Abstract
Rhizobia described so far belong to three distinct phylogenetic branches within the α-2 subclass of Proteobacteria. Here we report the discovery of a fourth rhizobial branch involving bacteria of the Methylobacterium genus. Rhizobia isolated from Crotalaria legumes were assigned to a new species, “Methylobacterium nodulans,” within the Methylobacterium genus on the basis of 16S ribosomal DNA analyses. We demonstrated that these rhizobia facultatively grow on methanol, which is a characteristic of Methylobacterium spp. but a unique feature among rhizobia. Genes encoding two key enzymes of methylotrophy and nodulation, the mxaF gene, encoding the α subunit of the methanol dehydrogenase, and the nodA gene, encoding an acyltransferase involved in Nod factor biosynthesis, were sequenced for the type strain, ORS2060. Plant tests and nodA amplification assays showed that “M. nodulans” is the only nodulating Methylobacterium sp. identified so far. Phylogenetic sequence analysis showed that “M. nodulans” NodA is closely related to Bradyrhizobium NodA, suggesting that this gene was acquired by horizontal gene transfer.
Symbioses between leguminous plants and soil bacteria commonly referred to as rhizobia are of considerable environmental and agricultural importance since they are responsible for most of the atmospheric nitrogen fixed on land. Rhizobia are able to elicit on most of the 18,000 species of the Leguminosae family the formation of specialized organs, called nodules, in which they reduce atmospheric nitrogen to ammonia to the benefit of the plant. Nodule formation is controlled by extracellular bacterial signal molecules, called Nod factors, which are recognized by the host plant (21, 34). The rhizobial species described so far are very diverse and do not form an evolutionary homogenous clade. They belong to three distinct branches within the α-2 subclass of Proteobacteria and are phylogenetically intertwined with non-symbiotic bacteria (40) (Fig. 1). A first large branch groups the genera Rhizobium, Sinorhizobium, Mesorhizobium, and Allorhizobium with Agrobacterium, a pathogenic bacterium of plants. A second branch contains the genus Bradyrhizobium together with photosynthetic free-living Rhodopseudomonas, whereas the third branch includes the genus Azorhizobium as well as the chemiautotroph Xanthobacter. Each rhizobial species has a defined host range, varying from very narrow, as in the case of Azorhizobium caulinodans (6), to very broad, as in the case of Sinorhizobium sp. strain NGR234 (30). Symbionts of legumes exhibiting ecological and agronomic potential should be characterized prior to their use in sustainable agriculture and environment management.
FIG. 1.
Unrooted phylogenetic tree showing the different rhizobial branches, including the new “M. nodulans” in the α subdivision of Proteobacteria. The tree was constructed by using the neighbor-joining method from almost full-length 16S rDNA sequences. The GenBank/EMBL accession numbers are as follows (the first letters of the genus and species are given in parentheses): D 12790 (Pr), D12797 (Mh), X67229 (Ml), L38825 (Mmed), X67224 (Ar), X67234 (Rt), U29386 (Rl), U28916 (Re), Y17047 (Au), X67225 (Av), X67223 (At), X67226 (Rg), X67222 (Sm), X68390 (Ss), X68387 (St), X67231 (Sf), X94198 (Xag), X94201 (Xau), X94199 (Xf), D11342 (Ac), U35000 (Be), M65248 (Af), L11661 (Nw), S46917 (Bd), D25312 (Rp), D12781 (Bj), U69637 (Bl), D32226 (Mo), D32225 (Mmes), D32227 (Mrad), D32229 (Mrhodi), D32230 (Mz), D32228 (Mrhode), D32224 (Me), D32236 (Msp), and AF220763 (Mn).
Crotalaria spp. are herbs and shrubs of the subfamily Papilionoideae; it is the largest plant genus in Africa. More than 500 species commonly occur in diverse climatological situations, from semidesert to rain forests and high mountains (1, 29). Some Crotalaria spp. are of great agronomic interest since they are used as green manure to improve soil fertility or control nematode populations in infested soils (4, 20). Characterization of a collection of rhizobia isolated from various Crotalaria species revealed two very distinct groups of symbiotic bacteria, a group of broad-host-range rhizobia related to Bradyrhizobium and a group of highly specific rhizobia of unknown taxonomic status (33).
We now report that the latter group of highly specific Crotalaria rhizobia belong to the Methylobacterium genus and assign them to a new species, for which we propose the name “M. nodulans.” These Methylobacterium strains thus constitute a novel and fourth group of nitrogen-fixing legume-symbiotic bacteria. We demonstrated that “M. nodulans” is a facultative methylotroph, which is a unique property among rhizobia.
MATERIALS AND METHODS
Bacterial strains.
Strains isolated from Crotalaria spp. are listed in Table 1. Methylobacterium rhodesianum LMG6086, Methylobacterium organophilum LMG6083, Methylobacterium extorquens LMG4250, Methylobacterium rhodinum LMG2275, Methylobacterium zatmanii LMG6087, Methylobacterium mesophilicum LMG5275, and Methylobacterium sp. strains LMG6378, LMG6085, and LMG6380 were from the collection of the Universiteit Gent (5). Sinorhizobium meliloti RCR2011, Sinorhizobium medicae A321, Sinorhizobium fredii USDA205, Sinorhizobium terangae ORS1009, Rhizobium leguminosarum bv. viciae 248, Rhizobium etli CFN42, Rhizobium tropici CIAT899, Methylobacterium ciceri UPMCa-7, Methylobacterium loti NZP2213, Bradyrhizobium japonicum USDA110, Bradyrhizobium elkanii USDA61, Allorhizobium undicola ORS995, and Azorhizobium caulinodans ORS571 were from our collection. The growth medium for Methylobacterium strains, including “M. nodulans” ORS2060 and ORS1917, was M72 (5) supplemented with 50 mM methanol. The complete medium for other strains was YM (37). Nodulating Methylobacterium strains were grown at 37°C; other strains were grown at 30°C.
TABLE 1.
Crotalaria rhizobia used in this study
| Strain(s)a | Host plant |
|---|---|
| “M. nodulans” | |
| ORS1917, ORS1991, ORS2060T | C. podocarpa |
| ORS1924, ORS1928, ORS1937, ORS2030, ORS2092 | C. perrottetii |
| ORS2026, ORS2045, ORS2076 | C. glaucoides |
| Bradyrhizobium sp. | |
| ORS1810 | C. lathyroides |
| ORS1816 | C. hyssopifolia |
| ORS1813 | C. hyssopifolia |
| ORS1929 | C. comosa |
| ORS2077 | C. retusa |
| ORS2088 | C. goreensis |
All strains are described by Samba et al. (33).
DNA technology.
Genomic DNA was prepared by using the method of Chen and Kuo (7). Plasmid DNA was isolated with a Miniprep kit (Promega, Charbonières, France). PCR products were purified with a QIAquick gel extraction kit (Qiagen, Courtaboeuf, France). Restriction endonucleases and ligase were used according to the manufacturer's specifications (Roche, Meylan, France, or Eurogentec, Seraing, Belgium). For Southern blot hybridization, restricted DNA was blotted to positively charged nylon membranes by the alkali transfer procedure and hybridized with digoxigenin (DIG)-dUTP using the DIG labeling kit supplied by Roche.
DNA amplification, sequencing, and analysis.
The primers used for DNA amplification and sequencing are described in Table 2. Nearly full-length 16S ribosomal DNA (rDNA) was amplified using the universal eubacterial 16S rDNA primers FGPS6 and FGPS1509 (28). To perform 16S rDNA PCR-restriction fragment length polymorphism analysis, 1,500-bp PCR products were digested with Sau96I, HinfI, MspI, and HaeIII and restriction fragments were analyzed by horizontal agarose gel electrophoresis using Metaphor agarose (FMC Bioproducts, Hellerup, Denmark). The 1,500-bp fragments of ORS2060 and ORS1924 were sequenced by using the primers FGPS6, FGPS1509, 16S-370f, 16–1080r, 16S-870f, and 16S-1924r; 555-bp sequences homologous to the mxaF gene were amplified from Crotalaria rhizobia and sequenced by using the nondegenerate primers f1003 and r1561 (26). For rhizobial species, a fragment of about 440 bp homologous to mxaF was amplified and sequenced by using the degenerate primers mxaf916 and mxar1360. Two pairs of primers, nodAfbrad/nodArbrad and nodA1f/nodAb1r, were tested for nodA amplification of reference Methylobacterium strains (LMG6086, LMG6083, LMG4250, LMG2275, LMG6087, LMG5275, LMG6378, LMG6085, and LMG6380). nodA amplification and sequencing of ORS2060 were performed using three pairs of primers, nodAfbrad/nodArbrad, nodboxuniv2/nodArbrad, and nodAfbrad/NodB76r.
TABLE 2.
Primers used for DNA amplification and sequencing
| Primer name | Primer sequence | Target gene | Reference |
|---|---|---|---|
| FGPS6 | 5′-GGA GAG TTA GAT CTT GGC TCA G-3′ | 16S rRNA | 28 |
| FGPS1509 | 5′-AAG GAG GGG ATC CAG CCG CA-3′ | 16S rRNA | 28 |
| 16S-370f | 5′-CCT GGG GAG TAC GGT CGC AAG-3′ | 16S rRNA | This study |
| 16S1080r | 5′-GGG ACT TAA CCC AAC ATC T-3′ | 16S rRNA | This study |
| 16S-870f | 5′-CCT GGG GAG TAC GGT CGC AAG-3′ | 16S rRNA | This study |
| 16S-1924r | 5′-GGC ACG AAG TTA GCC GGG GC-3′ | 16S rRNA | This study |
| f1003 | 5′-GCG GCA CCA ACT GGG GCT GGT-3 | mxaF | 26 |
| r1561 | 5′-GGG CAG CAT GAA GGG CTC CC-3′ | mxaF | 26 |
| mxaf916 | 5′-GGC GAC AAC AAG TGG WCS ATG-3′ | mxaF/xoxF | This study |
| mxar1360 | 5′-ART CCA TRC ARA YGT GGT T-3′ | mxaF/xoxF | This study |
| xoxFr | 5′-CCG GAA CGG CTC GTA RTC CA-3′ | xoxF | This study |
| nodAfbrad | 5′-GTY GAG TGG AGS STK CGC TGG G-3′ | nodA | This study |
| nodArbrad | 5′-TCA CAR CTC KGG CCC GTT CGG-3′ | nodA | This study |
| nodA1f | 5′-TGC RGT GGA ARN TRB VYT GGG-3′ | nodA | This study |
| nodAb1r | 5′-GGN CCG TCR TCR AAS GTC ARG TA-3′ | nodA | This study |
| nodboxuniv2 | 5′-ATC NAA ACA AWN RAT TTT AC-3′ | nod box | This study |
| NodB76r | 5′-GGR TKN GGN CCR TCR TCR AAN GT-3′ | nodB | This study |
PCR amplification was performed with a Perkin-Elmer model 2400 thermocycler in a 25-μl (total volume) reaction mixture containing 50 ng of genomic DNA, each deoxynucleotide triphosphate (200 μM), primers (0.8 μM each), MgCl2 (1.5 mM), 1.25 U of Taq DNA polymerase (Gibco BRL, Cergy Pontoise, France), and the buffer supplied with the enzyme. A touchdown PCR (12) was performed for primer pairs nodAfbrad/nodArbrad and nodA1f/nodAb1r (annealing temperature from 65 to 55°C in 20 cycles), primer pairs nodboxuniv2/nodArbrad and nodAfbrad/NodB76r (annealing temperature from 60 to 45°C in 30 cycles), and mxaf916/mxar1360 (annealing temperature from 60 to 50°C in 20 cycles). A standard PCR method was used for primer pairs FGPS6/FGPS1509 (60°C annealing temperature) and f1003/r1561 (55°C annealing temperature). Sequencing reactions were performed with the ABI Prism BigDye Terminator Cycle sequence kit (Applied Biosystems, Foster City, Calif.) and analyzed on an Applied Biosystems model 310 DNA sequencer. Sequences were aligned by using the PILEUP program (11). Phylogenetic trees were constructed by the neighbor-joining method (32), and a bootstrap confidence analysis was performed on 1,000 replicates to determine the reliability of the tree topology obtained (13).
Rhizobial mxaF-homologous partial sequences are available upon request.
Construction and screening of a genomic library of ORS2060.
To obtain a genomic library of the ORS2060 strain, total DNA was subjected to partial digestion with Sau3AI and dephosphorylated with an alkaline phosphatase treatment. Fragments were then ligated to SuperCos I XbaI/BamHI arms (Stratagene, La Jolla, Calif.) as instructed by the manufacturer. Ligated DNA was packaged by using the Gigapack III Gold packaging extract (Stratagene). Standard methods were used for titrating and cosmid propagation using Escherichia coli XL1-MR as the host. Approximately 2,000 white colonies were picked individually into 96-well microtiter plates containing Luria-Bertani medium plus kanamycin (50 μg/ml), grown overnight at 37°C, and stored with 30% glycerol at −80°C. Screening of mxaF-containing cosmids was performed by DNA amplification using the primer pair f1003/r1561. A selected clone, pSTM217, was confirmed by hybridization with an mxaF probe constructed by DIG labeling the ORS2060 555-bp mxaF internal fragment.
Methanol utilization tests.
Cells were grown for 48 h to mid-log phase in minimum mineral medium M72 (5) supplemented with pyruvate (10 mM) and yeast extract (0.5g/liter). Bacterial suspensions were diluted in M72 medium to an optical density of 0.05, and one of the following compounds was added: methanol (MeOH) (10, 50, 100, or 500 mM), pyruvate (10 mM), or succinate (10 mM). Growth was monitored by measuring optical density at 620 nm. MeOH dosage in culture supernatants was performed as described previously (39) except that KMnO4 was replaced by alcohol oxidase (EC 1.1.3.13) at 0.1 U/ml (Sigma, L'Isle d'Abeau, France).
Plant tests.
Plant cultivation and nodulation tests were carried out as described previously (23), with the following modifications: seeds were superficially sterilized with concentrated sulfuric acid for 35 min (Crotalaria podocarpa), 30 min (Crotalaria perrottetii), 15 min (Crotalaria comosa), 40 min (C. goreensis), or 25 min (Crotalaria ochroleuca). Effectiveness was estimated by visual observation of plant vigor and foliage color of 30-day-old plants.
Fresh nodules were observed under an Olympus SHZ 10 stereomicroscope. Sections of 80-μm thickness were made using a Leica VT1000S Vibratome. Microscopic preparations were examined without further staining with an Olympus Provis microscope.
Bacteriochlorophyll detection.
The presence of bacteriochlorophyll a in “M. nodulans” ORS2060 was checked as described previously (22).
Nucleotide sequence accession numbers.
Accession numbers for 16S rRNA and the mxaF and nodA genes are as follows: AF220762 (ORS1924 16S rRNA gene), AF220763 (ORS2060 16S rRNA gene), AF220764 (ORS2060 mxaF gene), and AF266748 (ORS2060 nodA gene). Accession numbers for rhizobial partial mxaF-homologous sequences are AF304307 to AF304313.
RESULTS
Bacteria that specifically nodulate Crotalaria belong to the Methylobacterium genus.
Rhizobia isolated from Crotalaria glaucoides, C. perrottetii, and C. podocarpa were previously shown to be highly specific, since they effectively nodulated only these three species (33). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis grouped almost all the strains into three related electrophoretic clusters separated from other rhizobial species (33).
To determine the bacterial genus to which they belong, 16S rDNA analysis was performed on 11 representative strains belonging to the previously identified sodium dodecyl sulfate-polyacrylamide gel electrophoresis clusters (Table 1). All strains tested gave identical patterns by 16S rDNA PCR-restriction fragment length polymorphism analysis, showing that the strains form a very homogenous group. The 16S rRNA genes of two strains, ORS2060 isolated from C. podocarpa and ORS1924 isolated from C. perrottetii, were sequenced and shown to be identical (accession numbers AF220763 and AF220762, respectively). Phylogenetic 16S rDNA sequence analysis revealed that this group was distinct from the three main branches containing all known rhizobial species. Surprisingly, the specific Crotalaria symbionts belonged to the Methylobacterium lineage of the α-Proteobacteria, thus constituting a fourth phylogenetic rhizobium branch (Fig. 1). Sequence similarities with the different Methylobacterium species described so far ranged from 93.64% (Methylobacterium radiotolerans) to 94.52% (M. extorquens) and was 95.16% with its closest phylogenetic neighbor, Methylobacterium sp. strain F48. These ranges are comparable with those found between most Methylobacterium species, thus demonstrating that the specific Crotalaria rhizobia phylogenetically belong to the Methylobacterium genus.
Bacteria of the new species “M. nodulans” are facultative aerobic methylotrophs.
Methylobacterium spp. oxidase methanol via a key periplasmic enzyme, methanol dehydrogenase, which belongs to the family of the pyrroloquinoline quinone (PQQ)-linked enzymes that are known as quinoproteins (2, 3). Methanol dehydrogenase is an α2β2 tetramer noncovalently bound to PQQ (15). The structural gene for the α subunit of the methanol dehydrogenase encoded by mxaF is well conserved among gram-negative methylotrophic bacteria (24). Therefore, to evaluate the presence of methanol oxidation genes in Crotalaria rhizobia, we performed PCR amplifications using nondegenerate primers f1003 and r1561, defined from conserved parts of mxaF genes (26). These primers indeed produced an amplification product of the expected size for 9 of the 11 specific Crotalaria strains tested. The 555-bp PCR product obtained from the representative strain ORS2060 was homologous to mxaF genes. The full-length mxaF gene was obtained from a genomic library of ORS2060 probed with the 555-bp PCR product (see Materials and Methods for details). The corresponding nucleotide sequence contained a single extended 1,890-bp open reading frame (accession number AF220764) encoding a 629-amino-acid protein that exhibits 88% identity with the MxaF proteins of M. extorquens (accession number M31108) and M. organophilum (accession number M22629). A putative ribosome binding site was identified upstream from the proposed ATG start codon. The first 93 nucleotides of the structural gene encode a typical signal sequence for secretion (38). These results indicated that Crotalaria-nodulating Methylobacterium strains did contain, as all other Methylobacterium strains, the structural gene, mxaF, required for methanol oxidation.
In order to directly assess the methylotrophic properties of Methylobacterium species from Crotalaria, the strains were then tested for their ability to use methanol (50 mM) as the sole carbon source in liquid culture. Growth was compared to that of M. extorquens LMG4250 and Bradyrhizobium sp. isolated from Crotalaria species. No growth was observed for Bradyrhizobium strains over a 10-day period (Fig. 2). By contrast, all Methylobacterium strains from Crotalaria grew on this substrate, with generation times ranging from 9.5 to 40 h. The fastest growth rates were obtained with strains ORS2060 (Fig. 2) and ORS1917 (9.5-h doubling time) and were similar to the growth rate of M. extorquens LMG4250 on MeOH (50 mM) (7.5-h doubling time). Similar growth was noted on either pyruvate or succinate as the sole carbon source. Growth was obtained at an MeOH concentration up to 500 mM.
FIG. 2.
Methanol utilization by “M. nodulans” ORS2060. Bradyrhizobium sp. strain ORS1810 (33) was used as a negative control. Shown is growth of ORS2060 (black circles) and ORS1810 (black squares) on minimum mineral medium 72 containing MeOH as the sole carbon source. MeOH concentrations in supernatants of cultures of ORS2060 (empty circles) and ORS1810 (empty squares) are also shown.
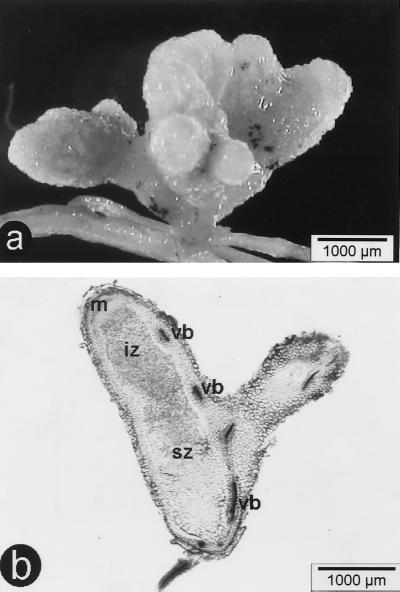
To confirm that the same bacterium isolated from Crotalaria exhibits both nodulation ability and methylotrophic properties, strain ORS2060 was grown on methanol, repurified from a single colony, and inoculated onto C. podocarpa and C. perrottetii. The bacteria formed nitrogen-fixing indeterminate nodules (Fig. 3). Single colonies reisolated from the nodules retained the ability to grow on methanol, clearly demonstrating that this new Methylobacterium strain is able both to grow on one-carbon (C1) compounds and to effectively nodulate legumes.
FIG. 3.
Nodules of C. perrottetii inoculated with “M. nodulans” ORS2060. (a) Multilobed fresh nodule; (b) unstained longitudinal section displaying the classical structure of indeterminate nodules with an apical meristematic zone (m), an infection zone (iz), a senescent zone (sz), and peripheral vascular bundles (vb).
Proposal of a new species was warranted by the low 16S rDNA sequence similarity values between Methylobacterium strains isolated from Crotalaria and other Methylobacterium strains. The unique symbiotic properties of these bacteria (see below) led us to propose the name “Methylobacterium nodulans.” Methylobacterium spp., which are mostly isolated from water and leaf surface microflora, are known in the literature as pink-pigmented facultative methylotrophs (19). However, “M. nodulans” strains are not pigmented and the type strain, ORS2060, did not exhibit the characteristic bacteriochlorophyll absorption peak at 766 nm. This could account for their adaptation to the soil rather than to the phyllosphere or water.
“M. nodulans” is the only nodulating Methylobacterium species identified to date.
The Methylobacterium genus has never been reported to contain nodulating bacteria. However, to be certain that nodulation is not a general but hitherto undetected Methylobacterium feature, we tested representative strains of several Methylobacterium species (M. rhodesianum, M. organophilum, M. extorquens, M. rhodinum, M. mesophilicum, M. zatmanii, M. radiotolerans, and “M. nodulans”) and a few Methylobacterium sp. strains (LMG6378, LMG6085, and LMG6380) by inoculation on Crotalaria species nodulated by broad-host-range Bradyrhizobium (C. comosa, C. goreensis, and C. ochroleuca) or by “M. nodulans” (C. perrottetii). None of them nodulated, except “M. nodulans.”
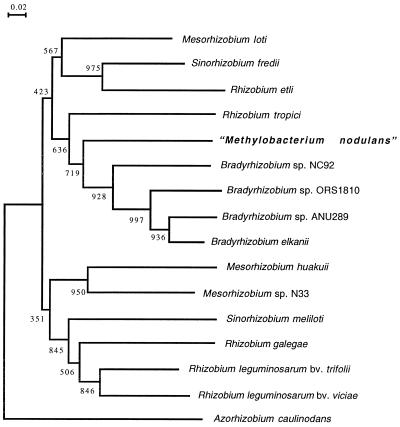
Specific rhizobial infection and nodulation of legumes is mainly controlled by a set of bacterial nodulation genes involved in the production of lipochitooligosaccharides (Nod factors) that act as morphogenic signal molecules on specific legume hosts (10, 36). Structural nodABC genes encoding key enzymes in Nod factor biosynthesis are present in all rhizobia. We thus looked for the presence of the nodA gene in the Methylobacterium representative strains listed above by PCR amplifications using two pairs of degenerate primers defined from conserved parts of NodA sequences. None of these strains responded positively, except “M. nodulans.” The full-length nodA sequence of “M. nodulans” ORS2060 was determined after PCR amplification. The open reading frame, which probably corresponds to nodA, is 642 nucleotides long (accession number AF266748). Sequence similarity with the different complete rhizobial NodA protein sequences available in databases ranged from 53.06% (A. caulinodans) to 74.11% (B. elkanii USDA94). Phylogenetic analysis of available NodA proteins showed that “M. nodulans” is grouped with Bradyrhizobium spp. (Fig. 4).
FIG. 4.
Phylogenetic tree based on full-length NodA sequences construced by using the neighbor-joining method. Bootstrap values (from 1,000 replications) are indicated. The GenBank/EMBL accession numbers are as follows (the first letters of the genus and species are given in parentheses): L18897 (Ac), 106241 (Ml), M73699 (Sf), M58625 (Re), X98514 (Rt), AF266748 (Mn), U33192 (BspNC92), U04609 (BspANU289), U04609 (Be), AJ249353 (Mh), U53327 (MspN33), M11268 (Sm),X87578 (Rg), and X01650 (Rl). The NodA sequence of Bradyrhizobium sp. strain ORS1810 was kindly provided by T. Stepkowski.
Methylotrophy is not a common feature of rhizobia.
We tested a collection of rhizobia belonging to various genera and species (S. meliloti, S. medicae, S. fredii, S. terangae, R. leguminosarum bv. viciae, R. etli, R. tropici, M. ciceri, M. loti, B. japonicum, B. elkanii, A. undicola, and A. caulinodans; see Materials and Methods) for their ability to grow on MeOH (50 mM) as the sole carbon source in solid and liquid culture. No growth of these strains was observed within 6 days. We also used PCR amplification to investigate the possible presence of a mxaF-homologous gene in the rhizobial strains. No amplification could be obtained using primers f1003 and r1561. When degenerate primers defined from conserved parts of both MxaF and MxaF homologs from α-Proteobacteria were used, all strains were found to contain a 440-bp sequence homologous to mxaF. These sequences however exhibited higher homology to two mxaF homologs, mxaF′ from M. extorquens (72 to 80% amino acid identity) and xoxF from Paracoccus denitrificans (72 to 83% amino acid identity) than to mxaF from M. extorquens (50 to 53% amino acid identity). xoxF and mxaF′ are thought to encode an alternative PQQ-dependent dehydrogenase (8, 18). Phylogenetic analysis further revealed that the rhizobial protein sequences are clustered with XoxF and MxaF′ and separated from the methanol dehydrogenase MxaF from α- and β-Proteobacteria (Fig. 5). We thus conclude that the sequences found in rhizobia probably do not correspond to a bona fide mxaF ortholog.
FIG. 5.
Phylogenetic tree based on about 140-amino-acid MxaF-homologous sequences constructed by using the neighbor-joining method. Bootstrap values (from 1,000 replications) are indicated. Cluster 1 groups only MxaF sequences with the following GenBank/EMBL accession numbers (the first letters of the genus and species are given in parentheses): M17339 (Pd), AB004097 (Hm), M22629 (Mo), M31108 (Me), AF220764 (Mn), U41040 (Mm), and AF184915 (Msp). Cluster 2 groups the rhizobial sequences (accession numbers given in Materials and Methods) together with XoxF from P. denitrificans (U34346) and MxaF′ from M. extorquens (U72662). MxaF from Bacillus sp. (M65004) was used as the root. The topology of the tree constructed with the full-length MxaF-homologous sequences (thus excluding the rhizobial sequences) is similar. (α), member of α-Proteobacteria; (β), member of β-Proteobacteria.
DISCUSSION
The rhizobial species described so far belong to three distinct 16S rRNA branches within α-Proteobacteria, including nonsymbiotic bacteria such as the plant pathogen Agrobacterium and the human and animal pathogen Afipia. The discovery of a fourth rhizobial phylogenetic branch, constituted by bacteria of the Methylobacterium genus, thus confirms and extends the polyphyletic origin of rhizobia within the α subclass of Proteobacteria (40). It is noteworthy that although rhizobia have been studied for more than 100 years, symbionts of less than 50 of the 750 known legume genera have been fully characterized. Therefore, it is quite likely that much greater rhizobial diversity will be discovered by characterizing symbionts of unexplored legumes and by focusing on legumes of unexplored areas.
Bacterial nodulation (nod) genes have been shown to play a central role in the molecular dialog between the plant and the bacterium, leading to plant recognition, infection, and nodulation (10, 34, 36). The presence of structural nodABC genes in all rhizobia indicates the unique origin of these genes, which could have been disseminated among Proteobacteria via self-transmissible plasmids (25) or other mechanisms allowing transfer of “symbiotic islands” (35). The presence of the nodA gene in “M. nodulans” is consistent with nodulation data and suggests that this bacterium is able to establish symbiosis by the same molecular mechanisms as other rhizobia. The NodA phylogenetic analysis confirms the monophyletic origin of this structural common nod gene (Fig. 4). The deduced absence of nodA in nonsymbiotic Methylobacterium spp. together with the close phylogenetic relationship between “M. nodulans” and Bradyrhizobium NodA proteins suggests that “M. nodulans” has acquired nodulation properties by lateral gene transfer.
Bacteria of the Methylobacterium genus are facultative methylotrophs capable of growing on one-carbon compounds such as formate, formaldehyde, and methanol as the sole source of carbon and energy, as well as on a wide range of multicarbon substrates (16, 17). They constitute one of the major methylotrophic branches in the α-2 subclass of Proteobacteria. It should be noted that other branches, including the three rhizobial branches described to date (Fig. 1), also contain methylotrophs. Indeed, Xanthobacter species are all autotrophic methylotrophs (27), most Rhodopseudomonas species grow photosynthetically with methanol (31), and a methyl bromide-utilizing methylotroph closely related to Rhizobium species was recently identified (9). Methylobacterium species are known to occur in man-made environments such as drinking water supplies, swimming pools, and hospital washbasins, where they often become highly resistant to chlorine. They are also widespread in natural environments, including soil, dust, air, and fresh water, wherever one-carbon compounds are abundant. Because of their ability to metabolize various plant decomposition compounds such as methylated compounds, methylotrophic bacteria play an important ecological role in the environmental carbon cycle. Although Methylobacterium strains have been frequently found on plant tissues (19), there is no previous evidence of symbiotic association of such microorganisms with plants. We demonstrated here that a group of Methylobacterium strains, identified as the new species “M. nodulans,” are able to form nitrogen-fixing nodules on the roots of leguminous plants. However, this case may not be unique since we recently learned that CB376, a nodulating photosynthetic strain from L. bainesii (14), could be classified in the Methylobacterium genus on the basis of its 16S rRNA sequence (W. Heumann, personal communication).
A polyphyletic origin of rhizobia versus a monophyletic origin of common nodulation genes suggests that rhizobia have evolved through acquisition of nodulation functions in different bacterial branches susceptible to adaptation to different legume environments. “M. nodulans” is a facultative methylotroph, a unique property among rhizobia, raising the question of the role of methylotrophy in Crotalaria-“M. nodulans” symbiosis. We are currently starting a genetic analysis to understand why Methylobacterium strains are specifically associated with some particular legumes.
ACKNOWLEDGMENTS
We thank D. Fleischmann, W. Heumann, J. Dénarié, and P. Normand for helpful discussions.
A.S. is indebted to IRD for a doctoral grant.
REFERENCES
- 1.Allen O N, Allen E K. The Leguminosae, a source book of characteristics, uses, and nodulation. Madison: University of Wisconsin Press; 1981. [Google Scholar]
- 2.Anthony C. Bacterial oxidation of methane and methanol. Adv Microbiol Physiol. 1986;27:113–210. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- 3.Anthony C, Ghosh M, Blake C C F. The structure and function of methanol dehydrogenase and related PQQ-containing quinoproteins. Biochem J. 1994;304:665–674. doi: 10.1042/bj3040665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker M, Johnson D E. The role of legume fallows in intensified upland rice-based systems of West Africa. Nutr Cycling Agroecosyst. 1999;1:71–81. [Google Scholar]
- 5.Belgian Co-ordinated Collection of Micro-organisms/Laboratorium voor Microbiologie. Bacteria. Catalogue. Ghent, Belgium: Universiteit Gent; 1998. [Google Scholar]
- 6.Boivin C, Ndoye I, Lortet G, Ndiaye A, De Lajudie P, Dreyfus B. The Sesbania root symbionts Sinorhizobium saheli and S. teranga bv. sesbaniae can form stem nodules on Sesbania rostrata, although they are less adapted to stem nodulation than Azorhizobium caulinodans. Appl Environ Microbiol. 1997;63:1040–1047. doi: 10.1128/aem.63.3.1040-1047.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W P, Kuo T T. A simple and rapid method for the preparation of Gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdova L, Lidstrom M E. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology. 1997;143:1729–1736. doi: 10.1099/00221287-143-5-1729. [DOI] [PubMed] [Google Scholar]
- 9.Connell Hancock T L, Costello A M, Lidstrom M E, Oremland R S. Strain IMB-1, a novel bacterium for the removal of methyl bromide in fumigated agricultural soils. Appl Environ Microbiol. 1998;64:2899–2905. doi: 10.1128/aem.64.8.2899-2905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dénarié J, Debellé F, Promé J-C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Don R H, Cox P T, Wainwright B, Baker K, Mattick J S. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:424–429. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Fleischmann D, Kramer D. Photosynthetic rhizobia. Biochim Biophys Acta. 1998;1364:17–36. doi: 10.1016/s0005-2728(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh M, Anthony C, Harlos K, Goodwin M G, Blake C C F. The refined structure of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens at 1.94 A. Structure. 1995;3:177–187. doi: 10.1016/s0969-2126(01)00148-4. [DOI] [PubMed] [Google Scholar]
- 16.Green P N, Bousfield I J. Emendation of Methylobacterium Patt, Cole and Hanson 1976; Methylobacterium rhodinum (Heumann 1962) comb. nov. corrig.; Methylobacterium radiotolerans (Ito and Lizuka 1971) comb. nov. corrig.; and Methylobacterium mesophilicum (Austin and Goodfellow 1979) comb. nov. Int J Syst Bacteriol. 1983;33:875–877. [Google Scholar]
- 17.Green P N. The genus Methylobacterium. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. New York, N.Y: Springer-Verlag; 1992. pp. 2342–2349. [Google Scholar]
- 18.Harms N, Ras J, Koning S, Reijnders W N M, Stouthamer A H, Van Spanning R J M. Genetics of C1 metabolism regulation in Paracoccus denitrificans. In: Lidstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 126–132. [Google Scholar]
- 19.Holland M A. Methylobacterium and plants. Recent Res Dev Plant Physiol. 1997;1:207–212. [Google Scholar]
- 20.Huang C S, Tenente R C V, Silva F C C, Lara J A R. Effect of Crotalaria spectabilis and two nematicides, on numbers of Meloidogyne incognita and Helicotylenchus. Nematologica. 1981;27:1–5. [Google Scholar]
- 21.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome J-C, Denarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 22.Lorquin J, Molouba F, Dreyfus B L. Identification of the carotenoid pigment canthaxanthin from photosynthetic Bradyrhizobium strains. Appl Environ Microbiol. 1997;63:1151–1154. doi: 10.1128/aem.63.3.1151-1154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lortet G, Méar N, Lorquin J, Dreyfus B, de Lajudie P, Rosenberg C, Boivin C. Nod factor thin-layer chromatography profiling as a tool to characterize symbiotic specificity of rhizobial strains: application to Sinorhizobium saheli, S. teranga and Rhizobium sp. strains isolated from Acacia and Sesbania. Mol Plant-Microbe Interact. 1996;9:736–747. [Google Scholar]
- 24.Machlin S M, Hanson R S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J Bacteriol. 1988;170:4739–4747. doi: 10.1128/jb.170.10.4739-4747.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Romero E, Caballero-Mellado J. Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci. 1996;15:113–140. [Google Scholar]
- 26.McDonald I R, Kenna E M, Murrell J C. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer W G, Croes L M, Jenni B, Lehmicke L G, Lidstrom M E, Dijkhuizen L. Characterization of Xanthobacter strains H4–14 and 25a and enzyme profiles after growth under autotrophic and heterotrophic conditions. Arch Microbiol. 1990;153:360–367. doi: 10.1007/BF00249006. [DOI] [PubMed] [Google Scholar]
- 28.Normand P, Cournoyer B, Nazaret S, Simonet P. Analysis of a ribosomal RNA operon in the actinomycete Frankia. Gene. 1992;111:119–124. doi: 10.1016/0378-1119(92)90612-s. [DOI] [PubMed] [Google Scholar]
- 29.Polhill R M. Crotalaria in Africa and Madagascar. Rotterdam, The Netherlands: Balkema, A. A.; 1982. [Google Scholar]
- 30.Pueppke S G, Broughton W J. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant-Microbe Interact. 1999;12:293–318. doi: 10.1094/MPMI.1999.12.4.293. [DOI] [PubMed] [Google Scholar]
- 31.Sahm H, Cox R B, Quayle J R. Metabolism of methanol by Rhodopseudomonas acidophila. J Gen Microbiol. 1976;94:313–322. doi: 10.1099/00221287-94-2-313. [DOI] [PubMed] [Google Scholar]
- 32.Saitou R R, Nei M. A neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;44:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Samba R T, de Lajudie P, Gillis M, Neyra M, Spencer-Barreto M M, Dreyfus B. Diversity of rhizobia nodulating Crotalaria spp. from Senegal. Symbiosis. 1999;27:259–268. [Google Scholar]
- 34.Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan J T, Ronson C W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent J M. A manual for the pratical study of root-nodule bacteria. Oxford, United Kingdom: Blackwell Scientific Publications; 1970. [Google Scholar]
- 38.Von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 39.Wood P J, Siddiqui I R. Determination of methanol and its application to measurement of pectin ester content and pectin methyl esterase activity. Anal Biochem. 1971;39:418–428. doi: 10.1016/0003-2697(71)90432-5. [DOI] [PubMed] [Google Scholar]
- 40.Young J P W, Haukka K E. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]