Abstract
Glioma is a common primary aggressive tumor with limited clinical treatment. Recently, growing research suggests that gut microbiota is involved in tumor progression, and several probiotics can inhibit tumor growth. However, evidence for the effect of probiotics on glioma is lacking. Here, we found that Bifidobacterium (B.) lactis combined with Lactobacillus (L.) plantarum reduced tumor volume, prolonged survival time and repaired the intestinal barrier damage in an orthotopic mouse model of glioma. Experiments demonstrated that B. lactis combined with L. plantarum suppressed the PI3K/AKT pathway and down-regulated the expression of Ki-67 and N-cadherin. The glioma-inhibitory effect of probiotic combination is also related to the modulation of gut microbiota composition, which is characterized by an increase in relative abundance of Lactobacillus and a decrease in some potential pathogenic bacteria. Additionally, probiotic combination altered fecal metabolites represented by fatty acyls and organic oxygen compounds. Together, our results prove that B. lactis combined with L. plantarum can inhibit glioma growth by suppressing PI3K/AKT pathway and regulating gut microbiota composition and metabolites in mice, thus suggesting the potential benefits of B. lactis and L. plantarum against glioma.
Keywords: glioma, gut microbiota, probiotics, PI3K/Akt signal pathway, bacterial metabolites
Introduction
Glioma, the most frequent primary intracranial neoplasm, represents 81% of the malignant tumors of the central nervous system. According to the 2021 World Health Organization classification, glioma is divided into four grades (grade 1 to grade 4). Unfortunately, more than 50% of the glioma patients developed glioblastoma (GBM, grade 4), which is the most malignant glioma with a mean survival time of 8 months (Ostrom et al., 2021). At present, the clinical treatment strategy of glioma is mainly maximum safe surgical resection, followed by radiotherapy and chemotherapy (temozolomide, TMZ). However, the therapy of glioma is challenged by its diffusion and invasion, which leads to high recurrence rates (Xu et al., 2020).
The complicated cross-talk between gut microbiota and tumors has been substantiated in numerous studies. Actually, polymorphic microbiomes have been identified as a new hallmark of cancer in the latest research (Hanahan, 2022). Gut dysbiosis, generally induced by oxidative stress, bacteriocins, etc., can disrupt intestinal integrity, promote bacterial translocation and induce inflammation. Inflammatory factors and bacterial metabolites can increase susceptibility to cancer and may reduce the permeability of the blood–brain barrier (BBB) and into the brain (Carlessi et al., 2021). Noteworthily, the alteration of gut microbiota composition caused by long-term antibiotic administration promoted glioma growth through regulating brain immune responses in mice (D’alessandro et al., 2020). Certain bacteria, such as Helicobacter pylori, Bacteroides fragilis and Clostridium, have been reported to drive tumorigenesis in the gastrointestinal tract through modulating autophagy, inflammation, oxidative stress or systemic immune responses (Matson et al., 2021). In addition, bacterial metabolites, such as short chain fatty acids (SCFA), lipopolysaccharide (LPS), serotonin, gamma-aminobutyric acid (GABA), etc., can affect brain function through stimulating the vagal nerve or passing the BBB (D’alessandro et al., 2021).
Probiotics, as naturally occurring bacteria that are extensively observed in human and animal intestine, have shown their potential against various diseases including cancer and neurodegenerative diseases. Lactobacillus and Bifidobacterium are common probiotics with positive effects in the prevention and treatment of cancers. Probiotics can repress tumor development through secreting antioxidant enzymes, regulating cell cycle associated proteins and inducing apoptosis (Mohseni et al., 2021). In the recent research, B. bifidum was markedly enriched in the patients who positively responded to the programmed cell death protein 1 (PD-1) blockade, and studies demonstrated that oral gavage of B. bifidum inhibited colon tumor growth in mice through stimulating CD8+ T cells (Lee et al., 2021). Recently, a study showed that gut microbiota composition was different between normal and glioma mice, the abundance of Lactobacillus decreased at an early stage and then gradually increased. After temozolomide treatment, Akkermansia and Bifidobacterium were enriched in glioma mice (Li et al., 2021). Fecal metabolites were also altered in glioma patients and mice, with glioma patient having a reduced levels of norepinephrine and 5-hydroxyindoleaceic acid (serotonin metabolite). Interestingly, the effects of glioma on fecal metabolites were abrogated after temozolomide treatment (Dono et al., 2020). Administration of L. plantarum can increase serotonin and dopamine level in the striatum of germ-free mice, and selective serotonin reuptake inhibitors can inhibit proliferation and induce apoptosis in glioma cell lines (D’alessandro et al., 2021). A study has shown that a ketogenic diet can slightly increase the survival of glioma mice through altering gut microbiota composition (McFarland et al., 2017). The supernatant of kefir, a commercial probiotic product, has dose-dependent cytotoxicity effect on U87 GBM cell line (Fatahi et al., 2021).
We aimed to investigate the effect of specific probiotic strains on glioma growth in GL261 orthotopic mouse model by gavage treatment with B. lactis or L. plantarum individually or in combination. Our previous experiments found that the combined application of B. lactis and L. plantarum was the most effective in reducing tumor volume of glioma-bearing mice. In this study, our results demonstrate that B. lactis combined with L. plantarum suppressed glioma development, at least partly by modulating phosphoinositide 3-kinase (PI3K)/AKT pathway, gut microbiota and metabolites. Collectively, these results reveal that B. lactis and L. plantarum might display potential efficacy against glioma.
Materials and methods
Cell culture
Cell line GL261 was bought from BeNa Culture Collection (Beijing, China) and cultured in DMEM-H/F12 media (Gibco, MA, United States) containing 10% fetal bovine serum (Gibco, MA, United States) and 1% penicillin/streptomycin (Invitrogen, MA, United States) at 37°C, 5% CO2 (Wang et al., 2020).
Bacterial preparation
Bifidobacterium lactis (BI516) and L. plantarum (LP-Onlly) freeze-dried powders were purchased from Shanghai Onlly Company (Shanghai, China) and suspended in sterile saline to a concentration of 4 × 109 CFU/0.2 ml (B. lactis: 2 × 109 CFU, L. plantarum: 2 × 109 CFU). The microbial number was counted using cell viable counting method, and bacteria was identified using molecular biological means by Guangdong Detection Center of Microbiology (Guangzhou, China). The detailed test results are shown in Supplementary Tables S1–S3, Sequence S1 and raw data is included in Data Sheet 1–4.
Design of animal experiments
Male C57BL/6 mice (4 weeks) were purchased from Chengdu Dashuo Laboratory Animal Company (Chengdu, China) and housed at 22 ± 2°C under a 12 h: 12 h light–dark cycle in specific pathogen-free facilities. All animal experiments followed the regulations of ethics committee of the Experimental Animal Administration of Sichuan University. Two batches of C57BL/6 mice were used: one batch was used for survival curve (n = 7), and another batch was used for indicator detection (n = 11). The mice in each batch were randomly divided into three groups: sham, model (Mod) and Mod-L. plantarum (LP) + B. lactis (BL) group. The Mod-LP + BL group was daily gavaged with 200 μl probiotic mixture of B. lactis (2 × 109 CFU) and L. plantarum (2 × 109 CFU) till the end of experiment. The sham and Mod group were daily gavaged with 200 μl sterile saline (Van Zyl et al., 2018). Stereotaxic surgery was performed on mice 10 days after the first gavage. The mice were intraperitoneally anesthetized with 4% chloral hydrate (66 mg/kg). Then, 1 × 105 GL261 cells (in 1 μl PBS) were stereotactically injected into the right striatum in the Mod and Mod-LP + BL groups and the stereotaxic coordinates to the bregma were 1.0 mm anterior, + 1.5 mm lateral, and 3.5 mm ventral (Lam et al., 2018). The sham group was injected with 1 μl PBS. The mice used for indicator detection were sacrificed 28 days after inoculation.
Clinical score and behavioral testing
Clinical scores and behavioral testing were daily assessed on the last 6 days before sacrifice based on previous research (Helgers et al., 2020). Detailed clinical scoring criterion is included in Supplementary Table S4. Open-field test: Locomotor activity was measured in an open-field apparatus (40 cm × 40 cm × 40 cm). The mouse was placed in the corner of the box and the spontaneous activity was recorded for 5 min using a video camera located above the box. The total motion distance and number of line crossings were assessed by EthoVision XT software (Zheng et al., 2016). Beam balance test: Motor coordination and balance were assessed by beam balance test and scoring criteria was as previously described (Supplementary Table S5; Chen et al., 2001).
Magnetic resonance imaging
In vivo cerebral magnetic resonance imaging (MRI) was performed on a small animal 7 T MRI scanner (Time Medical, Shanghai, China). All mice were anesthetized with 2% inhaled isoflurane. Images were collected using a 2D T2-weighted sequence (axial orientation): Repetition time = 3,500 ms, echo time = 54 ms, number of averages = 4, matrix = 256 × 256, 6 slices, slice thickness = 1 mm. Tumor volumes were calculated using ITK-SNAP program (Agliardi et al., 2021).
Fecal samples and tissue collection
Fresh fecal pellets were collected and immediately frozen at −80°C on days 25 to 27 after inoculation. All mice were anesthetized with 4% chloral hydrate (66 mg/kg) and intracardially perfused with cold sterilized saline at the end of experiments. Brain, ileum and colon tissues were collected, part tissues were snap-frozen in liquid nitrogen and stored at −80°C, and part tissues were fixed with 4% paraformaldehyde and embedded in paraffin (Yang et al., 2020).
Histological and immunohistochemical analyses
Paraffin-embedded tissues were cut into 3-μm-thick slides. Hematoxylin and eosin (HE) staining was conducted according to standard histological protocols. For immunohistochemistry (IHC), slides were, respectively, incubated with the following primary antibodies from Abcam (Cambridge, United Kingdom) after blocking: Ki-67 (ab16667), Occludin (ab216327), ZO-1 (ab221547). Quantification of IHC was performed by counting immunostaining-positive cells (Ki-67) and calculating positive areas (Occludin, ZO-1) using Image Pro Plus software (Chen et al., 2020).
Reverse transcription quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Thermo fisher scientific, MA, United States) and reverse-transcribed with the RevertAid First Strand cDNA Synthesis Kit (Thermo fisher scientific, MA, United States). Real-time-polymerase chain reaction was conducted according to previous research and data was processed by 2−ΔΔCT method (Yang et al., 2020). The primer sequences for target genes are listed in Supplementary Table S6.
Western blot
Total proteins were extracted with RIPA buffer (Beyotime, Shanghai, China) following the manufacturer’s instructions. The protein lysate was electrophoresed and transferred to a PVDF membrane (Bio-Rad, CA, United States). The membrane was, respectively, incubated with the following primary antibodies from CST (Shanghai, China): p-PI3K (Tyr458/Tyr199; 1:1000; 4,228), PI3K (1:1000; 4,257), p-AKT (Ser473; 1:2000; 4,060), AKT (1:1000; 4,691), PTEN (1:1000; 9,559), survivin (1:1000; 2,808), FoxO1 (1:1000; 2,880), Bad (1:1000; 9,292), E-Cadherin (1:1000; 3,195) and N-Cadherin (1:1000; 13,116). The proteins were detected with ECL Plus Reagent (Beyotime, Shanghai, China) and quantified using ImageJ software (Yang et al., 2020).
Gut microbial analysis
Fecal Genomic DNA was Extracted with a Fecal DNA Extraction kit (TIANGEN, Beijing, China) Following the manufacturer’s Instructions. Amplification of the Near-Full-Length 16S Rrna Genes was Performed With the 27F/1390R Primers. Next, the Constructed Library was Sequenced on the Pacbio Sequel Platform. The High-Quality Reads Were Analyzed With QIIME Software (Stearns et al., 2011). The Qualified Sequences Were Clustered Into Operational Taxonomic Units (OTUs) at 97% Similarity. Raw Data for the 16S Rrna Sequences Were Submitted to the Genbank Databases Under Accession Number PRJNA863912.
Next-generation sequencing
RNA-seq was performed based on previous research (Wang et al., 2019). Briefly, mRNA was purified from total RNA and used to construct RNA-seq library with NEBNext® Ultra™ RNA Library Prep Kit (NEB, MA, United States). Paired-end mRNA sequencing was carried out on Noves6000 platform. Clean reads were mapped to the mouse reference genome (GRCm38.p6) using HISAT2. Differentially expressed genes (DEGs) were selected with|fold change (FC)| > 1.5 and value of p < 0.05 using edgeR. Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) analysis were performed using the clusterProfiler package. The RNA-seq raw data have been deposited in GenBank database with the accession number of PRJNA863751.
Untargeted metabolomics by GC–MS
Metabolic extracts from fecal were analyzed by gas chromatography–mass spectrometry (GC–MS) based on previous research (He et al., 2019). Fecal samples were handled with extract solvent (methanol-chloroform, 3:1, containing internal standard). The quality control samples were prepared by mixing an equal aliquot of all collected samples. GC–MS analyses were performed on Agilent 7,890 system with DB-5MS capillary column (30 m × 250 μm × 0.25 μm). Samples (1 μl) were injected in splitless mode. The injector, transfer line and ion source temperatures were 280°C, 280°C and 250°C, respectively. The oven temperature program was 50°C for 1 min, then raised to 310°C at 10°C/min, and held steady for 8 min. Raw GC–MS data were processed with ChromaTOF software (LECO). The raw data for GC–MS have been submitted to the Metabolights database under accession number MTBLS5563.
Statistics analysis
Statistical analyses were carried out with the SPSS 25.0 software. Log-rank test was used for Kaplan–Meier survival curves. Statistical difference between 2 groups was identified using a two-tailed Student’s t-test. For multiple comparisons, two-way ANOVA followed by LSD post hoc test was performed. Differences at p < 0.05 were considered significant.
Results
Bifidobacterium lactis combined with Lactobacillus plantarum inhibit glioma growth in mice
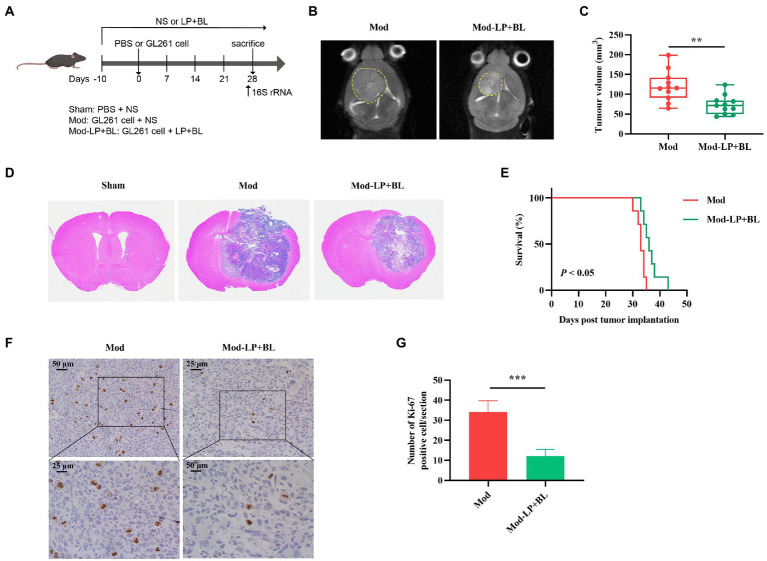
To determine whether B. lactis combined with L. plantarum can inhibit glioma growth in mice, we treated glioma-bearing mice with B. lactis and L. plantarum via daily oral gavage (Figure 1A). Tumor volume of glioma-bearing mice was evaluated by cerebral T2-weighted MRI sequence on day 28 post-inoculation (Figure 1B). Quantitative analysis showed that compared with the Mod group, tumor volume of the Mod-LP + BL group was declined (120.36 ± 38.69 mm3 vs. 73.25 ± 23.88 mm3, p < 0.01; Figure 1C). HE staining of glioma tissues substantiated this point (Figure 1D). As shown in Figure 1E, B. lactis combined with L. plantarum slightly prolonged the median survival time of glioma mice (33.0 days vs. 37.0 days, p < 0.05). To examine whether probiotics regulate tumor proliferation, IHC staining for Ki-67 was performed, and the number of Ki-67 positive cells was decreased in the Mod-LP + BL group compared to the Mod group (Figures 1F,G).
Figure 1.
Bifidobacterium lactis and Lactobacillus plantarum inhibit glioma growth in mice. (A) Experimental design. (B) Representative T2-weighted MRI images and (C) tumor volume quantification of glioma mice receiving either PBS or probiotic combination at day 28 post tumor implantation, n = 11, two-tailed Student’s t-test. (D) Representative HE-stained coronal brain sections. (E) Kaplan–Meier survival curves, n = 7, Log-rank test. (F) Representative Ki-67-stained sections and (G) numbers of Ki-67 positive cells of brain tissues, scale bars, 50 μm (upper) or 25 μm (lower). n = 6. Two-tailed Student’s t-test. Mod, Model. LP, Lactobacillus plantarum; BL, Bifidobacterium lactis; NS, normal saline. *p < 0.05, **p < 0.01, ***p < 0.001.
Effects of Bifidobacterium lactis and Lactobacillus plantarum on neurobehavior in glioma mice
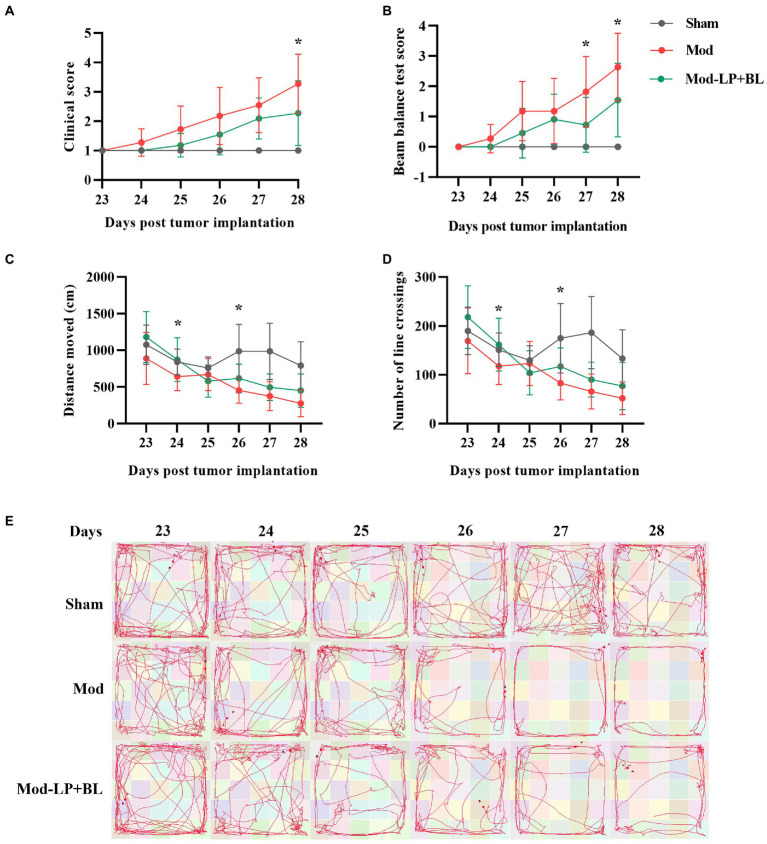
To explore the influences of B. lactis and L. plantarum on neurobehavior in glioma mice, we performed clinical scoring and behavioral testing. With the tumor growth, the locomotor ability and motor coordination of the mice in the Mod group deteriorated rapidly on the 23rd to 28th day after inoculation compared with the sham group, and probiotic mix alleviated this symptom. Compared with the Mod group, the clinical score and beam balance test score of the Mod-LP + BL group were decreased (day 27 and day 28: p < 0.05; Figures 2A,B). Open-field test demonstrated that distance moved and number of line crossings of the Mod group continuously declined compared with the sham group, and probiotic mix slowed down this trend (day 24 and day 26: p < 0.05; Figures 2C–E).
Figure 2.
Effects of B. lactis and L. plantarum on neurobehavior in glioma mice. (A) Clinical score and (B) beam balance test score of mice at day 23 to 28 post tumor implantation. (C) Distance moved and (D) number of line crossings in the open field of mice at day 23 to 28 post tumor implantation. (E) Representative images of the movement track of mice in the open-field test. Mod, Model. LP, Lactobacillus plantarum; BL, Bifidobacterium lactis. Two-way ANOVA. *p < 0.05. n = 11.
Bifidobacterium lactis combined with Lactobacillus plantarum repair intestinal barrier damage in glioma mice
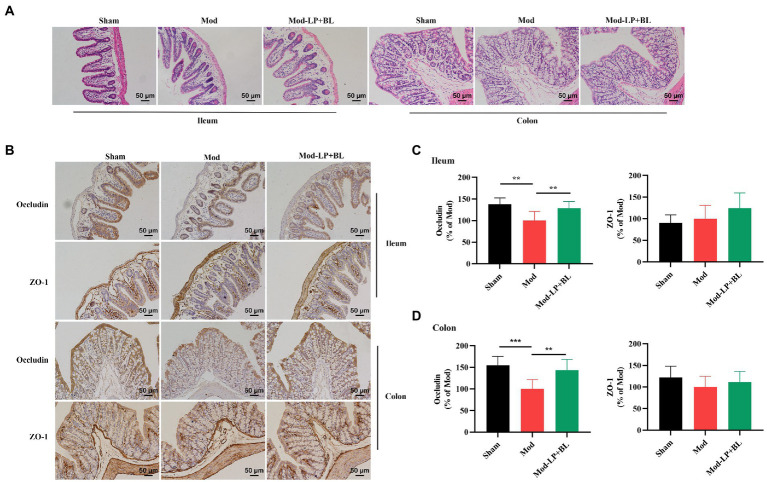
The intestinal defense system plays a pivotal role in preventing the bacterial overgrowth and mucosal immune response. Intercellular tight junctions, which are constituted by Occludin, claudin, zonula occludens (ZO) and junctional adhesion, are the major determinants of the intestinal physical barrier (Suzuki, 2020). HE staining did not show obvious abnormality in ileum and colon tissues of mice in each group (Figure 3A). To explore the effect of probiotics on intestinal barrier in glioma mice, we detected the expression of tight junction proteins. IHC staining showed that protein expression of Occludin in ileum and colon tissues was down-regulated in the Mod group compared with the Sham group (Ileum, p < 0.01; colon, p < 0.001), and probiotic combination up-regulated Occludin expression in ileum and colon tissues (Ileum, p < 0.01; colon, p < 0.01; Figures 3B–D).
Figure 3.
B. lactis and L. plantarum repair intestinal barrier damage in glioma mice. Representative (A) H&E, (B) ZO-1 and Occludin stained sections of ileum and colon tissues harvested from mice, scale bars, 50 μm. Quantification of ZO-1 and Occludin immunopositive area in (C) ileum and (D) colon tissues, two-way ANOVA. Mod, Model. LP, Lactobacillus plantarum; BL, Bifidobacterium lactis. *p < 0.05, **p < 0.01, ***p < 0.001. n = 6.
Bifidobacterium lactis combined with Lactobacillus plantarum inhibit PI3K/AKT pathway in glioma mice
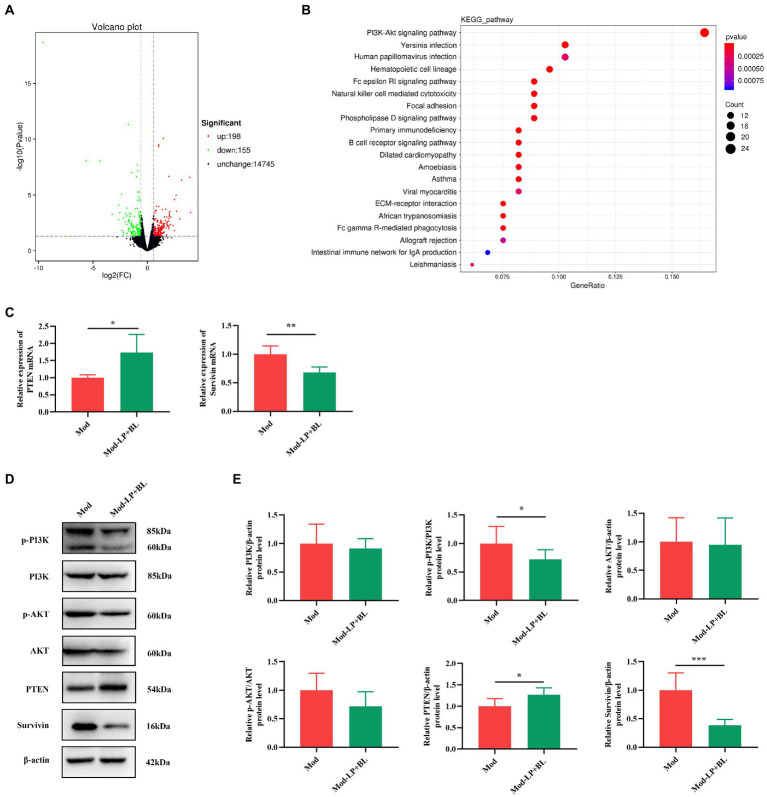
To delve the potential mechanism of the inhibitory effect of probiotics on glioma, RNA-seq was performed on the glioma tissues of mice. DEGs between the Mod and Mod-LP + BL groups were identified based on |FC| > 1.5 and value of p <0.05, among which 198 were up-regulated and 155 were down-regulated (Figure 4A). Then, a transcriptional heatmap was constructed to compare the expression profiles of DEGs between different groups, and the comparison showed significantly transcriptomic alterations in the Mod-LP + BL group compared with the Mod group (Supplementary Figure S1). GO analysis of DEGs revealed enrichment in cell adhesion, extracellular space and calcium ion binding (Supplementary Figure S2). KEGG pathway enrichment analysis predicted that PI3K-AKT signaling pathway involved in the regulation of probiotics on glioma (Figure 4B).
Figure 4.
B. lactis combined with L. plantarum inhibit PI3K/AKT pathway in glioma mice. (A) Volcano plot of DEGs of glioma tissues between the Mod and Mod-LP + BL groups. n = 3. (B) KEGG pathway enrichment analysis of DEGs. (C) The mRNA expression of PTEN and survivin in glioma tissues. n = 6. Two-tailed Student’s T-test. (D) Representative immunoblots and (E) quantitative analysis of p-PI3K, p-AKT, PTEN and survivin in glioma tissues. n = 6. Two-tailed Student’s T-test. Mod, Model. LP, Lactobacillus plantarum; BL, Bifidobacterium lactis. *p < 0.05, **p < 0.01, ***p < 0.001.
PI3K/AKT pathway, as the most enriched pathway identified by KEGG analysis, plays a crucial role in tumor initiation and progression. As a negative regulator of PI3K signals, the loss of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) results in PI3K pathway activation (Lee et al., 2018). Abnormal activation of PI3K and its downstream signals (Bad (Bui et al., 2018), FoxO1 (Yan and Huang, 2019), survivin (Yang et al., 2017), etc.) can promote carcinogenesis. To assess the activation of PI3K/AKT pathway, q-PCR and western blot of glioma tissues were performed. As shown in Figure 4C, probiotic combination up-regulated the mRNA expression of PTEN and down-regulated the mRNA expression of survivin in glioma tissues (PTEN, p < 0.05; survivin, p < 0.01; Figure 4C). At the protein level, probiotic combination down-regulated p-PI3K and survivin (p-PI3K, p < 0.01; survivin, p < 0.001; Figures 4D,E), and up-regulated PTEN expression in glioma tissues (p < 0.05; Figures 4D,E). Nevertheless, the protein expression of Bad and FoxO1 did not change significantly (Supplementary Figure S3).
Effects of Bifidobacterium lactis and Lactobacillus plantarum on the migration and invasion-related genes in glioma mice
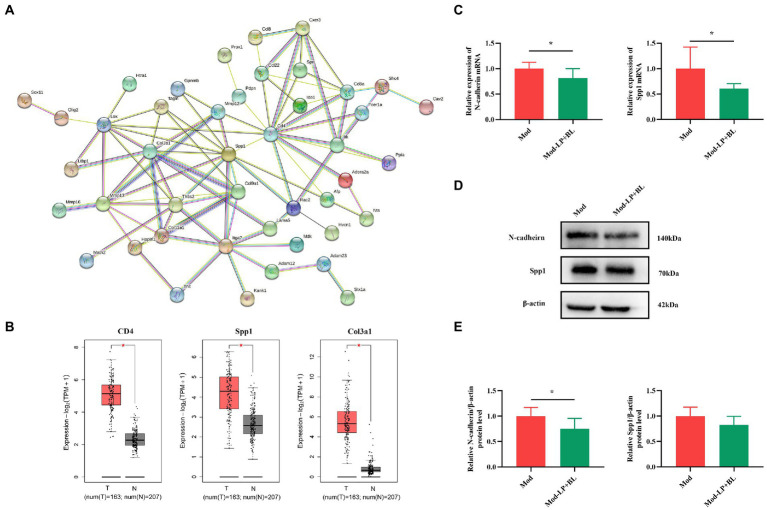
To predict interactions between candidate genes at the protein level, candidate genes were selected by taking the intersection of DEGs and glioma-associated genes, and a protein–protein interaction (PPI) network was constructed using the STRING database. The PPI network consisted of 44 nodes and 89 edges, and Cd4, Spp1 and Col3a1 were identified as the top three hub genes (Figure 5A). Subsequently, the cancer genome atlas (TCGA) and the genotype-tissue expression (GTEx) datasets were analyzed by GEPIA to evaluate the differential expression and prognostic value of the three hub genes. Compared with normal tissues, Cd4, Spp1 and Col3a1 expression were significantly increased in glioma tissues (Cd4, p < 0.05; Spp1, p < 0.05; Col3a1, p < 0.05; Figure 5B), but their expression levels were not correlated with overall survival (Supplementary Figure S4).
Figure 5.
Effects of B. lactis and L. plantarum on the migration and invasion-related genes in glioma mice. (A) PPI network. (B) Differential expression of CD4, Spp1 and Col3a1 in normal and glioma tissues from TCGA and GTEx datasets. (C) mRNA expression of N-cadherin and Spp1 in glioma tissues in Mod and Mod-LP + BL groups. n = 6, two-tailed Student’s T-test. (D) Representative immunoblots and (E) quantitative analysis of N-cadherin and Spp1 in glioma tissues. n = 6, two-tailed Student’s T-test. Mod, Model. LP, Lactobacillus plantarum; BL, Bifidobacterium lactis. T, tumor. N, normal. *p < 0.05, **p < 0.01, ***p < 0.001.
PPI network showed that genes related to tumor migration and invasion were different in the Mod and Mod-LP + BL groups. Osteopontin (OPN/Spp1), a potential biomarker for glioblastoma, could induce glioma cell migration and invasion. Experiments demonstrated that selective knockdown of osteopontin displayed an anti-tumoral activity in a GBM model (Lamour et al., 2010). Tumor invasion and metastasis are generally associated with epithelial-mesenchymal transition, which is a cellular process accompanied with the loss of epithelial marker (E-cadherin, etc.) and increase in mesenchymal markers (N-cadherin, vimentin, etc.) (Pastushenko and Blanpain, 2019). q-PCR was conducted to prove that mRNA expression of Spp1 and CDH2/N-cadherin are down-regulated by probiotic combination in glioma mice (Spp1, p < 0.05; N-cadherin, p < 0.05; Figure 5C). Western blot showed that probiotic combination suppressed N-cadherin protein expression (p < 0.05), but did not statistically affect the protein expression of Spp1 (Figures 5D,E) in glioma tissues. Additionally, the mRNA and protein expression of CDH1/E-cadherin did not alter in the Mod-LP + BL group compared with the Mod group (Supplementary Figure S5). The original images of western blot are shown in Supplementary Figure S6.
Bifidobacterium lactis combined with Lactobacillus plantarum altered the gut microbiota composition in glioma mice
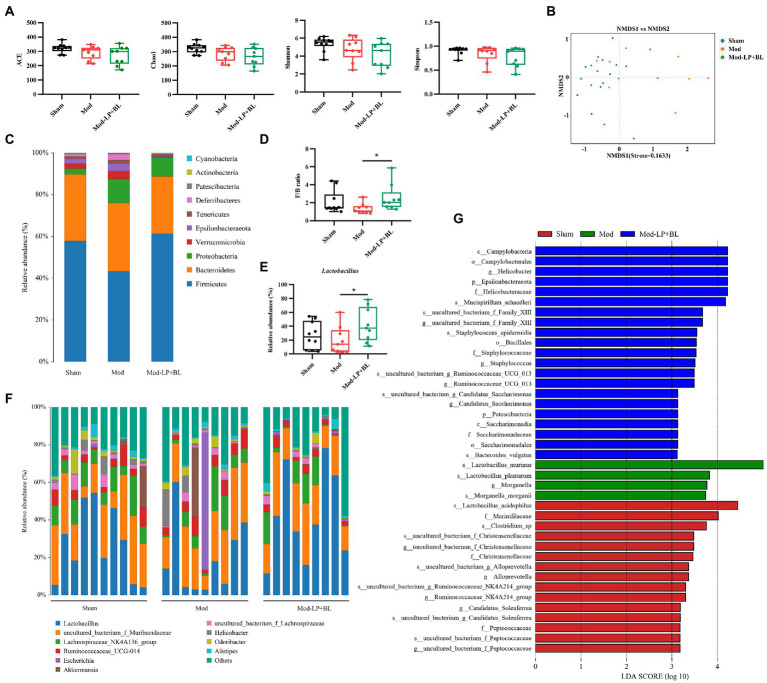
To compare the alteration of the gut microbiota composition in the three treatment groups, 16S rRNA sequencing was conducted on feces from mice. The α-diversity of gut microbiota was analyzed by four metrics: ACE, Chao1, Simpson and Shannon index, and results showed that B. lactis combined with L. plantarum did not affect the α-diversity of gut microbiota in glioma mice (Figure 6A). For β diversity, the nonmetric multidimensional scaling (NMDS), based on binary-jaccard distance, exposed insignificant clustering of gut microbiota composition between glioma and normal mice, but probiotic combination conspicuously affect the composition of gut microbiota in glioma mice (Figure 6B). As shown in Figure 6C, gut microbiota in the three groups was dominated by Bacteroidetes and Firmicutes at the phylum level. A lower abundance of Firmicutes was observed in the Mod group compared with the Sham group, and probiotic combination increased the abundance of Firmicutes and decreased the abundance of Bacteroidetes, leading to a rising in the ratio of Firmicutes/Bacteroidetes (F/B) compared with the Mod group (2.54 ± 1.48 vs. 1.35 ± 0.58, p < 0.05; Figure 6D). Genus-level analysis exhibited that Lactobacillus, the predominant bacterium in the three groups, was reduced in glioma mice compared with healthy mice, whereas oral gavage of probiotics increases the relative abundance of Lactobacillus (42.04% ± 24.26% vs. 19.52% ± 19.71%, p < 0.05; Figures 6E,F). To further identify the specific microbiota, linear discriminant analysis effect size (LEfSe) was performed and results showed that numerous pathogenic bacteria were enriched in the Mod group, including Helicobacter and Staphylococcus (Figure 6G).
Figure 6.
B. lactis combined with L. plantarum altered the gut microbiota composition in glioma mice. (A) Comparison of α-diversity of gut microbiota including ACE, Chao1, Simpson and Shannon index between the three groups. Wilcoxon rank-sum test. (B) NMDS analysis based on binary-jaccard distance, stress = 0.1633. Bar charts of the gut microbiota composition at the (C) phylum and (F) genus-level in the three groups. (D) F/B ratio, two-way ANOVA. (E) Relative abundance of Lactobacillus in the three groups, two-way ANOVA. (G) LDA score of enriched bacterial taxa, LDA > 3.0. Mod, Model. LDA, Linear discriminant analysis. LP, Lactobacillus plantarum; BL, Bifidobacterium lactis. *p < 0.05. n = 9–10.
Effects of Bifidobacterium lactis and Lactobacillus plantarum on microbial metabolites in glioma mice
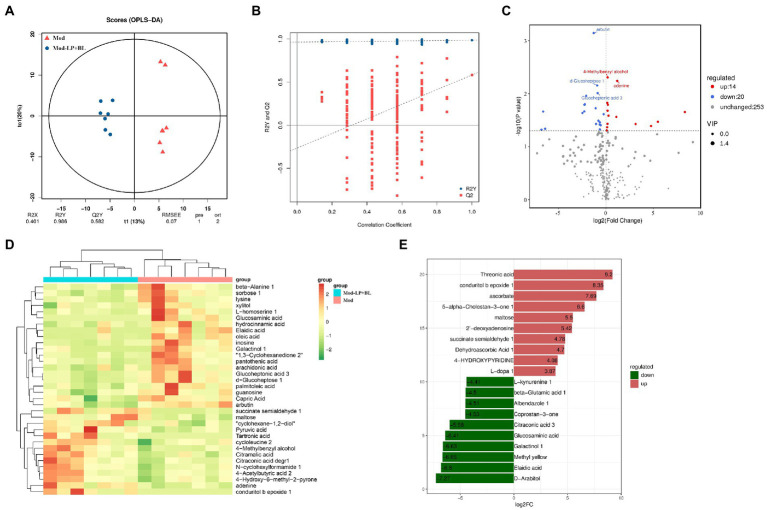
Untargeted metabolomics by GC–MS was conducted on stool collected from mice in the Mod and Mod-LP + BL groups. Supervised orthogonal projections to latent structures discriminant analysis (OPLS-DA) was performed to compare the difference between the two groups. The representative OPLS-DA score plot indicated distinct separation between the Mod and Mod-LP + BL groups (R2Y = 0.986; Q2Y = 0.582; Figure 7A), and permutation tests further validated the constructed OPLS-DA model (Figure 7B). In addition, variable importance in projection (VIP) score was calculated, which is a measure of estimating the importance of each metabolite in the OPLS-DA model. With VIP > 1 and p < 0.05, 14 up-regulated metabolites and 20 down-regulated metabolites were identified from feces of the Mod and Mod-LP + BL groups (Figure 7C). Unsupervised analysis of all differential metabolites clearly discriminated two clusters, one gathered glioma mice treated with probiotics and another gathered glioma mice treated with PBS (Figure 7D). The primary differences were the markedly increased/decreased level of threonic acid (FC = 9.2), conduritol b epoxide 1 (FC = 8.35), ascorbate (FC = 7.69), D − Arabitol (FC = −7.27) and Elaidic acid (FC = −6.8; Figure 7E). Analysis of metabolites via the human metabolome database showed that differential metabolites mainly enriched in fatty acyls and organooxygen compounds (Supplementary Figure S7).
Figure 7.
Effects of B. lactis and L. plantarum on microbial metabolites in glioma mice. (A) OPLS-DA score plot and (B) permutation tests of OPLS-DA models. (C) Differential fecal metabolites volcano plot of the Mod and Mod-LP + BL groups. (D) Unsupervised hierarchical heatmap of differential fecal metabolites between the Mod and Mod-LP + BL groups. (E) Fold change of differential metabolites.
Combined analysis of metabolomics and microbiome
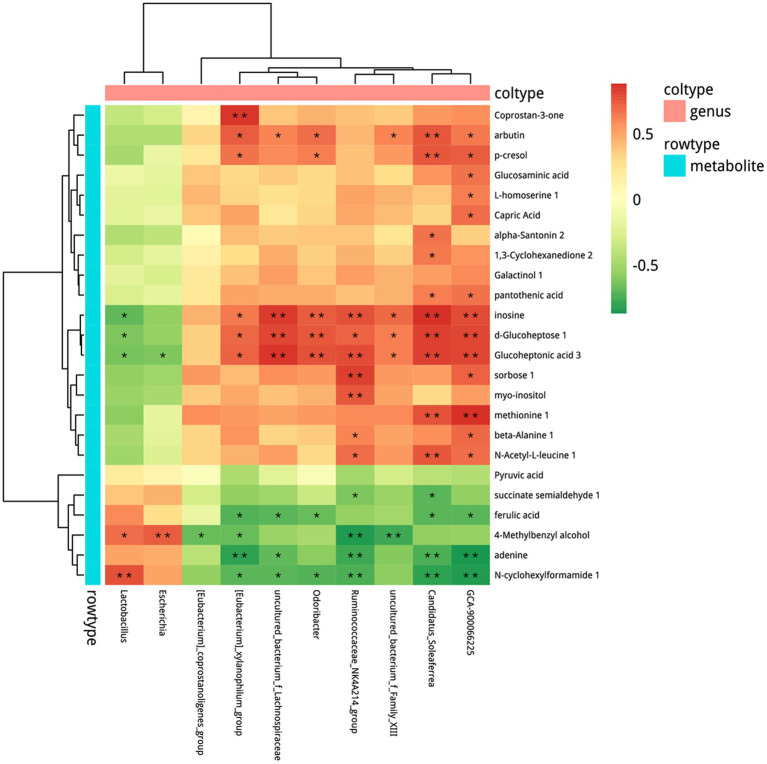
Based on the alteration in microbial composition and the metabolites, we combined microbiome with metabolomics to explore the possible relationship between microbes and metabolites. Correlation analysis between OTU and all differential fecal metabolites was conducted (Supplementary Figure S8) and the results are shown in Figure 8 with p < 0.05 at the genus level. Microbes such as Lactobacillus, were significantly increased after probiotics treatment and showed a positive correlation with N-cyclohexylformamide 1, 4-methylbenzyl alcohol, and a negative correlation with glucoheptonic acid 3, d-glucoheptose 1, inosine.
Figure 8.
Combined analysis of metabolomics and microbiome. Spearman’s rank correlation. *p < 0.05, **p < 0.01. n = 5–6.
Discussion
Recently, numerous researches have focused on the cross-interaction between gut microbiota and glioma (D’alessandro et al., 2021). Diverse studies have shown the distinct separation of gut microbiota between normal and glioma individuals both in humans and mice (Patrizz et al., 2020; Li et al., 2021). Notably, gut dysbiosis, caused by antibiotic supplements, can promote tumor growth in glioma mice via affecting immune system (D’alessandro et al., 2020; Fan et al., 2022). Therefore, we treated glioma mice with probiotics to determine whether probiotics can repress glioma progression. Our experiments show that B. lactis combined with L. plantarum can suppress glioma growth through regulating PI3K/AKT pathway and gut microbiota composition in mice. Additionally, the inhibitory effect was also associated with the alteration of fecal metabolites. Our results indicate the potential protective effects of B. lactis and L. plantarum against glioma.
The PI3K/AKT signaling pathway plays a crucial role in regulating cellular functions comprising cell survival, proliferation, growth and metabolism. Thorough molecular studies have identified RTK/RAS/PI3K (88%) as one of the significantly altered pathways in human GBMs (Colardo et al., 2021). Epidermal growth factor receptor (EGFR), a type of receptor tyrosine kinases, can recruit and active PI3K/AKT pathway. More than 50% GBMs have EGFR amplifications or mutations (Oprita et al., 2021), and PIK3CA mutation (15%) and PTEN deletion can also active PI3K/AKT pathway (Colardo et al., 2021). Actually, accumulating evidence has demonstrated that probiotics, including B. lactis and L. plantarum, can down-regulate hyperactivated PI3K/AKT pathway in numerous diseases both in vivo and in vitro, such as colorectal cancer, gastric cancer and aging (Mohseni et al., 2021). In the present study, L. plantarum induced apoptosis in a gastric cancer cell line (AGS) through up-regulating PTEN, Bax, TLR4 and down-regulating Akt (Maleki-Kakelar et al., 2020). Experiments showed that B. animalis subsp. lactis can prevent the Salmonella infection through suppressing PI3K/Akt signal pathway (Huang and Huang, 2016). Our study shows that B. lactis combined with L. plantarum down-regulated the protein expression of p-PI3K, down-regulated the protein and mRNA expression of survivin, and up-regulated the protein and mRNA expression of PTEN in glioma mice. Noteworthily, Helicobacter (H.) pylori infection can induce reactive oxygen species- (ROS-) mediated DNA damage and activate PI3K/Akt pathway in gastric cancer both in vivo and in vitro (Xie et al., 2018). After infection with Staphylococcus aureus, the mammary glands of mice showed high levels of PI3K, AKT, and p-NF-κB (Han et al., 2020). Researches have depicted that staphylococcal superantigen can induce toxicity by activating PI3K/Akt pathway (Krakauer et al., 2010; Krakauer, 2012). Our results show that B. lactis combined with L. plantarum decreased the abundance of Staphylococcus and Helicobacter, therefore, we speculated that the down-regulation of PI3K/AKT pathway may be partly associated with the decrease in Staphylococcus and Helicobacter.
A recent study showed that increased abundance of Bacteroidetes and decreased abundance of Firmicutes were observed in glioma mice compared with health mice, and the decreased abundance of Bacteroidetes was related to delayed glioma progression (Fan et al., 2022). Our results show an analogous trend, with probiotics reducing the abundance of Bacteroidetes and raising the abundance of Firmicutes. After oral gavage of B. lactis and L. plantarum, abundance of L. plantarum was increased but B. lactis did not colonize. Numerous studies have shown that both colonized and non-colonized probiotics can benefit the host (Xiao et al., 2003; Plaza-Diaz et al., 2019; Van Zyl et al., 2020). We speculated that the antitumor effect of B. lactis in glioma mice can also be exerted by allowing it to stay in the intestine for a certain period of time each day. The findings of Se-Hoon et al. validated our guesses, daily gavage of different Bifidobacterium strains reduced tumor burden in mice. However, these strains did not colonize, and the results showed that the abundance of Bifidobacterium strains increased 4 h after gavage, but decreased significantly after 24 h (Lee et al., 2021). Additionally, B. lactis and L. plantarum decreased the abundance of several potential pathogenic bacteria. Uncontrolled proliferation of pathogens can contribute to dysbiosis and further lead to decreased permeability of intestinal mucosal barrier (Iacob and Iacob, 2019). Previous researches have shown that H. pylori can disrupt claudin-4 and claudin-5 through activating myosin light-chain kinase. Actually, proteases of the high temperature requirement A (HtrA), which are broadly expressed by various bacteria, can cleave the junction factors such as Occludin and claudin-8 (Backert et al., 2018). In this research, protein expression of Occludin in ileum and colon tissues were down-regulated in glioma mice, which means intestinal barrier was damaged. B. lactis combined with L. plantarum repaired the intestinal barrier damage in glioma mice.
Accumulating evidence demonstrates that microbial metabolites are involved in development of various tumors. Some microbiota-derived metabolites, such as SCFA (acetate, propionate, butyrate, etc.) can suppress tumor growth, while other metabolites, such as secondary bile acids can promote tumor development (Liu et al., 2020; Mirzaei et al., 2021; Zhang et al., 2021). Interestingly, microbial metabolites can penetrate the damaged intestinal barrier, even cross the BBB into the brain, modulating brain functions (Carlessi et al., 2021). In addition, bacterial metabolites can affect the vagal nerve by stimulating enteroendocrine cells or provoking enteric nervous system, regulating brain functions (D’alessandro et al., 2021). A study demonstrated that Lactobacillus rhamnosus can increase GABA levels in the brain (cortical regions) and reduce depression- and anxiety-related behavior in mice, while the behavioral and neurochemical effects were not observed in vagotomized mice (Bravo et al., 2011). In our research, we performed untargeted GC–MS analysis of fecal metabolites and detected a total of 253 metabolites, including 14 up-regulated metabolites and 20 down-regulated metabolites. Numerous substances with antitumor activity such as ascorbate were identified as up-regulated metabolites, and potential carcinogen such as elaidic acid and methyl yellow were identified as down-regulated metabolites. Ascorbate is an antioxidant that scavenges ROS and prevents DNA damage, and its anticancer effects have been found in a variety of tumors (Pawlowska et al., 2019). A study showed that high dose ascorbate can cause double stranded breaks and aggregate in S-phase in GBM cell lines (Castro et al., 2017). Elaidic acid, a trans fatty acids, can promote metastasis of tumor cell lines by activating EGFR (Kishi et al., 2018).
Taken together, our results indicate that B. lactis combined with L. plantarum repressed glioma development at least partly by regulating PI3K/AKT pathway and the gut microbiota. Therefore, B. lactis and L. plantarum might be regarded as promising candidates for the treatment of glioma.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA863912 (16S rRNA genes) and PRJNA863751 (RNA-seq).
Ethics statement
The studies involving animal subjects were reviewed and approved by the ethics committee of Experimental Animal Administration of Sichuan University.
Author contributions
LW: methodology, investigation, formal analysis and writing-original draft preparation, writing-review and editing. SL, HF, MH, and JX: methodology, supervision and writing-review and editing. JD and FP: conceptualization, project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by National Natural Science Foundation of China (no.82003879 and U19A2010) the Key Project of Science and Technology Department of Sichuan Province (no. 2020YFS0053; 2021YFS0044), and Youth Talent Promotion Project of China Association for Science and Technology (CACM-2020-QNRC1-01) and the Open Research Fund of Chengdu University of Traditional Chinese Medicine Key Laboratory of Systematic Research of Distinctive Chinese Medicine Resources in Southwest China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.986837/full#supplementary-material
References
- Agliardi G., Liuzzi A. R., Hotblack A., De Feo D., Núñez N., Stowe C. L., et al. (2021). Intratumoral IL-12 delivery empowers CAR-T cell immunotherapy in a pre-clinical model of glioblastoma. Nat. Commun. 12, 444. doi: 10.1038/s41467-020-20599-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Bernegger S., Skórko-Glonek J., Wessler S. (2018). Extracellular Htr A serine proteases: An emerging new strategy in bacterial pathogenesis. Cell. Microbiol. 20:e12845. doi: 10.1111/cmi.12845, PMID: [DOI] [PubMed] [Google Scholar]
- Bravo J. A., Forsythe P., Chew M. V., Escaravage E., Savignac H. M., Dinan T. G., et al. (2011). Ingestion of lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U. S. A. 108, 16050–16055. doi: 10.1073/pnas.1102999108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui N. L., Pandey V., Zhu T., Ma L., Basappa, Lobie P. E. (2018). Bad phosphorylation as a target of inhibition in oncology. Cancer Lett. 415, 177–186. doi: 10.1016/j.canlet.2017.11.017, PMID: [DOI] [PubMed] [Google Scholar]
- Carlessi A. S., Borba L. A., Zugno A. I., Quevedo J., Réus G. Z. (2021). Gut microbiota-brain axis in depression: the role of neuroinflammation. Eur. J. Neurosci. 53, 222–235. doi: 10.1111/ejn.14631, PMID: [DOI] [PubMed] [Google Scholar]
- Castro M. L., Carson G. M., Mcconnell M. J., Herst P. M. (2017). High dose ascorbate causes both genotoxic and metabolic stress in glioma cells. Antioxidants (Basel) 6, 58. doi: 10.3390/antiox6030058, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Jin D., Huang S., Wu J., Xu M., Liu T., et al. (2020). Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 469, 456–467. doi: 10.1016/j.canlet.2019.11.019, PMID: [DOI] [PubMed] [Google Scholar]
- Chen J., Sanberg P. R., Li Y., Wang L., Lu M., Willing A. E., et al. (2001). Intravenous administration of human umbilical cord blood reduces behavioral deficits after stroke in rats. Stroke 32, 2682–2688. doi: 10.1161/hs1101.098367, PMID: [DOI] [PubMed] [Google Scholar]
- Colardo M., Segatto M., Di Bartolomeo S. (2021). Targeting RTK-PI3K-mTOR axis in gliomas: an update. Int. J. Mol. Sci. 22, 4899. doi: 10.3390/ijms22094899, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’alessandro G., Antonangeli F., Marrocco F., Porzia A., Lauro C., Santoni A., et al. (2020). Gut microbiota alterations affect glioma growth and innate immune cells involved in tumor immunosurveillance in mice. Eur. J. Immunol. 50, 705–711. doi: 10.1002/eji.201948354, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’alessandro G., Lauro C., Quaglio D., Ghirga F., Botta B., Trettel F., et al. (2021). Neuro-signals from gut microbiota: perspectives for brain Glioma. Cancers (Basel) 13:2810. doi: 10.3390/cancers13112810, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dono A., Patrizz A., Mccormack R. M., Putluri N., Ganesh B. P., Kaur B., et al. (2020). Glioma induced alterations in fecal short-chain fatty acids and neurotransmitters. CNS. Oncologia 9, Cns57. doi: 10.2217/cns-2020-0007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Su Q., Chen J., Wang Y., He S. (2022). Gut microbiome alterations affect Glioma development and Foxp3 expression in tumor microenvironment in mice. Front. Oncol. 12:836953. doi: 10.3389/fonc.2022.836953, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatahi A., Soleimani N., Afrough P. (2021). Anticancer activity of kefir on Glioblastoma cancer cell as a new treatment. Int. J. Food Sci. 2021, 8180742–8180745. doi: 10.1155/2021/8180742, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Li X., Liu J., Zou Z., Luo L., Wu R., et al. (2020). Bta-miR-223 targeting CBLB contributes to resistance to Staphylococcus aureus mastitis Through the PI3K/AKT/NF-κB pathway. Front Vet Sci 7, 529. doi: 10.3389/fvets.2020.00529, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (2022). Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46. doi: 10.1158/2159-8290.Cd-21-1059, PMID: [DOI] [PubMed] [Google Scholar]
- He Z., Wang M., Li H., Wen C. (2019). GC-MS-based fecal metabolomics reveals gender-attributed fecal signatures in ankylosing spondylitis. Sci. Rep. 9, 3872. doi: 10.1038/s41598-019-40351-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgers S. O. A., Talbot S. R., Riedesel A. K., Wassermann L., Wu Z., Krauss J. K., et al. (2020). Body weight algorithm predicts humane endpoint in an intracranial rat glioma model. Sci. Rep. 10, 9020. doi: 10.1038/s41598-020-65783-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F. C., Huang S. C. (2016). The different effects of probiotics treatment on salmonella-induced interleukin-8 response in intestinal epithelia cells via PI3K/Akt and NOD2 expression. Benef. Microbes. 7, 739–748. doi: 10.3920/bm2015.0181, PMID: [DOI] [PubMed] [Google Scholar]
- Iacob S., Iacob D. G. (2019). Infectious threats, the intestinal barrier, and its Trojan horse: dysbiosis. Front. Microbiol. 10:1676. doi: 10.3389/fmicb.2019.01676, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S., Fujiwara-Tani R., Luo Y., Kawahara I., Goto K., Fujii K., et al. (2018). Pro-metastatic signaling of the trans fatty acid elaidic acid is associated with lipid rafts. Oncol. Lett. 15, 4423–4426. doi: 10.3892/ol.2018.7817, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer T. (2012). PI3K/Akt/mTOR, a pathway less recognized for staphylococcal superantigen-induced toxicity. Toxins (Basel) 4, 1343–1366. doi: 10.3390/toxins4111343, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer T., Buckley M., Issaq H. J., Fox S. D. (2010). Rapamycin protects mice from staphylococcal enterotoxin B-induced toxic shock and blocks cytokine release in vitro and in vivo. Antimicrob. Agents Chemother. 54, 1125–1131. doi: 10.1128/aac.01015-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam F. C., Morton S. W., Wyckoff J., Vu Han T. L., Hwang M. K., Maffa A., et al. (2018). Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat. Commun. 9, 1991. doi: 10.1038/s41467-018-04315-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour V., Le Mercier M., Lefranc F., Hagedorn M., Javerzat S., Bikfalvi A., et al. (2010). Selective osteopontin knockdown exerts anti-tumoral activity in a human glioblastoma model. Int. J. Cancer 126, 1797–1805. doi: 10.1002/ijc.24751, PMID: [DOI] [PubMed] [Google Scholar]
- Lee Y. R., Chen M., Pandolfi P. P. (2018). The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 19, 547–562. doi: 10.1038/s41580-018-0015-0, PMID: [DOI] [PubMed] [Google Scholar]
- Lee S. H., Cho S. Y., Yoon Y., Park C., Sohn J., Jeong J. J., et al. (2021). Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 6, 277–288. doi: 10.1038/s41564-020-00831-6, PMID: [DOI] [PubMed] [Google Scholar]
- Li X. C., Wu B. S., Jiang Y., Li J., Wang Z. F., Ma C., et al. (2021). Temozolomide-induced changes in gut microbial composition in a mouse model of brain Glioma. Drug Des. Devel. Ther. 15, 1641–1652. doi: 10.2147/dddt.S298261, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Song X., Khan S., Li Y., Guo Z., Li C., et al. (2020). The gut microbiota at the intersection of bile acids and intestinal carcinogenesis: an old story, yet mesmerizing. Int. J. Cancer 146, 1780–1790. doi: 10.1002/ijc.32563, PMID: [DOI] [PubMed] [Google Scholar]
- Maleki-Kakelar H., Dehghani J., Barzegari A., Barar J., Shirmohamadi M., Sadeghi J., et al. (2020). Lactobacillus plantarum induces apoptosis in gastric cancer cells via modulation of signaling pathways in Helicobacter pylori. Bioimpacts 10, 65–72. doi: 10.34172/bi.2020.09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V., Chervin C. S., Gajewski T. F. (2021). Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology 160, 600–613. doi: 10.1053/j.gastro.2020.11.041, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland B., Dees K., Melo N., Fehling S., Gibson S., Yan Z., et al. (2017). Exth-30. Therapeutic benefit of a ketogenic diet through altered gut microbiota in a mouse model of glioma. Neuro Oncol. 19, vi78. doi: 10.1093/neuonc/nox168.322 [DOI] [Google Scholar]
- Mirzaei R., Afaghi A., Babakhani S., Sohrabi M. R., Hosseini-Fard S. R., Babolhavaeji K., et al. (2021). Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 139:111619. doi: 10.1016/j.biopha.2021.111619, PMID: [DOI] [PubMed] [Google Scholar]
- Mohseni A. H., Casolaro V., Bermúdez-Humarán L. G., Keyvani H., Taghinezhad S. S. (2021). Modulation of the PI3K/Akt/mTOR signaling pathway by probiotics as a fruitful target for orchestrating the immune response. Gut Microbes 13, 1–17. doi: 10.1080/19490976.2021.1886844, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprita A., Baloi S. C., Staicu G. A., Alexandru O., Tache D. E., Danoiu S., et al. (2021). Updated insights on EGFR signaling pathways in Glioma. Int. J. Mol. Sci. 22:587. doi: 10.3390/ijms22020587, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom Q. T., Cioffi G., Waite K., Kruchko C., Barnholtz-Sloan J. S. (2021). CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2014-2018. Neuro Oncol. 23, iii1–iii105. doi: 10.1093/neuonc/noab200, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastushenko I., Blanpain C. (2019). EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226. doi: 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Patrizz A., Dono A., Zorofchian S., Hines G., Takayasu T., Husein N., et al. (2020). Glioma and temozolomide induced alterations in gut microbiome. Sci. Rep. 10, 21002. doi: 10.1038/s41598-020-77919-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowska E., Szczepanska J., Blasiak J. (2019). Pro- and antioxidant effects of vitamin C in cancer in correspondence to its dietary and pharmacological concentrations. Oxidative Med. Cell. Longev. 2019, 7286737–7286718. doi: 10.1155/2019/7286737, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza-Diaz J., Ruiz-Ojeda F. J., Gil-Campos M., Gil A. (2019). Mechanisms of action of probiotics. Adv. Nutr. 10, S49–S66. doi: 10.1093/advances/nmy063, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns J. C., Lynch M. D., Senadheera D. B., Tenenbaum H. C., Goldberg M. B., Cvitkovitch D. G., et al. (2011). Bacterial biogeography of the human digestive tract. Sci. Rep. 1, 170. doi: 10.1038/srep00170, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. (2020). Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim. Sci. J. 91:e13357. doi: 10.1111/asj.13357, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zyl W. F., Deane S. M., Dicks L. M. T. (2018). In vivo bioluminescence imaging of the spatial and temporal colonization of Lactobacillus plantarum 423 and enterococcus mundtii ST4SA in the intestinal tract of mice. BMC Microbiol. 18:171. doi: 10.1186/s12866-018-1315-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zyl W. F., Deane S. M., Dicks L. M. T. (2020). Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 12:1831339. doi: 10.1080/19490976.2020.1831339, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang X., Tan Z., Su Y., Liu J., Chang M., et al. (2019). Human ESC-derived expandable hepatic organoids enable therapeutic liver repopulation and pathophysiological modeling of alcoholic liver injury. Cell Res. 29, 1009–1026. doi: 10.1038/s41422-019-0242-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang F., Zhong J., Zhu T., Zheng Y., Zhao T., et al. (2020). Using apelin-based synthetic notch receptors to detect angiogenesis and treat solid tumors. Nat. Commun. 11, 2163. doi: 10.1038/s41467-020-15729-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S.-D., De Zhang Z., Lu H., Jiang S. H., Liu H. Y., Wang G. S., et al. (2003). Multicenter, randomized, controlled trial of heat-killedLactobacillus acidophilus LB in patients with chronic diarrhea. Adv. Ther. 20, 253–260. doi: 10.1007/BF02849854, PMID: [DOI] [PubMed] [Google Scholar]
- Xie C., Yi J., Lu J., Nie M., Huang M., Rong J., et al. (2018). N-Acetylcysteine reduces ROS-mediated oxidative DNA damage and PI3K/Akt pathway activation induced by Helicobacter pylori infection. Oxidative Med. Cell. Longev. 2018, 1874985. doi: 10.1155/2018/1874985, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Tang L., Li X., Fan F., Liu Z. (2020). Immunotherapy for glioma: current management and future application. Cancer Lett. 476, 1–12. doi: 10.1016/j.canlet.2020.02.002, PMID: [DOI] [PubMed] [Google Scholar]
- Yan Y., Huang H. (2019). Interplay among PI3K/AKT, PTEN/FOXO and AR signaling in prostate cancer. Adv. Exp. Med. Biol. 1210, 319–331. doi: 10.1007/978-3-030-32656-2_14, PMID: [DOI] [PubMed] [Google Scholar]
- Yang C., Wang M. H., Zhou J. D., Chi Q. (2017). Upregulation of miR-542-3p inhibits the growth and invasion of human colon cancer cells through PI3K/AKT/survivin signaling. Oncol. Rep. 38, 3545–3553. doi: 10.3892/or.2017.6054, PMID: [DOI] [PubMed] [Google Scholar]
- Yang X., Yu D., Xue L., Li H., Du J. (2020). Probiotics modulate the microbiota-gut-brain axis and improve memory deficits in aged SAMP8 mice. Acta Pharm. Sin. B 10, 475–487. doi: 10.1016/j.apsb.2019.07.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., An Y., Qin X., Wu X., Wang X., Hou H., et al. (2021). Gut microbiota-derived metabolites in colorectal cancer: The bad and the challenges. Front. Oncol. 11:739648. doi: 10.3389/fonc.2021.739648, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Zeng B., Zhou C., Liu M., Fang Z., Xu X., et al. (2016). Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol. Psychiatry 21, 786–796. doi: 10.1038/mp.2016.44, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA863912 (16S rRNA genes) and PRJNA863751 (RNA-seq).