Abstract
MicroRNAs (miRNAs) are non-coding RNAs which are essential post-transcriptional gene regulators in various neuronal degenerative diseases and playact a key role in these physiological progresses. Neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, multiple sclerosis, and, stroke, are seriously threats to the life and health of all human health and life kind. Recently, various studies have reported that some various miRNAs can regulate the development of neurodegenerative diseases as well as act as biomarkers to predict these neuronal diseases conditions. Endogenic miRNAs such as miR-9, the miR-29 family, miR-15, and the miR-34 family are generally dysregulated in animal and cell models. They are involved in regulating the physiological and biochemical processes in the nervous system by targeting regulating different molecular targets and influencing a variety of pathways. Additionally, exogenous miRNAs derived from homologous plants and defined as botanmin, such as miR2911 and miR168, can be taken up and transferred by other species to be and then act analogously to endogenic miRNAs to regulate the physiological and biochemical processes. This review summarizes the mechanism and principle of miRNAs in the treatment of some neurodegenerative diseases, as well as discusses several types of miRNAs which were the most commonly reported in diseases. These miRNAs could serve as a study provided some potential biomarkers in neurodegenerative diseases might be an ideal and/or therapeutic targets for neurodegenerative diseases. Finally, the role accounted of the prospective exogenous miRNAs involved in mammalian diseases is described.
Graphical abstract
1. Listing a large number of neural-related miRNAs and sorting out their pathways.
2. Classify and sort miRNAs according to their mechanism of action.
3. Demonstrating the effects of up-regulation or down-regulation of each miRNAs on the nervous system.
Keywords: MicroRNAs, Small RNA, Neurodegenerative diseases, Mechanisms
Introduction
MicroRNAs (miRNAs) are non-coding single-stranded chain RNA molecules with 21 to 25 nucleotides. They play an indispensable and irreplaceable role in regulating all events such as timing development, cell proliferation, asymmetric development, dendritic ridge development, early embryonic development, tumor development, stem cell differentiation, and antiviral effects. Meanwhile, microRNAs also play an important role in pathogenic mechanisms, such as oxidative stress (Cabezas et al. 2019). When free radicals are produced excessively and cannot be cleared in time, they will cause oxidative damage to nervous tissues (Abrahams et al. 2019). In 1998, Fire and his team (Fire et al. 1998) demonstrated that purified double-stranded RNAs can efficiently and specifically block the expression of target genes, a phenomenon called RNA interference. By 1999, Baulcombe (Hamilton and Baulcombe 1999) had shown that the miRNA let-7miRNA family was also involved in plant gene silencing, and proved that miRNAs are widely distributed in various animal cells. Moreover, miR-31 is believed to be a target of BCL6 and is also thought to reduce cell damage by inhibiting polycystin 1 (PKD1) expression. Inhibition of BCL6 mitigated oxidative stress–induced neuronal injury by targeting the miR-31/PKD1 axis (Wei et al. 2021). Mitochondrial malfunction can lead to promote oxidative stress, which can then lead to neuronal death and neurodegenerative disorders. miR-142a-5p induces mitochondrial dysfunction, mitochondrial autophagy, and apoptosis by targeting mitofusin-1 and can serve as an important regulator of denervation-induced skeletal muscle atrophy (Yang et al. 2020). Based on the extensive body of miRNA research, so far, the related researchers have established theories regarding the roles of miRNAs, and these related researches have been gradually carried out. Furthermore, as an authoritative database of miR-genes, miRBase serves as an authoritative database that has made it more convenient for us researchers to study various miRNA genes (Griffiths-Jones et al. 2006). With the deepening of miRNAs research, the role of miRNAs in neurodegenerative diseases has been explored (Mohr and Mott 2015). Although these studies have provided several available miRNAs as biomarkers in neurodegenerative disease, the detailed function as well as the potential regulatory mechanism of miRNAs that contribute to the pathogenesis of neurodegenerative diseases still remains unclear.
Neurodegenerative diseases would tend to worsen over time and are seriously a threat to human health. According to the World Health Organization, neurodegenerative diseases are likely to overtake cancer as the world’s second leading cause of death by 2040. Because the human nervous system is almost non-regenerative, the progression of the disease is irreversible (Alzheimer's 2016; Heemels 2016). According to previous research, inflammation has been linked to the pathophysiology of neurodegenerative disorders (Marogianni et al. 2020). When harmful substances are stimulated over a long period and become uncontrollable, the innate immune system can cause damage to the brain (Fakhoury 2016; Milo et al. 2020). Macrophage polarization comprises both M1 and M2 phenotypes, and it plays a crucial role in cell proliferation and the advancement of inflammatory disorders like cancer and multiple sclerosis. miR-9, miR-125b, miR-127, and miR-155 have been found to promote M1 polarization, while miR-124, miR-125a-5p, miR-223, miR-146a, miR-34a, miR-132, and let-7c have been demonstrated to generate promote M2 polarization in macrophages by targeting numerous transcription factors and signal transduction proteins. Inflammation-related disorders may benefit from miRNAs that affect macrophage polarization (Essandoh et al. 2016). Metabolic disorders also contribute to the disease. The accumulation of amyloid beta (Aβ) outside nerve cells to produce amyloid (senile) plaques to create senile plaques is a hallmark of AD, also known as amyloid plaques (O'Brien and Wong 2011; Qiang et al. 2017). The root cause of this protein disease and its associated neurodegeneration is a metabolic disorder (Haass and Selkoe 2007). Furthermore, different altered neurotransmitter metabolisms are also related to AD and its associated neurodegeneration (Li et al. 2020). Reduction of miR-96-5p in AD models decreases the Aβ42/Aβ40 ratios by upregulating the production of ATP binding cassette subfamily A member 1, implying that miR-96-5p is vital in managing the parts of Aβ production (M. Zhu et al. 2021a, b).
Excitatory toxins such as glutamate can also lead to neurodegenerative diseases (Beal 1992; Ikonomidou and Turski 1996). Glutamate toxicity occurs when the quantity of glutamate in the intercellular space is excessive high, resulting in neuronal degeneration, senescence, and even death (Meldrum 2000; Rajendra et al. 2004). miR-223, which is secreted by exosomes, targets glutamate receptors. The changes in miR-223 levels in the orbitofrontal cortex are positively correlated with inflammation, but negatively associated with GABAergic gene expression (Amoah et al. 2020). miR-124, through the Akt and mammalian target of rapamycin (mTOR) signaling pathways, upregulates the astrocytic glutamate transporter-1, which is the major mechanism that prevents extracellular glutamate overaccumulation in the central nervous system (CNS) after ischemic stroke. miR-124 upregulation can minimize the infarct size and increase neurological function recovery after ischemic stroke (W. Y. Huang et al. 2019a, b). Therefore, it seems to be critical to study the molecular mechanism and function of miRNAs involved in the neurodegenerative diseases.
In the current review, we briefly introduce and classify RNAs, focus on the mechanism and principle of miRNAs in the treatment of some neurodegenerative diseases, and summarize several types of miRNAs that are most effective in the treatment of diseases as well as the mechanisms and pathways worthy of further study. In summary, there is not only one mechanism leading to neurodegenerative diseases, but usually a common result of the action of multiple pathogenic principles and mechanisms contributes to a disease (Calin and Croce 2006; Jay et al. 2007; Lu et al. 2005). Thus, this study provided discusses the therapeutic potential of miRNA-mediated neurodegenerative diseases and biomarkers by analyzing and classifying the relationship between miRNAs and neurodegenerative diseases.
Small non-coding RNA
Non-coding RNAs are separately divided into long NC non-coding RNAs and small NC non-coding RNAs, moreover which are separated into miRNAs, small interfering RNAs (siRNAs), small nuclear RNAs (snRNAs), and Piwi-interacting RNAs (pi-RNAs) (Panni et al. 2020). They play an irreplaceable indispensable role in regulating all events such as timing development, cell proliferation, asymmetric development, dendritic ridge development, early embryonic development, tumor development, stem cell differentiation, and the antiviral responses (Higgs and Lehman 2015; Mattick and Makunin 2006; Re et al. 2014). To be more specific, siRNA is a two-stranded small interfering RNA. It is often involved in RNA interference, which regulates gene expression in a specific way (Kanasty et al. 2013; Nikam and Gore 2018). siRNA is often associated with the treatment of cancer diseases. As technology has progressed, some targeted drugs mediated by siRNA play an increasingly important role in the treatment of cancer (Jain et al. 2018; Kanasty et al. 2013; Nikam and Gore 2018). And snRNA is another small non-coding RNA distributed in the nucleoli of eukaryotic cells with conserved structural elements. snRNA does not exist in a free form; but rather, it binds to a protein to form a complex, a small ribonucleic protein particle (Kandels-Lewis and Seraphin 1993). The protein portion of snRNA has nuclease and ligase activity that clears transcription at the intron–exon junction and joins the two free ends (Chen and Wagner 2010; Lardelli and Lykke-Andersen 2020). Nuclear pre-snRNA exportation is a critical quality control mechanism for functional spliceosomes and is involved in RNA processing (Becker et al. 2019). pi-RNA also is a class of RNA that interacts with the Piwi protein, which is commonly found in germline stem cells (Blom-Dahl and Azpiazu 2018). It shows a distinctive localization type in the genome and plays an important role in the maintenance of stem cell function, gamete formation, and silencing of foreign transposons (Czech et al. 2018). Moreover, a new role for the piRNA/Piwi complex in cancer has been discovered (Y. Liu et al. 2019a, b).
Last but not least, in plants and animals, miRNAs are non-coding single-stranded RNA molecules with a length of about 22 nucleotides that are encoded by endogenous genes and are implicated in the control of post-transcriptional expression of genes (Bushati and Cohen 2007). This kind of miRNA forms a double-stranded complex with target messenger RNA (mRNA) or DNA through the complementary pairing of base pairings; this binding affects the modification, translation, transcription, and other processes of RNA, and blocks or inhibits the normal expression of genes (Ambros 2004; Cai et al. 2009). So far, hundreds of identical miRNAs have been found in different species, but some miRNAs differ significantly in expression levels in different tissues and at different developmental stages (Kozomara et al. 2019). Some miRNAs show strictly controlled differential spatial and temporal expression patterns and present a more rigorous one (Fabian and Sonenberg 2012; Rupaimoole and Slack 2017).
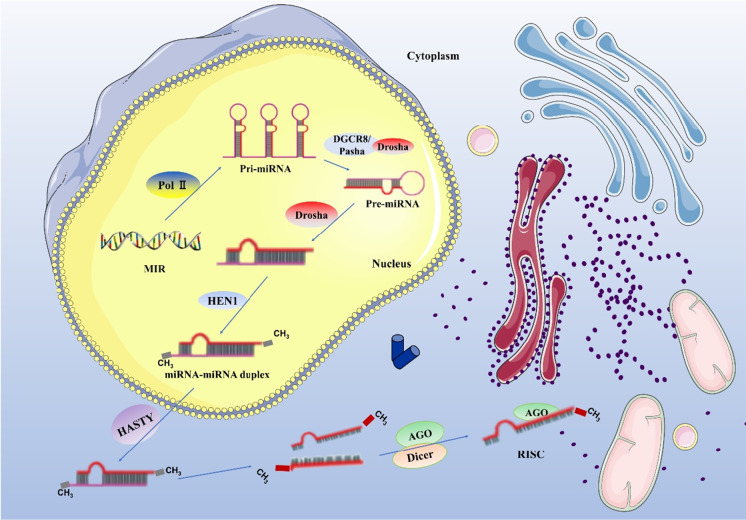
There are some microRNA genes in the organism, which generate pri-miRNA under the action of reverse transcriptase RNA POL II (polymerase II). The complementary regions of the molecule are combined in a rod-shaped structure, while the non-complementary regions are organized in a ring or chain structure. Under the action of the Drosha enzyme, the hairpin structure in pri-miRNA is cut out and pre-miRNA is generated. Under the action of HEN1, a (methyl transferase) protein, a methyl group is added to the 3′end of the double-stranded chain structure with a methyl group to avoid the degradation of the double-stranded chain structure, and miRNA–miRNA duplex molecules are generated. At this point, the molecule enters the cytoplasm from the nucleus, and the double strand is dissociated. One strand is degraded, and the other single strand becomes a mature miRNA, which is combined with an Argonaute protein to form the RNA-induced silencing complex (RISC) to regulate the transcription of the corresponding target gene (Fig. 1) (Cai et al. 2009; Kim 2005; Krol et al. 2010; Murchison and Hannon 2004).
Fig. 1.
miRNA biogenesis
Role of miRNAs
In nature, miRNAs are widely believed to exert their biological functions by forming an incorrect pairing with partial binding to the target mRNA’s 3′-untranslated region of target mRNAs, preventing the mRNA from being translated into the target protein, resulting in the degradation of the mRNA or reduction of its stability (Bartel 2004; Lewis et al. 2005). It is called post-transcriptional gene silencing (PTGS) which is the process by which microRNAs target bind with full complementary to mRNAs, leading to the cleavage and degradation of the target microRNAs. PTGS is effective only when the mRNA transcript is cleaved or degraded by RISC (Cogoni and Macino 2000; Sijen and Kooter 2000). microRNAs have multiple modes of action playing a biological role. The first PTGS is common in plants, where microRNAs can bind to target gene mRNAs in a completely and full complementary manner, acting similarly to siRNAs, and eventually leading to cleavage of the target mRNAs (Hamilton and Baulcombe 1999; Wassenegger and Pelissier 1998; Zhang et al. 2018). In animals, however, microRNAs are usually only partially complementary to target gene mRNAs when acting. When they bind, thus they prevent translation but without affecting the stability of the mRNAs (Kim et al. 2008). The third mode has both. Finally, miRNAs can combine both of the abovementioned modes of action, binding with full complementary to degrade mRNAs or binding with partial complementary to block translation. When microRNAs and target genes are fully complementary, they can directly target to cut the mRNA; when miRNAs and target genes are not complementary totally, the target gene’s translation is blocked (Correia de Sousa et al. 2019).
The role of miRNAs in nervous system development
miRNAs are required for neural stem cells and neural progenitor cells to develop. For example, miR-9 and miR-124 highly expressed in the brain are both affluent. In neural progenitor cells and neural stem cells, overexpression of miR-124 overexpression promotes neurogenesis by inhibiting the Toll-like receptor 4 pathway. Meanwhile, miR-124 can target the Tal1 axis and promote neuronal proliferation, and can also activate microglia by targeting vesicle-associated membrane protein 3. And miR-9 accelerates the differentiation process and increases glial and neuronal differentiation by targeting the nuclear receptor TLX. The maturation of neurons, the growth of neurites, and the occurrence of synapses integrate the functions of the CNS and jointly regulate the complex behavior of the body. Meanwhile, miRNA plays a critical role in all of these processes. For example, miR-132/miR-212 clusters are involved in dendritic growth and dendritic spine formation. And miR-134, which is specifically expressed in the brain, targets LIM kinase 1, thus inhibiting the growth of dendritic spines and increasing the synaptic excitatory area. The dicer enzyme is indispensable in the synthesis of miRNA synthesis (Song and Rossi 2017). When certain brain regions are knocked out and the Dicer enzyme is removed, miRNAs cannot be formed, which will eventually lead to the occurrence of neurodegenerative diseases (Chmielarz et al. 2017). For example, it would occur cerebellar degeneration and ataxia, when Dicer is knocked out by Pcp2 promoter–Cre recombinase in differentiated mature Purkinje cells (Schaefer et al. 2007), which may be related to the downregulation of miRNAs targeting spinal cerebellar ataxin 1, such as miR-130, miR-9, and miR-101 (Lee et al. 2008). As several factors influence the level of miRNA expression levels, such as transcription, maturation, and degradation, miRNAs play a role in various aspects of brain tissue growth and development (Diaz et al. 2014).
The expression level of miRNA expression is regulated by a fine regulatory mechanism to ensure that it is upregulated or downregulated at specific periods of biological development (J. Wang et al. 2020a, b). miRNA expression in the CNS is transient and regional (Bak et al. 2008). Many miRNAs expressed in the brain occur only during development, and the expression of some miRNAs varies with cell proliferation, regional differentiation, and pathway establishment during cortical formation (Shu et al. 2019). For example, in experiments to study the development of the mouse cortex, researchers found that miR-19b is expressed before birth; miR-128 is expressed after birth; and the expressions of miR-178, miR-125b, miR-9, and miR-131 reached their peak expression on embryonic day 21 of the embryonic stage, while miR-124a and miR-266 expressions increase during the embryonic stage development and remain in a stable state after birth, and miR-103 expression increases over time (Sempere et al. 2004).
The CNS of an embryo grows sequentially and spatially (de Lahunta et al. 2016). miRNAs may be involved in the regulation of temporal and spatial growth patterns of the embryonic CNS (Braoudaki and Lambrou 2015). For example, the Hox gene is involved in the anterior and posterior growth modes of brain tissue, and Hox gene expression is regulated by miRNAs in the Hox gene cluster, which controls the anterior and posterior growth modes of brain tissue (Mallo and Alonso 2013; Pearson et al. 2005).
The role of miRNAs in AD
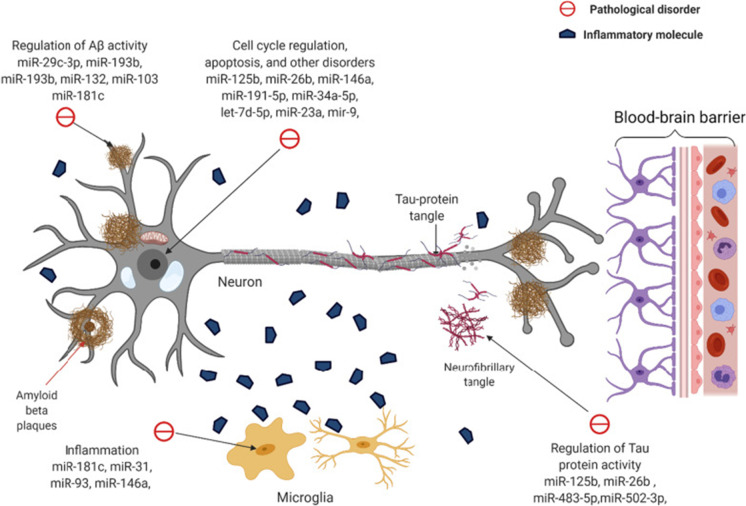
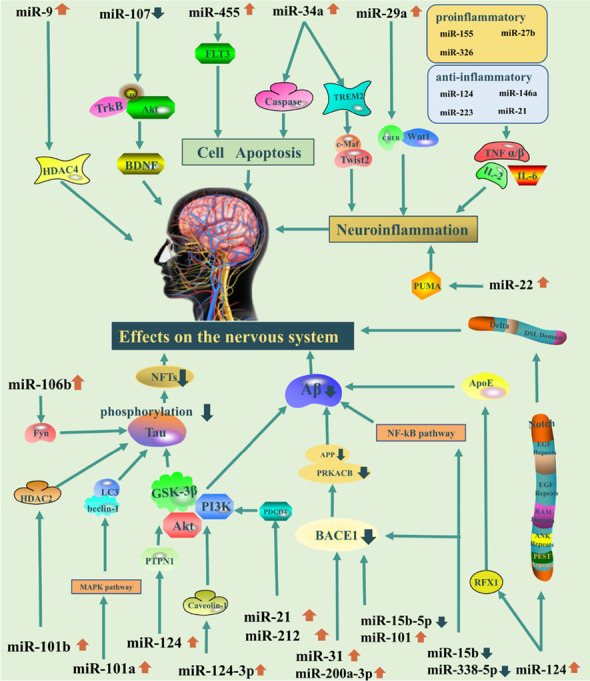
Neurodegenerative diseases are complex and progressive, diseases that progress and eventually lead to death. Many researchers have discovered that miRNAs are involved in the development and plasticity of the nervous system, as well as the occurrence and progression of neurodegenerative disorders (Quinlan et al. 2017). Aβ deposition is one of the main pathological symptoms of AD; these deposits consist of insoluble protein inclusions and plaques including Aβ42 and Aβ40 in AD. Numerous studies indicated that the Aβ could be a potential and effective biomarker to be used as a diagnostic strategy biomarker for AD. Meanwhile, tau is the major physiological protein which stabilizes microtubules in neuronal axons. Neuro axial degeneration could lead to increased release of tau from neurons in AD. Subsequently, dysfunctional tau assembles into the neurofibrillary tangles of the proximal axoplasm. Therefore, increased phosphorylated tau (p-tau181, p-tau217, and p-tau231) in the axoplasm and cerebrospinal fluid such as P-Tau181, 217, and 231 could be regarded as AD biomarkers (Quinlan et al. 2017). Neuro-inflammation, such as chronic inflammation and microglia, is also an important biomarker in AD disease. Many numbers of genes were are highly expressed in AD including sialic acid binding Ig-like lectin 3 (CD33), ephrin type-A receptor 1 (EPHA1), membrane-spanning 4-domains (MS4), triggering receptor expressed on myeloid cells 2 (TREM2), and ATP-binding cassette sub-family A member 7 (ABCA7) showed in Fig. 2. miRNAs involved in AD diseases have been demonstrated that it could regulate gene expression by binding to mRNA, either blocking its translation or leading to its degradation, or blocking to protein synthesis or leading to the degradation of targeted mRNA as well as the above biomarkers. Moreover, in encapsulation of the given stability of miRNAs due to its stability in body fluids, as well as regulation ability provided, researchers have speculated that they could possibly serve as biomarkers in AD (Sonntag 2010). Abundant expression dysregulation of microRNAs and their maladjustment could eventually lead to a variety of neurodegenerative diseases (Sonntag 2010). Herein, Table 1 and Fig. 2 present the role of miRNAs involved in the course of AD that affects processes critical to the processes for AD development and progression, including tau phosphorylation, Aβ metabolism, neuro-inflammation, and amyloid precursor protein (APP) production which were exhibited in the current study (Table 1 and Fig. 2).
Fig. 2.
The molecular mechanism of miRNAs on the pathology of AD.
Reproduced with permission from reference (Klyucherev et al. 2022). Copyright 2022 BMC Online Library
Table 1.
The relationship between different miRNAs and neurodegenerative diseases
| Article no | Mature miRNAs | Disease | Target | Pathways | Cells/mouse model | Reference |
|---|---|---|---|---|---|---|
| 1 | miR-15b | AD | BACE1/NF-κB | miR-15b↑ → APP and Aβ ↓ → AD ( −) | SH-SY5Y cell | Li and Wang (2018) |
| 2 | miR-15b-5p | AD | APP | miR-15b-5p↑ → APP and Aβ ↓ → AD ( −) | swAPP695-HEK293 cell | H. Y. Liu et al. (2019a, b) |
| 3 | miR-29c | AD | BACE1 | miR-29c ↓ → PKA/CREB ↑ → AD ( +) | SAMP8 mice | Yang et al. (2015) |
| 4 | miR-31 | AD | APP/BACE1 | miR-31↑ → Aβ ↓ → AD ( −) | 3xTg-AD mice | Barros-Viegas et al. (2020) |
| 5 | miR-9 | Ischemic stroke | HDAC4 |
miR-9 ↑ → HDAC4 ↓ → Ischemic stroke ( −) |
SH-SY5Y cell | Nampoothiri and Rajanikant (2019) |
| 6 | miR-200a-3p | AD | BACE1 and PRKACB | MiR-200a-3p↑ → Aβ Overproduction and Tau Hyperphosphorylation ↓ → AD ( −) | SAMP8 mice | Wang et al. (2019) |
| 7 | miR-338-5p | AD | BACE1/NF-κB |
miR-338-5p ↓ → Aβ ↓ → AD ( −) |
5XFAD transgenic (TG) mice | Qian et al. (2019) |
| 8 | miR-101 | AD/MCI | APP/RanBP9/Rab5 | miR-101↓ → AMPK Hyperphosphorylation ↑ → AD/MCI ( +) | pLSyn-miR-101 sponge mice | Barbato et al. (2020) |
| 9 | miR-101a | AD | MAPK | miR-101a ↑ → protein LC3 and beclin-1 ↓ → AD ( −) | SH-SY5Y cells、APPswe/PS1ΔE9 transgenic mice | Q. Li, Y. Wang et al. (2019) |
| 10 | miR-101b | AD | HDAC2/HNF-4A/AMPK | miR-101b ↑ → HDAC2 ↓ → AD ( −) | AD mice | D. Liu et al. (2017a, b) |
| 11 | miR-106b | AD | Fyn | miR-106 b ↑ → tau phosphorylation at Tyr18 ↓ → AD ( −) | SH-SY5Y cells | Liu et al. (2016) |
| 12 | miR-132 | AD/HD | BBB | miR-132 ↑ → VE-cadherin/β-Catenin ↑ → AD/HD ( −) | MCAO mice | Zuo et al. (2019) |
| 13 | miR-132 | AD/HD | C1q | miR-132 ↑ → C1q ↓ → AD/HD ( −) | APP/PS1 transgenic mice | N. Xu et al. (2019a, b) |
| 14 | miR-212/132 | AD | NOS1 | miR212 ↓ → SNO-GAPDH/SNO-Drp1/SNO-Cdk5 ↑ → AD ( +) | human neural cells | Y. Wang et al. (2017a, b) |
| 15 | miR-142 | MS | SOCS1/TGFBR1 | miR-142 ↑ → SOCS1/TGFBR1 ↓ → MS ( +) | EAE mice | Talebi et al. (2017) |
| 16 | miR-146 | AD | NF-κB | miR-146 ↑ → IL-1/TNF ↓ → AD ( +) | THP-1/U937/HL-60/WEHI-3 cell | Taganov et al. (2006) |
| 17 | miR-146a | AD | TRAF6/NF-κB | miR-146a ↑ → p62/Beclin1 ↑ → AD ( +) | HT-22 cells/C8-B4 cells/bEnd.3 cells | Fang et al. (2014); Kim et al. (2021) |
| 18 | miR-146a | AD | IRAK-1 | miR-146a ↑ → IRAK-1 ↓ → AD ( +) | HAG cells | Cui et al. (2010) |
| 19 | miR-155 | MS/EAE/ALS | IL-6/IL-1/TNF | miR-155 ↓ → IL-6/IL-1/TNF ↓ → MS/EAE/ALS ( −) | SOD1 mice | Butovsky et al. (2015) |
| 20 | miR-211 | MACO/I/R | PUMA | miR-211 ↑ → PUMA↓ → MACO/I/R ( −) | PC12 cell/MCAO mice | Liu et al. (2020) |
| 21 | miR-455-5p | I/R | FLT3 | miR-455 ↑ → FLT3 ↓ → I/R ( −) | I/R mice | Chen et al. (2020) |
| 22 | miR-21 | AD | PDCD4/PI3K/AKT/GSK-3β | miR-21 ↑ → PI3K/Akt/GSK3β↓ → AD ( −) | SH-SY5Y cells | Feng et al. (2018) |
| 23 | miR-124-3p | AD | PI3K/AKT/GSK-3β | miR-21 ↑ → Caveolin-1-PI3K/Akt/GSK3β↓ → AD ( −) | N2a/APP695swe cells | Kang et al. (2017) |
| 25 | miR-124 | AD | RFX1 | miR-124 ↓ → RFX1 ↑ → ApoE ↓ → AD ( −) | BV2 microglia cell | Feng et al. (2017) |
| 26 | miR-124 | AD | Delta in Notch Signaling Pathway | miR-124 ↑ → Delta ↓ → AD ( −) | AD flies | Kong et al. (2015) |
| 27 | miR-34a | AMD | TREM2 | miR-34a ↑ → TREM2 ↓ → AMD ( +) | C8B4 microglial (MG) Cells | Bhattacharjee et al. (2016) |
| 28 | miR-34a | AD | Caspase-2 | miR-34a ↑ → Caspase-2 ↓ → AD ( +) | SH-SY5Y cells | Q. Li, T. Liu et al. (2019a, b) |
| 29 | miR-22 | I/R | PUMA | miR-22 ↑ → PUMA↑ → I/R ( −) | PC12 cells | Jiao et al. (2020) |
| 30 | miR-29a | AD | Wnt1/CREB | miR-29a ↑ → inflammatory cytokines ↓ → AD ( −) | PBMCs | Sedighi et al. (2019) |
| 31 | miR-107 | AD | PDCD10 | miR-107 ↑ → PDCD10 ↓ → AD ( −) | the mouse model insulted by 6-OHDA | Sun et al. (2020) |
| 32 | miR-933 | AD | 27-OHC | miR-933 ↑ → 27-OHC↑ → inflammatory cytokines ↑ → AD ( +) | HMVEC | Dias et al. (2018) |
| 33 | miR-200b/c | AD | S6K1 | miR-200b/c ↑ → IRS-1pSer ↓ → AD ( −) | Tg2576 transgenic mice | Higaki et al. (2018) |
| 34 | miR-142-5p | AD | sGC/sGMP | miR-142-5p ↑ → sGC/sGMP ↓ → AD ( −) | SH-SY5Y cells | H. Xu et al. (2019a, b) |
| 35 | miR-30b | PD | SNCA | miR-30b ↑ → Bax/Bcl-2 ↓ → PD ( −) | SH-SY5Y cells | Shen et al. (2020) |
| 36 | miR-148b-3p | neuroinflammation | PIK3CA/Akt/NF-κB | miR-148b-3p ⬆ → PI3K/pAKt ↓ → neuroinflammation ( −) | Sprague Dawley rats | M. Wang et al. (2020a, b) |
| 37 | miR-34c | AD | SYT1 | miR-34c ↑ → ROS‐JNK‐p53 ↑ → AD ( −) | SAMP8 mice | Shi et al. (2020) |
| 38 | miR-125b | AD | GluN2A | miR-125b ↑ → GluN2A ↓ → AD (− | AD mice | Tang et al. (2019) |
| 39 | miR-125b | AD | DUSP6/PPP1CA/Bcl-W | miR-125b ↑ → Bcl-W/DUSP6/PPP1CA↓ → Tau Hyperphosphorylation ↑ → AD ( +) | AD mice | Banzhaf-Strathmann et al. (2014) |
| 40 | miR-142-5p | AD | Aβ42 | miR-142-5p ↑ → PSD-95 ↓ → AD ( +) | SH-SY5Y cells | Song and Kim (2017) |
| 41 | miR-6845-3p | AD | Aβ25-35 | miR-6845-3p ↓ → Aβ25-35 ↓ → AD ( −) | SH-SY5Y cells | Hu et al. (2018) |
| 42 | miR-26b | AD | Rb1 | miR-26b ↑ → p27/Kip1↓ → Cdk5 ↑ → Tau Hyperphosphorylation ↑ → AD ( +) | E18 Sprague Dawley rat | Absalon et al. (2013) |
| 43 | miR-138 | AD | RARA | miR-138 ↑ → RARA/GSK-3β↑ → Tau Hyperphosphorylation ↑ → AD ( +) | HEK293/tau cells | X. Wang et al. (2015a, b) |
| 44 | miR-922 | AD | UCHL1 | miR-922 ↑ → UCHL1 ↓ → Tau Hyperphosphorylation ↑ → AD ( +) | AD mice | Zhao et al. (2014) |
| 45 | miR-96 | VaD/AD | mTOR | miR-96 ↑ → mTOR ↓ → VaD/AD ( +) | CCH rat | Liu et al. (2018) |
| 46 | miR-10a | AD | BDNF-TrkB | miR-10a ↑ → BDNF-TrkB ↓ → AD ( +) | AD rat | Wu et al. (2018) |
| 48 | miR-206-3p | AD | BDNF | miR-206-3p ↑ → BDNF ↓ → AD ( +) | APP/PS1 mice | C. N. Wang et al. (2017a, b) |
| 49 | miR-140-5p | AD | ADAM10 | miR-104-5p ↑ → ADAM10 ↓ → AD ( +) | SHSY5Y/CHP212 cells | Akhter et al. (2018) |
| 50 | miR-937 | AD | BDNF | miR-937 ↑ → Brn-4 ↑ → Aβ↓ → AD ( −) | AD mice | Liu et al. (2015) |
| 51 | miR-144 | AD | NRF2/GSH | miR-144 ↑ → NRF2/GSH ↓ → AD ( +) | SHSY5Y cells | Zhou et al. (2017) |
| 52 | miR-433 | PD | FGF20 | miR-433 ↓ → FGF20 ↑ → alpha-synuclein↑ → PD ( +) | DCHG | G. Wang et al. (2008a, b) |
| 53 | miR-7/miR-153 | PD | α-syn | miR-7/miR-153↑ → α-syn ↓ → PD ( +) | Primary neuron cells | Doxakis (2010); Zhu, Wang, Qi et al. (2018) |
| 54 | miR-205 | PD | LRRK2 | miR-205↑ → LRRK2 ↓ → PD ( +) | G2019S (Y1699C mutation cells | Cho et al. (2013) |
| 55 | miR-599 | PD | LRRK2 | miR-599↑ → LRRK2 ↓ → PD ( +) | SH-SY5Y cells | Wu et al. (2019) |
| 56 | miR-29c | PD | PTEN | miR-29c↑ → PTEN ↓ → PD ( +) | PC12 cells | Zou et al. (2015) |
| 57 | miR-21 | PD | α-syn | miR-21↑ → α-syn ↑ → PD ( −) | SH-SY5Y cells | Su et al. (2016) |
| 58 | miR-181b | PD | PTEN | miR-181b↑ → PTEN ↓ → PD ( +) | PC12 cells | W. Li et al. (2018a, b) |
| 59 | miR-221 | PD | PI3K/AKT | miR-221↑ → PI3K/AKT ↑ → PD ( +) | Rats | Salama et al. (2020) |
| 60 | miR-100 | PD | PI3K/AKT | miR-100↑ → PI3K/AKT ↑ → PD ( +) | SH-SY5Y cells | (Peng et al. (2019) |
| 61 | miR-126 | PD | PI3K/AKT | miR-126↑ → PI3K/AKT ↓ → PD ( −) | Neuron cells | Kim et al. (2014) |
| 62 | miR-185 | PD | PI3K/AKT | miR-185↑ → PI3K/AKT ↑ → PD ( +) | Rats | Qin et al. (2021) |
| 63 | miR-410 | PD | PTEN/pAKT | miR-410↑ → PTEN/AKT ↑ → PD ( +) | SH-SY5Y and PC12 cells | Ge et al. (2019) |
| 64 | miR-34a | PD | Nrf2/Bax/caspase-3 | miR-34a↑ → Nrf2↓ → Bax/caspase-3 ↑ → PD ( −) | SH-SY5Y | Alural et al. (2015) |
| 65 | miR-626 | PD | Keap 1/Nrf2 | miR-626↑ → Keap 1↓ → Nrf2 ↑ → PD ( +) | Retinal pigment epithelium cells | Qin et al. (2019) |
| 66 | miR-153 | PD | Nrf2-HO-1 | miR-153↑ → Nrf2-HO-1 ↓ → PD ( −) | SH-SY5Y | Zhu et al. (2018b) |
| 67 | miR-27a/miR-27b | PD | PINK1 | miR-27a/miR-27b↑ → PINK1 ↓ → PD ( −) | Hela and M17 cells | Kim et al. (2016) |
| 68 | miR-212-5p | PD | SIRT2/p53 | miR-212-5p↑ → SIRT2 ↓ → p53 ↓ → PD ( +) | Rats | Sun et al. (2018) |
| 69 | miR-7 | PD | SIRT2/p53 | miR-7↑ → SIRT2 ↓ → p53 ↓ → PD ( +) | Dopaminergic neurons cells | Li et al. (2016) |
| 70 | miR-183 | PD | Bcl-2 | miR-183↑ → Bcl-2 ↑ → PD ( −) | Rats | J. X. Gao et al. (2019a, b) |
| 71 | miR133a | PD | Bax/caspase-3 | miR-133a↑ → Bax/caspase-3 ↓ → PD ( +) | PC12 | Lu et al. (2020) |
| 72 | miR-505 | PD | caspase-3 | miR-505↑ → caspase-3 ↑ → PD ( −) | SH-SY5Y | Zhu et al. (2018a, b) |
| 73 | miR-15b-5p | PD | caspase-3 | miR-15b-5p↑ → caspase-3 ↑ → PD ( −) | SH-SY5Y | J. Zhu et al. (2021a, b) |
( −): anesis, ( +): deterioration, ↓: descend, ↑: ascend.
The most frequent neurodegenerative illness is AD with clinical manifestations of AD including cognitive impairment, personality changes, and progressive loss of memory (Alzheimer's 2016). The pathological findings were degeneration of neurons in the hippocampus and cortex, neurofibrillary tangles, and “senile plaques” precipitated containing Aβ-amyloid (Lane et al. 2018). Studies have shown that apoptosis may be the main mechanism of neuronal degeneration in AD (Caccamo et al. 2017), and there are a large number of apoptotic programmed death neurons in the brain of AD patients. Some microRNAs related to apoptosis, such as the miR-16 and the miR-15, participate in the regulation of the human anti-apoptotic BCL-2 gene (Cimmino et al. 2005). There has also been speculation that the expressions of miR-9, miR-128, and miR-1256 are expressed significantly more in the brains of patients with AD which are significantly higher than the average expression levels in the brain tissues of the adult control group brains (Sethi and Lukiw 2009). The level of miR-107 expression is also highly linked to the severity of AD; its expression decreases as the disease has progresses (W. X. Wang et al. 2008a, b). Beta-secretase 1 (BACE1) is the enzyme that controls the rate of Aβ-amyloid formation, and miRNAs such as miR-328 and miR-298 can also regulate the expression of BACE1 to affect the production of Aβ-amyloid production. Specific locations on BACE1 can be recognized by the miR-328 and the miR-298, which regulate the expression of the BACE1 enzyme (Boissonneault et al. 2009).
The role of miRNAs in Parkinson’s disease
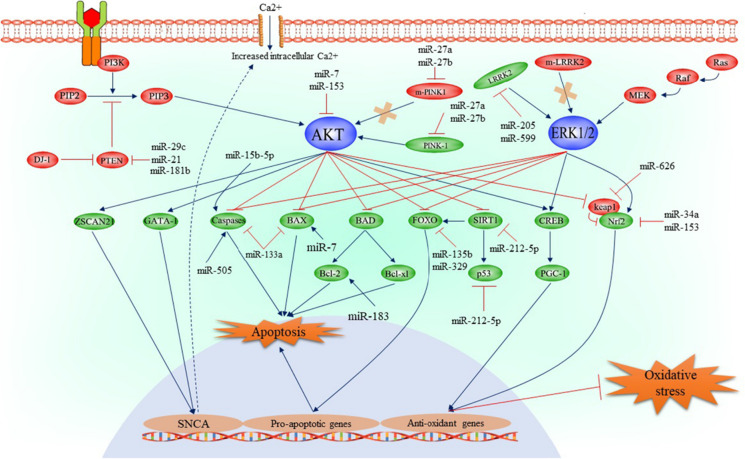
Parkinson’s disease (PD) is a widespread and complicated neurodegenerative illness caused by the death of dopamine neurons in the substantia nigra and manifests with a broad range of motor and non-motor symptoms. There are many biomarkers for PD, including clinical biomarkers such as rapid eye movement sleep behavior disorder, imaging biomarkers, inflammatory biomarkers, and biofluid-based biomarkers such as α-synuclein and extracellular vesicle. miRNAs are being investigated as potential biomarkers for the early diagnosis of PD and monitoring the pathological progression because they are stable and easily detectable in biofluids (Li and Le 2020). Table 1 shows various miRNAs with reported expression in different cells and rats with PD. The crucial factors involved in inducing neuronal death in PD are parkin, VPS35, DJ1, PTEN-induced kinase 1, and leucine-rich repeat kinase 2 (LRRK2); these proteins facilitate oxidative stress, inflammation, apoptosis, and dysregulated calcium homeostasis in neurons (Zimprich et al. 2004). The molecular mechanisms by which miRNAs contribute to the pathology of PD are summarized in Fig. 3.
Fig. 3.
The molecular mechanism of miRNAs on the pathology of PD.
Reproduced with permission from reference (Khezri et al. 2022). Copyright 2022 Springer Nature and Copyright Clearance Center
SNCA encodes α-synuclein, one of the main precipitating factors for PD. Increased α-synuclein can induce dopaminergic neuron dysfunction in the PD brain by triggering excessive oxidative stress and disrupting calcium homeostasis (Lashuel et al. 2002). A polymorphism of PITX3 is related to PD, while miR-133b participates in passive feedback regulation of Pitx3. When miR-133b expression is reduced, the balance is disrupted, which will eventually lead to PD. SNCA overexpression enhances the sensitivity of dopaminergic neurons to oxidative stress, which is one of the pathological changes of PD. Inhibition of miR-433 expression increases the expression of fibroblast growth factor 20 protein and ultimately leads to SNCA overexpression (Higgs and Lehman 2015). miR-7 and miR-153 participate in post-transcriptional regulation of SNCA, and changes in their expression lead to PD. The phosphoinositide 3-kinase (P13K)/AKT signaling pathway can also induce α-synuclein overexpression. Activated miR-7 and miR-153 can inhibit the P13K/AKT signaling pathway and are potential biomarkers involved in α-synuclein expression for PD (Fig. 3).
LRRK2 is regarded as one of most important proteins involved in the development of dopaminergic neurons; mutations in this gene can cause familial and sporadic PD diseases (Zimprich et al. 2004). miR-205 and miR-599 can decrease LRRK2 expression by targeting the 3′-untranslated region (UTR) of LRRK2 mRNA and attenuate MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine)-induced apoptosis in SH-SY5Y cells (Fig. 3). Moreover, miR-34a, miR-153, and miR-626 are associated with the pathologic mechanisms in PD by regulating oxidative stress and neuronal apoptosis. The P13K/AKT, LRRK2/ERK1/2, and P13K/PTEN signaling pathways are involved in neuronal apoptosis, oxidative stress, and other pathophysiological mechanisms in PD and are key regulators in the homeostasis of dopaminergic neurons. Many miRNAs have been reported and predicted to regulate neurodegeneration in PD, and these bioactive miRNAs may serve as a platform for potential biological markers and therapeutic targets in future.
The role of miRNAs in Huntington’s disease and other neurodegenerative diseases
Huntington’s disease (HD) is a malignant autosomal dominant neurological illness generally known as Huntington’s chorea, mainly due to the production of abnormal Huntington protein (Htt), resulting in the progressive death of nerve cells in the striatum and cortex (McColgan and Tabrizi 2018). Increased restrictive element-1 silencing transcription factor (REST) expression has been found in both HD mouse models and human HD brain tissue (Buckley et al. 2010; Johnson and Buckley 2009). REST plays a critical role in the regulation of neuronal genes; it is widely expressed throughout the body and exhibits especially strong expression in immature CNS cells. Its function is to suppress a large number of neuron-specific genes by attracting a multi-subunit complex of DNA regulatory motifs known as repressor 1 to a specific location (Buckley et al. 2010). It promotes neuroprotection by suppressing genes implicated in oxidative stress and amyloid toxicity and by fine-tuning genes involved in synaptic plasticity and normal aging (Hwang and Zukin 2018). Moreover, miRNAs that target the REST complex can significantly reduce other miRNAs, including miR-330, miR-29a, miR-132, and miR-124 (Yoo et al. 2011). In particular, miR-132 and miR-124 are decreased most significantly in HD (Johnson et al. 2008). miR-124 is highly expressed specifically in normal neurons, and its target gene is a component of REST (Lee et al. 2017). During embryonic CNS system development, miR-124 targets the 3′-UTR of synaptonemal complex protein 1 (SCPY1) to limit its expression (Visvanathan et al. 2007). In addition, miR-132 downregulation promotes axonal growth by inhibiting p250GA transcription in the cortex of patients with HD (Fukuoka et al. 2018). Furthermore, in vivo upregulation of REST alters the production of myriad miRNAs, including miR-9*, miR-9, and miR-124 (Bithell et al. 2009). To be more specific, miR-124 mainly inhibits the expression of non-neuronal genes in neurons, which is critical for neuronal development. In addition, miR-9 and miR-9* can regulate the expression of REST, thus affecting the manifestation of HD (Chang et al. 2017). These studies suggest that REST can directly inhibit gene expression and indirectly regulate the function of miRNAs and their target genes by influencing the expression of some miRNAs. Other miRNAs such as miR-9, miR-142, miR-155, and miR-22, among others, have been identified to contribute to the neurodegenerative disease including ischaemic stroke, IR, and MS (Table 1).
The role of endogenous miRNAs
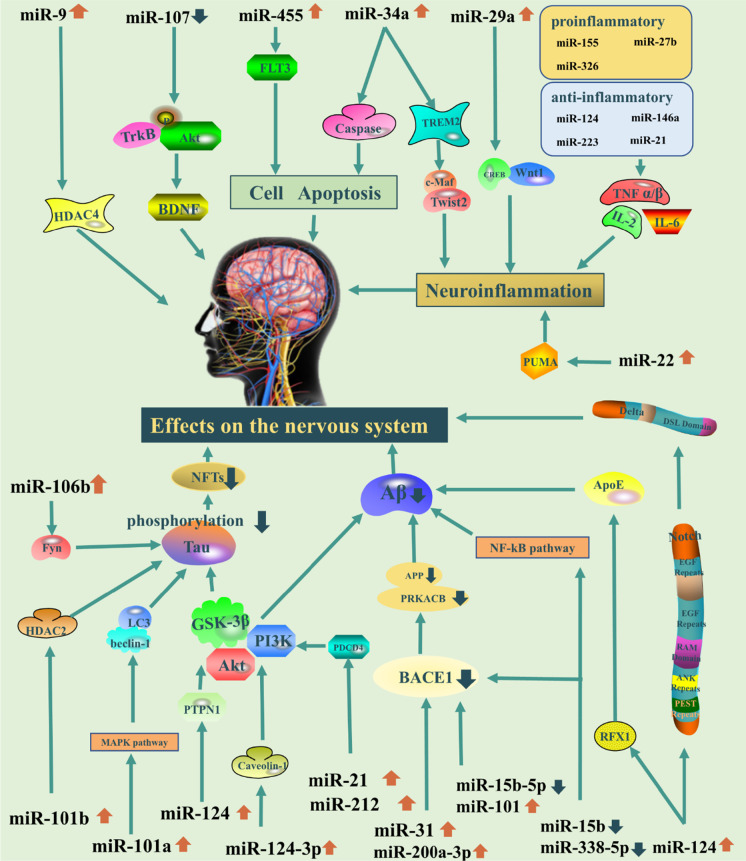
Researchers have examined the role of endogenous miRNAs in eight neurodegenerative diseases, including AD, amyotrophic lateral sclerosis (ALS), ataxia, dementia, HD, MS, PD, and prion diseases. Most studies have focused on AD to examine the relationship between miRNAs and neurodegenerative diseases. The changes in various miRNAs in animal models of different neurodegenerative diseases and their corresponding regulatory pathways are described Table 1. There is a dual effect of mRNAs on the nervous system. On the one hand, some miRNAs have protective effects. For example, in AD, miR-15b, miR-107, miR-388-5p, miR-200a-3p, miR-31, and miR-29c can directly act on the BACE1 (Barros-Viegas et al. 2020; Jiao et al. 2016; Li and Wang 2018; Qian et al. 2019; Wang et al. 2019; Zong et al. 2011), reducing Aβ accumulation, and play a protective role against AD. miR-15b can also act on nuclear transcription factor-kβ signal to play a regulatory role (Dai et al. 2014). Meanwhile, miR-132, miR-212, miR-101a/b, miR-155a-5p, miR-142a-5p, miR-455a-5p, and miR-146a-5p can inhibit tau phosphorylation through different pathways and regulatory pathways (D. Liu et al. 2017a, b; Sierksma et al. 2018; Smith et al. 2015). Specifically, miR-132 and miR-212 can inhibit tau phosphorylation by disrupting the balance of S-nitrosylation (Cha et al. 2019), while miR-101a/b regulates tau phosphorylation through two different pathways, histone deacetylase 2 and mitogen-activated protein kinases (D. Liu et al. 2017a, b). miR-155a-5p, miR-142a-5p, miR-455a-5p, and miR-146a-5p can regulate or directly act on tau protein by targeting glycogen synthase kinase-3β and the cyclin-dependent kinase 5 (CDK5)/PTEN/PPP2CA axis to inhibit its phosphorylation (Sierksma et al. 2018). On the other hand, some miRNAs can also damage the nervous system to some extent. For example, several miRNAs are pro-inflammatory, promoting neuroinflammation (Vezzani et al. 2011) and neuronal damage (Stephenson et al. 2018). The pro-inflammatory miR-155, miR-27b, and miR-326; the anti-inflammatory miR-21, miR-124, miR-146a, and miR-223 (Gaudet et al. 2018); and the pro- and anti-inflammatory let-7 family (Roush and Slack 2008) can regulate inflammation through the nuclear factor kappa B (NF-κβ) pathway in the brain (Fig. 4).
Fig. 4.
The regulatory mechanisms of miRNAs in CNS
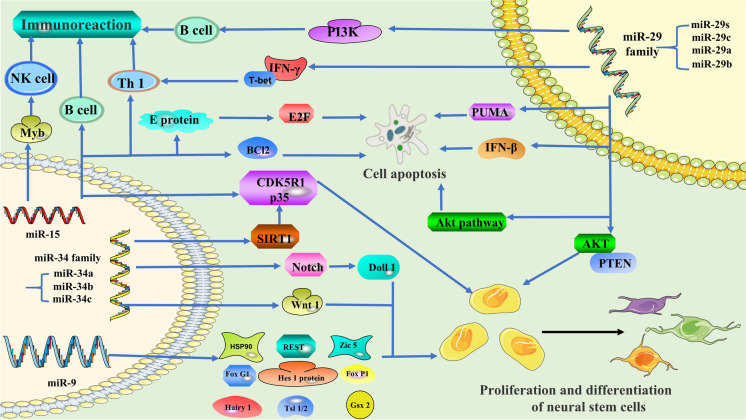
A review of the literature shows that many miRNAs are dysregulated in animal models of neurodegenerative diseases. miR-9, miR-15, the miR-29 family, and the miR-34 family have been studied most extensively (Fig. 5). For example, the occurrence of neurodegenerative diseases is related to cell proliferation and differentiation, and miR-9 is the key miRNA involved in regulating cell proliferation and differentiation (Khafaei et al. 2019). Neurodegenerative diseases involve extensive apoptosis, and the miR-34 family is one of the important miRNAs involved in regulating apoptosis. In clinical applications, the miR-34 family is often used for cancer treatment (Hermeking 2010). However, as a key miRNA regulating programmed cell death, the miR-34 family’s function in neurodegenerative illnesses should not be overlooked (Bazrgar et al. 2021). Pro-inflammatory, immune-mediated mechanisms are undeniably important in the pathogenesis and progression of neurodegenerative disease. Furthermore, miR-15 regulates cellular immunity and is involved in the onset and progression of neurodegenerative disorders (Hutter et al. 2021). Meanwhile, it is also involved in apoptosis, cell differentiation, and proliferation, and plays a wide role in various processes. The miR-29 family also participates in the regulation of various channels in the brain (Swahari et al. 2021), and plays a modulatory role within the immune reaction, multiplication, separation, and apoptosis of cells (Alizadeh et al. 2019; Sharma et al. 2021).
Fig. 5.
The regulatory mechanisms of critical miRNAs in CNS
In the following subsections, we summarize the regulatory pathways and mechanisms of action of these endogenous miRNAs in animal models of neurodegenerative diseases. The expression profile of miRNAs is summarized in Table 1, which also forms a general map to clarify the expression of these miRNAs in neurodegenerative diseases.
miR-9
miR-9 is highly expressed in the brain of developing and adult mammals (Sempere et al. 2004). miR-9 is indispensable to the proliferation and differentiation of neural stem cells (Tan et al. 2012). Indeed, suppression of miR-9 leads to decreased proliferation of neural progenitor cells and increased cell migration ability (Delaloy et al. 2010). Studies have revealed distinct sets of miRNAs in different species, and the differences in miR-9 may be partly responsible for the differences in brain shape, size, and function among vertebrates. This may be because miR-9 is closely related to the proliferation and differentiation of neural stem cells (Alwin Prem Anand et al. 2020). miR-9 can modulate these processes by interacting with the 3′-UTR of Hes1 mRNA and thus regulating Hes1 protein expression. When miR-9 is overexpressed, Hes1 protein is reduced, inducing neuronal differentiation and cell cycle termination (Tan et al. 2012). The foremost characteristic of miR-9 in neurogenesis is to downregulate hairy1 mRNA. The miR-9/hairy1 complex impacts apoptosis through p53 and proliferation through cyclin D/p27 (Bonev et al. 2011). miR-9 inhibits the expression of Gsh2 and Foxg1 proteins, enabling appropriate differentiation of Cajal–Retzius cells and preterm neurons, and can also affect neural development by playing a role in the neuronal cell cycle (Dajas-Bailador et al. 2012; Shibata et al. 2011). FoxP1 and ISL1/2, which are expressed in spinal motor neurons, are both assumed to be targets of miR-9. Therefore, miR-9 can impair the differentiation and axonal projection of spinal motor neurons by regulating FoxP1 protein levels (Otaegi et al. 2011).
miR-9 is strongly associated with the proliferation and differentiation of neurons, and when the homeostasis between miR-9 and transcription factors is disrupted, the proliferation, differentiation, and apoptosis of neurons are disturbed, leading to neurodegenerative diseases (Reddy et al. 2017). For example, a pathological feature of HD is transcriptional alterations in certain brain regions that are associated with abnormal activity of REST (Buckley et al. 2010). A prevailing view is that in neural progenitor cells, REST is essential for the inhibition of neuronal genes, and downregulation of REST during neuronal differentiation facilitates the acquisition and maintenance of neuronal phenotypes (Ballas et al. 2005). miR-9 controls dendritic development by targeting REST, and a vital function of miR-9 is to maintain low REST levels throughout neuronal maturation (Giusti et al. 2014). Studies have shown that REST silencing complex downregulation via miR-9 provides a double-negative feedback loop in the progression of HD; specifically, miR-9 and miR-9* target REST and core REST, respectively (Packer et al. 2008). Therefore, the dysregulation of and changes in miR-9 in corresponding targets in the populations of neuronal progenitor cells could be a common mechanism of neurodegenerative diseases.
The miR-29 family
The miR-29 family consists of clusters of miRNAs that produce miR-29a, miR-29b, miR-29c, and miR-29s, among others. They share parts of the same sequence and therefore the same target (Kriegel et al. 2012). T-bet and IFN expression are increased when miR-29 is lost. T-bet and IFN are transcription factors that work together to coordinate type 1 inflammatory responses. They are found in a variety of cell types, including antigen-presenting cells and lymphocytes. The immune system defends the body against a wide range of harmful microorganisms, but when the timing and duration of the expression of these immune factors are out of balance, autoimmune diseases can develop (Smith et al. 2012) (Steiner et al. 2011). The miR-29 family regulates Th1 differentiation and also maintains B cell survival and controls terminal differentiation through PI3K signal transduction (Hines et al. 2020), which is closely related to the immune response. In rat models of cerebral infarction, miR-29 contributes to the regulation of the Akt signaling pathway, which can suppress neuronal death (Rong et al. 2020). Different members of the miR-29 family are highly expressed in olfactory bulb neurons, the hippocampus, and cerebellar Purkinje cells. After temporary forebrain ischaemia, neurons in the hippocampal CA1 region continue to die in a delayed manner, which affects disease progression. By targeting PUMA, a member of the pro-apoptotic Bcl2 family, miR-29a causes hippocampal neuronal cell death and protects CA1 from delayed neuronal death after forebrain ischaemia (Ouyang et al. 2013). In mice, knockout of the miR-29a/b-1 cluster produces obvious characteristics of ataxia, with the main lesion site in the cerebellum; the major participants in the main cerebellar circuit are Purkinje cells. miR-29a/b-1 clusters show abundant expression in Purkinje cells. Kcnc3 and Hip1 proteins are significantly upregulated when the miR-29a/b-1 cluster is removed (Papadopoulou et al. 2015). Upregulation of Hip1 leads to N-methyl-d-aspartate receptor over-activation and increased excitotoxicity. Kcnc3 encodes Kv3.3, a voltage-gated sodium–potassium subunit crucial for Purkinje cell fast peaking and linked to spinocerebellar ataxia 13 (Hurlock et al. 2008). miR-29 also increases rat neural stem cell proliferation through the PTEN/AKT signaling pathway (Y. Gao et al. 2019a, b). IFN-β is broadly used as a factor in the immunoregulatory therapy of patients with MS. Several miRNAs are downregulated when IFN-β response genes are upregulated. Members of the miR-29 family, which is linked to apoptosis and IFN feedback loops, are among them (Hecker et al. 2013).
miR-15
Since the discovery of the miR-15/107 group, its role in humans has been increasingly studied. In vertebrate species, these miRNAs control the expression of genes involved in cell division, metabolism, the immune response, and apoptosis. Human cancer, cardiovascular disease, and neurological illnesses have all been linked to the miR-15/107 family (Parsi et al. 2015; Finnerty et al. 2010; Liu and Wang 2012). The miR-15/107 family regulates CDK5R1/p35, which is crucial for brain development and function and has been linked to a number of neurological disorders, including AD. Indeed, downregulation of the miR-15/107 family increases CDK5R1/p35 levels and, consequently, increases CDK5 activity, which is involved in the etiology of AD (Moncini et al. 2017). In addition to neural effects, miR-15 is also related to the immune response. miR-15 can help pre-B cells shift from proliferation to differentiation (Lindner et al. 2017); limit T cell cycle, survival, and memory T cell differentiation (Gagnon et al. 2019); and govern natural killer cell maturation by antagonizing Myb (Sullivan et al. 2015). When miR-15 regulates these immune cells, it regulates the immune response in the body. miR-15 can also regulate cell division and apoptosis. miR-15 and miR-16 have been identified as direct transcription targets of E2F1. Targeting cyclin E inhibits e2F-induced proliferation (Ofir et al. 2011). By targeting BCL2, miR-15 and miR-16 potentially cause apoptosis (Cimmino et al. 2005). Neurodegenerative diseases are often associated with neuroinflammation and abnormal apoptosis. Because miR-15 regulates the immune response and apoptosis, it may directly regulate some neural pathways and may also affect the occurrence of neurological diseases by regulating cellular immunity, division, or apoptosis.
The miR-34 family
The miR-34 family in mammals is made up of three processed miRNAs that are encoded by two separate genes: miR-34a is encoded by one transcript, while miR-34b and miR-34c share a transcript (L. Zhang et al. 2019a, b). miR-34a is abundant in mature and differentiated neurotransmitters in mice (Jauhari and Yadav 2019), whereas miR-34b/c is mostly found in lung tissues (Kim et al. 2019). p53 is required for miR-34 expression, and the stability of the p53 protein greatly increases miR-34 expression (Cortez et al. 2016). Feedback loops are formed between miR-34 expression and p53 protein levels during neurodevelopment (Jauhari et al. 2018). miR-34a contributes to the regulation of mouse neural stem cell proliferation by targeting SIRT1 and regulating the activity of p53. Increased expression of miR-34a can promote neurons and neurites to grow longer following mitosis in mouse neural stem cells. Decreased miR-34a expression inhibits neuronal differentiation (Aranha et al. 2011; Yamakuchi and Lowenstein 2009). TAp73, a member of the p53 family of proteins, is a transcription factor that acts on particular binding sites on the miR-34a promoter to stimulate the production of these miRNAs. Neuronal differentiation by TAp73 is mediated by miR-34a regulation of synaptic proteins (Agostini et al. 2011). miR-34a affects many aspects of cell physiology, regulating somatic reprogramming and neural differentiation by regulating the p53 protein level (Choi et al. 2011). The Notch ligand Dll1 is targeted by miR-34a when it downregulates Dll1 expression, negatively regulating cell proliferation and inducing MB cell apoptosis and neural differentiation (de Antonellis et al. 2011). In fruit flies, miR-34a modulates age-related events and long-term brain integrity, indicating a biological link between aging and neurodegeneration. The loss of this molecule can cause accelerated aging and delayed brain degeneration. miR-34a upregulation prolongs median lifespan and alleviates neurodegenerative changes caused by human pathogenic polyglutamine disease proteins (Liu et al. 2012). miR-34b/c also has a regulatory role in the nervous system, regulating Wnt1 and enhancing the differentiation of brain dopamine neurons (De Gregorio et al. 2018). Therefore, the miR-34 family is also significant in neurodegenerative illnesses.
The role of exogenous miRNAs
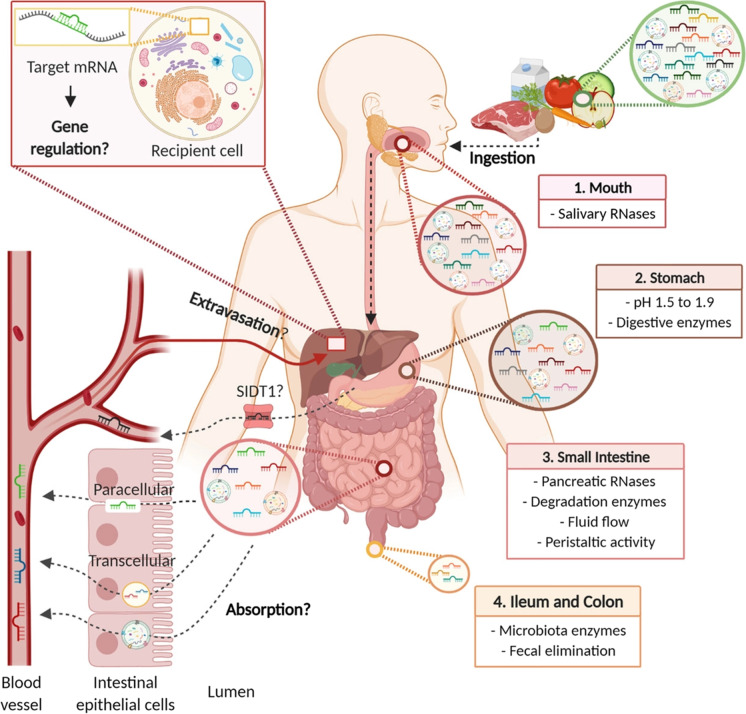
Recently, a growing body of literature has shown that some exogenous RNAs can be recognized by a myriad of receptors in host eukaryotic cells and that such small non-coding RNAs can be transmitted between different species to induce signal interference in a transboundary manner (Reniewicz et al. 2016). Just like microbes and hosts, symbiotic or pathogenic relationships between various organisms may be mediated by this pattern of cross-species communication (Liang et al. 2013). When exogenous miRNAs enter the host body, they can be delivered actively to recipient cells by being packaged selectively into microvesicles. Studies have shown that exogenous plant miRNAs can be obtained through ingestion of food; they can then be detected in serum, organs, and tissues of experimental animals (Link et al. 2019). In addition, some plant miRNAs can also be obtained through breast milk (Lukasik et al. 2017). Plant-derived miRNAs can be absorbed by gastrointestinal cells via SITT1, allowing them to reach the bloodstream and regulate the expression of endogenous mRNA (Zhang et al. 2012). Broccoli-derived miRNAs can withstand ordinary food processing and digestive conditions (Chapado et al. 2021), and fragments of plant miRNAs have also been detected in feces, blood, the gastrointestinal tract, and organs after plant RNA had been added directly to food particles (Liang et al. 2014).
Exogenous miRNAs can regulate the expression of target genes and the function of recipient cells. When exogenous miRNAs enter the human body, they seem to act similarly to endogenous miRNAs, participating in the host body’s management of life activities. By suppressing GSK3β-mediated NF-κB and transforming growth factor beta 1 (TGF-β1) pathways, soybean-derived gma-miR159a improves hepatic stellate cell activation and lowers inflammation (Yu et al. 2021). Moreover, cross-kingdom gene regulation by miRNAs has initiated the hot topic of the potential function of miRNAs on human or mice gene expression after oral administration.
Various miRNAs derived from plants are shown in Table 2 and their regulatory mechanisms on the human body are illustrated in Fig. 6. Among these miRNAs, miR168a, miR2911, and miR156a show high serum concentrations while miR166a shows moderate serum concentrations in mammals. Interestingly, miR2911 derived from honeysuckle presents antiviral activity against influenza A viruses (H1N1, H5N1, and H7N9) and SARS-CoV-2 in cells and mice (Zhou et al. 2020a). miR2911 is derived from plant 26S ribosomal RNA and has a high GC content; hence, it remains stable after decoction and oral administration. However, other exogenous miRNAs from honeysuckle such as miR166a and miR2910 are extensively degraded by boiling, and pre-miRNAs such as pre-miR168a and pre-miR156 are not detected in cooked rice or mouse serum while mature miR168a and miR156 levels are high. These results suggest that only mature miRNAs can be absorbed in the gut and that miRNAs derived from ribosome fragments could be more stable and abundant after decoction (Chen et al. 2021). These miRNAs derived from ribosome fragments are called atypical miRNAs because they have no precursor miRNAs. These miRNAs should receive additional research attention in the future. Other miRNAs have shown effective medicinal function, including miR159 as a breast cancer suppressor through targeting the TCF7 gene, and miR156a, miR168, and miR166a can reduce cytokine-induced monocytes adhesion and act as vasoprotective molecules by triggering low-density lipoprotein receptor adapter protein 1 (LDLRAP1) to treat cancer (Chin et al. 2016; Hou et al. 2018). The following subsections highlight the regulatory mechanisms of the main exogenous miRNAs in mammals.
Table 2.
The identified miRNAs in cross-kingdom gene regulation
| Article no | Dietary miRNAs | Original plants | Target/host | Disease/target gene | Reference |
|---|---|---|---|---|---|
| 1 | miR2911 | Honeysuckle | Human | Influenza A virus, SARS-CoV-2 | Zhou et al., (2020b); Zhou et al., (2015) |
| 2 | miR168 | Rice | Mice | LDLRAP1, Breast cancer | Lang et al. (2019) |
| 3 | miR166a | Oryza sativa | Human/mice/rat | LDLRAP1 | Zhang et al. (2012) |
| 4 | miR156a | Cabbage, spinach, lettuce | Human/HAEC cells | LDLRAP1 | Hou et al. (2018) |
| 5 | miR2910 | Populus euphratica | Human | JAK-STAT signal pathway, breast cancer | Y. C. Liu et al. (2017a, b) |
| 6 | miR159 | Flycine max Arabidopsis thaliana | Mice/cancer cells | TCF7, breast cancer | Chin et al. (2016) |
| 7 | miR14 | Curcuma longa | Human | Rheumatoid arthritis | Sharma et al. (2017) |
| 8 | miR5754 | Medicago truncatula | HCT116 cells | MALAT1, NEAT1 | Marzano et al. (2020) |
| 9 | miR4995 | Glycine max | HCT116 cells | MALAT1, NEAT1 | Marzano et al. (2020) |
| 10 | miR167e-5p | Moringa oleifera Lam | Mice | β-Catenin | Li et al. (2019a) |
| 11 | miR156 | Cabbage, spinach, lettuce | BxPc-3, AsPC-1, and PANC-1 cells | Wnt10b | M. Li, T. Chen, R. Wang et al. (2019) |
| 12 | miR471 | Lettuce | Mice | Hepatitis B virus (HBV) | S. Zhang et al. (2019a, b) |
| 13 | miR519 | Lettuce | Mice | Hepatitis B virus (HBV) | S. Zhang et al. (2019a, b) |
| 14 | miR5338 | Rape bee pollen | Rat | Mitochondrial fusion requires fusion protein 1 | Chen et al. (2018) |
Fig. 6.
The cross-kingdom regulatory mechanisms of plant miRNAs: the absorbing of miRNAs after oral administration (Del Pozo-Acebo et al. 2021).
Copyright 2021 The British Pharmacological Society
miR168a
miR168a is abundant in rice and is one of the most abundant exogenous plant miRNAs in the serum. Zhang et al. (2012) first reported that miR168 can be transferred into the human body and regulate the gene expression. It resists the gut tract conditions and reaches the serum and organs, and it could keep high levels where it could regulate the expression of LDLRAP1 mRNA (Lang et al. 2019). Specifically, miR168a binds to human/mouse LDLRAP1 mRNA to suppress its expression in the liver, reducing low-density lipoprotein elimination in mouse plasma. These findings imply that exogenous plant miRNAs obtained from food can affect mammalian target gene expression (Lang et al. 2019; Zhang et al. 2012).
miR168 can also participate in physiological and biochemical processes and play a regulatory role in the host. Furthermore, the miR168 family has 123 targets in the human transcriptome; 58% of these targets are cleaved and translation is inhibited for 41% of these targets. Among the 10 genes chosen at random are ATXN1, RPL34, ALS2, and AKAPI3, which are involved in transcription, cell transport, cell metabolism, and neurodegenerative disorders (Javed et al. 2017). Overall, miR-168 is highly conserved and can play a regulatory role after being absorbed by the body, and even participate in the regulation of some neurodegenerative diseases.
miR2911
miR2911 is a highly conserved miRNA that is abundant in honeysuckle and is stable after high-temperature frying (Zhou et al. 2020b). Most miRNAs such as miR166a, miR2914, and miR156 enriched in honeysuckle decoction are easily degraded after boiling, but miR2911 is stable. Although the reason underlying the high stability of miR2911 is still unclear, previous reports indicate that miR2911 is an atypical miRNA derived from ribosomal RNA (rRNA) and has a high GC content, which might contribute to its high stability. These results revealed that herb miRNAs derived from rRNA and with a high GC content might play an important role in traditional Chinese medicine. This eventuality requires additional research to exploit this unrecognized component.
A murine study reported that miR2911 could be detected in various organs and could pass through the mouse GI tract and be transferred to the bloodstream and lungs. Subsequently, SID1 transmembrane family member 1 in the gastric pit cell membrane can mediate uptake of dietary miRNAs, including miR2911, into cells (Fig. 6). miR2911 has various activities, including antiviral, antineoplastic, and anti-inflammatory. miR2911 can directly and adequately inhibit the replication of the H1N1 virus. In vitro and in vivo, miR2911 can suppress the replication of the H5N1 and H7N9 viruses, suggesting that it could be a unique product from nature that efficiently prevents viral infection (Zhou et al. 2015). miR2911 encoded by Lonicera japonica inhibits the replication of enterovirus 71 by targeting the VP1 gene (X. Li et al. 2018a, b), and directly inhibits the replication of varicella-zoster virus by targeting the IE62 gene (Y. Huang et al. 2019a, b). In addition to its antiviral effects, miR2911 can inhibit tumor growth by targeting TGF-β1 (Liu et al. 2021).
Exogenous miRNAs from food sources can enter the mammalian circulatory system and accumulate in organs. Once sin organs, these plant miRNAs can regulate target genes in a manner similar to endogenous miRNAs, thus affecting related physiological processes. Although a few exogenous miRNAs have been developed into molecules for drugs, miR2911 has not been exploited for this purpose. Hence, it is critical to find more new miRNAs similar to miR2911 with the excellent characteristics that could serve as therapeutics.
Discussion
miRNAs are involved in different types of neurodegenerative diseases and are upregulated or downregulated to different degrees in the brain of patients. Hence, they are receiving more and more attention as potential biomarkers of neurological diseases (J. Wang et al. 2015a, b). miR-155-5p, miR-9-5p, miR-146a-5p, miR-21-5p, miR-223-3p, miR-124-3p, and miR-132-3p are all frequently investigated miRNAs, in addition to miR-9, miR-15, the miR-29 family, and the miR-34 family (Juzwik et al. 2019). In neurodegenerative diseases, some of these miRNAs regulate immune responses in the brain (Neal and Richardson 2018); some regulate neuronal division, differentiation, and apoptosis (Lynam-Lennon et al. 2009); and some are associated with oxidative stress (Konovalova et al. 2019). Major pathways involved in neurodegenerative diseases, such as the NF-Kβ pathway, the Akt signaling pathway, and the Notch pathway, are closely related to miRNAs. In this review, we have discussed miRNAs in the context of neurodegenerative diseases. It is important to note that miRNAs do not play independent roles. Their functions overlap, suggesting that they are likely to work together in neurodegenerative diseases. We have systematically sorted and identified miRNAs related to neurodegenerative diseases. These miRNAs are involved in many common pathological mechanisms of neurodegenerative diseases, a fact that underscores the common pathways of disease pathogenesis.
miRNAs are highly conserved among species due to their hairpin structure, which enables them to be expressed stably in the body. In addition, the development and application of high-throughput sequencing technology has greatly improved the diagnostic sensitivity of miRNA profiling. The type and amounts of exosomes released by nerve cells are affected by pathological changes in the nervous system microenvironment, and miRNAs, which are the important parts of neuronal genetics, are often significantly altered, suggesting that miRNA precursors have considerable value as early diagnostic markers for neurodegenerative diseases. Wang (Wang & Zhang 2020) summarized several representative miRNAs that could be used as biomarkers in neurodegenerative diseases and proposed the merit and demerit of miRNAs as diagnostic markers for neurodegenerative diseases. Cheng et al. (2015) examined whether unique miRNAs might also have comparable diagnostic efficacy as conventional biomarkers and may be used as diagnostic biomarkers to mirror the development of AD. Therefore, circulating ex-miRNAs have great potential as biomarkers for the prognosis and analysis of neurodegenerative diseases.
Some miRNAs also show their considerable therapeutic potential for neurodegenerative diseases. Currently, there are two miRNA-based treatment strategies. The first is to use miRNA mimics to improve the expression of some miRNAs and to regulate the expression of their specific target proteins (Bouchie 2013). The second approach is to use miRNA inhibitors to block certain miRNAs when they are overexpressed by injecting complementary RNA sequences that bind to and inactivate the target miRNAs (Rupaimoole and Slack 2017). Drugs for neurodegenerative diseases need to cross the blood–brain barrier (BBB) to be effective; miRNAs, as small molecules of only about 20 bp, have the ability to cross the BBB. Targeted delivery of miRNAs is also a topic worth studying. miRNA-targeted agents with improved pharmacological activity and optimized pharmacokinetic properties are expected to be novel agents for the treatment of neurodegenerative diseases (Gupta et al. 2021). However, the application of miRNA-based therapies faces many difficulties. One is potential off-target effects (Seok et al. 2018). miRNAs should reach the right target without interfering with normal cell function and deliver drug molecules across the BBB to the right cells in the brain to act on their corresponding proteins. In addition, it is crucial to avoid activating the innate immune response when delivering this “foreign material” to the brain. Thus, there should be exploration of the potential impact of using exosomes from dendritic cells to carry nucleic acids and proteins to target cells (Zhang et al. 2021).
It is precisely because miRNAs are particularly important in the pathogenesis of neurodegenerative diseases that it is important to elucidate the unknown molecular mechanisms behind these diseases and to enable the use of these molecules as potential therapeutic interventions. It is possible to use miRNAs as biomarkers of neurodegenerative diseases or as new therapeutic agents. At the same time, it is important to understand disease occurrence and progression and its related molecular/cellular mechanisms, as well as the detailed role of miRNA networks in sporadic forms of neurodegenerative disease.
In addition, some exogenous miRNAs such as miR168 and miR2911 play a regulatory role by binding to their matching targets. Because these exogenous miRNAs can enter the human body and participate in regulatory processes, we speculate that some exogenous miRNAs can also cross the BB. It may even play a protective role in neurodegenerative diseases, but more studies are needed.
Acknowledgements
The authors thank the Wuhan University of Technology for permission to conduct this research, the supporting from the Shandong Chambroad Holding Group Co. LTD and Beijing Metalulu Botanmin Technology Co., LTD.
Abbreviations
- PRKACB
Protein kinase cAMP-activated catalytic subunit beta
- HDAC4
Histone deacetylase 4
- PKA
Protein kinase A
- CREB
Cyclic adenosine monophosphate response element-binding protein
- SAMP8
Senescence-accelerated mouse prone 8
- AMPK
Phosphorylation-dependent AMP-activated protein kinase
- MCI
Mild cognitive impairment
- HNF-4A
Hepatocyte nuclear factor 4α
- MCAO
Middle cerebral artery occlusion
- NOS1
Neuronal nitric oxide synthase
- MS
Multiple sclerosis
- EAE
Experimental autoimmune encephalomyelitis
- SOCS1
Suppressor of cytokine signaling
- TGFBR1
Transforming growth factor, beta receptor I
- OGD/R
Oxygen-glucose deprivation/reoxygenation
- I/R
Cerebral ischemia–reperfusion
- FLT3
Feline McDonough sarcoma-like tyrosine kinase 3
- CDK5
Calpain 1/p25/cyclin-dependent kinases 5
- STAT3
Signal transducer and activator of transcription 3
- Bim
Bcl-2-interacting mediator of cell death
- ERK
Extracellular signal-regulated kinase
- AMD
Age-related macular degeneration
- MG
Murine microglial
- PUMA
P53 upregulated modulator of apoptosis
- PBMCs
Peripheral blood mononuclear cells
- PDCD10
Programmed cell death 10
- 6-OHDA
6-Hydroxydopamine
- HMVEC
27 Hydroxycholesterols (27-OHC) on microvascular endothelial cell
- SH-SY5Y
Human neuroblastoma cells
- C1q
Primary protein of classical complement cascade
- RF1
Regulatory factor X1
- S6K1
Ribosomal protein S6 kinase B1
- IRS-1pSer
Phosphorylates insulin receptor substrate 1
- sGC
Soluble guanylate cyclase
- SNCA
Synuclein-alpha
- DUSP6
Dual specificity phosphatase 6
- PPP1CA
Protein phosphatase 1 catalytic subunit α isoform
- GSK-3β
Glycogen synthase kinase-3β
- CCH
Chronic cerebral hypoperfusion
- VaD
Vascular dementia
- mTOR
Mammalian target of rapamycin
- IRAK
Interleukin-1 receptor-associated kinase
- HAG cells
Human astroglial cells
- HSPC
Hematopoietic stem/progenitor cells
- PS1
Presenilin-1
- ADAM10
A disintegrin and metalloproteinase 10
- Nrf2
Nuclear factor-erythroid 2-related factor 2
- DCHG
Duke Center for Human Genetics
- BDNF
Brain-derived neurotrophic factor
- CDK5/PTEN/PPP2CA
Calpain 1/p25/cyclin-dependent kinases 5/phosphatase and tensin homolog deleted on chromosome ten/protein phosphatase 2 catalytic subunit alpha
- IFN-β
Interferon-β
- Dll1
Notch ligand delta-like 1
- LIMK-1
The p-Lin-11/Isl-1/Mec-3 kinases
- BACE1
Beta-site APP cleaving enzyme 1
- UTR
Untranslated region
- NC RNAs
Non-coding RNAs
- RISC
RNA-induced silencing complex
Author contribution
S.L. and T.S. conceived this study. Z.L., S.L., and T.S. designed and performed this research. S.L. and Z.L. wrote this review. Z.L. and T.S. improved the language. All authors reviewed this manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (No.32202857), the Fundamental Research Funds for the Central Universities (WUT: 2021IVA075) as well as the Project of Industry-school Cooperation and Collaborative Education (No.KGRCYJY2021005) and Grant (No.20212h0313) from Beijing Metalulu Botanmin Technology Co., LTD.
Data availability
All data generated or analyzed during this study are included in this published article.
Code availability
Not applicable.
Declarations
Ethics approval
All operation in this study was granted approval by the ethics board at the Wuhan University of Technology.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhixin Lei and Taolei Sun contributed equally to this paper.
Contributor Information
Zhixin Lei, Email: leizhixin@whut.edu.cn.
Taolei Sun, Email: suntl@whut.edu.cn.
References
- Abrahams S, Haylett WL, Johnson G, Carr JA, Bardien S. Antioxidant effects of curcumin in models of neurodegeneration, aging, oxidative and nitrosative stress: a review. Neuroscience. 2019;406:1–21. doi: 10.1016/j.neuroscience.2019.02.020. [DOI] [PubMed] [Google Scholar]
- Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. MiR-26b, upregulated in Alzheimer's disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci. 2013;33(37):14645–14659. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, di Val R, Cervo P, Melino G. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A. 2011;108(52):21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter R, Shao Y, Shaw M, Formica S, Khrestian M, Leverenz JB, Bekris LM. Regulation of ADAM10 by miR-140-5p and potential relevance for Alzheimer's disease. Neurobiol Aging. 2018;63:110–119. doi: 10.1016/j.neurobiolaging.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh M, Safarzadeh A, Beyranvand F, Ahmadpour F, Hajiasgharzadeh K, Baghbanzadeh A, Baradaran B. The potential role of miR-29 in health and cancer diagnosis, prognosis, and therapy. J Cell Physiol. 2019;234(11):19280–19297. doi: 10.1002/jcp.28607. [DOI] [PubMed] [Google Scholar]
- Alural B, Ozerdem A, Allmer J, Genc K, Genc S. Lithium protects against paraquat neurotoxicity by NRF2 activation and miR-34a inhibition in SH-SY5Y cells. Front Cell Neurosci. 2015;9:209. doi: 10.3389/fncel.2015.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlwinPremAnand A, Alvarez-Bolado G, Wizenmann A. MiR-9 and the midbrain-hindbrain boundary: a showcase for the limited functional conservation and regulatory complexity of microRNAs. Front Cell Dev Biol. 2020;8:586158. doi: 10.3389/fcell.2020.586158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer's A. 2016 Alzheimer's disease facts and figures. Alzheimers Dement. 2016;12(4):459–509. doi: 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Amoah SK, Rodriguez BA, Logothetis CN, Chander P, Sellgren CM, Weick JP, Mellios N. Exosomal secretion of a psychosis-altered miRNA that regulates glutamate receptor expression is affected by antipsychotics. Neuropsychopharmacology. 2020;45(4):656–665. doi: 10.1038/s41386-019-0579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranha MM, Santos DM, Sola S, Steer CJ, Rodrigues CM. miR-34a regulates mouse neural stem cell differentiation. PLoS ONE. 2011;6(8):e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121(4):645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Banzhaf-Strathmann J, Benito E, May S, Arzberger T, Tahirovic S, Kretzschmar H, Edbauer D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer's disease. EMBO J. 2014;33(15):1667–1680. doi: 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato C, Giacovazzo G, Albiero F, Scardigli R, Scopa C, Ciotti MT, Ruberti F. Cognitive decline and modulation of Alzheimer's disease-related genes after inhibition of microRNA-101 in mouse hippocampal neurons. Mol Neurobiol. 2020;57(7):3183–3194. doi: 10.1007/s12035-020-01957-8. [DOI] [PubMed] [Google Scholar]
- Barros-Viegas AT, Carmona V, Ferreiro E, Guedes J, Cardoso AM, Cunha P, Cardoso AL. miRNA-31 improves cognition and abolishes amyloid-beta pathology by targeting APP and BACE1 in an animal model of Alzheimer's disease. Mol Ther Nucleic Acids. 2020;19:1219–1236. doi: 10.1016/j.omtn.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bazrgar M, Khodabakhsh P, Prudencio M, Mohagheghi F, Ahmadiani A. The role of microRNA-34 family in Alzheimer's disease: a potential molecular link between neurodegeneration and metabolic disorders. Pharmacol Res. 2021;172:105805. doi: 10.1016/j.phrs.2021.105805. [DOI] [PubMed] [Google Scholar]
- Beal MF. Does impairment of energy metabolism result in excitotoxic neuronal death in neurodegenerative illnesses? Ann Neurol. 1992;31(2):119–130. doi: 10.1002/ana.410310202. [DOI] [PubMed] [Google Scholar]
- Becker D, Hirsch AG, Bender L, Lingner T, Salinas G, Krebber H. Nuclear Pre-snRNA export is an essential quality assurance mechanism for functional spliceosomes. Cell Rep. 2019;27(11):3199–3214 e3193. 10.1016/j.celrep.2019.05.031 [DOI] [PubMed]
- Bhattacharjee S, Zhao Y, Dua P, Rogaev EI, Lukiw WJ. microRNA-34a-mediated down-regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS One. 2016;11(3):e0150211. doi: 10.1371/journal.pone.0150211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bithell A, Johnson R, Buckley NJ. Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington's disease. Biochem Soc Trans. 2009;37(Pt 6):1270–1275. doi: 10.1042/BST0371270. [DOI] [PubMed] [Google Scholar]
- Blom-Dahl D, Azpiazu N. The Pax protein Eyegone (Eyg) interacts with the pi-RNA component Aubergine (Aub) and controls egg chamber development in Drosophila. Dev Biol. 2018;434(2):267–277. doi: 10.1016/j.ydbio.2017.12.012. [DOI] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284(4):1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20(1):19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31(7):577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- Braoudaki M, Lambrou GI. MicroRNAs in pediatric central nervous system embryonal neoplasms: the known unknown. J Hematol Oncol. 2015;8:6. doi: 10.1186/s13045-014-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley NJ, Johnson R, Zuccato C, Bithell A, Cattaneo E. The role of REST in transcriptional and epigenetic dysregulation in Huntington's disease. Neurobiol Dis. 2010;39(1):28–39. doi: 10.1016/j.nbd.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Cialic R, Krasemann S, Murugaiyan G, Fanek Z, Weiner HL. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann Neurol. 2015;77(1):75–99. doi: 10.1002/ana.24304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas R, Baez-Jurado E, Hidalgo-Lanussa O, Echeverria V, Ashrad GM, Sahebkar A, Barreto GE. Growth factors and neuroglobin in astrocyte protection against neurodegeneration and oxidative stress. Mol Neurobiol. 2019;56(4):2339–2351. doi: 10.1007/s12035-018-1203-9. [DOI] [PubMed] [Google Scholar]
- Caccamo A, Branca C, Piras IS, Ferreira E, Huentelman MJ, Liang WS, Oddo S. Necroptosis activation in Alzheimer's disease. Nat Neurosci. 2017;20(9):1236–1246. doi: 10.1038/nn.4608. [DOI] [PubMed] [Google Scholar]
- Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinforma. 2009;7(4):147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Cha DJ, Mengel D, Mustapic M, Liu W, Selkoe DJ, Kapogiannis D, Walsh DM. miR-212 and miR-132 are downregulated in neurally derived plasma exosomes of Alzheimer's patients. Front Neurosci. 2019;13:1208. doi: 10.3389/fnins.2019.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Wu YR, Chen CM. Down-regulation of miR-9* in the peripheral leukocytes of Huntington's disease patients. Orphanet J Rare Dis. 2017;12(1):185. doi: 10.1186/s13023-017-0742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapado LA, Martin-Hernandez R, Hernandez de la Red S, Tome-Carneiro J, Gil-Zamorano J, Ruiz-Roso MB, Davalos A. Connection between miRNA mediation and the bioactive effects of broccoli (Brassica oleracea var. italica): exogenous miRNA resistance to food processing and GI digestion. J Agric Food Chem. 2021;69(32):9326–9337. 10.1021/acs.jafc.1c04087 [DOI] [PubMed]
- Chen J, Wagner EJ. snRNA 3' end formation: the dawn of the Integrator complex. Biochem Soc Trans. 2010;38(4):1082–1087. doi: 10.1042/BST0381082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wu RZ, Zhu YQ, Ren ZM, Tong YL, Yang F, Dai GH. Study on the inhibition of Mfn1 by plant-derived miR5338 mediating the treatment of BPH with rape bee pollen. BMC Complement Altern Med. 2018;18(1):38. doi: 10.1186/s12906-018-2107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhu C, Jia W, Wang J, Gu L. MiR-455-5p attenuates cerebral ischemic reperfusion injury by targeting FLT3. J Cardiovasc Pharmacol. 2020;76(5):627–634. doi: 10.1097/FJC.0000000000000898. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhang F, Dong L, Wu H, Xu J, Li H, Zhang CY. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2021;31(3):247–258. doi: 10.1038/s41422-020-0389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Doecke JD, Sharples RA, Villemagne VL, Fowler CJ, Rembach A, Lifestyle Research G. Prognostic serum miRNA biomarkers associated with Alzheimer's disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry. 2015;20(10):1188–1196. doi: 10.1038/mp.2014.127. [DOI] [PubMed] [Google Scholar]
- Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu X, Wang SE. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26(2):217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielarz P, Konovalova J, Najam SS, Alter H, Piepponen TP, Erfle H, Domanskyi A. Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 2017;8(5):e2813. doi: 10.1038/cddis.2017.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, Cai H. MicroRNA-205 regulates the expression of Parkinson's disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013;22(3):608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, He L. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13(11):1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10(6):638–643. doi: 10.1016/s0959-437x(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' action through miRNA editing. Int J Mol Sci. 2019;20(24). 10.3390/ijms20246249 [DOI] [PMC free article] [PubMed]
- Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Welsh J\W. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst. 2016;108(1). 10.1093/jnci/djv303 [DOI] [PMC free article] [PubMed]
- Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J Biol Chem. 2010;285(50):38951–38960. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Munafo M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet. 2018;52:131–157. doi: 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhang W, Zhang H, Sun S, Yu H, Guo Y, Zhou Y. Modulation of HBV replication by microRNA-15b through targeting hepatocyte nuclear factor 1alpha. Nucleic Acids Res. 2014;42(10):6578–6590. doi: 10.1093/nar/gku260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci. 2012;15(5):697–699. doi: 10.1038/nn.3082. [DOI] [PubMed] [Google Scholar]
- de Antonellis P, Medaglia C, Cusanelli E, Andolfo I, Liguori L, De Vita G, Zollo M. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS One. 2011;6(9):e24584. doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lahunta A, Glass EN, Kent M. Embryonic development of the central nervous system. Vet Clin North Am Small Anim Pract. 2016;46(2):193–216. doi: 10.1016/j.cvsm.2015.10.011. [DOI] [PubMed] [Google Scholar]
- De Gregorio R, Pulcrano S, De Sanctis C, Volpicelli F, Guatteo E, von Oerthel L, Bellenchi GC. miR-34b/c regulates Wnt1 and enhances mesencephalic dopaminergic neuron differentiation. Stem Cell Reports. 2018;10(4):1237–1250. doi: 10.1016/j.stemcr.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo-Acebo L, de Las L, Hazas MC, Margolles A, Davalos A, Garcia-Ruiz A. Eating microRNAs: pharmacological opportunities for cross-kingdom regulation and implications in host gene and gut microbiota modulation. Br J Pharmacol. 2021;178(11):2218–2245. doi: 10.1111/bph.15421. [DOI] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6(4):323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias IHK, Brown CL, Shabir K, Polidori MC, Griffiths HR. miRNA 933 expression by endothelial cells is increased by 27-hydroxycholesterol and is more prevalent in plasma from dementia patients. J Alzheimers Dis. 2018;64(3):1009–1017. doi: 10.3233/JAD-180201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz NF, Cruz-Resendiz MS, Flores-Herrera H, Garcia-Lopez G, Molina-Hernandez A. MicroRNAs in central nervous system development. Rev Neurosci. 2014;25(5):675–686. doi: 10.1515/revneuro-2014-0014. [DOI] [PubMed] [Google Scholar]
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285(17):12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essandoh K, Li Y, Huo J, Fan GC. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46(2):122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19(6):586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Fakhoury M. Immune-mediated processes in neurodegeneration: where do we stand? J Neurol. 2016;263(9):1683–1701. doi: 10.1007/s00415-016-8052-0. [DOI] [PubMed] [Google Scholar]
- Fang J, Barker B, Bolanos L, Liu X, Jerez A, Makishima H, Starczynowski DT. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-kappaB gene network. Cell Rep. 2014;8(5):1328–1338. doi: 10.1016/j.celrep.2014.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CZ, Yin JB, Yang JJ, Cao L. Regulatory factor X1 depresses ApoE-dependent Abeta uptake by miRNA-124 in microglial response to oxidative stress. Neuroscience. 2017;344:217–228. doi: 10.1016/j.neuroscience.2016.12.017. [DOI] [PubMed] [Google Scholar]
- Feng MG, Liu CF, Chen L, Feng WB, Liu M, Hai H, Lu JM. MiR-21 attenuates apoptosis-triggered by amyloid-beta via modulating PDCD4/ PI3K/AKT/GSK-3beta pathway in SH-SY5Y cells. Biomed Pharmacother. 2018;101:1003–1007. doi: 10.1016/j.biopha.2018.02.043. [DOI] [PubMed] [Google Scholar]
- Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Takahashi M, Fujita H, Chiyo T, Popiel HA, Watanabe S, Hohjoh H. Supplemental treatment for Huntington's disease with miR-132 that is deficient in Huntington's disease brain. Mol Ther Nucleic Acids. 2018;11:79–90. doi: 10.1016/j.omtn.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JD, Kageyama R, Shehata HM, Fassett MS, Mar DJ, Wigton EJ, Ansel KM. miR-15/16 restrain memory T cell differentiation, cell cycle, and survival. Cell Rep. 2019;28(8):2169–2181 e2164. 10.1016/j.celrep.2019.07.064 [DOI] [PMC free article] [PubMed]
- Gao JX, Li Y, Wang SN, Chen XC, Lin LL, Zhang H. Overexpression of microRNA-183 promotes apoptosis of substantia nigra neurons via the inhibition of OSMR in a mouse model of Parkinson's disease. Int J Mol Med. 2019;43(1):209–220. doi: 10.3892/ijmm.2018.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Qiao H, Lu Z, Hou Y. miR29 promotes the proliferation of cultured rat neural stem/progenitor cells via the PTEN/AKT signaling pathway. Mol Med Rep. 2019;20(3):2111–2118. doi: 10.3892/mmr.2019.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet AD, Fonken LK, Watkins LR, Nelson RJ, Popovich PG. MicroRNAs: roles in regulating neuroinflammation. Neuroscientist. 2018;24(3):221–245. doi: 10.1177/1073858417721150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Yan Z, Zhu H, Zhao H. MiR-410 exerts neuroprotective effects in a cellular model of Parkinson's disease induced by 6-hydroxydopamine via inhibiting the PTEN/AKT/mTOR signaling pathway. Exp Mol Pathol. 2019;109:16–24. doi: 10.1016/j.yexmp.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Giusti SA, Vogl AM, Brockmann MM, Vercelli CA, Rein ML, Trumbach D, Refojo D. MicroRNA-9 controls dendritic development by targeting REST. Elife. 2014;3.10.7554/eLife.02755 [DOI] [PMC free article] [PubMed]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–144. 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed]
- Gupta A, Andresen JL, Manan RS, Langer R. Nucleic acid delivery for therapeutic applications. Adv Drug Deliv Rev. 2021;178:113834. doi: 10.1016/j.addr.2021.113834. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hecker M, Thamilarasan M, Koczan D, Schroder I, Flechtner K, Freiesleben S, Zettl UK. MicroRNA expression changes during interferon-beta treatment in the peripheral blood of multiple sclerosis patients. Int J Mol Sci. 2013;14(8):16087–16110. doi: 10.3390/ijms140816087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemels MT. Neurodegenerative diseases. Nature. 2016;539(7628):179. doi: 10.1038/539179a. [DOI] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Higaki S, Muramatsu M, Matsuda A, Matsumoto K, Satoh JI, Michikawa M, Niida S. Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer's disease models. PLoS ONE. 2018;13(5):e0196929. doi: 10.1371/journal.pone.0196929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PG, Lehman N. The RNA World: molecular cooperation at the origins of life. Nat Rev Genet. 2015;16(1):7–17. doi: 10.1038/nrg3841. [DOI] [PubMed] [Google Scholar]
- Hines MJ, Coffre M, Mudianto T, Panduro M, Wigton EJ, Tegla C, Koralov SB. miR-29 sustains B cell survival and controls terminal differentiation via regulation of PI3K signaling. Cell Rep. 2020;33(9):108436. doi: 10.1016/j.celrep.2020.108436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou D, He F, Ma L, Cao M, Zhou Z, Wei Z, Jiang X. The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J Nutr Biochem. 2018;57:197–205. doi: 10.1016/j.jnutbio.2018.03.026. [DOI] [PubMed] [Google Scholar]
- Hu L, Zhang R, Yuan Q, Gao Y, Yang MQ, Zhang C, Hu X. The emerging role of microRNA-4487/6845-3p in Alzheimer's disease pathologies is induced by Abeta25-35 triggered in SH-SY5Y cell. BMC Syst Biol. 2018;12(Suppl 7):119. doi: 10.1186/s12918-018-0633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WY, Jiang C, Ye HB, Jiao JT, Cheng C, Huang J, Shao JF. miR-124 upregulates astrocytic glutamate transporter-1 via the Akt and mTOR signaling pathway post ischemic stroke. Brain Res Bull. 2019;149:231–239. doi: 10.1016/j.brainresbull.2019.04.013. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu H, Sun X, Ding M, Tao G, Li X. Honeysuckle-derived microRNA2911 directly inhibits varicella-zoster virus replication by targeting IE62 gene. J Neurovirol. 2019;25(4):457–463. doi: 10.1007/s13365-019-00741-2. [DOI] [PubMed] [Google Scholar]
- Hurlock EC, McMahon A, Joho RH. Purkinje-cell-restricted restoration of Kv3.3 function restores complex spikes and rescues motor coordination in Kcnc3 mutants. J Neurosci. 2008;28(18):4640–4648. 10.1523/JNEUROSCI.5486-07.2008 [DOI] [PMC free article] [PubMed]
- Hutter K, Rulicke T, Drach M, Andersen L, Villunger A, Herzog S. Differential roles of miR-15a/16-1 and miR-497/195 clusters in immune cell development and homeostasis. FEBS J. 2021;288(5):1533–1545. doi: 10.1111/febs.15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JY, Zukin RS. REST, a master transcriptional regulator in neurodegenerative disease. Curr Opin Neurobiol. 2018;48:193–200. doi: 10.1016/j.conb.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidou C, Turski L. Neurodegenerative disorders: clues from glutamate and energy metabolism. Crit Rev Neurobiol. 1996;10(2):239–263. doi: 10.1615/critrevneurobiol.v10.i2.50. [DOI] [PubMed] [Google Scholar]
- Jain S, Pathak K, Vaidya A. Molecular therapy using siRNA: recent trends and advances of multi target inhibition of cancer growth. Int J Biol Macromol. 2018;116:880–892. doi: 10.1016/j.ijbiomac.2018.05.077. [DOI] [PubMed] [Google Scholar]
- Jauhari A, Yadav S. MiR-34 and MiR-200: regulator of cell fate plasticity and neural development. Neuromolecular Med. 2019;21(2):97–109. doi: 10.1007/s12017-019-08535-9. [DOI] [PubMed] [Google Scholar]
- Jauhari A, Singh T, Singh P, Parmar D, Yadav S. Regulation of miR-34 family in neuronal development. Mol Neurobiol. 2018;55(2):936–945. doi: 10.1007/s12035-016-0359-4. [DOI] [PubMed] [Google Scholar]
- Javed M, Solanki M, Sinha A, Shukla LI. Position based nucleotide analysis of miR168 family in higher plants and its targets in mammalian transcripts. Microrna. 2017;6(2):136–142. doi: 10.2174/2211536606666170215154151. [DOI] [PubMed] [Google Scholar]
- Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26(5):293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Kong L, Yao Y, Li S, Tao Z, Yan Y, Yang J. Osthole decreases beta amyloid levels through up-regulation of miR-107 in Alzheimer's disease. Neuropharmacology. 2016;108:332–344. doi: 10.1016/j.neuropharm.2016.04.046. [DOI] [PubMed] [Google Scholar]
- Jiao H, Chen R, Jiang Z, Zhang L, Wang H. miR-22 protect PC12 from ischemia/reperfusion-induced injury by targeting p53 upregulated modulator of apoptosis (PUMA) Bioengineered. 2020;11(1):209–218. doi: 10.1080/21655979.2020.1729321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Buckley NJ. Gene dysregulation in Huntington's disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11(3):183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008;29(3):438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Juzwik CA, S SD, Zhang Y, Paradis-Isler N, Sylvester A, Amar-Zifkin A, Fournier AE. microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog Neurobiol. 2019;182:101664. 10.1016/j.pneurobio.2019.101664 [DOI] [PubMed]
- Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S, Seraphin B. Involvement of U6 snRNA in 5' splice site selection. Science. 1993;262(5142):2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Kang Q, Xiang Y, Li D, Liang J, Zhang X, Zhou F, Li Y. MiR-124–3p attenuates hyperphosphorylation of Tau protein-induced apoptosis via caveolin-1-PI3K/Akt/GSK3beta pathway in N2a/APP695swe cells. Oncotarget. 2017;8(15):24314–24326. 10.18632/oncotarget.15149 [DOI] [PMC free article] [PubMed]
- Khafaei M, Rezaie E, Mohammadi A, Shahnazi Gerdehsang P, Ghavidel S, Kadkhoda S, Tavallaie M. miR-9: from function to therapeutic potential in cancer. J Cell Physiol. 2019 doi: 10.1002/jcp.28210. [DOI] [PubMed] [Google Scholar]
- Khezri MR, Yousefi K, Zolbanin NM, Ghasemnejad-Berenji M. MicroRNAs in the pathophysiology of Alzheimer's disease and Parkinson's disease: an overview. Mol Neurobiol. 2022;59(3):1589–1603. doi: 10.1007/s12035-022-02727-4. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lee Y, McKenna ND, Yi M, Simunovic F, Wang Y, Sonntag KC. miR-126 contributes to Parkinson's disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiol Aging. 2014;35(7):1712–1721. doi: 10.1016/j.neurobiolaging.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Fiesel FC, Belmonte KC, Hudec R, Wang WX, Kim C, Kim J. miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1) Mol Neurodegener. 2016;11(1):55. doi: 10.1186/s13024-016-0121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Kim EJ, Lee S, Tan X, Liu X, Park S, Ahn YH. MiR-34a and miR-34b/c have distinct effects on the suppression of lung adenocarcinomas. Exp Mol Med. 2019;51(1):1–10. doi: 10.1038/s12276-018-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Russell AE, Wang W, Gemoets DE, Sarkar SN, Simpkins JW, Brown CM. miR-146a dysregulates energy metabolism during neuroinflammation. J Neuroimmune Pharmacol. 2021 doi: 10.1007/s11481-021-09999-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyucherev TO, Olszewski P, Shalimova AA, Chubarev VN, Tarasov VV, Attwood MM, Schioth HB. Advances in the development of new biomarkers for Alzheimer's disease. Transl Neurodegener. 2022;11(1):25. doi: 10.1186/s40035-022-00296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Wu J, Zhang D, Wan C, Yuan L. The role of miR-124 in Drosophila Alzheimer's disease model by targeting delta in notch signaling pathway. Curr Mol Med. 2015;15(10):980–989. doi: 10.2174/1566524016666151123114608. [DOI] [PubMed] [Google Scholar]
- Konovalova J, Gerasymchuk D, Parkkinen I, Chmielarz P, Domanskyi A. Interplay between MicroRNAs and oxidative stress in neurodegenerative diseases. Int J Mol Sci. 2019;20(23). 10.3390/ijms20236055 [DOI] [PMC free article] [PubMed]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics. 2012;44(4):237–244. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lane CA, Hardy J, Schott JM. Alzheimer's disease. Eur J Neurol. 2018;25(1):59–70. doi: 10.1111/ene.13439. [DOI] [PubMed] [Google Scholar]
- Lang C, Karunairetnam S, Lo KR, Kralicek AV, Crowhurst RN, Gleave AP, Ingram JR. Common variants of the plant microRNA-168a exhibit differing silencing efficacy for human low-density lipoprotein receptor adaptor protein 1 (LDLRAP1) Microrna. 2019;8(2):166–170. doi: 10.2174/2211536608666181203103233. [DOI] [PubMed] [Google Scholar]
- Lardelli RM, Lykke-Andersen J. Competition between maturation and degradation drives human snRNA 3' end quality control. Genes Dev. 2020;34(13–14):989–1001. doi: 10.1101/gad.336891.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel HA, Petre BM, Wall J, Simon M, Nowak RJ, Walz T, Lansbury PT., Jr Alpha-synuclein, especially the Parkinson's disease-associated mutants, forms pore-like annular and tubular protofibrils. J Mol Biol. 2002;322(5):1089–1102. doi: 10.1016/s0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- Lee Y, Samaco RC, Gatchel JR, Thaller C, Orr HT, Zoghbi HY. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat Neurosci. 2008;11(10):1137–1139. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Im W, Ban JJ, Lee M, Jung KH, Lee SK, Kim M. Exosome-based delivery of miR-124 in a Huntington's Disease model. J Mov Disord. 2017;10(1):45–52. 10.14802/jmd.16054 [DOI] [PMC free article] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li T, Le W. Biomarkers for Parkinson's disease: how good are they? Neurosci Bull. 2020;36(2):183–194. doi: 10.1007/s12264-019-00433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Jiang Y, Wang Y, Yang S, Bi X, Pan X, Li W. MiR-181b regulates autophagy in a model of Parkinson's disease by targeting the PTEN/Akt/mTOR signaling pathway. Neurosci Lett. 2018;675:83–88. doi: 10.1016/j.neulet.2018.03.041. [DOI] [PubMed] [Google Scholar]
- Li X, Huang Y, Sun M, Ji H, Dou H, Hu J, Chen L. Honeysuckle-encoded microRNA2911 inhibits Enterovirus 71 replication via targeting VP1 gene. Antiviral Res. 2018;152:117–123. doi: 10.1016/j.antiviral.2018.02.015. [DOI] [PubMed] [Google Scholar]
- Li M, Chen T, Wang R, Luo JY, He JJ, Ye RS, Zhang YL. Plant MIR156 regulates intestinal growth in mammals by targeting the Wnt/beta-catenin pathway. Am J Physiol Cell Physiol. 2019;317(3):C434–C448. doi: 10.1152/ajpcell.00030.2019. [DOI] [PubMed] [Google Scholar]
- Li Q, Liu T, Yang S, Zhang Z. Upregulation of miR-34a by inhibition of IRE1alpha has protective effect against abeta-induced injury in SH-SY5Y cells by targeting caspase-2. Oxid Med Cell Longev. 2019;2019:2140427. doi: 10.1155/2019/2140427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang H. miR-15b reduces amyloid-beta accumulation in SH-SY5Y cell line through targetting NF-kappaB signaling and BACE1. Biosci Rep. 2018;38(6). 10.1042/BSR20180051. [DOI] [PMC free article] [PubMed]
- Li Q, Wang Y, Peng W, Jia Y, Tang J, Li W, Yang J. MicroRNA-101a regulates autophagy phenomenon via the MAPK pathway to modulate Alzheimer's-associated pathogenesis. Cell Transplant. 2019;28(8):1076–1084. doi: 10.1177/0963689719857085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RJ, Liu Y, Liu HQ, Li J. Ketogenic diets and protective mechanisms in epilepsy, metabolic disorders, cancer, neuronal loss, and muscle and nerve degeneration. J Food Biochem. 2020;44(3):e13140. doi: 10.1111/jfbc.13140. [DOI] [PubMed] [Google Scholar]
- Li S, Lv X, Zhai K, Xu R, Zhang Y, Zhao S, Lou J. MicroRNA-7 inhibits neuronal apoptosis in a cellular Parkinson's disease model by targeting Bax and Sirt2. Am J Transl Res. (2016;8(2):993–1004. https://www.ncbi.nlm.nih.gov/pubmed/27158385 [PMC free article] [PubMed]
- Li M, Chen T, He JJ, Wu JH, Luo JY, Ye RS, Zhang YL. Plant MIR167e-5p inhibits enterocyte proliferation by targeting beta-catenin. Cells. 2019a;8(11). 10.3390/cells8111385 [DOI] [PMC free article] [PubMed]
- Liang H, Zen K, Zhang J, Zhang CY, Chen X. New roles for microRNAs in cross-species communication. RNA Biol. 2013;10(3):367–370. doi: 10.4161/rna.23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhu Y, Sun B, Shao Y, Jing A, Wang J, Xiao Z. Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr. 2014;2(4):380–388. doi: 10.1002/fsn3.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Lohmuller M, Kotkamp B, Schuler F, Knust Z, Villunger A, Herzog S. The miR-15 family reinforces the transition from proliferation to differentiation in pre-B cells. EMBO Rep. 2017;18(9):1604–1617. 10.15252/embr.201643735 [DOI] [PMC free article] [PubMed]
- Link J, Thon C, Schanze D, Steponaitiene R, Kupcinskas J, Zenker M, Link A. Food-derived Xeno-microRNAs: influence of diet and detectability in gastrointestinal tract-proof-of-principle study. Mol Nutr Food Res. 2019;63(2):e1800076. doi: 10.1002/mnfr.201800076. [DOI] [PubMed] [Google Scholar]
- Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Bonini NM. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang C, Wang X, Xu S. Therapeutic effects of transplantation of As-MiR-937-expressing mesenchymal stem cells in murine model of Alzheimer's disease. Cell Physiol Biochem. 2015;37(1):321–330. doi: 10.1159/000430356. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhao J, Lu G. miR-106b inhibits tau phosphorylation at Tyr18 by targeting Fyn in a model of Alzheimer's disease. Biochem Biophys Res Commun. 2016;478(2):852–857. doi: 10.1016/j.bbrc.2016.08.037. [DOI] [PubMed] [Google Scholar]
- Liu D, Tang H, Li XY, Deng MF, Wei N, Wang X, Zhu LQ. Targeting the HDAC2/HNF-4A/miR-101b/AMPK pathway rescues tauopathy and dendritic abnormalities in Alzheimer's disease. Mol Ther. 2017;25(3):752–764. doi: 10.1016/j.ymthe.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Chen WL, Kung WH, Huang HD. Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genomics. 2017;18(Suppl 2):112. doi: 10.1186/s12864-017-3502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Liu P, Wang Z, Fang S, Liu Y, Wang J, Wang L. Inhibition of microRNA-96 ameliorates cognitive impairment and inactivation autophagy following chronic cerebral hypoperfusion in the rat. Cell Physiol Biochem. 2018;49(1):78–86. doi: 10.1159/000492844. [DOI] [PubMed] [Google Scholar]
- Liu HY, Fu X, Li YF, Li XL, Ma ZY, Zhang Y, Gao QC. miR-15b-5p targeting amyloid precursor protein is involved in the anti-amyloid eflect of curcumin in swAPP695-HEK293 cells. Neural Regen Res. 2019;14(9):1603–1609. doi: 10.4103/1673-5374.255979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, Xu W. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18(1):123. doi: 10.1186/s12943-019-1052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Miao Y, Zhang L, Xu X, Luan Q. MiR-211 protects cerebral ischemia/reperfusion injury by inhibiting cell apoptosis. Bioengineered. 2020;11(1):189–200. doi: 10.1080/21655979.2020.1729322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xu M, Yan L, Wang Y, Zhou Z, Wang S, Dong L. Honeysuckle-derived microRNA2911 inhibits tumor growth by targeting TGF-beta1. Chin Med. 2021;16(1):49. doi: 10.1186/s13020-021-00453-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang Y. [Cellular function of microRNA-15 family]. Sheng Li Xue Bao. 2012;64(1):101–106. https://www.ncbi.nlm.nih.gov/pubmed/22348968 [PubMed]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu W, Lin J, Zheng D, Hong C, Ke L, Wu X, Chen P. Overexpression of microRNA-133a inhibits apoptosis and autophagy in a cell model of Parkinson's disease by downregulating Ras-related C3 botulinum toxin substrate 1 (RAC1). Med Sci Monit. 2020;26:e922032. 10.12659/MSM.922032 [DOI] [PMC free article] [PubMed] [Retracted]
- Lukasik A, Brzozowska I, Zielenkiewicz U, Zielenkiewicz P. Detection of plant miRNAs abundance in human breast milk. Int J Mol Sci. 2017;19(1). 10.3390/ijms19010037 [DOI] [PMC free article] [PubMed]
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84(1):55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- Mallo M, Alonso CR. The regulation of Hox gene expression during animal development. Development. 2013;140(19):3951–3963. doi: 10.1242/dev.068346. [DOI] [PubMed] [Google Scholar]
- Marogianni C, Sokratous M, Dardiotis E, Hadjigeorgiou GM, Bogdanos D, Xiromerisiou G. Neurodegeneration and inflammation-an interesting interplay in Parkinson's disease. Int J Mol Sci. 2020;21(22). 10.3390/ijms21228421 [DOI] [PMC free article] [PubMed]
- Marzano F, Caratozzolo MF, Consiglio A, Licciulli F, Liuni S, Sbisa E, Catalano D. Plant miRNAs reduce cancer cell proliferation by targeting MALAT1 and NEAT1: a beneficial cross-kingdom interaction. Front Genet. 2020;11:552490. doi: 10.3389/fgene.2020.552490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. 10.1093/hmg/ddl046 [DOI] [PubMed]
- McColgan P, Tabrizi SJ. Huntington's disease: a clinical review. Eur J Neurol. 2018;25(1):24–34. doi: 10.1111/ene.13413. [DOI] [PubMed] [Google Scholar]
- Meldrum BS. Glutamate as a neurotransmitter in the brain: review of physiology and pathology. J Nutr. 2000;130(4S Suppl):1007S–1015S. doi: 10.1093/jn/130.4.1007S. [DOI] [PubMed] [Google Scholar]
- Milo R, Korczyn AD, Manouchehri N, Stuve O. The temporal and causal relationship between inflammation and neurodegeneration in multiple sclerosis. Mult Scler. 2020;26(8):876–886. doi: 10.1177/1352458519886943. [DOI] [PubMed] [Google Scholar]
- Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncini S, Lunghi M, Valmadre A, Grasso M, Del Vescovo V, Riva P, Venturin M. The miR-15/107 family of microRNA genes regulates CDK5R1/p35 with implications for Alzheimer's disease pathogenesis. Mol Neurobiol. 2017;54(6):4329–4342. doi: 10.1007/s12035-016-0002-4. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16(3):223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Nampoothiri SS, Rajanikant GK. miR-9 upregulation integrates post-ischemic neuronal survival and regeneration in vitro. Cell Mol Neurobiol. 2019;39(2):223–240. doi: 10.1007/s10571-018-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal M, Richardson JR. Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. Biochim Biophys Acta Mol Basis Dis. 2018;1864(2):432–443. doi: 10.1016/j.bbadis.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikam RR, Gore KR. Journey of siRNA: clinical developments and targeted delivery. Nucleic Acid Ther. 2018;28(4):209–224. doi: 10.1089/nat.2017.0715. [DOI] [PubMed] [Google Scholar]
- O'Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer's disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir M, Hacohen D, Ginsberg D. MiR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol Cancer Res. 2011;9(4):440–447. doi: 10.1158/1541-7786.MCR-10-0344. [DOI] [PubMed] [Google Scholar]
- Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31(3):809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Xu L, Lu Y, Sun X, Yue S, Xiong XX, Giffard RG. Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to forebrain ischemia. Glia. 2013;61(11):1784–1794. doi: 10.1002/glia.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28(53):14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech. 2020;1863(6):194417. doi: 10.1016/j.bbagrm.2019.194417. [DOI] [PubMed] [Google Scholar]
- Papadopoulou AS, Serneels L, Achsel T, Mandemakers W, Callaerts-Vegh Z, Dooley J, De Strooper B. Deficiency of the miR-29a/b-1 cluster leads to ataxic features and cerebellar alterations in mice. Neurobiol Dis. 2015;73:275–288. doi: 10.1016/j.nbd.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Parsi S, Smith PY, Goupil C, Dorval V, Hebert SS. Preclinical evaluation of miR-15/107 family members as multifactorial drug targets for Alzheimer's disease. Mol Ther Nucleic Acids. 2015;4:e256. doi: 10.1038/mtna.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Peng T, Liu X, Wang J, Liu Y, Fu Z, Ma X, Lu H. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson's disease via regulating miR-100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif Cells Nanomed Biotechnol. 2019;47(1):2764–2774. doi: 10.1080/21691401.2019.1636805. [DOI] [PubMed] [Google Scholar]
- Qian Q, Zhang J, He FP, Bao WX, Zheng TT, Zhou DM, Peng GP. Down-regulated expression of microRNA-338-5p contributes to neuropathology in Alzheimer's disease. FASEB J. 2019;33(3):4404–4417. doi: 10.1096/fj.201801846R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Yau WM, Lu JX, Collinge J, Tycko R. Structural variation in amyloid-beta fibrils from Alzheimer's disease clinical subtypes. Nature. 2017;541(7636):217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin LX, Tan JQ, Zhang HN, Tang JG, Jiang B, Shen XM, Wang CY. Preliminary study of hsa-miR-626 change in the cerebrospinal fluid of Parkinson's disease patients. J Clin Neurosci. 2019;70:198–201. doi: 10.1016/j.jocn.2019.08.082. [DOI] [PubMed] [Google Scholar]
- Qin X, Zhang X, Li P, Wang M, Yan L, Pan P, Bao Z. MicroRNA-185 activates PI3K/AKT signalling pathway to alleviate dopaminergic neuron damage via targeting IGF1 in Parkinson's disease. J Drug Target. 2021;29(8):875–883. doi: 10.1080/1061186X.2021.1886300. [DOI] [PubMed] [Google Scholar]
- Quinlan S, Kenny A, Medina M, Engel T, Jimenez-Mateos EM. MicroRNAs in neurodegenerative diseases. Int Rev Cell Mol Biol. 2017;334:309–343. doi: 10.1016/bs.ircmb.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Rajendra W, Armugam A, Jeyaseelan K. Neuroprotection and peptide toxins. Brain Res Brain Res Rev. 2004;45(2):125–141. doi: 10.1016/j.brainresrev.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Re A, Joshi T, Kulberkyte E, Morris Q, Workman CT. RNA-protein interactions: an overview. Methods Mol Biol. 2014;1097:491–521. doi: 10.1007/978-1-62703-709-9_23. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Williams J, Smith F, Bhatti JS, Kumar S, Vijayan M, Reddy AP. MicroRNAs, aging, cellular senescence, and Alzheimer's disease. Prog Mol Biol Transl Sci. 2017;146:127–171. doi: 10.1016/bs.pmbts.2016.12.009. [DOI] [PubMed] [Google Scholar]
- Reniewicz P, Zyzak J, Siednienko J. The cellular receptors of exogenous RNA. Postepy Hig Med Dosw (online) 2016;70:337–348. doi: 10.5604/17322693.1199987(KomorkowereceptoryegzogennegoRNA.). [DOI] [PubMed] [Google Scholar]
- Rong W, Yang L, Li CY, Wu XT, Zhou ZD, Zhu WL, Yan Y. MiR-29 inhibits neuronal apoptosis in rats with cerebral infarction through regulating Akt signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(2):843–850. 10.26355/eurrev_202001_20068 [DOI] [PubMed]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- Salama RM, Abdel-Latif GA, Abbas SS, El Magdoub HM, Schaalan MF. Neuroprotective effect of crocin against rotenone-induced Parkinson's disease in rats: interplay between PI3K/Akt/mTOR signaling pathway and enhanced expression of miRNA-7 and miRNA-221. Neuropharmacology. 2020;164:107900. doi: 10.1016/j.neuropharm.2019.107900. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204(7):1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedighi M, Baluchnejadmojarad T, Fallah S, Moradi N, Afshin-Majdd S, Roghani M. Klotho ameliorates cellular inflammation via suppression of cytokine release and upregulation of miR-29a in the PBMCs of diagnosed Alzheimer's disease patients. J Mol Neurosci. 2019;69(1):157–165. doi: 10.1007/s12031-019-01345-5. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok H, Lee H, Jang ES, Chi SW. Evaluation and control of miRNA-like off-target repression for RNA interference. Cell Mol Life Sci. 2018;75(5):797–814. doi: 10.1007/s00018-017-2656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459(2):100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Sharma A, Sahu S, Kumari P, Gopi SR, Malhotra R, Biswas S. Genome-wide identification and functional annotation of miRNAs in anti-inflammatory plant and their cross-kingdom regulation in Homo sapiens. J Biomol Struct Dyn. 2017;35(7):1389–1400. doi: 10.1080/07391102.2016.1185381. [DOI] [PubMed] [Google Scholar]
- Sharma S, Pavlasova GM, Seda V, Cerna KA, Vojackova E, Filip D, Mraz M. miR-29 modulates CD40 signaling in chronic lymphocytic leukemia by targeting TRAF4: an axis affected by BCR inhibitors. Blood. 2021;137(18):2481–2494. doi: 10.1182/blood.2020005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YF, Zhu ZY, Qian SX, Xu CY, Wang YP. miR-30b protects nigrostriatal dopaminergic neurons from MPP(+)-induced neurotoxicity via SNCA. Brain Behav. 2020;10(4):e01567. doi: 10.1002/brb3.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Zhang K, Zhou H, Jiang L, Xie B, Wang R, Xu S. Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer's Disease. Aging Cell. 2020;19(3):e13125. doi: 10.1111/acel.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31(9):3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu P, Wu C, Liu W, Ruan X, Liu C, Hou L, Peng X. The spatiotemporal expression pattern of microRNAs in the developing mouse nervous system. J Biol Chem. 2019;294(10):3444–3453. doi: 10.1074/jbc.RA118.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierksma A, Lu A, Salta E, Vanden Eynden E, Callaerts-Vegh Z, D'Hooge R, De Strooper B. Deregulation of neuronal miRNAs induced by amyloid-beta or TAU pathology. Mol Neurodegener. 2018;13(1):54. doi: 10.1186/s13024-018-0285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Kooter JM. Post-transcriptional gene-silencing: RNAs on the attack or on the defense? BioEssays. 2000;22(6):520–531. doi: 10.1002/(SICI)1521-1878(200006)22:6<520::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, Mavrikis Cox G, Whitacre CC. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189(4):1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PY, Hernandez-Rapp J, Jolivette F, Lecours C, Bisht K, Goupil C, Hebert SS. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum Mol Genet. 2015;24(23):6721–6735. doi: 10.1093/hmg/ddv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Kim YK. Identification of the role of miR-142-5p in Alzheimer's disease by comparative bioinformatics and cellular analysis. Front Mol Neurosci. 2017;10:227. doi: 10.3389/fnmol.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Rossi JJ. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J. 2017;474(10):1603–1618. doi: 10.1042/BCJ20160759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag KC. MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res. 2010;1338:48–57. doi: 10.1016/j.brainres.2010.03.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35(2):169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson J, Nutma E, van der Valk P, Amor S. Inflammation in CNS Neurodegenerative Diseases. Immunology. 2018;154(2):204–219. doi: 10.1111/imm.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Yang X, Lou J. Geniposide reduces alpha-synuclein by blocking microRNA-21/lysosome-associated membrane protein 2A interaction in Parkinson disease models. Brain Res. 2016;1644:98–106. doi: 10.1016/j.brainres.2016.05.011. [DOI] [PubMed] [Google Scholar]
- Sullivan RP, Leong JW, Schneider SE, Ireland AR, Berrien-Elliott MM, Singh A, Fehniger TA. MicroRNA-15/16 antagonizes Myb to control NK cell maturation. J Immunol. 2015;195(6):2806–2817. doi: 10.4049/jimmunol.1500949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Han X, Li X, Song Q, Lu M, Jia M, Hu G. MicroRNA-212-5p prevents dopaminergic neuron death by inhibiting SIRT2 in MPTP-induced mouse model of Parkinson's disease. Front Mol Neurosci. 2018;11:381. doi: 10.3389/fnmol.2018.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang T, Xiu W, Cao W, He M, Sun W, Zhao W. MiR-107 overexpression attenuates neurotoxicity induced by 6-hydroxydopamine both in vitro and in vivo. Chem Biol Interact. 2020;315:108908. doi: 10.1016/j.cbi.2019.108908. [DOI] [PubMed] [Google Scholar]
- Swahari V, Nakamura A, Hollville E, Stroud H, Simon JM, Ptacek TS, Deshmukh M. MicroRNA-29 is an essential regulator of brain maturation through regulation of CH methylation. Cell Rep. 2021;35(1):108946. doi: 10.1016/j.celrep.2021.108946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebi F, Ghorbani S, Chan WF, Boghozian R, Masoumi F, Ghasemi S, Noorbakhsh F. MicroRNA-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J Neuroinflammation. 2017;14(1):55. doi: 10.1186/s12974-017-0832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SL, Ohtsuka T, Gonzalez A, Kageyama R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells. 2012;17(12):952–961. doi: 10.1111/gtc.12009. [DOI] [PubMed] [Google Scholar]
- Tang H, Ma M, Wu Y, Deng MF, Hu F, Almansoub H, Zhu LQ. Activation of MT2 receptor ameliorates dendritic abnormalities in Alzheimer's disease via C/EBPalpha/miR-125b pathway. Aging Cell. 2019;18(2):e12902. doi: 10.1111/acel.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzani A, French J, Bartfai T, Baram TZ. The role of inflammation in epilepsy. Nat Rev Neurol. 2011;7(1):31–40. doi: 10.1038/nrneurol.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21(7):744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang L. Circulating exosomal miRNA as diagnostic biomarkers of neurodegenerative diseases. Front Mol Neurosci. 2020;13:53. doi: 10.3389/fnmol.2020.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82(2):283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28(5):1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu JT, Tan L, Tian Y, Ma J, Tan CC, Tan L. Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci Rep. 2015;5:9522. doi: 10.1038/srep09522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tan L, Lu Y, Peng J, Zhu Y, Zhang Y, Sun Z. MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 2015;589(6):726–729. doi: 10.1016/j.febslet.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Wang CN, Wang YJ, Wang H, Song L, Chen Y, Wang JL, Jiang B. The anti-dementia effects of Donepezil involve miR-206-3p in the hippocampus and cortex. Biol Pharm Bull. 2017;40(4):465–472. doi: 10.1248/bpb.b16-00898. [DOI] [PubMed] [Google Scholar]
- Wang Y, Veremeyko T, Wong AH, El Fatimy R, Wei Z, Cai W, Krichevsky AM. Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer's disease. Neurobiol Aging. 2017;51:156–166. doi: 10.1016/j.neurobiolaging.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu J, Wang Q, Jiang H, Zeng L, Li Z, Liu R. MicroRNA-200a-3p mediates neuroprotection in Alzheimer-related deficits and attenuates amyloid-beta overproduction and tau hyperphosphorylation via coregulating BACE1 and PRKACB. Front Pharmacol. 2019;10:806. doi: 10.3389/fphar.2019.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao Y, Lu X, Wang T, Li S, Kong X, Wang L. MicroRNAs and nervous system diseases: network insights and computational challenges. Brief Bioinform. 2020;21(3):863–875. doi: 10.1093/bib/bbz032. [DOI] [PubMed] [Google Scholar]
- Wang M, Suo L, Yang S, Zhang W. CircRNA 001372 Reduces inflammation in propofol-induced neuroinflammation and neural apoptosis through PIK3CA/Akt/NF-kappaB by miRNA-148b-3p. J Invest Surg. 2020b;1-11.10.1080/08941939.2020.1771639 [DOI] [PubMed]
- Wassenegger M, Pelissier T. A model for RNA-mediated gene silencing in higher plants. Plant Mol Biol. 1998;37(2):349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- Wei P, Chen H, Lin B, Du T, Liu G, He J, You C. Inhibition of the BCL6/miR-31/PKD1 axis attenuates oxidative stress-induced neuronal damage. Exp Neurol. 2021;335:113528. doi: 10.1016/j.expneurol.2020.113528. [DOI] [PubMed] [Google Scholar]
- Wu BW, Wu MS, Guo JD. Effects of microRNA-10a on synapse remodeling in hippocampal neurons and neuronal cell proliferation and apoptosis through the BDNF-TrkB signaling pathway in a rat model of Alzheimer's disease. J Cell Physiol. 2018;233(7):5281–5292. doi: 10.1002/jcp.26328. [DOI] [PubMed] [Google Scholar]
- Wu Q, Xi DZ, Wang YH. MicroRNA-599 regulates the development of Parkinson's disease through mediating LRRK2 expression. Eur Rev Med Pharmacol Sci. 2019;23(2):724–731. 10.26355/eurrev_201901_16886 [DOI] [PubMed]
- Xu H, Yue C, Chen L. Post-transcriptional regulation of soluble guanylate cyclase that governs neuropathic pain in Alzheimer's disease. J Alzheimers Dis. 2019;71(4):1331–1338. doi: 10.3233/JAD-190743. [DOI] [PubMed] [Google Scholar]
- Xu N, Li AD, Ji LL, Ye Y, Wang ZY, Tong L. miR-132 regulates the expression of synaptic proteins in APP/PS1 transgenic mice through C1q. Eur J Histochem. 2019b;63(2). 10.4081/ejh.2019.3008 [DOI] [PMC free article] [PubMed]
- Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8(5):712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Yang G, Song Y, Zhou X, Deng Y, Liu T, Weng G, Pan S. MicroRNA-29c targets beta-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol Med Rep. 2015;12(2):3081–3088. doi: 10.3892/mmr.2015.3728. [DOI] [PubMed] [Google Scholar]
- Yang X, Xue P, Chen H, Yuan M, Kang Y, Duscher D, Chen Z. Denervation drives skeletal muscle atrophy and induces mitochondrial dysfunction, mitophagy and apoptosis via miR-142a-5p/MFN1 axis. Theranostics. 2020;10(3):1415–1432. doi: 10.7150/thno.40857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WY, Cai W, Ying HZ, Zhang WY, Zhang HH, Yu CH. Exogenous plant gma-miR-159a, identified by miRNA library functional screening, ameliorated hepatic stellate cell activation and inflammation via inhibiting GSK-3beta-mediated pathways. J Inflamm Res. 2021;14:2157–2172. doi: 10.2147/JIR.S304828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Zhang CY. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yun Z, Gong L, Qu H, Duan X, Jiang Y, Zhu H. Comparison of miRNA evolution and function in plants and animals. Microrna. 2018;7(1):4–10. doi: 10.2174/2211536607666180126163031. [DOI] [PubMed] [Google Scholar]
- Zhang L, Liao Y, Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res. 2019;38(1):53. doi: 10.1186/s13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sang X, Hou D, Chen J, Gu H, Zhang Y, Hong Z. Plant-derived RNAi therapeutics: a strategic inhibitor of HBsAg. Biomaterials. 2019;210:83–93. doi: 10.1016/j.biomaterials.2019.04.033. [DOI] [PubMed] [Google Scholar]
- Zhang S, Cheng Z, Wang Y, Han T. The risks of miRNA therapeutics: in a drug target perspective. Drug Des Devel Ther. 2021;15:721–733. doi: 10.2147/DDDT.S288859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZB, Wu L, Xiong R, Wang LL, Zhang B, Wang C, Chen SD. MicroRNA-922 promotes tau phosphorylation by downregulating ubiquitin carboxy-terminal hydrolase L1 (UCHL1) expression in the pathogenesis of Alzheimer's disease. Neuroscience. 2014;275:232–237. doi: 10.1016/j.neuroscience.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, Zhang CY. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25(1):39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhao L, Zheng J, Wang K, Deng H, Liu P, Mu H. MicroRNA-144 modulates oxidative stress tolerance in SH-SY5Y cells by regulating nuclear factor erythroid 2-related factor 2-glutathione axis. Neurosci Lett. 2017;655:21–27. doi: 10.1016/j.neulet.2017.06.045. [DOI] [PubMed] [Google Scholar]
- Zhou LK, Zhou Z, Jiang XM, Zheng Y, Chen X, Fu Z, Yi Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6(1):54. doi: 10.1038/s41421-020-00197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LK, Zhou Z, Jiang XM, Zheng Y, Chen X, Fu Z, Yi Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6:54. doi: 10.1038/s41421-020-00197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wang S, Liang Y, Xu X. Inhibition of microRNA-505 suppressed MPP+-induced cytotoxicity of SHSY5Y cells in an in vitro Parkinson's disease model. Eur J Pharmacol. 2018;835:11–18. doi: 10.1016/j.ejphar.2018.07.023. [DOI] [PubMed] [Google Scholar]
- Zhu J, Xu X, Liang Y, Zhu R. Downregulation of microRNA-15b-5p targeting the Akt3-mediated GSK-3beta/beta-catenin signaling pathway inhibits cell apoptosis in Parkinson's disease. Biomed Res Int. 2021;2021:8814862. doi: 10.1155/2021/8814862. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu M, Jia L, Jia J. Inhibition of miR-96-5p may reduce Abeta42/Abeta40 ratio via regulating ATP-binding cassette transporter A1. J Alzheimers Dis. 2021;83(1):367–377. doi: 10.3233/JAD-210411. [DOI] [PubMed] [Google Scholar]
- Zhu J, Wang S, Qi W, Xu X, Liang Y. Overexpression of miR-153 promotes oxidative stress in MPP(+)-induced PD model by negatively regulating the Nrf2/HO-1 signaling pathway. Int J Clin Exp Pathol. 2018b;11(8):4179–4187. https://www.ncbi.nlm.nih.gov/pubmed/31949812 [PMC free article] [PubMed]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Zong Y, Wang H, Dong W, Quan X, Zhu H, Xu Y, Qin C. miR-29c regulates BACE1 protein expression. Brain Res. 2011;1395:108–115. doi: 10.1016/j.brainres.2011.04.035. [DOI] [PubMed] [Google Scholar]
- Zou H, Ding Y, Shi W, Xu X, Gong A, Zhang Z, Liu J. MicroRNA-29c/PTEN pathway is involved in mice brain development and modulates neurite outgrowth in PC12 cells. Cell Mol Neurobiol. 2015;35(3):313–322. doi: 10.1007/s10571-014-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Lu J, Manaenko A, Qi X, Tang J, Mei Q, Hu Q. MicroRNA-132 attenuates cerebral injury by protecting blood-brain-barrier in MCAO mice. Exp Neurol. 2019;316:12–19. doi: 10.1016/j.expneurol.2019.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Not applicable.