Graphical abstract
Keywords: Optic nerve, Multiple sclerosis, Deep learning, CNN, MRI, Optic neuritis
Abbreviations: ON, Optic neuritis; CNN, Convolutional neural network; MS, Multiple sclerosis; MRI, Magnetic resonance imaging; SVM, Support vector machine; RF, Random forest; ROC, Receiver operating characteristic
Highlights
-
•
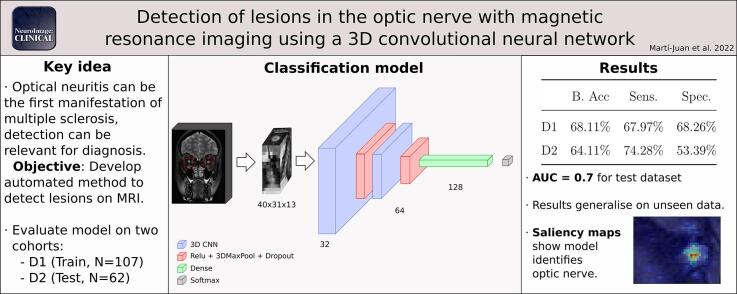
We developed a 3D Convolutional neural network trained with optic nerve images to automatically detect lesions.
-
•
Results obtained are evaluated on two cohorts and comparable to results by trained radiologists using a single image.
-
•
Interpretation of the network using saliency maps show that the model correctly identifies the optic nerve.
Abstract
Background
Optic neuritis (ON) is one of the first manifestations of multiple sclerosis, a disabling disease with rising prevalence. Detecting optic nerve lesions could be a relevant diagnostic marker in patients with multiple sclerosis.
Objectives
We aim to create an automated, interpretable method for optic nerve lesion detection from MRI scans.
Materials and Methods
We present a 3D convolutional neural network (CNN) model that learns to detect optic nerve lesions based on T2-weighted fat-saturated MRI scans. We validated our system on two different datasets (N = 107 and 62) and interpreted the behaviour of the model using saliency maps.
Results
The model showed good performance (68.11% balanced accuracy) that generalizes to unseen data (64.11%). The developed network focuses its attention to the areas that correspond to lesions in the optic nerve.
Conclusions
The method shows robustness and, when using only a single imaging sequence, its performance is not far from diagnosis by trained radiologists with the same constraint. Given its speed and performance, the developed methodology could serve as a first step to develop methods that could be translated into a clinical setting.
1. Introduction
Optic neuritis (ON) is an acute inflammation of the optic nerve (De Lott et al., 2022) that results in visual loss. It has a prevalence of between 1 and 5 per 100,000 (Preziosa et al., 2016) on the general population, and can be the first manifestation of inflammatory central nervous system diseases such as Multiple Sclerosis (MS) (Rodríguez-Acevedo et al., 2022). MS is a disabling autoimmune condition of the central nervous system (Thompson et al., 2018) characterized by inflammation, demyelinatization and neurodegeneration in the brain and the spinal chord that can cause a great variety of neurological manifestations, including visual impairment. It has a prevalence of 35.9 per 100,000 in the general population, and it has been rising worldwide over the last decade (up to 30% from 2013 to 2020 (Walton et al., 2020)). ON is the first manifestation of MS in 15 to 20% of the patients, and up to 40% of MS patients will present ON during the follow-up (Tintoré et al., 2005). In the 2017 update to the McDonald criteria for the diagnosis of MS (Thompson et al., 2018), presence of visible lesions in the optic nerve was not included as a relevant topography in spite of previous recommendations (Filippi et al., 2016), but the study of optic nerve pathology for MS diagnosis was deemed as a high research priority. As discussions about this topic continues (Brownlee et al., 2018, Wattjes et al., 2021, Rovira and Auger, 2021), interest in optic nerve lesions as a biomarker to diagnose and assess MS will continue to rise in the following years.
Magnetic Resonance Imaging (MRI) of the optic nerve can be used by radiologists to observe the optic nerve and diagnose any damage present (Gass and Moseley, 2000). However, conventional imaging sequences are not appropriate to capture the optic nerve, due to its small size, the composition of its surroundings (mainly fat, bone and cerebrospinal fluid), and distortions caused by eye movement during scan. For this reason, protocols such as short-tau inversion-recovery, fat-suppressed spin-echo (FAT-SAT) (Gass and Moseley, 2000, Faizy et al., 2019) are being used, which better capture the structure of the optic nerve and pathology-induced changes. Sensitivity of MRI to detect lesions in the optic nerve is high, specially on FAT-SAT (McKinney et al., 2013, De Lott et al., 2022), but the MRI interpretation by radiologists does not always coincide, due to scan quality, observer expertise, or clinical knowledge of the patient by the radiologist, among other factors (Bursztyn et al., 2019). In this context, the implementation of automatic analysis pipelines may help neuroradiologists to have a higher agreement. Schroeder et al., 2021 proposed an image processing automated tool to improve contrast of the optic nerve in a study with 60 patients, improving the specificity of lesion detection significantly, as well as the intra-observer agreement, while the sensitivity did not change. However, no other tools have been proposed for automated or semi-automated ON diagnosis at this time.
Machine learning is a set of tools and models that can learn from existing data to solve a specific problem (Yu et al., 2018). One example of such models are deep learning systems, composed of stacks of non-linear transformations (commonly named neural networks) that learn abstract representations of data and are used to solve specific tasks (LeCun et al., 2015). Deep learning models have been successfully used for a variety of medical imaging problems (Zhang et al., 2021) such as detection of diabetic retinopathy (Gulshan et al., 2016) or brain tumor segmentation (Havaei et al., 2017), and can be deployed as tools to assist clinicians and radiologists (McBee et al., 2018).
In this work, we present an automated pipeline to extract the optic nerve from T2-weighted FAT-SAT scans and train a 3D convolutional neural network (CNN) for the classification task. We evaluated our model on two different cohorts and compared it to two additional classification approaches to assess the robustness of the model. We also evaluated the interpretability of the network by computing saliency maps and evaluating if the network is able to locate the optic nerve. As far as we know, this is the first automated tool for detecting lesions in the optic nerve proposed in the literature. We believe that this model can serve as a stepping stone to develop tools that can be incorporated in the clinical routine of a neuroradiology department. The code is publicly available at:https://github.com/GerardMJuan/optic-nerve-3dcnn-ms.
2. Materials and methods
To design the experiments for this work, we have followed the checklist for Artificial Intelligence in Medical imaging described in (Mongan et al., 2020), to make sure that we include all the necessary information for the experiments to be robust and understandable.
2.1. Data
We have used two different retrospective cohorts of patients in this work, which we named D1 (used for training the model and hyperparameter search) and D2 (used for testing, unseen during training). The first one was acquired with a Trio scanner (Siemens, Erlangen, Germany) and is composed of 169 subjects, whereas the second cohort, acquired in a Prisma scanner (Siemens, Erlangen, Germany) has 62 subjects, with a lower percentage of patients with lesions (see Table 1), the latter being used for validation. Both cohorts included, among other sequences, a 3T tcoronal 2D T2 turbo spin-echo fat-suppressed (FAT-SAT) with identical acquisition parameters (Acquisition time = 3:46 min, 20 slices, voxel size 0.5 x 0.5 x 3.0 mm3 field of view = 180 mm, flip angle = 120°, TR = 4000 ms, TE = 84 ms), the difference between the cohorts being the model of the machine used. Data was taken from patients in clinical routine. standardized protocol FAT-SAT have been recently added to the clinical routine in our center, regardless of clinical presentation, and our cohort is formed from those scans. Both cohorts were labelled by compiling their medical records and a written report of an expert radiologist, to discern if optic nerve lesions were present in the scan, and revised by a separate radiologist. Lesion had to be clinically apparent to be labelled as one.
Table 1.
Description of the data used in the project. N refers to the number of patients, whereas Lesion and Healthy refer to optic nerve scans (2 scans per subject, corresponding to each eye). Sex is described as percentage of females in the dataset, and age is described as meanstd. CIS: Clinically isolated syndrome. RR: Relapsing remitting MS. PMS: Progressive MS, including primary and secondary progressive. NMOSD: Neuromyelitis optica spectrum disorder. MOGAD:. Uncategorized: Patients with no information regarding clinical status available.
| D1 | D2 | Total | |
|---|---|---|---|
| N | 107 | 62 | 169 |
| Lesion (Eye) | 72 | 14 | 86 |
| Healthy (Eye) | 142 | 110 | 252 |
| Sex | |||
| Age | |||
| CIS () | 72 | 40 () | 112 () |
| RR () | 6 | 10 () | 16 () |
| ON () | 22 | 4 () | 26 () |
| PMS () | 5 | 2 () | 7 () |
| NMOSD () | 1 | 0 | 1 () |
| MOGAD () | 0 | 2 () | 2 () |
| Uncategorized () | 1 | 4 () | 5 () |
a: Unequal variances t-test comparing the age between the two cohorts , .
Table 1 contains demographic and clinical information of the two datasets, as well as the number of optic nerve scans, divided by label.
2.2. Processing pipeline
Prior to training, scans were processed to remove irrelevant parts of the image and reduce its dimensionality, leaving only the optic nerve and surrounding area. We designed the following semiautomated method:
-
•
Image split: we split each scan along the midline, with each eye and optic nerve fully contained in each half.
-
•
Left eye flip: we mirror the left half so that both eyes have the same orientation.
-
•
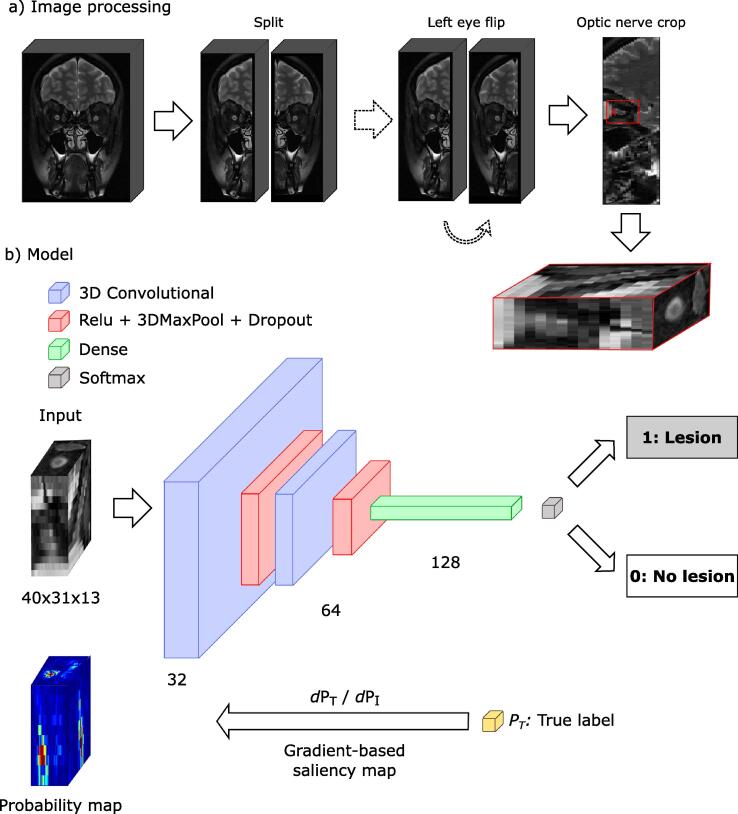
Optic nerve cropping: with a visual interface, we manually select the area where the optic connects with the eye globe. Then, an automatic crop is performed around the selected point. The size of the crop has been manually selected so that it has enough margin to contain the full optic nerve accounting for any possible inter-subject variation. The size of the final crop is 40 x 31 x 13 voxels. We used this final crop as input for all subsequent experiments.
Fig. 1a shows a diagram of the full processing pipeline.
Fig. 1.
Outline of the classification pipeline. a) Processing pipeline: image split, left eye flip and optic nerve cropping. (b) Architecture of the 3D convolutional neural network, from image to label, and saliency maps for interpretability. Filters drawn as 2D for easier representation.
2.3. Classification model
We implemented a 3D CNN for the classification task. It receives as input the 3D crops of the optic nerve described in the previous section and it is composed of two 3D convolutional layers with 64 and 128 filters, each one having a rectifying linear unit non-linearity layer, a max pooling layer to reduce dimensionality, and a dropout layer (Srivastava et al., 2014) with probability 0.1 to avoid overfitting to the training data and improve generalization. A final dense, fully connected layer with 256 weights leads to a softmax layer that outputs probabilities for the two possible outputs: presence of lesion or not. The network was trained using Adam optimizer (Kingma and Ba, 2017), which updates the weights of each layer of the network by minimizing a loss function, with a learning rate of . Our loss function is the binary cross-entropy between predicted and real labels, for N scans:
| (1) |
with being the label for scan i, and being the predicted lesion probability (i.e., if there is a lesion present or not) for that scan. By minimizing this expression, the network learns to assign the correct label to each scan. All the data were passed through the network 200 times (epochs), stopping early if the loss did not improve for 10 consecutive epochs. For a more in-depth explanation of each of the components of the network and its optimization, we refer the reader to (Deng, 2014). Fig. 1b shows a diagram of the network.
We also implemented two simpler classification models to compare the performance of the CNN model against two other approaches. The first model uses a Support Vector Machine (SVM) (Boser et al., 1992), whereas the second one uses a Random Forest (RF) (Breiman, 2001). Both models have as input all the voxels of the 3D crop of the optic nerve.
2.4. Data augmentation
Data augmentation is a popular procedure for training neural networks. It consists of adding random variations to the training data in each training iteration, improving the generalization of the model. It has been shown to improve the performance of neural networks, compared to not using it (Shorten and Khoshgoftaar, 2019). For this project, we used random recropping and random noise augmentation. The first procedure consists of performing 8 additional crops around the x and y axis of the selected voxel. Then, at training time, each crop can be selected randomly for each epoch. Regarding noise augmentation, for each epoch, there is 50% probability of adding white noise to each input image (with variance = 0.05). Those methods are use for the CNN. For the RF and SVM models, to make the comparison fairer, we included all the additional crops of the training sets as input to the models.
2.5. Experimental setup
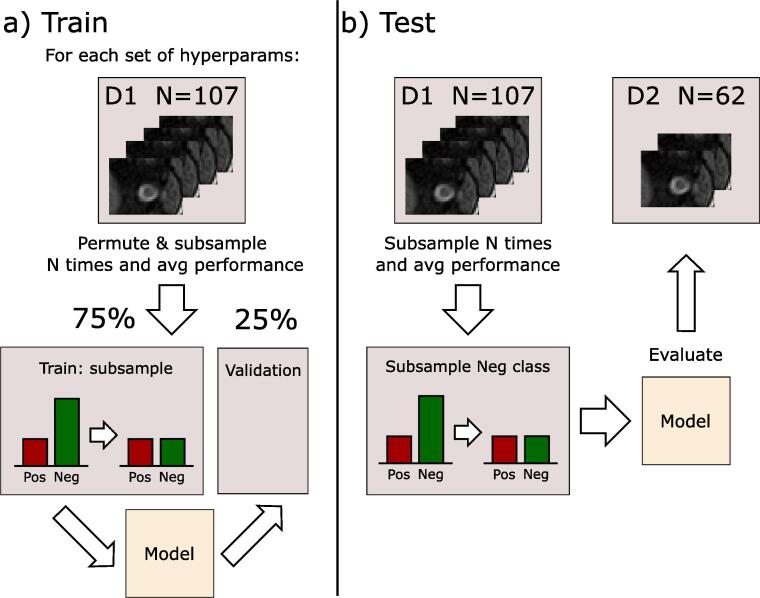
Given the unbalanced state of the dataset (see Table 1), we opted to do a permutation procedure to remove any existing biases caused by this unbalance. Over 200 iterations, we randomly separated the training dataset (D1) into training and validation (75/25 split) ensuring that subjects remain consistent across train/validation sets (i.e., both eyes of a subject go to either train or validation subsets), and subsampling the healthy subjects in the training partition to balance it. The performance of the network was then evaluated over the average of the results on the validation dataset of each iteration. Fig. 2(a) shows this training procedure.
Fig. 2.
Outline of the training procedure.
Hyperparameters of the network (dropout rate, number of layers, number of filters and learning rate) and of the SVM and RF models were set using an exhaustive search over the average performance of the model on the permutation using only the training set. Full set of hyperparameter space searched for the three models can be found in the Supplementary material.
After defining the final hyperparameters of the model, we used the testing set (D2) to evaluate the performance of the model on a cohort unseen by the network during training. Performance was evaluated by subsampling the negative class on the whole training set, over 200 iterations, and doing the final prediction by combining its results via averaging the resulting probabilities outputted by the model. This approach is shown in Fig. 2(b). The other two models (RF and SVM) were trained and evaluated following the same procedure as with the CNN model. The models are evaluated using accuracy, sensitivity (true positive rate), specificity (true negative rate) and balanced accuracy:
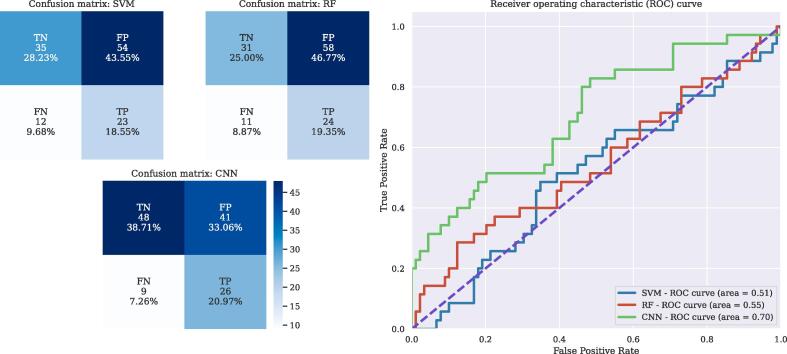
where TP, FP, TN and FP are true positives, false positives, true negatives and false positives, respectively. We also computed the Receiver operating characteristic (ROC) curve to visualize the behavior of the three models, by plotting the False Positive rate and the True Positive rate at different binary classification thresholds, as shown in Fig. 3.
Fig. 3.
(a) Confusion matrices for the three models, evaluated on the D2 (testing) cohort. (b) ROC curve over D2 for the three models. Legend of the figure includes Area under the Curve (AUC) for each model.
2.6. Network interpretation
The only output of the network is a probability value that tells us if the models detects a lesion in that image or not. However, we would like to have further information: for example, knowing which regions of the image were used by the network to reach its decision. To obtain this information, we generated saliency maps (Zhou et al., 2016) of the network for each input image. Saliency maps are probability maps over the input image that tells us which areas of the image are more important for the model to make a decision. There are different ways to implement saliency maps (see (Zhang and Zhu, 2018) for a comprehensive survey). We decided to implement gradient-based saliency maps: for a given input image I, we compute the derivatives of its true class probability w.r.t. I (Eq. 2). This is done by backpropagating the gradient over the whole network, akin to training the model, obtaining a map of weights over the original image that highlights the most important regions used by the model to reach . Fig. 1(b) shows a diagram of the saliency maps computation.
| (2) |
3. Results
Table 2 summarizes the average results on training, test and validation set after our experimental setup.
Table 2.
Table of results on the two datasets, D1 and D2. D1 results are presented as meanstd percentages over 200 iterations, as detailed in Section 2.5. Acc: Accuracy. B. Acc: Balanced Accuracy. Sens: Sensitivity. Spec: Specificity. SVM: Support vector machine. RF: random forest. 3D CNN: 3D convolutional neural network.
| SVM | RF | 3D CNN | ||
|---|---|---|---|---|
| D1 | Acc | |||
| B. Acc | ||||
| Sens. | ||||
| Spec. | ||||
| D2 | Acc | 44.35% | 46.77% | |
| B. Acc | 51.7% | 52.52% | ||
| Sens. | 68.57% | 65.71% | ||
| Spec. | 34.83% | 39.32% |
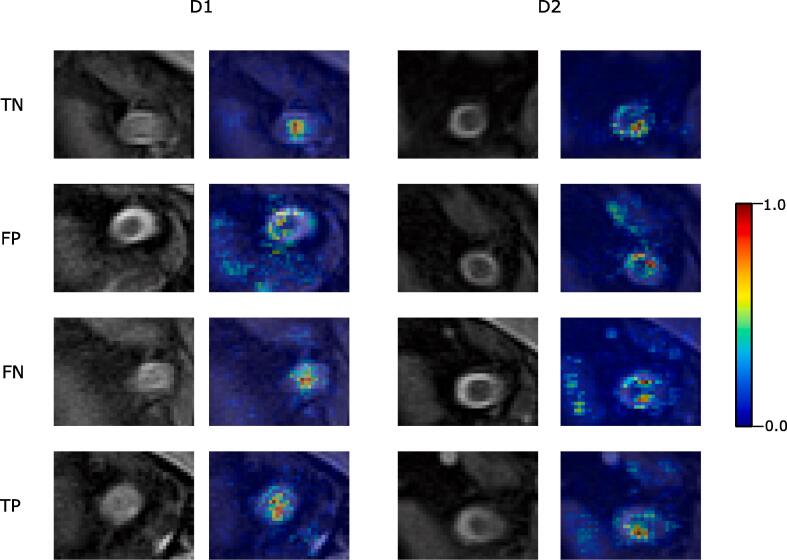
Fig. 4 shows four examples of scans correctly classified with and without optic nerve lesion for both TRIO and PRISMA datasets, and their corresponding saliency maps.
Fig. 4.
Coronal view of selected scans, for the two cohorts, and their corresponding saliency map generated by the network, by their characteristics (lesion or not) and their classification (correct or incorrect). TP: True positive (lesion). TN: True negative (no lesion). FN: False negative. FP: False positive.
4. Discussion
Detecting optic nerve lesions using MRI scans is highly relevant for MS diagnosis and other diseases linked with ON. In this paper, we have explored an automated method to detect such lesions and help clinicians make better informed decisions. As far as we know, this is the first automated approach for optic nerve lesion detection using MRI.
Our results show balanced accuracies of around 68% for validation, with similar sensitivity and specificity, so classification results are not affected by the uneven proportion of positive and negative labels. Results of the model are consistently better than the two other classification methods we compare to, obtaining results around mid 50%, meaning that those simpler models are not able to distinguish the presence/absence of optic nerve lesions, and that our approach correctly captures the signal from the images that lead to correct decisions.
When looking at the test set (D2), the performance of the CNN, while it slightly decreases compared to the training dataset (D1) (specially on the specificity), it has a similar balanced accuracy, albeit slightly lower (64%). The sensitivity is much higher, so the model has few FN but a sizeable number of FP. Given that those scans were completely unseen by the model during training, the results suggest that the model has not overfitted to the training data and it is able to generalize well. In contrast, the other two models show very poor performance, with balanced accuracies around 50% and bad specificities, which indicates that it assigns the same label to all the cohort, without learning to distinguish between them. Compared to the performance obtained in D1, SVM and RF do not work well on unseen data from another scanner. The confusion matrices of the three models show that the model overestimates the presence of lesion, with large amount of FP in the three models, although on a lower percentage in the CNN, while the number of FN remains low.
While the performance shows that the network is able to distinguish lesions well enough, we also wanted to explore what the network is learning to take such decisions. Saliency maps (Fig. 4) show that the network is able to correctly identify the zone of the optic nerve, giving more weight to the voxels that represents that area. This is consistent across scans and cohorts, with the optic nerve being in slightly different positions but the network being able to detect it. When the network fails, it normally is because it is confused by areas that are not the optic nerve (FP and FN in the figure). When the network correctly predicts the lesion, we do not appreciate significant differences between the areas observed for scans with and without lesions, or across cohorts.
While sensitivity of the model is not comparable to the one reported by trained neuroradiologists (80–94% within 30 days of symptoms (De Lott et al., 2022)), the performance of the model in D1 (68% balanced accuracy and 68% sensitivity) and D2 (74% sensitivity) makes us believe that the model is more comparable to the one reported by a radiologist unaware of the clinical symptoms and evaluating a single image, with a sensitivity of around 76%, (Bursztyn et al., 2019). Radiologists usually have access to a more diverse set of scans, repeated sequences, and information about the patient, while our model achieves the results with a single sequence. Given the speed of the model to process a single scan (around 20 s to preprocess the image with minimal human input and between 1 and 5 s to classify, depending on the machine), and the sequence used (short and easily acquired in clinical routine) the proposed system could serve as a stepping stone for future, more precise models that could be easily integrated in a diagnosis procedure with minimal costs. Also, a tool that provides another opinion could be useful to make better informed decisions when there is disagreement or doubt on patient assessment, as well as in conditions without experienced specialists in optic nerve or centers with lower resources.
Performance of the model, while better than the other methods we compare it to, is still behind diagnosis accuracy of trained radiologists. A clear limitation of the presented work is that the proportion of scans with a lesion is very small compared to healthy optic nerve. Our dataset, in fact, over-represents the actual incidence present in the regular MS population to be able to have enough samples to successfully train the model. Larger studies on patients affected with ON could help build better, more balanced datasets with which to build improved detection models. Another limitation of the data is that, while our two cohorts were taken using different machines, they are from the same manufacturer and with the same parameters. Testing the method on images taken with another machine, or with other sequences, would help clarify the robustness of the model. We also have no information about the treatments undergone by the patients, which could affect the ON and bias our results. All those limitations could be improved by designing a complete clinical study instead of a retrospective one.
The architecture of the neural network could also be improved. Approaches such as metric learning (Kaya and Bilge, 2019) could help building a better representation of the problem, transforming it from a binary classification task to a manifold learning approach, where the different subtypes of optic nerve lesions (De Lott et al., 2022) could be included. Also, simpler, more traditional 2D CNN using slices of the scan could be alternatives to our architecture and ought to be studied. Adding clinical symptoms to the input of the network could help the classification by adding extra context about the patient. Segmentation of the optic nerve could also help improve lesion detection and could be an interesting avenue to explore. For example, we could train a network to segment the optic nerve and use the saliency maps together with the segmentation to evaluate the prediction correctness, by looking at the percentage of pixel saliency probabilities outside the boundaries of the segmentation: if the network is looking at structures other than the optic nerve, it probably is a bad prediction. Finally, automated diagnostic models, including the one proposed, should be adapted to other sequences with improved optic nerve imaging (e.g. double inversion recovery sequences (Riederer et al., 2019)).
5. Conclusions
In this paper, we presented a first approach for automated optic nerve lesion detection based on MRI. We showed a simple and fast way to process the image, train a 3D CNN network and classify it. Performance and interpretation of the network indicates that it is able to learn to distinguish lesions in the optic nerve, and that it could be an important first step to develop systems that could support clinicians to make more informed diagnosis.
Disclosures
G Martí Juan has nothing to disclose. M Frías has nothing to disclose. A García Vidal has nothing to disclose. A Vidal-Jordana has received support for contracts Juan Rodes (JR16/00024) and receives research support from Fondo de Investigación en Salud (PI17/02162) from Instituto de Salud Carlos III, Spain; and has engaged in consulting and/or participated as speaker in events organized by Novartis, Roche, Biogen, and Sanofi. M Alberich has nothing to disclose. W Calderon has nothing to disclose. G Piella has nothing to disclose. O Camara has nothing to disclose. X Montalban has received speaking honoraria and travel expenses for participation in scientific meetings, has been a steering committee member of clinical trials or participated in advisory boards of clinical trials in the past years with Abbvie, Actelion, Alexion, Bayer, Biogen, Bristol-Myers Squibb/Celgene, EMD Serono, Genzyme, Hoffmann-La Roche, Immunic, Janssen Pharmaceuticals, Medday, Merck, Mylan, Nervgen, Novartis, Sandoz, Sanofi-Genzyme, Teva Pharmaceutical, TG Therapeutics, Excemed, MSIF and NMSS. J Sastre-Garriga has received grants and personal fees from Genzyme, personal fees from Almirall, Biogen, Celgene, Merck, Bayer, Biopass, Bial, Novartis, Roche and Teva, and he is European co-Editor of Multiple Sclerosis Journal and Scientific Director of Revista de Neurologia. A Rovira serves on scientific advisory boards for Novartis, Sanofi-Genzyme, Synthetic MR, TensorMedical, Roche, Biogen, Bristol, and OLEA Medical, and has received speaker honoraria from Bayer, Sanofi-Genzyme, Merck-Serono, Teva Pharmaceutical Industries Ltd, Novartis, Roche and Biogen. D Pareto has received a research contract from Biogen Idec.
Funding
This project was developed as a part of Gerard Martí-Juan ECTRIMS Research Fellowship Program 2021–2022. This study was partially supported by the Projects (PI18/00823, PI19/00950), from the Fondo de Investigación Sanitaria (FIS), Instituto de Salud Carlos III.
Ethical statement
Ethical approval for the project was done by the Vall d’Hebron ethical approval committee on 4/03/2022 (Proj. Number PR(IDI)124/2022).
CRediT authorship contribution statement
Gerard Martí-Juan: Conceptualization, Software, Validation, Writing - original draft, Visualization. Marcos Frías: Conceptualization, Methodology, Software, Validation, Investigation, Visualization. Aran Garcia-Vidal: Methodology, Software. Angela Vidal-Jordana: Investigation, Resources, Data curation, Writing - review & editing. Manel Alberich: Resources, Data curation. Willem Calderon: Writing - review & editing. Gemma Piella: Writing - review & editing. Oscar Camara: Writing - review & editing. Xavier Montalban: Resources, Funding acquisition. Jaume Sastre-Garriga: Resources, Writing - review & editing. Àlex Rovira: Investigation, Data curation, Resources, Supervision, Funding acquisition. Deborah Pareto: Conceptualization, Methodology, Formal analysis, Resources, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.nicl.2022.103187.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Boser B.E., Guyon I.M., Vapnik V.N. Proceedings of the 5th Annual ACM Workshop on Computational Learning Theory. 1992. A training algorithm for optimal margin classifiers; pp. 144–152. [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. doi: 10.1007/978-3-030-62008-0_35. [DOI] [Google Scholar]
- Brownlee W.J., Miszkiel K.A., Tur C., Barkhof F., Miller D.H., Ciccarelli O. Inclusion of optic nerve involvement in dissemination in space criteria for multiple sclerosis. Neurology. 2018;91:E1130–E1134. doi: 10.1212/WNL.0000000000006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursztyn L.L., De Lott L.B., Petrou M., Cornblath W.T. Sensitivity of orbital magnetic resonance imaging in acute demyelinating optic neuritis. Can. J. Ophthalmol. 2019;54:242–246. doi: 10.1016/j.jcjo.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lott L.B., Bennett J.L., Costello F. The changing landscape of optic neuritis: a narrative review. J. Neurol. 2022;269:111–124. doi: 10.1007/s00415-020-10352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, L., 2014. A tutorial survey of architectures, algorithms, and applications for deep learning. doi: 10.1017/ATSIP.2013.99.
- Faizy T.D., Broocks G., Frischmuth I., Westermann C., Flottmann F., Schönfeld M.H., Nawabi J., Leischner H., Kutzner D., Stellmann J.P., Heesen C., Fiehler J., Gellißen S., Hanning U. Spectrally fat-suppressed coronal 2D TSE sequences may be more sensitive than 2D STIR for the detection of hyperintense optic nerve lesions. Eur. Radiol. 2019;29:6266–6274. doi: 10.1007/s00330-019-06255-z. [DOI] [PubMed] [Google Scholar]
- Filippi, M., Rocca, M.A., Ciccarelli, O., De Stefano, N., Evangelou, N., Kappos, L., Rovira, A., Sastre-Garriga, J., Tintorè, M., Frederiksen, J.L., Gasperini, C., Palace, J., Reich, D.S., Banwell, B., Montalban, X., Barkhof, F., Group, M.S., 2016. MRI criteria for the diagnosis of multiple sclerosis: MAGNIMS consensus guidelines. The Lancet. Neurology 15, 292–303. URL:https://pubmed.ncbi.nlm.nih.gov/26822746 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4760851/, doi: 10.1016/S1474-4422(15)00393-2. [DOI] [PMC free article] [PubMed]
- Gass A., Moseley I.F. The contribution of magnetic resonance imaging in the differential diagnosis of optic nerve damage. J. Neurol. Sci. 2000;172:17–22. doi: 10.1016/S0022-510X(99)00272-5. [DOI] [PubMed] [Google Scholar]
- Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P.C., Mega J.L., Webster D.R. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. URL:https://doi.org/10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- Havaei M., Davy A., Warde-Farley D., Biard A., Courville A., Bengio Y., Pal C., Jodoin P.M., Larochelle H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. URL:https://doi.org/10.1016/j.media.2016.05.004 arXiv:1505.03540. [DOI] [PubMed] [Google Scholar]
- Kingma D.P., Ba J. 2017. Adam: A method for stochastic optimization. arXiv:1412.6980. [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Kaya Mahmut, Bilge Hasan Sakir. Deep Metric Learning: A Survey. Symmetry (Basel). 2019;11(9):1066. [Google Scholar]
- McBee M.P., Awan O.A., Colucci A.T., Ghobadi C.W., Kadom N., Kansagra A.P., Tridandapani S., Auffermann W.F. Deep Learning in Radiology. Acad. Radiol. 2018;25:1472–1480. doi: 10.1016/j.acra.2018.02.018. URL:https://doi.org/10.1016/j.acra.2018.02.018. [DOI] [PubMed] [Google Scholar]
- McKinney A.M., Lohman B.D., Sarikaya B., Benson M., Lee M.S., Benson M.T. Accuracy of routine fat-suppressed FLAIR and diffusion-weighted images in detecting clinically evident acute optic neuritis. Acta radiol. 2013;54:455–461. doi: 10.1177/0284185112471797. URL:https://doi.org/10.1177/0284185112471797. [DOI] [PubMed] [Google Scholar]
- Mongan J., Moy L., Kahn C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiology. Artificial Intelligence. 2020;2 doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preziosa, P., Comi, G., Filippi, M., 2016. Optic neuritis in multiple sclerosis. doi: 10.1212/WNL.0000000000002869. [DOI] [PubMed]
- Riederer I., Mühlau M., Hoshi M.M., Zimmer C., Kleine J.F. Detecting optic nerve lesions in clinically isolated syndrome and multiple sclerosis: double-inversion recovery magnetic resonance imaging in comparison with visually evoked potentials. J. Neurol. 2019;266:148–156. doi: 10.1007/s00415-018-9114-2. URL:https://doi.org/10.1007/s00415-018-9114-2. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Acevedo B., Rovira A., Vidal-Jordana A., Moncho D., Pareto D., Sastre-Garriga J. Optic neuritis: aetiopathogenesis, diagnosis, prognosis and management. Revista de neurologia. 2022;74:93–104. doi: 10.33588/rn.7403.2021473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira À., Auger C. Beyond McDonald: updated perspectives on MRI diagnosis of multiple sclerosis. Expert Review of Neurotherapeutics. 2021;21:895–911. doi: 10.1080/14737175.2021.1957832. URL:https://doi.org/10.1080/14737175.2021.1957832. [DOI] [PubMed] [Google Scholar]
- Schroeder A., Van Stavern G., Orlowski H.L., Stunkel L., Parsons M.S., Rhea L., Sharma A. Detection of optic neuritis on routine brain MRI without and with the assistance of an image postprocessing algorithm. Am. J. Neuroradiol. 2021;42:1130–1135. doi: 10.3174/ajnr.A7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorten C., Khoshgoftaar T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data. 2019;6 doi: 10.1186/s40537-019-0197-0. URL:https://doi.org/10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting Nitish. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G., Correale J., Fazekas F., Filippi M., Freedman M.S., Fujihara K., Galetta S.L., Hartung H.P., Kappos L., Lublin F.D., Marrie R.A., Miller A.E., Miller D.H., Montalban X., Mowry E.M., Sorensen P.S., Tintoré M., Traboulsee A.L., Trojano M., Uitdehaag B.M., Vukusic S., Waubant E., Weinshenker B.G., Reingold S.C., Cohen J.A. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17:162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Baranzini S.E., Geurts J., Hemmer B., Ciccarelli O. Multiple sclerosis. The Lancet. 2018;391:1622–1636. doi: 10.1016/S0140-6736(18)30481-1. URL:https://doi.org/10.1016/S0140-6736(18)30481-1. [DOI] [PubMed] [Google Scholar]
- Tintoré M., Rovira A., Rio J., Nos C., Grivé E., Téllez N., Pelayo R., Comabella M., Montalban X. Is optic neuritis more benign than other first attacks in multiple sclerosis? Annals of Neurology. 2005;57:210–215. doi: 10.1002/ana.20363. [DOI] [PubMed] [Google Scholar]
- Walton, C., King, R., Rechtman, L., Kaye, W., Leray, E., Marrie, R.A., Robertson, N., La Rocca, N., Uitdehaag, B., van der Mei, I., Wallin, M., Helme, A., Angood Napier, C., Rijke, N., Baneke, P., 2020. Rising prevalence of multiple sclerosis worldwide: Insights from the Atlas of MS, third edition. Mult. Scler. J. 26, 1816–1821. doi: 10.1177/1352458520970841. [DOI] [PMC free article] [PubMed]
- Wattjes M.P., Ciccarelli O., Reich D.S., Banwell B., de Stefano N., Enzinger C., Fazekas F., Filippi M., Frederiksen J., Gasperini C., Hacohen Y., Kappos L., Li D.K., Mankad K., Montalban X., Newsome S.D., Oh J., Palace J., Rocca M.A., Sastre-Garriga J., Tintoré M., Traboulsee A., Vrenken H., Yousry T., Barkhof F., Rovira À., Rocca M.A., Tintore M., Rovira A. 2021 MAGNIMS-CMSC-NAIMS consensus recommendations on the use of MRI in patients with multiple sclerosis. Lancet Neurol. 2021;20:653–670. doi: 10.1016/S1474-4422(21)00095-8. [DOI] [PubMed] [Google Scholar]
- Yu K.H., Beam A.L., Kohane I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018;2:719–731. doi: 10.1038/s41551-018-0305-z. URL:https://doi.org/10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- Zhang, Q.s., Zhu, S.c., 2018. Visual interpretability for deep learning: a survey. Front. Inf. Technol. Electron. Eng. 19, 27–39. URL:https://doi.org/10.1631/FITEE.1700808, doi: 10.1631/FITEE.1700808.
- Zhang Y., Gorriz J.M., Dong Z. Deep learning in medical image analysis. J Imaging. 2021;7:NA. doi: 10.3390/jimaging7040074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR) 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.