/S1464793105006822. [DOI] [PubMed] [Google Scholar]
- 53.Rankin AM, Galbreath KE, Teeter KC Signatures of adaptive molecular evolution in American pikas (Ochotona princeps) Journal of Mammalogy. 2017;98(4):1156–1167. doi: 10.1093/jmammal/gyx059. [DOI] [Google Scholar]
- 54.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al g: Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Research. 2019;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Revell LJ Phytools: an R package for phylogenetic comparative biology (and other things) Methods in Ecology and Evolution. 2012;3(2):217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 56.Revelle W. 2020. psych: Procedures for Personality and Psychological Research, v=2.0. 12. Evanston, Illinois, USA: Northwestern University,https://CRAN.R-project.org/package=psych.
- 57.Sato M, Nakazawa M, Usui T, Tanimoto N, Abe H, Ohguro H Mutations in the gene coding for guanylate cyclase-activating protein 2 (GUCA1B gene) in patients with autosomal dominant retinal dystrophies. Graefe's Archive for Clinical and Experimental Ophthalmology. 2005;243(3):235–242. doi: 10.1007/s00417-004-1015-7. [DOI] [PubMed] [Google Scholar]
- 58.Scala M, Brigati G, Fiorillo C, Nesti C, Rubegni A, Pedemonte M, et al Novel homozygous TSFM pathogenic variant associated with encephalocardiomyopathy with sensorineural hearing loss and peculiar neuroradiologic findings. Neurogenetics. 2019;20(3):165–172. doi: 10.1007/s10048-019-00582-5. [DOI] [PubMed] [Google Scholar]
- 59.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sikes RS, Gannon WL Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy. 2011;92(1):235–253. doi: 10.1644/10-MAMM-F-355.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silva RS, Salles MV, Motta FL, Sallum JMF Retinitis pigmentosa due to Rp1 biallelic variants. Scientific Reports. 2020;10(1):1603. doi: 10.1038/s41598-020-58243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 63.Smeitink JAM, Elpeleg O, Antonicka H, Diepstra H, Saada A, Smits P, et al Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. The American Journal of Human Genetics. 2006;79(5):869–877. doi: 10.1086/508434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith AT, Johnston CH, Alves PC, Hackländer K. 2018. Lagomorphs: Pikas, Rabbits, and Hares of the World. Baltimore: Johns Hopkins University Press.
- 65.Smith AT, Smith HJ, Wang XG, Yin XC, Liang J Social behavior of the steppe-dwelling black-lipped pika (ochotona curzoniae) Acta Theriologica Sinica. 1986;6(1):13–32. [Google Scholar]
- 66.Smith AT, Xie Y. 2009. A guide to the Mammals of China. Changsha: Hunan Education Press. (in Chinese)
- 67.Sokolov VE, Ivanitskaya EY, Gruzdev VV, Heptner VG. 2009. Lagomorphs: Mammals of Russia and Adjacent Regions. Boca Raton: CRC Press.
- 68.Song J, Smaoui N, Ayyagari R, Stiles D, Benhamed S, MacDonald IM, et al High-throughput retina-array for screening 93 genes involved in inherited retinal dystrophy. Investigative Ophthalmology & Visual Science. 2011;52(12):9053–9060. doi: 10.1167/iovs.11-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sukhotin A, Kovalev A, Sokolov E, Sokolova IM Mitochondrial performance of a continually growing marine bivalve, Mytilus edulis, depends on body size. Journal of Experimental Biology. 2020;223(13):jeb226332. doi: 10.1242/jeb.226332. [DOI] [PubMed] [Google Scholar]
- 71.Suyama M, Torrents D, Bork P PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research. 2006;34(S2):W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang XY, Liang D, Jin W, Tang MK, Shalayiwu, Liu SY, et al Out of Tibet: genomic perspectives on the evolutionary history of extant pikas. Molecular Biology and Evolution. 2020;37(6):1577–1592. doi: 10.1093/molbev/msaa026. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y Ochotona gaoligongensis-a new species. Zoological Research. 1988:9. [Google Scholar]
- 75.Wang YX. 2003. Taxonomic list and distribution of mammal species and subspecies in China. Beijing: China Forestry Publishing House (in Chinese)
- 76.Wieden HJ, Gromadski K, Rodnin D, Rodnina MV Mechanism of elongation factor (EF)-Ts-catalyzed nucleotide exchange in EF-Tu - Contribution of contacts at the guanine base. Journal of Biological Chemistry. 2002;277(8):6032–6036. doi: 10.1074/jbc.M110888200. [DOI] [PubMed] [Google Scholar]
- 77.Wilson DE, Reeder DM. 1993. Mammal Species of the World: A Taxonomic and Geographic Reference. 2nd ed. Washington: Smithsonian Press.
- 78.Wilson DE, Reeder DM. 2005. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd ed. Baltimore: Johns Hopkins University Press.
- 79.Wilson MC, Smith AT The pika and the watershed: the impact of small mammal poisoning on the ecohydrology of the Qinghai-Tibetan Plateau. Ambio. 2015;44(1):16–22. doi: 10.1007/s13280-014-0568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang ZH PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 81.Yu L, Wang GD, Ruan J, Chen YB, Yang CP, Cao X, et al Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nature Genetics. 2016;48(8):947–952. doi: 10.1038/ng.3615. [DOI] [PubMed] [Google Scholar]
- 82.Yu Y, Blair C, He XJ RASP 4: ancestral state reconstruction tool for multiple genes and characters. Molecular Biology and Evolution. 2020;37(2):604–606. doi: 10.1093/molbev/msz257. [DOI] [PubMed] [Google Scholar]
- 83.Zachos J, Pagani M, Sloan L, Thomas E, Billups K Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292(5517):686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 84.Zhang C, Rabiee M, Sayyari E, Mirarab S ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19(S6):153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang HF. 2021. DNA short-insert library construction protocol for illumina HiSeq 2500/4000/X ten or NovaSeq.
- 86.Zhang QJ, Zulfiqar F, Xiao XS, Riazuddin SA, Ahmad Z, Caruso R, et al. 2007. Severe retinitis pigmentosa mapped to 4p15 and associated with a novel mutation in the PROM1 gene. Human Genetics, 122(3–4): 293–299.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.