Abstract
The human gut microbiota protects the host from invading pathogens and the overgrowth of indigenous opportunistic species via a process called colonization resistance. Here, we investigated the antagonistic activity of human gut bacteria towards Candida albicans, an opportunistic fungal pathogen that can cause severe infections in susceptible individuals. Coculture batch incubations of C. albicans in the presence of faecal microbiota from six healthy individuals revealed varying levels of inhibitory activity against C. albicans. 16S rRNA gene amplicon profiling of these faecal coculture bacterial communities showed that the Bifidobacteriaceae family, and Bifidobacterium adolescentis in particular, were most correlated with antagonistic activity against C. albicans. Follow-up mechanistic studies performed under anaerobic conditions confirmed that culture supernatants of Bifidobacterium species, particularly B. adolescentis, inhibited C. albicans in vitro. Fermentation acids (FA), including acetate and lactate, present in the bifidobacterial supernatants were important contributors to inhibitory activity. However, increasing the pH of both bacterial supernatants and mixtures of FA reduced their anti-Candida effects, indicating a combinatorial effect of prevailing pH and FA. This work, therefore, demonstrates potential mechanisms underpinning gut microbiome-mediated colonization resistance against C. albicans, and identifies particularly inhibitory components such as bifidobacteria and FA as targets for further study.
Keywords: human gut microbiota, bifidobacteria, colonization resistance, Candida albicans, short chain fatty acids, lactate, pH
This study showed that faecal microbiota vary in their ability to inhibit the fungal pathogen Candida albicans, and identified bifidobacteria, and their fermentation acids, as inhibitory gut microbiota components.
Introduction
The human colon harbours a diverse microbiota that is dominated by obligate anaerobic bacteria (Pasolli et al. 2019, Whitman et al. 1998). The main energy sources for these gut microbes are nondigestible carbohydrates that resist digestion in the small intestine and become available for bacterial fermentation in the proximal colon (Flint et al. 2015). These substrates are fermented by the gut microbiota to produce short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate, and other fermentation acids (FA) such as lactate (Cummings 1981). SCFAs provide the host with up to 5%–10% of their total daily energy requirement (Mortensen and Clausen 1996), and positively impact intestinal and systemic host health (Cummings 1981, Koh et al.2016).
The intestinal microbiota also contributes to host health by bolstering resistance against colonization of the gut by pathogens (Bohnhoff et al. 1964, Buffie et al. 2015). This phenomenon, termed colonization resistance, can prevent pathogens from establishing and replicating in the gut, or from reaching the densities required to invade deeper tissues and cause overt disease (Bohnhoff et al. 1964). Colonization resistance is multifactorial, involving mechanisms such as the direct production of antimicrobial compounds (Donia and Fischbach 2015, Rea et al. 2010), competition for adhesion receptors on the gut epithelium (Ventura et al. 2016), and direct competition for niches and nutrients required for the growth of competing pathogenic bacteria (Deriu et al. 2013, Freter et al. 1983, Maltby et al. 2013, Wilson and Perini 1988). Additional mechanisms of colonization resistance include the creation of a less favourable gut environment, e.g. by lowering the luminal pH through the production of SCFAs (Cherrington et al. 1991, Rivera-Chávez et al. 2016, Roe et al. 2002), or depleting free molecular oxygen, which can prevent overgrowth and virulence gene expression of some pathogenic microbes (Marteyn et al. 2011, Rivera-Chávez et al. 2016). Furthermore, human gut commensals are instrumental in the training and modulation of the host immune system (Kau et al. 2011, Thaiss et al. 2016), inducing the release of host antimicrobial compounds (Cash et al. 2006, Fan et al. 2015), and in stimulating epithelial barrier reinforcement and repair (Geirnaert et al. 2017, Rossi et al. 2015). Importantly, microbiota-mediated colonization resistance can be weakened by various environmental factors and insults, such as Western-style diet (Martinez-Medina et al. 2014), antibiotic therapy (Bohnhoff et al. 1964, Vollaard et al. 1992), and acute and chronic inflammatory conditions (Carroll et al. 2012, Stecher et al. 2007).
Candida albicans is a diploid polymorphic fungus and a common opportunistic pathogen of humans, with an estimated annual incidence of 700 000 cases of Candida bloodstream infections globally (Guinea 2014). In susceptible patient cohorts, including premature infants and those undergoing chemo- or immune-therapy, organ or stem cell transplants, or abdominal surgery or trauma, C. albicans infections can be particularly devastating, with mortality rates of 46%–75% following systemic spread, even with antifungal drug interventions (Brown et al. 2012). The incidence of C. albicans infections has increased in vulnerable subjects over the past few decades (Low and Rotstein 2011) alongside the emergence of other clinically important Candida spp., such as C. auris (Heaney et al. 2020, Pfaller et al. 2000). Furthermore, a significant increase of isolates with resistance to common antifungal agents has been observed (Whaley et al. 2016).
Despite the pathogenic potential of C. albicans, it exists harmlessly in the gastrointestinal tract (GIT) of 40%–80% of healthy individuals in Western countries, predominantly in the yeast form, and with cell counts that do not typically exceed 104–5 colony forming units (CFU)/g faeces (Harnett et al. 2017, Mason et al. 2012, Nash et al. 2017, Neville et al. 2015, Odds et al. 1989). The GIT is therefore a natural reservoir of C. albicans (Hube 2004, Odds 2010) but, in health, its overgrowth is suppressed by the gut microbiota via colonization resistance (Fan et al. 2015, Kennedy and Volz 1985a). However, conditions such as weakened immunity, increased permeability of the intestinal mucosal barrier, and/or perturbation of microbiota-mediated colonization resistance via receipt of broad-spectrum antibiotics can favour C. albicans pathogenesis (d'Enfert et al. 2020, León et al. 2009, Samonis et al. 1994,). Furthermore, systemic candidiasis is often reported to derive from a preceding expansion of Candida spp. in the GIT and subsequent translocation from the intestinal niche into the bloodstream (Miranda et al. 2009, Zhai et al. 2020). GIT colonization by C. albicans is, therefore, a major risk factor for systemic candidiasis (Pittet et al. 1994).
Given the importance of the intestinal niche as a reservoir for systemic dissemination, and the known suppressive effects of the indigenous microbiota on the colonization of the gut by C. albicans in health (Fan et al. 2015), we here assessed the potential of the human gut microbiota, and individual gut anaerobe species, to suppress the growth of this opportunistic pathogen in vitro. We identified specific bacterial isolates, including Bifidobacterium adolescentis, in faecal samples of healthy individuals that inhibit C. albicans growth in vitro, and revealed the involvement of gut bacterial FA and pH in this process. These findings enhance current knowledge on potential mechanisms of colonization resistance against C. albicans in the human gut, and suggest targets for further studies that aim to utilize the gut microbiota as a source of novel therapies with antagonistic activity against this opportunistic fungal pathogen.
Materials and methods
Ethics
Faecal sample collections used for isolation of human gut anaerobes, and for coculture experiments with C. albicans, were approved by the Ethical Review Panel of the Rowett Institute under study number 5946. No donors had received antibiotic treatment for at least 6 months prior to faecal donation.
Cultivation of C. albicans strain SC5314
Candida albicans strain SC5314 (Gillum et al. 1984) was prepared by plating 2–10 µl of frozen glycerol stock on YPD plates [1% w/v yeast extract (Oxoid LP0021, Basingstoke, UK), 2% w/v mycological peptone (Oxoid LP0040), 2% w/v d-glucose, and 2% w/v agar No. 2 (Oxoid LP0012)] and incubating at 30°C for 48 h. A single colony was transferred from the Petri dish into NGY broth (0.1% yeast extract (Oxoid LP0021), 0.1% neopeptone (Difco, Franklin Lakes, NJ, USA), and 0.4% w/v d-glucose; MacCallum et al. 2006) and incubated at 30°C, with shaking at 200 rpm, overnight. The concentration of C. albicans cells in suspension (cells/ml) was estimated by counting using a haemocytometer. Yeast growth was assessed by measuring optical density of the cultures at a wavelength of 600 nm using a spectrophotometer. For determination of C. albicans CFUs in samples, cells were plated on Sabouraud dextrose agar (SDA; 4% w/v d-glucose, 1% w/v mycological peptone, and 2% w/v agar No. 2, pH 5.6).
Batch cocultures of C. albicans and mixed faecal microbiota from healthy donors
Cocultures of C. albicans and mixed faecal microbiota were performed in duplicate for each faecal donor in anaerobically sealed Wheaton bottles containing complex anaerobic medium. The medium contained (amounts given are for 1 l): oat spelt xylan (0.6 g; Sigma-Aldrich, St. Louis, MO, USA), pectin (citrus, 0.6 g; Sigma-Aldrich), amylopectin (0.6 g; Sigma-Aldrich), arabinogalactan (larch, 0.6 g; Sigma-Aldrich), potato starch (5.0 g; Sigma-Aldrich), inulin (0.6 g; Sigma-Aldrich), porcine mucin (0.5 g; Sigma-Aldrich), casein hydrolysate (0.5 g; Fluka, Charlotte, NC, USA), peptone water (0.5 g; Oxoid), K2HPO4 (2.0 g; BDH, Dubai, UAE), NaHCO3 (0.2 g; Sigma-Aldrich), NaCl (4.5 g; Fisher Scientific), MgSO4 · 7H2O (0.5 g; BDH), CaCl2 · 2H2O (0.45 g; Sigma-Aldrich), FeSO4 · 7H2O (0.005 g; Hopkin & Williams, UK), haemin (0.01 g; Sigma-Aldrich), bile salts (0.05 g, Oxoid), 0.1% w/v resazurin (0.6 ml), antifoam A (Y-30, 0.5 ml; Sigma-Aldrich), and dH2O to 1 l. In addition, the medium was supplemented with filter-sterilized reducing solution to ensure anaerobic conditions (0.5 g cysteine, 3.0 g NaHCO3, and dH2O to 40 ml). The pH was adjusted to 6.5 (using HCl and NaOH, as appropriate) before dispensing the medium (50 ml aliquots) into Wheaton bottles anaerobically and autoclaving. After autoclaving, Wheaton bottles were supplemented with 100 µl mineral solution (150 mg EDTA, 60 mg FeSO4 · 7H2O, 3.0 mg ZnSO4 · 7H2O, 0.9 mg MnCl2 · 7H2O, 9.0 mg boric acid, 6.0 mg CoCl2 · 6H2O, 0.3 mg CuCl2 · 2H2O, 0.6 mg NiCl2 · 6H2O, 0.9 mg NaMoO4 · 2H2O, and dH2O to 300 ml), 70 µl vitamin solution (0.2 g menadione, 0.4 g biotin, 0.4 g pantothenate, 2.0 g nicotinamide, 0.1 g vitamin B12, 0.8 g thiamine, 1.0 g p-aminobenzoic acid, and dH2O to 200 ml), 155 µl of a SCFA solution (17 ml acetic acid, 6ml propionic acid, 1 ml n-valeric acid, 1 ml iso-valeric acid, 1 ml iso-butyric acid, and 5 ml butyric acid) and 153 µl of a solution containing additional medium components (2 μg folic acid, 2000 μg inositol, 400 μg niacin, 400 μg pyridoxine HCl, 200 μg riboflavin, 100 μg potassium iodide, and 200 μg ferric chloride and dH2O to 1 l).
Candida albicans cells from an overnight culture grown in YPD broth were washed in sterile PBS, counted using a haemocytometer, and inoculated into 50 ml anaerobic media in Wheaton bottles at a final concentration of 5 × 106 cells/ml (except for one pilot experiment where the inoculum was 5 × 105 cells/ml, see ‘Results’ section for more details). Faecal samples were obtained from six different donors and slurries (10% w/v faeces) were prepared in gentleMACS™ M tubes (Miltenyi Biotech, Auburn, CA, USA) by homogenization in anaerobic PBS (PBS containing 0.05% cysteine). Faecal homogenates were centrifuged at 500 x g for 5 min and the liquid faecal component was injected into the Wheaton bottles using a sterile syringe to give a 0.02% faecal suspension at baseline. The inoculated Wheaton bottles were incubated at 35°C for 48 h with gentle shaking at 75 rpm. Measurements of C. albicans CFUs were carried out at t = 0, 24, and 48 h by plating 10-fold serial dilutions on SDA plates supplemented with 34 µg/ml chloramphenicol. CFUs were counted after aerobic incubation at 30°C for 2–3 days.
16S rRNA gene amplicon sequencing of cocultured incubation samples
The faecal inocula from healthy donors used in the coculture experiments, and from the two biological replicate samples collected after 24 and 48 h of incubation with C. albicans, were analyzed by Illumina MiSeq-based 16S rRNA gene profiling, targeting the V1–V2 region of the gene. Genomic DNA was extracted using the FastDNATM SPIN Kit for Soil (MP Biomedicals, Irvine, CA, USA) following the manufacturer's instructions. Barcoded fusion primers containing adaptors for downstream Illumina MiSeq sequencing MiSeq-27F (5′-AATGATACGGCGACCACCGAGATCTACACTATGGTAATTCCAGMGTTYGATYMTGGCTCAG-3′) and MiSeq-338R (5′-CAAGCAGAAGACGGCATACGAGAT-barcode-AGTCAGTCAGAAGCTGCCTCCCGTAGGAGT-3′) were used for PCR amplification of 16S rRNA genes from extracted DNA. PCR was performed using Q5 Taq polymerase (New England Biolabs, Ipswich, MA), with the following cycling conditions: 98°C for 2 min; followed by 20 cycles at 98°C for 30 s, 50°C for 30 s, and 72°C for 90 s; with a final extension at 72°C for 5 min. Each sample was amplified in quadruplicate; the four reactions were pooled, and PCR products were ethanol precipitated to generate a single PCR amplicon tube per sample. These PCR products were then quantified using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA), and a sequencing master mix was prepared by mixing the samples in equimolar amounts, which was then sequenced at the Centre for Genome-Enabled Biology and Medicine (CGEBM) at the University of Aberdeen (Aberdeen, UK). For sequencing, an Illumina MiSeq machine was used, with 2 × 250 bp read length. The raw output sequence data are available from the European Nucleotide Archive, under the project accession number PRJEB48351. Individual sample accession numbers are given in Table S1 (Supporting Information).
Analysis of 16S rRNA gene amplicon data
The raw read data in fastq format were analyzed using the open-source software Mothur (Schloss et al. 2009). For both of the timepoints after coculture, the two experimental replicates were pooled into single samples for final analyses as no statistically significant differences were detected between replicates. Briefly, contigs were created using the make.contigs command and low quality contigs (such as with length < 280 or > 470 bases, containing at least one ‘N’, and polymeric stretches > 7 bases) were filtered out using screen.seqs. The contigs were aligned against the SILVA reference (https://www.arb-silva.de/; Quast et al. 2013), and operational taxonomic units (OTUs) were generated at a 97% similarity cut-off level, with a preclustering step of diffs = 3 to reduce the impact of sequencing errors. Chimera removal software was not used as abundant OTUs corresponding to bifidobacteria were mistaken for chimeric sequences. Instead, the split.abund command was used to filter out low-abundance sequences that appeared less than 10 times in the dataset. All samples were rarefied to 9171 reads for subsequent comparative analyses. Samples derived from the D1 and D3 faecal inocula samples generated far fewer reads than this, so were excluded from the final analyses. Taxonomic classifications were assigned to each OTU by mapping against the RDP reference database (Cole et al. 2014). Taxonomies for selected OTUs were also validated by manually checking representative sequences using BLAST searches against the NCBI nucleotide database (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the Ribosomal Database Project (Cole et al. 2014, Johnson et al. 2008). Alpha-diversity measures, and phylotype analyses at the phylum, family and genus levels were carried out using Mothur. The final OTU table, phylum, family, genus, and alpha-diversity results for each sample are shown in Table S1 (Supporting Information). The faecal and enriched microbial community coculture samples were assigned to the categories ‘benign’ or ‘antagonistic’ according to the extent of the inhibition shown against C. albicans. Putative biomarkers at different taxonomic levels that correlated with antagonistic activity against C. albicans were assessed using LEfSe (Segata et al. 2011), as implemented in Mothur.
Culturing of human gut anaerobes
The gut anaerobes tested in the current study included isolates from the Rowett Institute (Aberdeen, UK) strain collection or purchased from DSMZ (Braunschweig, Germany) (Table S2, Supporting Information). The isolates were revived from stocks, anaerobically, in Hungate tubes containing M2GSC medium supplemented with 10% v/v clarified bovine rumen fluid (Bryant 1972, Miyazaki et al. 1997). Inoculated cultures were incubated at 37°C in a static 5% CO2 incubator overnight (NuAire, Plymouth, MN, USA). Cell growth was monitored by measuring optical density at 650 nm (OD650) using a spectrophotometer (Novaspec II, Amersham BioSciences UK Ltd., Little Chalfont, UK).
Some of the anaerobic bacteria tested for anti-Candida activity in this study were newly isolated from the stool samples of two consenting adults (D3 and DM1). For each donor, 10-fold serial faecal dilutions were prepared in M2 medium (Hobson 1969) with no added carbon source. Each preparation was then used to inoculate five different agar plates: fastidious anaerobe agar (FAA, LAB M Ltd, Heywood, UK) supplemented with 5% v/v horse blood and 0.5% w/v menadione; FAA supplemented with 5% v/v horse blood; brain heart infusion (BHI, Oxoid); M2GSC (Miyazaki et al. 1997); and M2GSC supplemented with 0.5% w/v haemin and 0.5% w/v menadione. The plates were incubated in an anaerobic cabinet (Don Whitley Scientific, Bingley, UK) for 48 h. In parallel, faecal dilutions were preincubated in M2-AXOS diluting broth [M2 supplemented with 0.2% w/v arabinoxylan oligosaccharides (Cargill, Wayzata, MN, USA)] before streaking. After 4 d of incubation, single colonies were selected and picked onto duplicate agar plates of the same type of culture medium on which they were first grown. Half of these duplicate plates were left to grow in the anaerobic cabinet, while the remaining plates were incubated aerobically, at 37°C, for up to 48 h. At the end of the incubation, the growth on anaerobic plates was compared with that on the aerobic counterparts to screen for strictly anaerobic isolates. Single colonies were picked from plates that only showed anaerobic growth and then grown in Hungate tubes containing either M2GSC medium supplemented with 0.5% w/v haemin and 0.5% w/v menadione, fastidious anaerobe broth supplemented with 5% v/v horse blood, and 0.5% w/v menadione, or BHI broth. DNA was extracted from the collected cultures using the FastDNATM SPIN Kit for Soil (MP Biomedicals) and 16S rRNA genes were amplified using universal bacterial primers (7F- AGAGTTTGATYMTGGCTCAG and 1510R- ACGGYTACCTTGTTACGACTT; Satokari et al. 2001) and Sanger sequenced (Eurofins Genomics) for taxonomic identification using BLAST (Johnson et al. 2008), and the Ribosomal Database Project Classifier (Cole et al. 2014). Culturing conditions used to obtain each of the novel isolates are shown in Table S3 (Supporting Information).
Inhibition of C. albicans growth by gut bacterial supernatants and gut bacterial FA
In order to assess the effect of individual gut bacterial isolates on the growth of C. albicans strain SC5314, anaerobes of interest (Table S2, Supporting Information) were cultured in tubes with anaerobic M2GSC medium at 37°C overnight. The individual culture supernatants were then collected after centrifugation at 658 × g for 10 min. The supernatants were filter-sterilized by passing through 0.2 μm syringe-driven filter units (Millex, Merck Millipore Ltd, Kenilworth, NJ, USA) to remove residual bacterial cells. Candida albicans cells pregrown in NGY to an OD600 of 0.8–0.95 were diluted 1 in 100 in fresh NGY medium and 100 μl was transferred to wells of 96-well microtitre plates (CoStar, Washington, WA, USA). The C. albicans suspensions were incubated with an equal amount of filter-sterilized bacterial culture supernatant, or fresh NGY medium as a control, to assess the fungal growth, with technical replicates. The 96-well plates were incubated anaerobically in a temperature-controlled plate reader at 37°C (Epoch 2 Microplate Spectrophotometer, BioTek, Swindon, UK). For each test and technical replicate, the growth of C. albicans was calculated by subtracting the OD600 value at time 0 from that measured after 24 h (T24–T0). The percentage growth of the fungus in fresh NGY medium in the absence of bacterial supernatant was set as 100% growth reference for each repeat run, and uninoculated filter-sterilized M2GSC medium was used as a control.
The impact of gut bacterial FA on C. albicans growth was assessed by monitoring fungal growth in the presence of a mixed solution of 45 mM sodium acetate (Sigma-Aldrich), 15 mM lactate (Sigma-Aldrich), and 10 mM sodium formate (VWR BDH Chemicals, Merck), supplemented with 0.4% w/v glucose, in addition to individual acids plus 0.4% w/v glucose. The pH of all solutions or NGY medium was adjusted using 1 M NaOH and 1 M HCl, as appropriate, to 4, 5, 6, or 7, and checked using a pH meter (Denver Instrument, Denver, CO, USA).
Quantification of FA in gut bacterial culture supernatants using gas chromatography
The culture supernatants of the tested gut bacterial isolates were analyzed by capillary gas chromatography (GC) to quantify the production of FA. To determine the concentrations of SCFAs and lactate, the samples were first derivatized as described elsewhere (Richardson et al. 1989). Briefly, 1 ml of a culture supernatant was placed in a Sorvall screw-capped tube and 50 μl of 0.1 M 2-ethylbutyric acid was added as an internal standard. Concentrations of derivatized fatty acids were determined after a double step extraction of organic acids in 0.5 ml of HCl and 2 ml of diethyl ether per sample, and quantification of their tertiary butyldimethylsisyl (t-BDMS) derivatives using capillary GC apparatus (Agilent 6890; Agilent Technologies, Santa Clara). A total of two technical replicates of an external standard (acetic acid, propionic acid, iso-butyric acid, n-butyric acid, iso-valeric acid, n-valeric acid, sodium formate, lithium lactate, and sodium succinate) were analyzed alongside the samples in each GC run to assess quality of the extraction.
Statistical analyses
The nonparametric Kruskal–Wallis test, followed by Dunn’s post hoc test, was used to analyze data from assays on the inhibition of C. albicans growth by gut bacterial supernatants, and to compare C. albicans growth in the absence and presence of gut anaerobe supernatants, using Prism v8.4.1 (GraphPad, San Diego, CA, USA). To test for associations between % C. albicans growth and the gut bacterial culture supernatants, a Spearman correlation was computed using Prism v8.4.1 (GraphPad). Exact P-values obtained using the Spearman correlation test were corrected using the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli (false discovery rate approach, with Q = 5%). Parameters included the OD of microbial cultures, pH, and FA levels (acetate, formate, and lactate; separately and combined), as quantified in the culture supernatants using GC.
Results
Inhibitory activity of cultivated faecal microbiota on C. albicans growth varies between faecal donors
To establish whether the gut microbiota from different individuals vary in their ability to suppress the growth of C. albicans, we performed coculturing experiments in batch culture, where C. albicans SC5314 cells were incubated for up to 48 h alongside faecal inocula from six healthy adults. The cocultures were performed under anaerobic conditions in a complex growth medium designed to mimic the human colon environment. The viability of C. albicans cells was assessed by determining CFUs following plating onto SDA medium plus chloramphenicol at 0 h and after 24 h and 48 h incubations with or without homogenized faecal inocula.
An initial experiment was conducted with a stool sample from a single healthy volunteer (Donor 1). As shown in Fig. 1(A), the coculture of C. albicans (inoculated at 5 × 105 cells/ml) with faecal material from Donor 1 showed a clear reduction in the fungal CFUs after 44 h incubation. However, viable cell counts were also reduced at the end of the control incubation when C. albicans was grown alone (Fig. 1, black lines), albeit the reduction was lower than that observed in coculture. Subsequent experiments, assessing the impact of faecal inocula from five additional donors were, therefore, performed using 10 times more C. albicans cells (inoculated at 5 × 106 cells/ml), which was sufficient to maintain significant C. albicans CFUs throughout the experiments (Fig. 1B). In the control samples, without the faecal inoculum, C. albicans CFUs remained relatively constant throughout the 48 h incubation period, with counts around 2.5 × 106 CFU/ml, indicating that the colon-mimicking growth medium and anaerobic conditions did not kill C. albicans (Fig. 1B, black lines). The experiment also revealed that the faecal microbiota from different individuals affected C. albicans viable counts to markedly differing degrees after 44 h of coculture (Fig. 1B, orange, red, green, brown, and blue lines). The faecal inoculum from Donor 5 resulted in the strongest inhibitory effect on C. albicans growth, with a 1000-fold (3-log) reduction of Candida CFUs at the end of the incubation period (1 × 103 CFU/ml). Cocultures with faecal inocula from Donors 3, 4, and 6 also resulted in a decrease in C. albicans CFUs (between 4- and 20-fold decrease). In contrast, the faecal inoculum from Donor 2 resulted in no effect on C. albicans growth, which was comparable with that of the no faecal inoculum control, suggesting that the gut bacteria cultured from the faecal inoculum of this individual did not impair the fungal survival under the tested conditions. We conclude that the cultivated faecal samples from healthy individuals differed in their ability to inhibit the survival of C. albicans.
Figure 1.
Faecal inocula from healthy donors resulted in varying killing activity against C. albicans cells in vitro. Candida albicans was cultured with faecal inocula from six different individuals (Donor 1–6), or with no faecal inocula as controls (black lines). Each data point (diamonds) represents C. albicans CFU/ml at sampled time points, while the line connects the means at each time point, calculated from two independent CFU measurements. Data were transformed to Log10 (y-axis). (A) Candida albicans was inoculated into the anaerobic medium at a density of 5 × 105 cells/ml. (B) Candida albicans was inoculated into the anaerobic medium at a concentration of 5 × 106 cells/ml.
Variance in faecal microbiota composition may impact colonization resistance against C. albicans
The differing extent of C. albicans growth inhibition observed in cocultures with faecal inocula from different donors might result from differences in the cultured gut microbiota species composition and, consequently, their metabolic activities. Therefore, we used 16S rRNA gene amplicon-based sequence profiling to analyze the bacterial communities present in the initial faecal inocula from the different donors and in the coculture batch samples after 1 and 2 days of incubation. The analysis revealed that, as anticipated, at the OTU level, the initial faecal inoculum samples contained the highest alpha diversity, which then became reduced as certain bacterial taxa were selectively enriched during coincubation (Fig. 2A; Table S1, Supporting Information).
Figure 2.
Bacterial community analysis of faecal samples and cocultures with varying inhibitory activity against C. albicans. (A) Bray–Curtis dendrogram of faecal inocula and subsequent cocultures with C. albicans. (B) Proportional family-level composition of Donor 2 (‘benign’) and Donor 5 (‘antagonistic’) faecal samples after 48 h coculture with C. albicans in anaerobic, colon-mimicking, medium.
We classified the cultured faecal samples into different groups according to the observed impact on C. albicans growth in the batch coculture. Specifically, Donor 5 was defined as ‘antagonistic’ as the faecal inoculum from this donor resulted in the strongest inhibitory effect, as were Donors 1, 3, 4, and 6 (all > 85% C. albicans inhibition). The Donor 2 inoculum was classified as ‘benign’ since coincubation had comparatively little effect on C. albicans survival in vitro (12% inhibition).
The nonparametric analysis of molecular variance (AMOVA) test implemented in the Mothur software package (Schloss et al. 2009) was first used to compare the bacterial compositions of the cultivated ‘benign’ and ‘antagonistic’ samples (D2 v D1, 3, 4, 5, and 6) at days 1 and 2 combined, and revealed a statistically significant difference between the two groups (P = .02).
We next used LEfSe (Segata et al. 2011) to identify taxa that were associated with either the ‘antagonistic’ (D1, 3, 4, 5, and 6) or ‘benign’ status (D2). The analysis indicated that the Bifidobacteriaceae family (P = .032), and more specifically, B. adolescentis (P = .032) and Bifidobacterium longum derived OTUs (P = .032) belonging to the Gram-positive Actinobacteria phylum correlated with samples exerting the strongest antagonistic activity against C. albicans (Fig. 2B; Tables S4 and S5, Supporting Information). In contrast, the Coriobacteriaceae family (P = .032) and its constituent species Collinsella aerofaciens (P = .026; hereon, indicated as Co. aerofaciens), also belonging to the Actinobacteria phylum, together with Clostridiaceae (P = .031) and Clostridium neonatale (P = .026) from the Firmicutes phylum, correlated with the greatly reduced antagonistic activity against C. albicans (Fig. 2B; Tables S4 and S5, Supporting Information).
Culture supernatants of specific human gut isolates inhibit C. albicans growth under anaerobic conditions
Having correlated the presence of bifidobacteria in the cultivated faecal samples with antagonistic activity against C. albicans using the 16S rRNA gene-based analysis, we next attempted to verify this finding by testing a panel of 37 common and dominant gut bacterial strains for inhibition of C. albicans growth in vitro. The original source of each bacterial isolate is shown in Table S2 (Supporting Information). A subset of the tested gut anaerobes was newly isolated for the purpose of this study from stool samples of healthy volunteers (see ‘Materials and Methods’ section for details of isolation steps), while other isolates were from the existing Rowett Institute strain collection or purchased from DSMZ. The species selected for these tests were also representative of the main phyla inhabiting the human gut (Table S2, Supporting Information). The bacterial isolates of interest therefore belonged to the phyla Firmicutes (nine strains belonging to the family Lachnospiraceae, four Eubacteriaceae, one Peptostreptococcaceae, three Clostridiaceae, six Ruminococcaceae, and one Oscillospiraceae), Actinobacteria (B. adolescentis, selected for analysis as this species was correlated with antagonist activity in coculture with C. albicans, and two Coriobacteriaceae), Bacteroidetes (five Bacteroidaceae, one Porphyromonadaceae, and two Prevotellaceae), and one Proteobacteria (Enterobacteriaceae).
We reasoned that the inhibitory effects of gut microbes upon C. albicans might be mediated, at least in part, by secreted factors or metabolites. Therefore, in order to assess the putative in vitro inhibitory activity of the selected gut bacterial isolates, each species (Fig. 3) was grown individually in M2GSC liquid medium overnight. Then, filter-sterilized culture supernatant was incubated with an overnight liquid culture of C. albicans under anaerobic conditions for 24 h. Candida albicans biomass was assessed using optical density (OD600) measurements. The percentage growth of the fungus alone in fresh NGY medium, without exposure to bacterial supernatants, was set as 100% reference for each repeat run, and uninoculated M2GSC medium was used as a control.
Figure 3.
Impact of culture supernatants from individual human gut anaerobe strains on C. albicans growth under anaerobic conditions. The whisker boxplot represents % C. albicans growth (T24–T0) when incubated with pure culture supernatants from human gut isolates. The growth of C. albicans alone in fresh NGY medium (black) was monitored via six technical replicates per test (total n = 54). Strains are grouped by family and colour-coded: dark red for Lachnospiraceae; red for Eubacteriaceae; orange for Ruminococcaceae; purple for Erysipelotrichia; blue for Bifidobacteriaceae; light blue for Coriobacteriaceae; green for Bacteroidaceae; light green for Porphyromonadaceae; lilac for Prevotellaceae; and turquoise for Enterobacteriaceae. The cross represents the mean, while the central horizontal line shows the median of six technical replicates per strain (except for ‘Uncharacterized’ sp. D3F10, n = 17; Coprococcus catus GD/7 and Lachnospira eligens DSM 3376T, n = 12; R. bromii DM137M, n = 11; B. adolescentis L2-32, n = 24; andBacteroides fragilis DSM 2151T, n = 11). The Kruskal–Wallis test revealed a highly significant difference between the effects of different supernatants (P< .0001), and Dunn’s post hoc identified multiple gut anaerobes whose culture supernatants significantly inhibited C. albicans growth compared to the C. albicans-only control, as indicated in the figure.
The experiments revealed that the different supernatants varied widely in their effect on C. albicans growth (Fig. 3). Most of the isolates tested, including Co. aerofaciens DSM 3979T, which was correlated with ‘benign’ status in the earlier sequence-based profiling analysis, did not inhibit C. albicans growth. Of note, however, Co. aerofaciens strain DM124M showed a mild inhibitory effect (P< .01; Fig. 3), suggesting that the activity observed may be strain specific. In contrast, the Blautia wexlerae D3M23, Faecalitalea cylindroides DM1E8M, Prevotella copri D3E4B, and Intestinibacter bartlettii 80/4 isolates showed more notable inhibitory effects (average inhibition in the range of 55%–60%, P< .0001). Importantly, B. adolescentis L2-32 was identified as the strongest antagonist among all of the strains tested (63.6% average inhibition, Fig. 3; P< .0001). This was consistent with the 16S rRNA gene-based analysis described above, which had associated bifidobacteria with inhibition of C. albicans in the coculture experiments. Incubation with the bacterial growth medium alone (M2GSC) appeared to promote the growth of C. albicans slightly, although the effect was not statistically significant (Fig. 3), likely due to the presence of glucose in the medium, which C. albicans assimilates for growth.
Because of the strong inhibitory impact displayed by the B. adolescentis strain L2-32 supernatant, combined with the previously identified correlation of this species with strong antagonism against C. albicans in the coculture faecal incubation experiments described above, and the fact that this species is commonly detected in faeces from healthy adults (Matsuki et al. 2004), we next decided to focus on Bifidobacterium isolates and, in particular, on B. adolescentis, in more detail.
Supernatants from specific Bifidobacterium strains inhibited C. albicans growth under anaerobic conditions
To investigate whether different species of bifidobacteria inhibited the growth of C. albicans in vitro, four different bifidobacterial species, including one B. animalis, four B. adolescentis, two B. bifidum, and six B. longum strains, all isolated from the faeces of healthy humans (Table S2, Supporting Information), were screened for inhibition of C. albicans growth using the anaerobic assay described above. As Co. aerofaciens was correlated with less inhibitory effects on C. albicans in the faecal coculture work, we also included supernatants from one strain of this species in these experiments for comparative purposes. The supernatants of all bifidobacteria species tested resulted in 20%–80% C. albicans growth inhibition (relative to C. albicans-only growth in fresh NGY medium), except for B. animalis T1-817, which had no inhibitory activity (Fig. 4). In agreement with the earlier experiments, supernatants from three out of four B. adolescentis strains (L2-32, L2-52, and L2-78) most strongly inhibited C. albicans growth (P< .001; 68%–78% fungal inhibition compared to the no supernatant controls; Fig. 4). In contrast, the type strain B. adolescentis DSM 20083T did not show a strong inhibitory effect, further indicating that the inhibitory activities may be strain-specific. Supernatants from B. bifidum T2-126 and T2-106 cultures were also significantly antagonistic against C. albicans in the anaerobic assay (P< .01 and P< .001, with 42%–49% fungal growth inhibition compared to the control, respectively). Finally, all representatives of the B. longum species tested showed a consistent, nonsignificant, mild inhibitory effect of approximately 20%–30% (Fig. 4).
Figure 4.
Impact of bifidobacterial and Co. aerofaciens culture supernatants on C. albicans growth under anaerobic conditions. The whisker boxplot represents % C. albicans growth (T24–T0) after incubation with culture supernatants from Bifidobacterium spp. or Co. aerofaciens strains isolated from healthy human donors. The crosses and central horizontal lines represent the mean and median, respectively, of six technical replicates per strain or for the C. albicans-only control (black). Strains are colour-coded by species. The Kruskal–Wallis test revealed a highly significant difference between samples (P< .0001), and Dunn’s post hoc test identified specific bifidobacterial isolates that exerted a significant inhibitory effect on C. albicans growth compared to the control.
The inhibitory activity of bifidobacterial supernatants on C. albicans growth correlated with FA production and acidic pH
Having determined that culture supernatants from certain Bifidobacterium species from the human gut exert inhibitory activity against C. albicans, we next investigated the potential mechanisms underlying this phenomenon. As anticipated, quantification of the FA in the bifidobacterial supernatants used in the anaerobic assay revealed that the main organic acids produced by these strains were acetate, lactate, and formate (Table 1). Bifidobacterium adolescentis L2-32 produced the highest levels of the FA (38.1 mM acetate, 9.9 mM lactate, and 4.2 mM formate), followed by B. adolescentis L2-52 (20.7 mM acetate, 8.2 mM lactate, and 4.7 mM formate), and B. adolescentis L2-78 (31.2 mM acetate, 11.4 mM lactate, and 6.2 mM formate; Table 1). The bifidobacterial strains producing the highest total concentrations of these FA, therefore, also displayed the strongest antagonistic activity against C. albicans (Fig. 5). In contrast, we detected low concentrations of organic acids in noninhibitory strain supernatants, such as those from B. animalis T1-817 and from the B. longum strains (Table 1), suggesting that the inhibitory capacity of certain human gut bifidobacteria might be associated with the release of primary metabolites into the supernatant.
Table 1.
Total and individual FA concentrations in the culture supernatants of bifidobacterial strains. In addition to FA profiles for each of the tested bifidobacterial, and Co. aerofaciens, strain supernatants, the corresponding supernatant pH, and % C. albicans growth (T24–T0; n = 6 technical replicates) are also shown. Table also shows Spearman correlation results, indicating that total FA, acetate, lactate and pH were all significantly associated with C. albicans inhibition.
| Strain | % C. albicans growth | Acetate (mM) | Lactate (mM) | Formate (mM) | Total FA | pH |
|---|---|---|---|---|---|---|
| B. adolescentis L2-32 | 22.00 | 37.17 | 8.7 | 8.34 | 54.21 | 5.07 |
| B. adolescentis L2-52 | 32.43 | 20.67 | 8.2 | 4.69 | 33.56 | 5.3 |
| B. adolescentis L2-78 | 30.26 | 31.21 | 11.42 | 6.23 | 48.86 | 5.28 |
| B. adolescentis DSM 20083T | 71.13 | 6.05 | 3.1 | 2.16 | 11.3 | 6.36 |
| B. animalis T1-817 | 100.48 | 7.64 | 0 | 3.81 | 11.45 | 6.76 |
| B. bifidum T2-126 | 51.30 | 13.84 | 2.77 | 3.1 | 19.71 | 6.47 |
| B. bifidum T2-106 | 58.17 | 16.22 | 4.07 | 3.12 | 23.41 | 6.4 |
| B. longum T2-150 | 74.74 | 13.83 | 4.12 | 3.24 | 21.19 | 6.4 |
| B. longum T2-97 | 64.68 | 17.62 | 5.45 | 2.53 | 25.6 | 6.29 |
| B. longum L2-40 | 65.94 | 14.62 | 4.32 | 1.71 | 20.64 | 6.24 |
| B. longum T2-159 | 67.03 | 14.02 | 4.59 | 1.55 | 20.17 | 6.57 |
| B. longum T2-133 | 68.17 | 12.95 | 3.87 | 3.58 | 20.41 | 6.47 |
| B. longum DSM 20219T | 82.52 | 8.74 | 4.22 | 1.64 | 14.6 | 6.29 |
| Co. aerofaciens DSM 3979T | 76.43 | 0.46 | 6.52 | 5.19 | 12.16 | 6.61 |
| M2GSC medium | 101.21 | 9.87 | 0 | 0.72 | 10.59 | 6.65 |
| Spearman coefficient | −0.87 | −0.62 | −0.46 | −0.82 | 0.78 | |
| Corrected P-value (two-tailed) | .0001 | .0043 | .0175 | .0002 | .0003 | |
Figure 5.
The inhibitory effect of Bifidobacterium and Co. aerofaciens isolates positively correlated with total concentration of FA and lower supernatant pH. Spearman correlation revealed that C. albicans inhibition was strongly associated with the FA concentration (A) and pH (B) of the bifidobacterial culture supernatants. Dots are colour-coded according to bacterial species, as per the key in the figure. P-values were corrected using the Benjamini, Krieger, and Yekutieli false discovery rate approach.
To assess whether the inhibitory activity observed in the anaerobic assay was associated with the production of FA, we performed Spearman’s coefficient analysis by plotting the % growth of C. albicans vs. the total amount of FA in the gut bacteria supernatants. We observed a strong positive correlation between total FA levels and fungal growth suppression (r = –0.82; Fig. 5A). Similarly, we noted a strong negative correlation between pH and C. albicans growth, with the lower pH correlating with reduced fungal growth (r = 0.78; Fig. 5B). We also calculated Spearman’s correlation coefficients for the main individual FA produced by the Bifidobacterium strains (Table 1). This analysis revealed that acetate, lactate, and formate concentrations were all significantly associated with C. albicans inhibition.
Sensitivity of C. albicans to individual and combined FA, and pH extremes, under anaerobic growth conditions
We next tested the effect of individual and mixed FA solutions, at concentrations analogous to the previously observed highly inhibitory B. adolescentis supernatants (40–50 mM acetate, 10–15 mM lactate, and 10 mM formate), on C. albicans growth in the anaerobic assay. The FA mixture containing acetate, lactate, and formate significantly reduced C. albicans growth compared to the control over the incubation period (mean fungal inhibition of 38%, P< .001; Fig. 6A). Similarly, the individual FA showed a consistent suppressive effect on C. albicans growth (mean fungal inhibition of approximately 35% compared to controls), despite formate and lactate being added at lower concentrations than acetate (Fig. 6A). This may be related to the fact that lactate and formate are stronger acids (pKa around 3.8) than acetate (pKa of 4.8). However, of note, the extent of inhibition exerted by the individual and mixed FA solutions was inferior to the impact on fungal growth displayed by B. adolescentis L2-32 supernatants in the same test (Fig. 6A). This suggests the potential existence of additional inhibitory factors in the supernatant.
Figure 6.
Impact of single and combined FA, as well as pH, on C. albicans growth under anaerobic conditions. (A) Individual FA and a mixed acid solution at concentrations detected in the most inhibitory (B. adolescentis L2-32) supernatant (40 mM acetate, 10 mM lactate, and 10 mM formate) were tested for their impact on the growth of C. albicans. The whisker boxplot includes the mean and median of six technical replicates as crosses and horizontal lines, respectively. The Kruskal–Wallis test indicated strong differences between the observed values (P< .0001); Dunn’s post hoc test revealed concentration-dependent inhibitory effects of the individual FA, with a particularly strong effect of 15 mM lactate and 10 mM formate, compared to the C. albicans-only control. (B) Effect of pH on C. albicans growth, under anaerobic conditions. pH values were adjusted by modifying NGY culture medium before filter-sterilization. The whisker boxplots show mean and median of four technical replicates. The Kruskal–Wallis test indicated significant differences between the observed values (P = .0024); Dunn’s post hoc testing indicated significant differences in fungal growth between the medium with unadjusted pH (pH 6.3, black), and pH 2 and pH 10. Significance values: ****P< .0001, *** P< .001, ** P< .01, and * P< .05.
We then assessed the sensitivity of C. albicans to pH, by incubating the fungus in NGY culture medium adjusted to pH values ranging from 2 to 10. In contrast to the FA-based tests, pH values within the normal range of those detected in the lower GIT seemed to have little impact on C. albicans growth when tested as the sole variable (Fig. 6B). Indeed, fungal growth was only significantly decreased at extreme pH values, particularly at pH 2 (P< .001) and at pH 10 (P< .05), compared to the fungal growth in unadjusted NGY medium (Fig. 6B). This indicated that the suppression of C. albicans growth observed in the presence of culture supernatants is not driven solely by pH.
Inhibition of C. albicans by bifidobacterial supernatants was mediated via the combined effects of pH and SCFAs
To further uncover the mechanisms underpinning the inhibitory capacity of the B. adolescentis strains tested, we next set up an anaerobic assay to study the effect of the following individual stressors on C. albicans growth: pH alone, exposure to a mixed solution of FA [45 mM acetate, 15 mM lactate, and 10 mM formate, to mimic the concentrations determined in the most inhibitory (B. adolescentis L2-32) supernatant], and bacterial culture supernatants. To better understand the combinatorial role of FA concentration and pH, we conducted the tests at different controlled pH values, in the range from 4 to 7, adjusting either the medium, or the test solution/supernatant.
Consistent with the previous observations (Fig. 6), C. albicans was highly resilient to the pH range tested under anaerobic conditions (Fig. 7). Critically though, altering the pH significantly impacted the inhibitory activity of the tested supernatants, and the FA mix. In all cases, these treatments were most inhibitory at the lowest pH tested (pH 4), and progressively lost potency against C. albicans as the pH increased (Fig. 7). This indicated that pH and FA combine to produce an inhibitory effect on C. albicans.
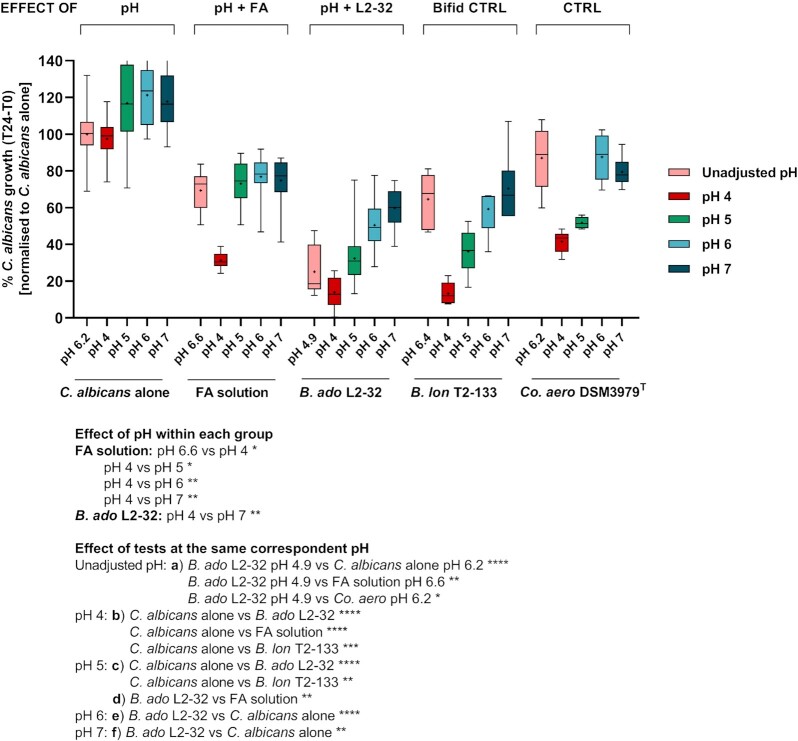
Figure 7.
Cumulative impact of pH and FA on C. albicans growth under anaerobic conditions. The whisker boxplot shows C. albicans growth when tested at different controlled pH values, adjusting either the medium or the test solution/supernatant in the range from pH 4 to 7, under anaerobic conditions. Crosses and central horizontal lines represent the mean and median, respectively, of 12 technical replicates per test (n = 32 for C. albicans alone at pH 6.2, n = 18 for FA solution at all tested pH values, n = 24 for both B. adolescentis L2-32 and Co. aerofaciens DSM 3979T at all tested pH values). The Kruskal–Wallis test indicated highly significant differences between groups (P< .0001); Dunn’s post hoc test, comparing the observations against each other, indicated significant differences within each group at different pH values (colour-coded as per legend) and between groups at the same corresponding pH, indicated separately in the Figure legend as ‘effect of pH’ and ‘effect of tests at the same correspondent pH’, respectively. Significance: ****P< .0001, *** P< .001, ** P < .01, and * P< .05. B. ado, Bifidobacterium adolescentis; B. lon, Bifidobacterium longum; Co. aero, Collinsella aerofaciens.
Of note, the antagonistic effect of the B. adolescentis L2-32 supernatant was significantly more pronounced than that of the SCFA solution at pH 5, as well as to that of a solution with an unadjusted pH value (Fig. 7), again suggesting that the bacterial supernatant might contain additional, but currently unidentified, inhibitory factors.
Discussion
Candida albicans is a major clinical challenge because of high mortality in susceptible patients, emerging resistance against antifungal and sanitizing agents, and the limited availability of additional therapeutic options (Pfaller et al. 2000). Alternative strategies to reduce carriage and dissemination of C. albicans in the gut should, therefore, be explored. The healthy intestinal microbiota is an appealing source of novel treatments, considering the well-established role it plays in protecting against systemic candidiasis by hindering fungal expansion and pathogenic initiation in the gut (Kennedy and Volz 1985a,b).
It is noteworthy that the level of C. albicans inhibition observed in some of our faecal coculture experiments (Fig. 1) were much greater than those observed with single-species supernatants (Fig. 3), indicating that the cumulative activities of the gut microbiota may be more powerful inhibitors of C. albicans than individual constituent species. Nonetheless, we also showed that the degree of inhibition from mixed microbial communities is highly variable, strongly indicating that some gut microbiota species are likely to be inherently more inhibitory against C. albicans than others. However, the gut microbiota is extremely complex and it is currently largely unknown which components are most likely to be potent inhibitors of C. albicans in the gut. We demonstrate here that B. adolescentis culture supernatants exert strong inhibitory activity against C. albicans under anaerobic conditions in vitro, and identified an inhibitory effect of secreted bacterial FA and prevailing pH on C. albicans growth. These observations were in agreement with our DNA sequence-based analysis correlating the presence of B. adolescentis with the inhibition of C. albicans in mixed coculture with faecal microbiota samples, under conditions mimicking the human colonic environment.
The Bifidobacterium genus is dominant in the colon of breast-fed infants (Khonsari et al. 2016, Yatsunenko et al. 2012) and it also accounts for approximately 5% of the microbiota in adults, of which the species B. adolescentis is a prevalent representative (Reuter 1963). Importantly, B. adolescentis is also enriched following consumption of resistant starch (Ze et al. 2012), and produces high amounts of organic acids as a result of carbohydrate fermentation (Table 1). Despite the relatively low proportional abundance of this genus in the total microbiota in adults, it has potential health benefits for the host (Fukuda et al. 2011, Rivière et al. 2014, Rossi et al. 2005). Aside from FA production, bifidobacteria have also been demonstrated to induce the anti-inflammatory cascade (Lammers et al. 2003, Meng et al. 2016), and improve colonization resistance against common food-borne pathogens such as E. coli O157: H7 and Salmonella enterica serovar Typhimurium (Fukuda et al. 2011, Makras and De Vuyst 2006, Ventura et al. 2016). In addition, B. adolescentis colonizes the epithelial mucus layer and may therefore out-compete pathogens for adhesion sites on the gut epithelium (Tan et al. 2016, Ventura et al. 2016), potentially impacting the biofilm formation that can be an important virulence factor in C. albicans (Gulati and Nobile 2016).
Importantly, previous work in immunosuppressed gnotobiotic mice showed that supplementation with the bifidobacterial species B. infantis and B. lactis suppressed C. albicans growth in the intestines of the mice, and reduced systemic candidiasis (Wagner et al. 1998). Bifidobacteria were also recently predicted as major antagonists against C. albicans in an in silico model of intermicrobial interactions in the human gut (Mirhakkak et al. 2020). Bifidobacteria such as B. adolescentis may, therefore, be promising candidates for novel microbiota-based therapeutics aimed at enhancing colonization resistance. Several clinical trials have reported some efficacy of probiotic supplementation of Bifidobacterium and Lactobacillus spp. in reducing C. albicans intestinal colonization and preventing invasive fungal sepsis in infants following antibiotic treatment (Romeo et al. 2011, Roy et al. 2014). Furthermore, because B. adolescentis is a common member of the adult gut microbiota (present in up to 83% of healthy adults; Junick and Blaut 2012, Matsuki et al. 1999) and responds to changes in the diet, the growth and metabolic activities of this species could potentially be modulated in vivo by prebiotic supplementation.
Aside from bifidobacteria, other gut bacterial taxa are also likely worthy of further study. For example, we also observed inhibitory effects against C. albicans by a number of other gut bacterial species (Fig. 3). Wider screening of gut bacterial isolates is, therefore, highly likely to identify additional candidates with pronounced anti-Candida activity. In contrast, we also identified bacterial supernatants with little effect on C. albicans growth, such as those derived from Flavonifractor plautii and Hungatella hathewayi. This is consistent with reports that the relative abundances of these two bacterial species are correlated with C. albicans levels in faecal samples from cancer patients (Mirhakkak et al. 2020). Our results also highlight that different strains of the same gut bacterial species may have varying impacts on C. albicans growth (Fig. 3). Better understanding of the mechanistic basis for some of the putative interactions, both beneficial and detrimental, between specific gut bacteria and C. albicans may help to prioritize candidates for further study as potential novel therapeutics.
Although specific members of the gut microbiota appear to be particularly inhibitory against C. albicans it must be acknowledged, however, that the in vitro-based inhibitions observed here may not be mirrored in vivo, where the inhibitory gut anaerobes must simultaneously compete with the dense and complex wider gut microbiota. Interactions between members of the gut microbiota might enhance or restrict any secreted anti-Candida activities via mechanisms such as competitive exclusion of the putatively beneficial gut bacteria, or suppression of metabolic activity. Further development of any single-species therapeutics will, therefore, require extensive testing for efficacy in vivo. Indeed, work by Maldonado-Gómez et al. (2016), using B. longum as an exemplar probiotic, has shown quite clearly that the engraftment success of such probiotics depends in large part on niches being available for colonization, and that this is strongly dependent on the baseline microbiota at time of probiotic consumption. This may mean that efficacy of supplemented therapeutics will vary between individuals.
Furthermore, relatively little is currently known about the potential health impacts of the vast majority of gut anaerobes. There is also some evidence to suggest that consumption of single-species therapies can impact on wider microbiota dynamics after supplementation into the gut ecosystem, and might actually delay microbiota reconstitution following perturbation episodes (Suez et al. 2018). Therefore, in addition to efficacy testing, extensive safety testing will likely also be required, particularly if these gut bacteria were to be used in the immunocompromised individuals who are typically at greatest risk of severe/systemic C. albicans infection.
A key mechanistic result of the current study is demonstrating the combinatorial effect of FA and pH on the growth of C. albicans. Our findings are consistent with previous work indicating that the protonated form of weak acids freely permeate and accumulate inside microbial cells, causing dissipation of the proton motive force (Axe and Bailey 1995), triggering energetically expensive stress responses (Henriques et al. 1997), and perturbation of essential metabolic reactions (Cottier et al. 2015, Jacobsen et al. 2018, Lourenço et al. 2018). Gut microbiota FA are, therefore, thought to play important roles in limiting C. albicans intestinal colonization in vivo (Guinan et al. 2019, Huang et al. 2011), and a decrease in caecal SCFA concentrations following antibiotic treatment is associated with increased C. albicans load in the faeces in mouse models (Bohnhoff et al. 1964, Guinan et al.2019).
In agreement with our observations presented here, C. albicans was shown to be susceptible to formate (Mirhakkak et al. 2020) and acetate, at concentrations of over 30 mM, in vitro, and the effect is aggravated by microaerophilic conditions (Lourenço et al. 2018). Further, acetate inhibits hyphal morphogenesis of C. albicans, and this inhibition of hyphae formation could, therefore, reduce fungal translocation through the epithelial barrier (Guinan et al. 2019). In contrast, previous work has shown that lactate does not impair fungal growth at concentrations tested in our study, even at low pH values, and under aerophilic/microaerophilic conditions (Lourenço et al. 2018). Indeed, lactate is a potential energy source for C. albicans under hypoxic conditions, and is known to induce sustained fungal resistance to osmotic and cell wall stress, via cell wall remodelling (Ene et al. 2012a,b, 2015). Nonetheless, substantial lactate release (up to approximately 110 mM), among other factors, is postulated to contribute to lactic acid bacteria-mediated colonization resistance to C. albicans in the vaginal tract (Köhler et al. 2012, Zangl et al.2020).
Importantly, the total FA and acetate concentrations that C. albicans cells were exposed to in this study are physiologically relevant for regions of the human GIT such as the proximal colon (Cummings and Macfarlane 1991). Indeed, total SCFA levels may reach up to 200 mM in the proximal colon (Cummings and Macfarlane 1991), suggesting that inhibition of C. albicans growth mediated by total FA may be greater than indicated by our pure culture studies, and may represent a key mechanism of colonization resistance to this opportunistic fungus. In contrast, the concentrations of formate and lactate detected here in the bifidobacterial culture supernatants appear to be slightly higher than those detected in human faecal samples, where they do not usually exceed 5–10 mM, as they are absorbed by the host or utilized by other bacteria (Duncan et al. 2007, Hove et al. 1994). Additionally, the finding that supernatants were often more inhibitory than defined FA mixtures (Figs 6 and 7) suggests that additional, but currently unidentified, antifungal compounds may be produced by some gut anaerobes. This may be a worthwhile avenue for further study.
Conclusions
In this in vitro study we showed that the degree of C. albicans growth inhibition by mixed human faecal microbiota communities can vary greatly between individual faecal donors. Specific components, such as B. adolescentis, were identified as being more antagonistic against C. albicans than other tested gut microbiota species. Inhibitory activity was predominantly driven by the release of FA, and the concomitant drop in ambient pH. The potential for altering the gut microbiota composition, e.g. by consumption of probiotics such as B. adolescentis, or increasing in vivo SCFA concentrations by consumption of dietary fibres such as resistant starch, are worthy of further study to determine whether these can bolster colonization resistance against C. albicans in the gut.
Supplementary Material
Acknowledgements
We thank Dr Donna M. MacCallum for critical reading of the manuscript, the Centre for Genome-Enabled Biology and Medicine at the University of Aberdeen for carrying out the 16S rRNA gene sequencing, and Donna Henderson for the GC analysis of bacterial fermentation acids. The authors also acknowledge the support of the Maxwell computer cluster funded by the University of Aberdeen.
Contributor Information
Liviana Ricci, Rowett Institute, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom; CIBIO - Department of Cellular, Computational and Integrative Biology, University of Trento, Trento, 38123, Italy.
Joanna Mackie, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom.
Gillian E Donachie, Rowett Institute, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom.
Ambre Chapuis, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom.
Kristýna Mezerová, Department of Microbiology, Faculty of Medicine and Dentistry, Palacký University Olomouc, Olomouc, 77515, Czech Republic.
Megan D Lenardon, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom; School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW 2052, Australia.
Alistair J P Brown, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom; MRC Centre for Medical Mycology, University of Exeter, Exeter, EX4 4QD, United Kingdom.
Sylvia H Duncan, Rowett Institute, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom.
Alan W Walker, Rowett Institute, University of Aberdeen, Aberdeen, AB25 2ZD, United Kingdom.
Authors’ contributions
A.W.W., S.H.D., A.J.P.B., and M.D.L. conceived of the research and designed the experiments. J.M., G.E.D., and A.C. carried out the batch coculture experiments and analyzed the resulting data. L.R. performed the rest of the experiments and analyzed the data. K.M. and L.R. isolated novel gut bacterial strains that were used in these experiments. L.R., S.H.D., and A.W.W. wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors have no conflicts of interest to declare.
Funding
Initial studies were funded from a Wellcome Institutional Strategic Support Fund (ISSF) Seed Corn Award (105625/Z/14/Z). Thereafter, the research was funded by the Scottish Government’s Rural and Environment Science and Analytical Services (RESAS) division. A.J.P.B. was supported by programme grants from the UK Medical Research Council (MR/M026663/1 and MR/M026663/2) and by the Medical Research Council Centre for Medical Mycology (MR/N006364/1 and MR/N006364/2).
References
- Axe D, Bailey J. Transport of lactate and acetate through the energized cytoplasmic membrane of Escherichiacoli. Biotechnol Bioeng. 1995;47:8–19. [DOI] [PubMed] [Google Scholar]
- Bohnhoff M, Miller CP, Martin WR. Resistance of the mouse's intestinal tract to experimental Salmonella infection. Factors which interfere with the initiation of infection by oral inoculation. J Exp Med. 1964;120:805–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR. et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165–78. [DOI] [PubMed] [Google Scholar]
- Bryant MP. Commentary on the hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–8. [DOI] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridiumdifficile. Nature. 2015;517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll IM, Ringel-Kulka T, Siddle JP. et al. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Mot. 2012;24:521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham C V, Behrendt CL. et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington CA, Hinton M, Pearson GR. et al. Short-chain organic acids at ph 5.0 kill Escherichiacoli and Salmonella spp. without causing membrane perturbation. J Appl Bacteriol. 1991;70:161–5. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA. et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottier F, Tan ASM, Chen J. et al. The transcriptional stress response of Candidaalbicans to weak organic acids. G3 Genes Genomes Genet. 2015;5:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70:443–59. [DOI] [PubMed] [Google Scholar]
- Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22:763–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert C, Kaune A-K, Alaban L-R. et al. The impact of the fungus-host-microbiota interplay upon Candidaalbicans infections: current knowledge and new perspectives. FEMS Microbiol Rev. 2020;45:fuaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M. et al. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;116:1477–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Belenguer A, Holtrop G. et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Adya AK, Wehmeier S. et al. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 2012a;14:1319–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Heilmann CJ, Sorgo AG. et al. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candidaalbicans. Proteomics. 2012b;12:3164–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Walker LA, Schiavone M. et al. Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. MBio. 2015;6:e00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Coughlin LA, Neubauer MM. et al. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candidaalbicans colonization. Nat Med. 2015;21:808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP. et al. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74:13–22. [DOI] [PubMed] [Google Scholar]
- Freter R, Brickner H, Botney M. et al. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun. 1983;39:676–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase Ket al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. [DOI] [PubMed] [Google Scholar]
- Geirnaert A, Calatayud M, Grootaert C. et al. Butyrate-producing bacteria supplemented in vitro to Crohn's disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Nature. 2017;7:11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum A, Tsay E, Kirsch D. Isolation of the Candidaalbicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–82. [DOI] [PubMed] [Google Scholar]
- Guinan J, Wang S, Hazbun TR. et al. Antibiotic-induced decreases in the levels of microbial-derived short-chain fatty acids correlate with increased gastrointestinal colonization of Candidaalbicans. Sci Rep. 2019;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect. 2014;20:5–10. [DOI] [PubMed] [Google Scholar]
- Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18:310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnett J, Myers SP, Rolfe M. Significantly higher faecal counts of the yeasts Candida and Saccharomyces identified in people with coeliac disease. Gut Pathog. 2017;9:28484520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney H, Laing J, Paterson L. et al. The environmental stress sensitivities of pathogenic Candida species, including Candidaauris, and implications for their spread in the hospital setting. Med Mycol. 2020;58:744–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques M, Quintas C, Loureiro-Dias M. Extrusion of benzoic acid in Saccharomycescerevisiae by an energy-dependent mechanism. Microbiology. 1997;143:1877–83. [DOI] [PubMed] [Google Scholar]
- Hobson PN. Rumen bacteria. Methods Microbiol. 1969;3B:133–49. [Google Scholar]
- Hove H, Norgard Andersen I, Mortensen PB. Fecal DL-lactate concentrations in 100 gastrointestinal patients. Scand J Gastroenterol. 1994;29:255–9. [DOI] [PubMed] [Google Scholar]
- Huang CB, Alimova Y, Myers TM. et al. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol. 2011;56:650–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube B. From commensal to pathogen: stage- and tissue-specific gene expression of Candidaalbicans. Curr Opin Microbiol. 2004;7:336–41. [DOI] [PubMed] [Google Scholar]
- Jacobson A, Lam L, Rajendram M. et al. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe. 2018;24:296–307.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y. et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junick J, Blaut M. Quantification of human fecal Bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl Environ Microbiol. 2012;78:2613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW. et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Volz PA. Ecology of Candidaalbicans gut colonization: inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect Immun. 1985a;49:654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Volz PA. Effect of various antibiotics on gastrointestinal colonization and dissemination by Candidaalbicans. Med Mycol. 1985b;23:265–73. [DOI] [PubMed] [Google Scholar]
- Khonsari S, Suganthy M, Burczynska B. et al. A comparative study of bifidobacteria in human babies and adults. Biosci Microbiota Food Heal. 2016;35:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P. et al. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–45. [DOI] [PubMed] [Google Scholar]
- Köhler GA, Assefa S, Reid G. Probiotic interference of Lactobacillusrhamnosus GR-1 and Lactobacillusreuteri RC-14 with the opportunistic fungal pathogen Candidaalbicans. Infect Dis Obstet Gynecol. 2012;2012:636474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers KM, Brigidi P, Vitali B. et al. Immunomodulatory effects of probiotic bacteria DNA: IL-1 and IL-10 response in human peripheral blood mononuclear cells. FEMS Immunol Med Microbiol. 2003;38:165–72. [DOI] [PubMed] [Google Scholar]
- León C, Ruiz-Santana S, Saavedra Pet al. Usefulness of the "Candida score" for discriminating between Candida colonization and invasive candidiasis in non-neutropenic critically ill patients: a prospective multicenter study. Crit Care Med. 2009;37:1624–33. [DOI] [PubMed] [Google Scholar]
- Lourenço A, Pedro NA, Salazar SB. et al. Effect of acetic acid and lactic acid at low pH in growth and azole resistance of Candidaalbicans and Candidaglabrata. Front Microbiol. 2018;9:3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low CY, Rotstein C. Emerging fungal infections in immunocompromised patients. F1000 Med Rep. 2011;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Findon H, Kenny CC. et al. Different consequences of ACE2 and SWI5 gene disruptions for virulence of pathogenic and nonpathogenic yeasts. Infect Immun. 2006;74:5244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makras L, De Vuyst L. The in vitro inhibition of Gram-negative pathogenic bacteria by bifidobacteria is caused by the production of organic acids. Int Dairy J. 2006;16:1049–57. [Google Scholar]
- Maldonado-Gómez MX, Martínez I, Bottacini F. et al. Stable engraftment of Bifidobacteriumlongum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–26. [DOI] [PubMed] [Google Scholar]
- Maltby R, Leatham-Jensen MP, Gibson T. et al. Nutritional basis for colonization resistance by human commensal Escherichiacoli strains HS and nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS ONE. 2013;8:e53957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteyn B, Scorza FB, Sansonetti PJ. et al. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol. 2011;13:171–6. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Denizot J, Dreux N. et al. Western diet induces dysbiosis with increased E. coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63:116–24. [DOI] [PubMed] [Google Scholar]
- Mason KL, Downward JRE, Mason KD. et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80:3371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Watanabe K, Fujimoto J. et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70:167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Watanabe K, Tanaka R. et al. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Zhu W, Ganguli K. et al. Anti-inflammatory effects of Bifidobacteriumlongum subsp infantis secretions on fetal human enterocytes are mediated by TLR-4 receptors. Am J Physiol Liver Physiol. 2016;311:G744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda LN, van der Heijden IM, Costa SF. et al. Candida colonisation as a source for candidaemia. J Hosp Infect. 2009;72:9–16. [DOI] [PubMed] [Google Scholar]
- Mirhakkak MH, Schäuble S, Klassert TE. et al. Metabolic modeling predicts specific gut bacteria as key determinants for Candidaalbicans colonization levels. ISME J. 2020;15:1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Martin J., Marinsek-Logar R. et al. Degradation and utilization of xylans by the rumen anaerobe Prevotellabryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe. 1997;3:373–81. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Clausen MR. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol. 1996;31:132–48. [DOI] [PubMed] [Google Scholar]
- Nash AK, Auchtung TA, Wong MC. et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 2017;5:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville BA, d'Enfert C, Bougnoux M-E. Candida albicans commensalism in the gastrointestinal tract. Munroe C (ed.), FEMS Yeast Res. 2015;15:fov081. [DOI] [PubMed] [Google Scholar]
- Odds FC, Kibbler CC, Walker E. et al. Carriage of Candida species and C. albicans biotypes in patients undergoing chemotherapy or bone marrow transplantation for haematological disease. J Clin Pathol. 1989;42:1259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Molecular phylogenetics and epidemiology of Candidaalbicans. Fut Microbiol. 2010;5:67–79. [DOI] [PubMed] [Google Scholar]
- Pasolli E, Asnicar F, Manara S. et al. Extensive unexplored human microbiome diversity revealed by over 150 000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Jones RN, Doern G V. et al. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997-1998. Antimicrob Agents Chemother. 2000;44:747–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet D, Monod M, Suter PM. et al. Candida colonization and subsequent infections in critically ill surgical patients. Ann Surg. 1994;220:751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea MC, Sit CS, Clayton E. et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridiumdifficile. Proc Natl Acad Sci. 2010;107:9352–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G. Comparative studies on the bifidus flora in the feces of infants and adults. Zentralbl Bakteriol Orig. 1963;191:486–507. [PubMed] [Google Scholar]
- Richardson AJ, Calder AG, Stewart CS. et al. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett Appl Microbiol. 1989;9:5–8. [Google Scholar]
- Rivera-Chávez F, Zhang LF, Faber F. et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière A, Moens F, Selak M. et al. The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl Environ Microbiol. 2014;80:204–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe AJ, O'Byrne C, McLaggan D. et al. Inhibition of Escherichiacoli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology. 2002;148:2215–22. [DOI] [PubMed] [Google Scholar]
- Romeo MG, Romeo DM, Trovato L. et al. Role of probiotics in the prevention of the enteric colonization by Candida in preterm newborns: incidence of late-onset sepsis and neurological outcome. J Perinatol. 2011;31:63–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Corradini C, Amaretti A. et al. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol. 2005;71:6150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O, Khan MT, Schwarzer M. et al. Faecalibacterium prausnitzii strain HTF-F and its extracellular polymeric matrix attenuate clinical parameters in DSS-induced colitis. Riedel CU (ed.), PLoS ONE. 2015;10:e0123013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Chaudhuri J, Sarkar D. et al. Role of enteric supplementation of probiotics on late-onset sepsis by Candida species in preterm low birth weight neonates: a randomized, double blind, placebo-controlled trial. N Am J Med Sci. 2014;6:50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonis G, Anastassiadou H, Dassiou Met al. Effects of broad-spectrum antibiotics on colonization of gastrointestinal tracts of mice by Candida albicans. Antimicrob Agents Chemother. 1994;38:602–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokari RM, Vaughan EE, Akkermans ADL. et al. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW. et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. Waldor M (ed.), PLoS Biol. 2007;5:e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J, Zmora N, Zilberman-Schapira G. et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–23.e16. [DOI] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N. et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci. 2016;113:E8141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zmora N, Levy M. et al. The microbiome and innate immunity. Nature. 2016;535:65–74. [DOI] [PubMed] [Google Scholar]
- Ventura M, Montes SA, Ruiz L. et al. High iron-sequestrating bifidobacteria inhibit enteropathogen growth and adhesion to intestinal epithelial cells in vitro. Front Microbiol. 2016;7:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollaard EJ, Clasener HAL, Janssen AJHM. Co-trimoxazole impairs colonization resistance in healthy volunteers. J Antimicrob Chemother. 1992;30:685–91. [DOI] [PubMed] [Google Scholar]
- Wagner RD, Warner T, Pierson C. et al. Biotherapeutic effects of Bifidobacterium spp. on orogastric and systemic candidiasis in immunodeficient mice. Rev Iberoam Micol. 1998;15:265–70. [PubMed] [Google Scholar]
- Whaley SG, Berkow EL, Rybak JM. et al. Azole antifungal resistance in Candidaalbicans and emerging non-albicansCandida species. Front Microbiol. 2016;7:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Perspective prokaryotes: the unseen majority. Proc Natl Acad Sci. 1998;95:6578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridiumdifficile by the colonic microflora. Infect Immun. 1988;56:2610–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ. et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangl I, Pap IJ, Aspöck C. et al. The role of Lactobacillus species in the control of Candida via biotrophic interactions. Microbial Cell. 2020;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, Duncan SH, Louis P. et al. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Ola M, Rolling T. et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med. 2020;26:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.