Abstract
Genetic ancestry inference can be used to stratify patient cohorts and to model pharmacogenomic variation within and between populations. We provide a detailed guide to genetic ancestry inference using genome-wide genetic variant datasets, with an emphasis on two widely used techniques: principal components analysis (PCA) and ADMIXTURE analysis. PCA can be used for patient stratification and categorical ancestry inference, whereas ADMIXTURE is used to characterize genetic ancestry as a continuous variable. Visualization methods are critical for the interpretation of genetic ancestry inference methods, and we provide instructions for how the results of PCA and ADMIXTURE can be effectively visualized.
Keywords: Admixture, Genetic ancestry inference, Pharmacogenomics, Health disparities, Genetic variants, Population-specific drug efficacy
1. Introduction
Pharmacogenomic variants that mediate patients’ response to medications often show large allele frequency differences among population groups [1, 2]. These allele frequency differences have important implications for treatment decisions, with population-specific effects observed for drug efficacy, dosage, and toxicity. Indeed, there are numerous examples of racial and ethnic differences in drug response, many of which can be attributed to allele frequency differences in pharmacogenomic variants [3-7]. It has been observed that up to 20% of newly approved drugs show distinct racial and ethnic response profiles, and differences of this kind can lead to group-specific treatment recommendations issued by the FDA [8].
Nevertheless, it should be stressed that race and ethnicity are socially ascribed characteristics, based on shared origins, culture, heritage, and social experiences. Race and ethnicity are not biological categories and are therefore imprecise proxies for genetic diversity [9]. Genetic ancestry, on the other hand, is a characteristic of the genome. Genetic ancestry measures individuals’ biogeographical origins, based on correlated allele frequency differences among ancestral source populations [10]. Genetic ancestry can be defined independently of the social dimensions of race and ethnicity, and it can be characterized objectively and with precision, as either a categorical or a continuous variable. Accordingly, pharmacogenomic variation among populations is better modeled with genetic ancestry as opposed to race and ethnicity.
The aim of this chapter is to provide a practical guide to genetic ancestry inference for pharmacogenetic researchers who may wish to stratify their study cohorts based on patterns of genetic diversity rather than, or in addition to, the more commonly used social categories of race and ethnicity. In light of the increasing availability of large-sale genomic datasets, we focus on genetic ancestry inference methods that make use of genome-wide genetic variant data, including whole-genome sequences, whole exome sequences, and whole-genome genotypes. We provide detailed protocols for two commonly used methods – principal components analysis (PCA) and ADMIXTURE analysis – and we emphasize visualization methods given their importance for large-scale data analysis and interpretation. PCA yields a high-level overview of the patterns of genetic diversity found in a genomic dataset and can be used to delineate genetic ancestry categories [11]. ADMIXTURE can be used to characterize genetic ancestry as a continuous variable, providing fractional estimates of ancestry components for each genomic sample [12].
2. Materials
In order to perform genetic ancestry inference, users will need access to (1) a unix/linux operating system, (2) the Conda package manager and environment management system, (3) program installation files, (4) all necessary program dependencies, and (5) appropriately formatted genomic variant data. We provide an overview of the operating system, the package manger, and the genomic data formats that are needed for both of the genetic ancestry inference methods described here. We also provide details on the installation of the R studio package, which can be used to visualize the results of the genetic ancestry inference.
2.1. Operating Systems
Scientific computing, including genetic ancestry inference, is generally conducted in the command line interface provided by unix/linx operating systems. There are numerous unix/linux operating systems available, many of which are provided free of charge. We recommend the freely available RedHat or Ubuntu Linux operating systems, and all protocols described here can be successfully executed in one of those operating systems.
2.2. Package Manager and Environment Management System
Installation and execution of scientific software packages often requires a specific environment along with a number of dependencies, i.e., other programs or libraries. Thus, environment specification and dependency installation is a rate-limiting step for the use of scientific software, including genetic ancestry inference packages. Conda is a freely available software package and environment management system that allows users to install and update software packages and their dependencies. Use of Conda can save a great deal of time and effort, allowing users to focus on software execution without the need for source code compilation. It should be noted that not all genetic ancestry inference software is made available through Conda, users may have to install and compile source files for some packages, but the tools described here can all be installed from Conda.
Conda version 4.9.2 https://repo.anaconda.com/miniconda/
2.3. PLINK
The program PLINK v1.90b6.21 64-bit, which can be used for PCA, is distributed through Conda.
2.4. ADMIXTURE
The program ADMIXTURE version 1.3.0 is distributed through Conda.
2.5. Genomic Variant Data
There are numerous sources of genomic variant data, and users can use their own appropriately formatted data to conduct the genetic ancestry inference analyses described here. The 1000 Genomes Project provides freely available human genomic variant data for <2000 individuals from 26 worldwide populations 13. Data are distributed in the variant call format (VCF) and can be downloaded as individual chromosome files. We recommend using a single small chromosome, e.g., chromosome 22, to get started with the analyses described here. Whole-genome analyses can be performed by concatenating all chromosome-specific VCF files.
- http://1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/phase3_liftover_nygc_dir/
- phase3.chr22.GRCh38.GT.crossmap.vcf.gz
- phase3.chr22.GRCh38.GT.crossmap.vcf.gz.tbi
2.6. Genomic Sample Information
The genetic ancestry inference protocols presented here entail the analysis of a subset of samples from four populations from the 1000 Genomes Project. Users will need to use the metadata on the population origins of the genomic variant samples in order to extract samples from those four populations.
2.7. Programming
The R studio version 4.0.3 integrated development environment (IDE) is used to visualize the results of genetic ancestry inference.
3. Methods
3.1. Software Installation
All of the necessary software listed in the Materials section should be installed, starting with the RedHat or Ubuntu operating system followed by the Conda package manager and environment management system. RedHat is often used for shared computer servers, whereas Ubuntu is recommended for laptop or personal computer use. Conda versions of individual programs can then be installed using the links provided in the Materials section. Finally, the R studio package for results visualization should be installed using the link provided in the Materials section. Instructions for software installation are provided in Notes 1–4.
3.2. Download Genomic Variant Data
Users can use their own genomic variant data as long as it is in the appropriate variant call format (VCF) https://samtools.github.io/hts-specs/VCFv4.1.pdf, or they can download human genome variant data from the 1000 Genomes Project using the link provided in the Materials section. See Note 5.
3.3. Download Genomic Sample Information
Metadata on the population origins of the genomic variants samples from the 1000 Genomes Project can be downloaded using the link in the Materials section. See Note 6.
3.4. Extract Samples from the Four Populations to Be Analyzed
Colombian in Medellin, Colombia [CLM], Iberian Populations in Spain [IBS], Peruvian in Lima Peru [PEL], and Yoruba in Ibadan, Nigeria [YRI]. Please note that the “$” symbol refers to the start of the Linux command line, after which the commands should be entered and executed as shown.
-
Extract sample identifiers for the four populations.
$ cut -f1,4 igsr_samples.tsv ∣ grep -e “CLM” -e “IBS” -e "PEL" -e "YRI" > samplesToPops.tsv
-
Extract the genomic variant data corresponding to the sample identifiers for the four populations.
$ cut -f1 samplesToPops.tsv > sampleIDs.tsv
$ plink --vcf phase3.chr22.GRCh38.GT.crossmap.vcf.gz --keep-allele-order --keep-fam sampleIDs.tsv --make-bed --out 1000Genomes.4Pops. Chr22.GRCh38
The last command creates the three PLINK format files, which are needed to run PCA in PLINK:
1000Genomes.4Pops.Chr22.GRCh38.bed
1000Genomes.4Pops.Chr22.GRCh38.bim
1000Genomes.4Pops.Chr22.GRCh38.fam
3.5. Linkage Disequilibrium (LD) Pruning
Perform linkage disequilibrium (LD) pruning to yield a reduced set of unlinked genetic variants. See Note 7.
1. Filter variants with minor allele frequency of 1% and perform LD pruning.
$ plink --bfile 1000Genomes.4Pops.Chr22.GRCh38 --keep-allele-order --maf0.01 --indep-pairwise 500 5 0.25 --out 1000Genomes.4Pops.Chr22.GRCh38
This command creates the file: 1000Genomes.4Pops.Chr22.GRCh38.prune.in with 20,936 variants.
$ plink --bfile 1000Genomes.4Pops.Chr22.GRCh38 --keep-allele-order --extract 1000Genomes.4Pops.Chr22.GRCh38.prune.in --make-bed --out 1000Genomes.4Pops.Chr22.GRCh38.Pruned
This command creates the files:
1000Genomes.4Pops.Chr22.GRCh38.Pruned.bed
1000Genomes.4Pops.Chr22.GRCh38.Pruned.bim
1000Genomes.4Pops.Chr22.GRCh38.Pruned.fam
3.6. PCA Analysis
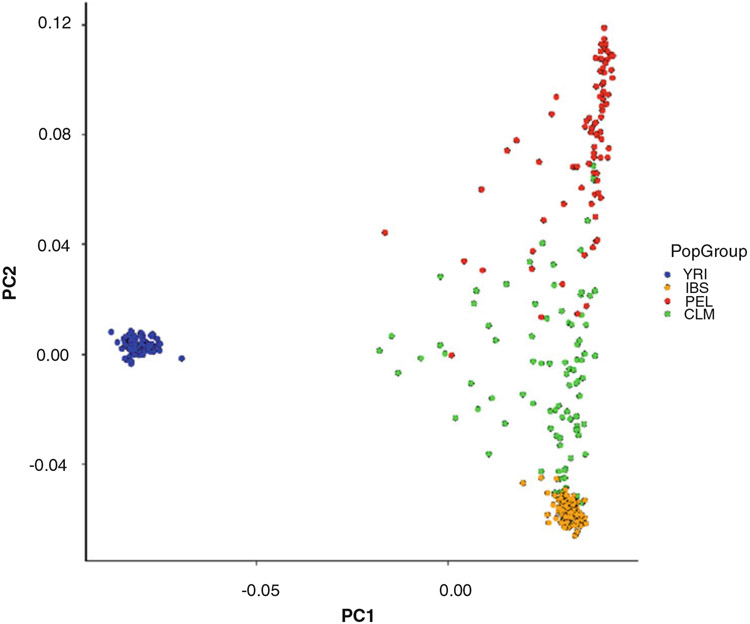
Run PCA analysis to characterize the genetic relationships among the samples (see Fig. 1). See Note 8.
Fig. 1.
Principal components analysis (PCA) of four human populations: Colombia (CLM – green), Peru (PEL – red), Spain (IBS – orange), Yoruba (YRI – blue). The first two principal components (PCs) are shown
$ plink --bfile 1000Genomes.4Pops.Chr22.GRCh38.Pruned --pca --out 1000Genomes.4Pops.Chr22.GRCh38.Pruned.PCA
This command creates the PCA results files, which will be subsequently visualized in R studio:
1000Genomes.4Pops.Chr22.GRCh38.Pruned.PCA.eigenval
1000Genomes.4Pops.Chr22.GRCh38.Pruned.PCA.eigenvec
3.7. Visualize PCA Results
Visualize PCA results in R studio.
-
Install R packages needed for visualization.
install.packages("dplyr")
install.packages("ggplot2")
install.packages("reshape2")
-
Configure and import the libraries.
options(scipen=100, digits=3)
library(‘dplyr’)
library(‘ggplot2’)
-
Read and process the eigenvector PCA output file and extract the top two PCs.
eigenvec <- read.table(‘1000Genomes.4Pops.Chr22.GRCh38.Pruned.PCA.eigenvec’, header = TRUE, sep = ’’)
rownames(eigenvec) <- eigenvec[,2]
eigenvec <- eigenvec[,3:ncol(eigenvec)]
colnames(eigenvec) <- paste(‘PC’, c(1:20), sep = ’’)
eigenvec$IndividualID = row.names(eigenvec)
eigenvec = eigenvec[c("IndividualID", "PC1", "PC2")]
row.names(eigenvec) <- NULL
-
Visually inspect the eigenvector file.
head(eigenvec)
IndividualID PC1 PC2 1 HG01113 0.0336 −0.00201 2 HG01119 0.0384 0.02310 3 HG01121 0.0371 0.02146 4 HG01122 0.0277 0.02503 5 HG01124 0.0283 −0.02970 6 HG01125 0.0293 −0.03324 -
Merge the results with population group data.
individualToPopGroupData = read.table(‘samplesToPops.tsv’, col.names = c("IndividualID", "PopGroup"), header = FALSE)
combinedPCsPopGroups = join(eigenvec, individualToPopGroupData)
head(combinedPCsPopGroups)
IndividualID PC1 PC2 PopGroup 1 HG01113 0.0336 −0.00201 CLM 2 HG01119 0.0384 0.02310 CLM 3 HG01121 0.0371 0.02146 CLM 4 HG01122 0.0277 0.02503 CLM 5 HG01124 0.0283 −0.02970 CLM 6 HG01125 0.0293 −0.03324 CLM -
Define population colors.
colors <- c("CLM" = "green", "IBS" = "orange", "PEL" = "red", "YRI" = "blue",)
-
Plot the PCA results for the top two PCs.
Options(repr.plot.width= 12, repr.plot.height=12)
ggplot(combinedPCsPopGroups, aes(x=PC1, y=PC2, fill=PopGroup)) + scale_fill_manual(values=colors) +
geom_point(size = 6, pch = 21, color= "black") + theme_classic() + theme(axis.title=element_text(size=18))
3.8. Admixture
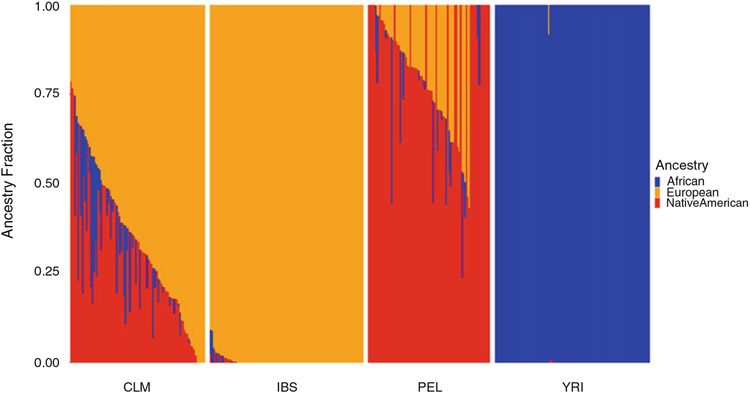
Run ADMIXTURE analysis to characterize patterns of genetic ancestry and admixture among the samples (see Fig. 2). See Note 9.
Fig. 2.
ADMIXTURE plot of four human populations: Colombia (CLM), Spain (IBS), Peru (PEL), and Yoruba (YRI). Each column is an individual, and for each individual the ancestry fraction for each of three continental population groups is shown: African (blue), European (orange), and Native American (red)
$ admixture 1000Genomes.4Pops.Chr22.GRCh38.Pruned.bed 3-j4
This command creates the files:
1000Genomes.4Pops.Chr22.GRCh38.Pruned.3.Q
1000Genomes.4Pops.Chr22.GRCh38.Pruned.3.P
3.9. Visualize ADMIXTURE Results
Visualize ADMIXTURE results using R studio.
-
Install R packages needed for visualization.
install.packages("dplyr")
install.packages("ggplot2")
install.packages("reshape2")
-
Import libraries.
library("dplyr")
library("ggplot2")
library("reshape2")
-
Get individual identifiers and individual population labels for all samples.
individualToPopGroupData = read.table(‘samplesToPops.tsv’, col.names = c("IndividualID", "PopGroup"), header = FALSE)
allCombinedFAMData = read.table(‘1000Genomes.4Pops.Chr22.GRCh38.Pruned.fam’, header = FALSE, col. names = c("FamilyID", "IndividualID", "N1", "N2", "N3", "N4"))
allCombinedFAMData = data.frame("IndividualID" = all-CombinedFAMData[ , 2:2])
-
Visually inspect the results.
head(individualToPopGroupData)
IndividualID PopGroup 1 HG01250 CLM 2 HG01255 CLM 3 HG01274 CLM 4 HG01279 CLM 5 HG01123 CLM 6 HG01130 CLM head(allCombinedFAMData)
IndividualID
1 HG01112 2 HG01113 3 HG01119 4 HG01121 5 HG01122 6 HG01124 -
Assign population labels to samples from ADMIXTURE output.
allCombinedFAMandPopGroupData = join(allCombined-FAMData, individualToPopGroupData)
head(allCombinedFAMandPopGroupData)
IndividualID PopGroup 1 HG01112 CLM 2 HG01113 CLM 3 HG01119 CLM 4 HG01121 CLM 5 HG01122 CLM 6 HG01124 CLM -
Extract ancestry estimates obtained from ADMIXTURE.
admixtureAncestryEstimates = read.table("1000Genome-s.4Pops.Chr22.GRCh38.Pruned.3.Q", header = FALSE)
head(admixtureAncestryEstimates)
V1 V2 V3 1 0.00001 0.00001 1.000 2 0.00001 0.33841 0.662 3 0.00001 0.49758 0.502 4 0.00001 0.49336 0.507 5 0.06223 0.48273 0.455 6 0.02341 0.16010 0.816 -
Combine individual ancestry estimates with individual identifiers and population labels.
combinedAncestryEstimatesData = cbind(allCombinedFA-MandPopGroupData, admixtureAncestryEstimates)
head(combinedAncestryEstimatesData)
IndividualID PopGroup V1 V2 V3 1 HG01112 CLM 0.00001 0.00001 1.000 2 HG01113 CLM 0.00001 0.33841 0.662 3 HG01119 CLM 0.00001 0.49758 0.502 4 HG01121 CLM 0.00001 0.49336 0.507 5 HG01122 CLM 0.06223 0.48273 0.455 6 HG01124 CLM 0.02341 0.16010 0.816 -
Characterize population ancestry means.
options(scipen = 10000)
combinedAncestryEstimatesData %>% group_by(PopGroup) %>% summarise_at(vars(V1, V2, V3), funs(mean))
PopGroup V1 V2 V3 CLM 0.0709 0.273122 0.65601 IBS 0.0028 0.001514 0.99569 PEL 0.0343 0.819700 0.14604 YRI 0.9991 0.000161 0.00078 -
Rename the columns and assign proper population labels to each cluster.
combinedAncestryEstimatesData = combinedAncestryEstimatesData %>% dplyr::rename("European" = "V3", "African" = "V1", "NativeAmerican" = "V2",)
head(combinedAncestryEstimatesData)
IndividualID PopGroup African NativeAmerican European 1 HG01112 CLM 0.00001 0.00001 1.000 2 HG01113 CLM 0.00001 0.33841 0.662 3 HG01119 CLM 0.00001 0.49758 0.502 4 HG01121 CLM 0.00001 0.49336 0.507 -
Check average ancestry for each population.
options(scipen = 10000)
combinedAncestryEstimatesData %>% group_by(PopGroup) %>% summarise_at(vars(European, African, NativeAmerican), funs(mean))
PopGroup European African NativeAmerican <fct> <dbl> <dbl> <dbl> CLM 0.65601 0.0709 0.273122 IBS 0.99569 0.0028 0.001514 PEL 0.14604 0.0343 0.819700 YRI 0.00078 0.9991 0.000161 -
Reformat the data in one column for graphs.
combinedAncestryEstimatesDataSorted = arrange(combinedAncestryEstimatesData, European, African, NativeAmerican, group_by = PopGroup)
row.names(combinedAncestryEstimatesDataSorted) <- NULL
combinedAncestryEstimatesDataSorted$index= as.numeric(row-names(combinedAncestryEstimatesDataSorted))
combinedAncestryEstimatesDataSortedMelt = melt(data = combinedAncestryEstimatesDataSorted, id.vars = c("IndividualID", "PopGroup", "European", "African", "NativeAmerican", "index"), measure.vars = c("European", "African", "NativeAmerican"))
colnames(combinedAncestryEstimatesDataSortedMelt)[7] <- ‘Ancestry’
colnames(combinedAncestryEstimatesDataSortedMelt)[8] <- ‘AncestryFraction’
print(combinedAncestryEstimatesDataSortedMelt[c(‘IndividualID’, "PopGroup", "Ancestry", "AncestryFraction", "index")])
-
Define the ancestry colors.
colors <- c("African" = "blue", "European" = "orange", "NativeAmerican" = "red")
-
Render the ADMIXTURE plot.
options(repr.plot.width=16, repr.plot.height=8)
ggplot(data=combinedAncestryEstimatesDataSortedMelt, aes(x=as.character(index), y=AncestryFraction, fill=Ancestry)) +
geom_bar(stat= "identity", width=1) + facet_grid(cols = vars(PopGroup), scales = "free", space = "free", drop = TRUE, switch= "both") +
scale_fill_manual(values=colors) +
labs(y= "Percentage Ancestry Estimate", title = "Admixture plot for four population groups from 1KGP", subtitle = "Groups: IBS, PEL, YRI, & CLM") +
theme(axis.title.x=element_blank(), axis.text.x=element_blank(), axis.ticks.x=element_blank(), panel.spacing.x=unit(1, "lines"),
strip.background = element_blank(), panel.background = element_blank(), axis.text=element_text(size=12),
axis.title=element_text(size=18), strip.text.x = element_text(size = 18), title = element_text(size = 20))
4. Notes
-
Operating system installation
We recommend using the Ubuntu operating system if users are working on their own laptop. Instructions for installing Ubuntu on a laptop can be found at https://ubuntu.com/tutorials/install-ubuntu-desktop#1-overview.
-
Conda installation
Conda can be installed using the Linux commands shown below, following the series of prompts after the first command:
$ bash Miniconda3-py38_4.9.2-Linux-x86_64.sh
$ conda create --name myenv
$ conda activate myenv
Please note that the “$” symbol refers to the start of the Linux command line, after which the commands should be entered and executed as shown.
-
PLINK and ADMIXTURE installation
The Conda versions of the programs PLINK and ADMIXTURE can be installed using the Linux commands shown below.
$ conda install -c sjnewhouse plink
$ conda install -c bioconda admixture
-
R studio
R studio can be installed following the instructions in the link provided in the Materials section, using the *.deb file corresponding to the Ubuntu operating system.
-
1000 Genomes Project human variant data
The 1000 Genomes Project human genome variant data can be downloaded using the following commands:
Please note that the wget command and the following ftp address should be entered on a single line.
-
The 1000 Genomes Project human genome sample
The 1000 Genomes Project human genome sample metadata can be downloaded using the “Download the list” button on the link provided in the Materials section. It should be noted that the sample metadata file igsr_samples.tsv will be downloaded into the user’s browser default download directory and will need to be transferred to the Linux working directory where the analyses are to be conducted.
-
Linkage disequilibrium (LD) pruning
Linkage disequilibrium (LD) pruning yields a reduced subset of genetic variants that are in approximate linkage equilibrium with each other (i.e., unlinked variants). The parameter values for the --indep-pairwise flag define how LD pruning proceeds. The first value (500) refers to the size of the genomic window, the second value (5) is the window step size, and the third value (0.25) is the variance inflation factor, which is used to measure the extent of linkage between pairs of variants. Details on LD pruning in PLINK can be found at https://zzz.bwh.harvard.edu/plink/summary.shtml#prune.
-
PCA analysis
Principal components analysis (PCA) is a technique that is used to reduce the dimensionality of large datasets and is ideal for the analysis of high-dimension genomic variant data. PCA analyzes the genomic variant covariance matrix to create uncorrelated, orthogonal variables that maximize the variance in the data – the principal components (PCs). Visualizing genomic data in PC-space provides an intuitive way to evaluate the genetic relationships among samples. The PLINK PCA program yields 20 PCs, and we visualize the first two PCs here.
-
ADMIXTURE
The program ADMIXTURE characterizes each individual with respect to the proportion of the genome that is made up of K theoretical ancestral source populations. The value of K can be chosen a priori, based on knowledge of the populations under analysis, or ADMIXTURE can be run across a series of K values to determine which value fits the data best. The samples analyzed here are from Africa (YRI), Europe (IBS), and the Americas (CLM and PEL). Since these populations are characterized primarily by African, European, and Native American continental ancestry fractions, we chose a value of K = 3 for the analysis. The value of 3 after the file name in the command line corresponds to the K-value, and the −j 4 flag corresponds to the number of threads to use.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Distinguished Scholars Program (DSP) to LMR and the Division of Intramural Research (DIR) of the National Institute on Minority Health and Health Disparities (NIMHD) at NIH (1ZIAMD000016 and 1ZIAMD000018).
References
- 1.Nagar SD, Conley AB, Jordan IK (2020) Population structure and pharmacogenomic risk stratification in the United States. BMC Biol 18:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagar SD, Moreno AM, Norris ET et al. (2019) Population pharmacogenomics for precision public health in Colombia. Front Genet 10:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachtiar M, Lee CG (2013) Genetics of population differences in drug response. Curr Genet MedRep 1:162–170 [Google Scholar]
- 4.Bjornsson TD, Wagner JA, Donahue SR et al. (2003) A review and assessment of potential sources of ethnic differences in drug responsiveness. J Clin Pharmacol 43:943–967 [DOI] [PubMed] [Google Scholar]
- 5.Chen ML (2006) Ethnic or racial differences revisited: impact of dosage regimen and dosage form on pharmacokinetics and pharmacodynamics. Clin Pharmacokinet 45:957–964 [DOI] [PubMed] [Google Scholar]
- 6.Huang SM, Temple R (2008) Is this the drug or dose for you? Impact and consideration of ethnic factors in global drug development, regulatory review, and clinical practice. Clin Pharmacol Ther 84:287–294 [DOI] [PubMed] [Google Scholar]
- 7.Yasuda SU, Zhang L, Huang SM (2008) The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther 84:417–423 [DOI] [PubMed] [Google Scholar]
- 8.Ramamoorthy A, Pacanowski MA, Bull J et al. (2015) Racial/ethnic differences in drug disposition and response: review of recently approved drugs. Clin Pharmacol Ther 97:263–273 [DOI] [PubMed] [Google Scholar]
- 9.Yudell M, Roberts D, DeSalle R et al. (2016) Science and society. Taking race out of human genetics. Science 351:564–565 [DOI] [PubMed] [Google Scholar]
- 10.Borrell LN, Elhawary JR, Fuentes-Afflick E et al. (2021) Race and genetic ancestry in medicine – a time for reckoning with racism. N Engl J Med 384:474–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price AL, Patterson NJ, Plenge RM et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909 [DOI] [PubMed] [Google Scholar]
- 12.Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]