Abstract
The cloning of the gene encoding the KlGpa1p subunit was achieved by standard PCR techniques and by screening a Kluyveromyces lactis genomic library using the PCR product as a probe. The full-length open reading frame spans 1,344 nucleotides including the stop codon. The deduced primary structure of the protein (447 amino acid residues) strongly resembles that of Gpa1p, the G-protein α subunit from Saccharomyces cerevisiae involved in the mating pheromone response pathway. Nevertheless, unlike disruption of Gpa1 from S. cerevisiae, disruption of KlGpa1 rendered viable cells with a reduced capacity to mate. Expression of a plasmidic KlGpa1 copy in a ΔKlgpa1 mutant restores full mating competence; hence we conclude that KlGpa1p plays a positive role in the mating pathway. Overexpression of the constitutive subunit KlGpa1p(K364) (GTP bound) does not induce constitutive mating; instead it partially blocks wild-type mating and is unable to reverse the sterile phenotype of ΔKlgpa1 mutant cells. K. lactis expresses a second Gα subunit, KlGpa2p, which is involved in regulating cyclic AMP levels upon glucose stimulation. This subunit does not rescue ΔKlgpa1 cells from sterility; instead, overproduction of KlGpa2p slightly reduces the mating of wild-type cells, suggesting cross talk within the pheromone response pathway mediated by KlGpa1p and glucose metabolism mediated by KlGpa2p. The ΔKlgpa1 ΔKlgpa2 double mutant, although viable, showed the mating deficiency observed in the single ΔKlgpa1 mutant.
Saccharomyces cerevisiae cells respond to mating pheromones by inducing activation of a G protein coupled to serpentine receptors (reviewed in reference 17). The G protein, composed of Gα (Gpa1p), Gβ (Ste4p), and Gγ (Ste18p) subunits, activates a phosphorylation cascade that involves at least Ste20p, the mitogen-activated protein kinase module, and transcription activators (reviewed in reference 9) that finally induce growth arrest, shmoo formation, induction of genes required for membrane fusion, including Fus1, and diploid formation. Epistasis analysis has established that the mating response is mediated by the Ste4 and Ste18 gene products and that lack of Gpa1p in S. cerevisiae confers lethality and lack of either Ste4p or Ste18p gives rise to sterility (6, 29).
A second Gα (Gpa2p) subunit (19) was identified and was involved, along with Ras2p, in the activation of adenylyl cyclase to induce cyclic AMP (cAMP) production in response to glucose stimulus. Isolation of Gpr1p, the seven-transmembrane segment receptor coupled to Gpa2p, showed that the Gpr1p/Gpa2p pathway acts in parallel to the Ras/cAMP pathway monitoring nutrient signals (4, 30). It has been shown that Gpa2p regulates growth and pseudohyphal development generated by nitrogen starvation (14).
In S. cerevisiae Gpa1p and Gpa2p subunits play divergent and unrelated roles in signal transduction, since Gpa1p does not participate in the integration of nutrient signals and Gpa2p does not participate in the pheromone response pathway required for mating.
Kluyveromyces lactis is a heterothallic budding yeast that is essentially aerobic (reviewed in reference 28). Its life cycle resembles that of S. cerevisiae, and the haploid and diploid vegetative growth and sexual reproduction of the two are highly similar. K. lactis shows two mating partners that undergo sexual reproduction when they respond to sexual pheromones. Mata and Matα cells are able to mate with each other to produce transient diploids that sporulate to generate four spores that germinate to haploid clones of two different mating types (8). We hypothesize that an S. cerevisiae-like pheromone response pathway is conserved in K. lactis. In fact a gene encoding a protein highly homologous to the α mating factor of S. cerevisiae has been identified (3). This sexual pheromone is thought to activate a pathway that finally turns on transcription factor KlSte12p (31). This factor is thought to bind to pheromone response elements located in promoter regions of genes required for mating. We are interested in searching for homologue genes coding for components of the G protein putatively involved in transducing the pheromone stimulus. In particular, here we describe the characterization of a gene encoding a Gα subunit that participates in the mating pathway. Like S. cerevisiae, K. lactis expresses a second Gα subunit (Gpa2p) that is directly involved in regulation of cAMP levels in response to glucose stimulus (25).
MATERIALS AND METHODS
Strains, plasmids, and media.
K. lactis strains used in this study were WM37 (NRRL Y-1140) (Mata his3); MD2/1 (MATa argA lysA uraA) (obtained from H. Fukuhara); 12/8 (Mata argA lysA uraA), a segregant of the cross between WM37 and MD2/1 that was used as tester strain; 155 (Matα ade2 his3 uraA), a segregant of the cross between MD2/1 and KA5-6C (Mata ade2 his3 leu1), of unknown origin (obtained from A. Brunner); and 136 (Mata argA his3 uraA), a segregant of the cross between WM37 and MD2/1. Homozygotic his3 uraA diploids were freshly generated by crossing strain 155 with strain 136. S. cerevisiae strains used in this study were W303-1A (Mata ade2-1 his3 leu2 trp1 ura3 can1-100), W303-3B (Mata ade2-1 his3 leu2 trp1 ura3 can1-100 scg1::Ura3), and 70 (Matα thr3). Escherichia coli strain DHα5 was used to propagate plasmids. Phagemid pTZ18R was used to subclone DNA fragments for sequencing. Vector YEpKD, which contains the Ura3 marker, was constructed by replacing the 2μm replication origin of vector YEp352 (10) by replication origin pKD from vector KEp6 (obtained from H. Fukuhara). Vector YEpKDHis was constructed by replacing the Ura3 marker by the His3 marker in YEpKD. The multicopy plasmid YEpKDHis-KlGpa1 was constructed by subcloning a 2.2-kb PstI fragment carrying the complete KlGpa1 open reading frame (ORF) into YEpKDHis digested with the same enzyme. This places KlGpa1 under the control of its own promoter. YEpKDHis-KlGpa2 was constructed by subcloning a 1.9-kb SalI-BamHI fragment that carries the KlGpa2 ORF and its promoter into YEpKDHis digested with the same enzymes. Construction of a genomic DNA library from K. lactis was described previously (18). YPD medium contained 1% yeast extract, 1% Bacto peptone, and 2% glucose. SD medium contained 0.67% yeast nitrogen base without amino acids, 2% glucose, and 25 mg of the required amino acids/ml. Luria-Bertani medium supplemented with 50 mg of ampicillin/ml was used to grow recombinant bacteria.
PCR-mediated mutagenesis.
Construction of KlGpa1p(K364) was done by standard PCR amplification. The 5′ AATATCTACTTTTTTTAAGAATAGT 3′ oligodeoxynucleotide (from position 1077 to 1101), which replaces cytosine with adenine at position 1089 of the sense strand (thereby replacing N364 with K in the putative protein), was used as the backward primer in a reaction that yielded a 277-bp amplification product. This fragment was then used as the forward primer in a second reaction that yielded a 643-bp product containing the naturally occurring sites ClaI (position 1016) and BstEII (93 bp beyond the stop codon). The ClaI-BstEII fragment was then subcloned back into the original wild-type gene. PCR conditions were as follows: 94°C for 5 min; 50 cycles of 94°C for 30 s, 45°C for 45 s, and 72°C for 60 s; and a final extension of 10 min at 72°C.
Cloning and sequencing.
The screening of a K. lactis genomic DNA library was done using the PCR product as a probe labeled with [α-32P]dCTP by random-primer DNA-labeling systems (Life Technologies). Positive clones were mapped with restriction enzymes, and one of them, containing the full gene, was sequenced in an automatic sequencer (ABI Prism 310; Applied Biosystems) at the Molecular Biology Facilities of the Instituto de Fisiología Celular, Universidad Nacional Autónoma de México. The sequence was analyzed using the GCG program from the Wisconsin Sequence Analysis Package.
Gene disruption.
The Ura3 marker, obtained as an HpaI/BamHI fragment (filled-in HpaI), was inserted into KlGpa1 previously digested at the naturally occurring sites XhoI (position 770) and BamHI (position 914) (filled-in XhoI). The resulting plasmid was treated with BglII, which digests KlGpa1 at positions 377 and 1119, to give rise to a cassette that carries the Ura3 marker flanked by fragments of 138 and 218 bp of the Klgpa1 ORF. This cassette was used to transfect haploid and diploid strains. Disruption of the KlGpa2 allele with the Ura3 marker was described previously (25). Disruption of KlGpa2 with the His3 cassette was as follows. The KlGpa2 gene was opened with BglII at positions 348, 408, and 478. A 1.8-kb BamHI fragment containing the His3 gene was then inserted. This construction was used as the template in a PCR using primers directed against KlGpa2 at positions 145 (forward) and 691 (backward), giving rise to a cassette containing the His3 gene flanked by KlGpa2 sequences of 204 and 213 bp, respectively. This was used to transfect haploid and diploid strains. Disruptions were confirmed by Southern blotting techniques.
Southern blot and Northern blot analysis.
Chromosomal and plasmidic DNA blots as well as RNA blots were hybridized with a 32P-labeled probe; the probe was labeled with the randomly primed system kit (Gibco BRL). Blots were hybridized overnight at 55°C and washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) and twice with 1× SSC–0.1% SDS. All washes were done at 65°C for 10 min.
Mating assays.
A patch of cells of the strain to be tested was grown on a plate of selective medium for 24 h. The tester strain was grown as a lawn on a YPD plate for 24 h. Both strains were replica plated onto a YPD plate and incubated overnight at 30°C to allow cells to mate. Diploids were selected on SD medium by replica plating. For quantitative mating assays, strains to be tested were grown until mid-log phase in YPD medium. Then 106 cells were mixed with 106 cells of the tester strain, collected on a nitrocellulose membrane filter, placed on a YPD plate, and incubated overnight at 30°C. The cells from each filter were suspended in water, diluted, and plated on SD medium.
Two-hybrid interaction assays.
Assays of physical interaction were done with the LexA-B42 two-hybrid systems as described previously (7). S. cerevisiae Gpa1 and Ste4 genes were subcloned into pJG4-5 and pEG202 as described previously (21). KlGpa1 and KlGpa2 were amplified by PCR, introducing NcoI (position −1) and EcoRI (position −5)/BamHI (18 nucleotides after the stop codon) sites, respectively. PCR products were subcloned in frame into pEG202.
Other.
Expression of Fus1-LacZ fusion was done as described previously (21). Molecular biology procedures were performed as described by Sambrook et al. (24). Standard yeast genetics procedures were done as described by Sherman et al. (26).
Nucleotide sequence accession number.
The sequence obtained in this study has been assigned GenBank accession no. AF135552.
RESULTS
By means of PCR we were able to amplify two products belonging to genes encoding α subunits of heterotrimeric G proteins. We have already demonstrated that one of these genes, KlGpa2, encodes a subunit directly involved in the regulation of cAMP levels in K. lactis (25). Cells lacking KlGpa2p are viable but fail to respond to transient stimulus by glucose and the cAMP level drops significantly, indicating that the G protein containing KlGpa2p is involved in regulating the activity of adenylyl cyclase and participates in a pathway related to monitoring the nutrient status of the cell (25).
The other PCR product was used as a probe to screen a K. lactis genomic library, looking for the full-length gene. Two positive clones were obtained; one showed a 2.2-kb PstI fragment that contained the complete ORF, as inferred from Southern blot analysis. The full sequence of this fragment contains an ORF of 1,344 nucleotides including the stop codon, which codes for a putative protein of 447 amino acid residues with a molecular mass of 50,739 Da.
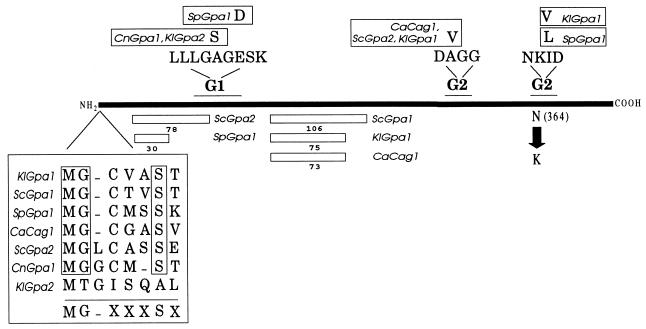
Analysis of the deduced primary structure of the protein showed a high degree of identity with the Gpa2p from K. lactis and with the Gpa proteins from S. cerevisiae. Close examination revealed 64% identity and 72% similarity with the Gpa1p from S. cerevisiae. Based on these criteria it could be that the cloned gene encodes the putative homologue of the Gα involved in the mating pathway of S. cerevisiae. The deduced primary structure of the protein (KlGpa1p) shows the characteristic structural domains conserved in Gα subunits from different organisms (Fig. 1). It shows the typical G1 and G2 regions involved in the binding and hydrolysis of the guanine nucleotide. It also shows the consensus amino terminus end (MG-XXXSXX) that has been identified as an N-myristoylation target in members of the mammalian Gαi family (11); however, unlike members of this family, KlGpa1p does not contain the cysteine residue at its carboxyl end that is ADP ribosylated by pertussis toxin (12), sharing this characteristic with the yeast Gα subunits known so far. Like Gpa1p from S. cerevisiae, KlGpa1p shows an extra internal fragment of 75 amino acid residues between the conserved G1 region and the first module of the G2 region (Fig. 1).
FIG. 1.
Primary structure of yeast Gα subunits. Rectangles below protein, inserts bigger than 29 amino acids. Amino acid sequences at the amino terminus are aligned, showing the consensus sequence for N-myristoylation. G1 and G2, conserved regions for guanine nucleotide binding and hydrolysis domains. Deviations from conserved sequences are boxed. N-to-K replacement at the G2 region that in S. cerevisiae confers the constitutive mating response is indicated. Kl, K. lactis; Sc, S. cerevisiae; Sp, S. pombe; Ca, Candida albicans; Cn, C. neoformans.
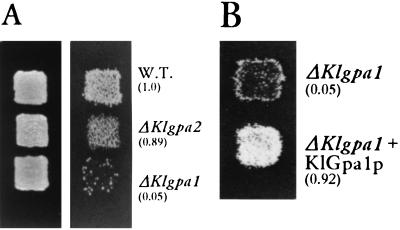
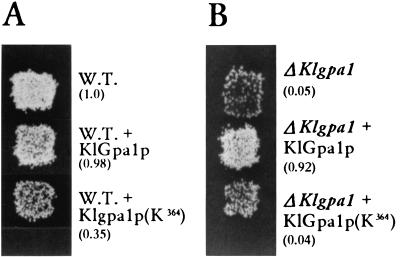
In the budding yeast S. cerevisiae, the heterotrimeric G protein composed of the Gpa1p (Gα), Ste4p (Gβ), and Ste18p (Gγ) subunits mediates pheromone-induced growth arrest. Activation of the G protein promoted by the binding of pheromone involves dissociation of the α subunit from the βγ dimer, which in turn activates a cascade signal that induces the expression of genes whose products are needed for conjugation (17). Lack of Gpa1p produces arrested cells with a transient ability to mate, indicating that Gpa1p plays a negative role in the response to the mating pheromone. We tested if lack of KlGpa1p has the same effect on the pheromone response pathway of K. lactis. Disruption of KlGpa1 of the strain 155 (Matα ade2 his3 uraA) was done by homologous recombination using a cassette containing the Ura3 marker flanked by fragments of 393 (from position 377 to 770) and 206 bp (from position 914 to 1119), respectively. Disruption of the gene and lack of its messenger were confirmed by Southern blot and Northern blot analyses, respectively (data not shown). Unlike what was found for disruption of Gpa1, which causes shmoo formation and lethality in S. cerevisiae (6), K. lactis haploid cells carrying the disrupted allele were viable (Fig. 2A) and no significant effect on morphology and doubling time was observed. Although in mating cultures wild-type K. lactis cells do not show visible shmoo morphology, we have observed that the number of budding cells is reduced two- to threefold, which suggests that growth arrest is induced by the mating partner. In addition, the disrupted strain had a dramatic impairment in mating (Fig. 2A) and forming diploids, showing a 20-fold reduction from the wild-type efficiency. This defect is totally reversed by expression of wild-type KlGpa1 under the control of its own promoter (Fig. 2B). Disruption of KlGpa1 in a Mata background (strain 12/8 [Mata argA lysA uraA]) has the same effect as that in the Matα cell, i.e., a 20- to 30-fold reduction of mating efficiency, and the level of mating of both disrupted strains (ΔKlgpa1 × ΔKlgpa1) is 0.001 of the wild-type level. Besides, after 72 h of incubation diploids appear in the selective medium.
FIG. 2.
(A) Effect of KlGpa1 and KlGpa2 disruption on growth properties and mating of K. lactis cells. Strain 155 (Matα ade2 his3 uraA), containing the wild-type (W.T.) or the disrupted alleles, was grown for 24 h on YPD plates (left) and then was replica plated to a YPD plate containing a lawn of the strain 12/8 (Mata argA lysA uraA) and incubated overnight at 30°C to allow cells to mate. Diploids were selected on SD medium supplemented with uracil (right). Numbers in parentheses, mating efficiencies relative to that of the wild type. Cells of the strain to be mated were combined with cells of the tester strain (12/8) on nitrocellulose membrane filters, placed on the surface of a YPD plate, and incubated overnight at 30°C. Cells were diluted and plated on SD medium. Mating efficiency is defined as the number of diploids divided by the number of haploids of the strain being tested. (B) The ΔKlgpa1 strain carrying either YEpKDHis or YEpKDHis-KlGpa1 was grown on YPD medium for 24 h. Patches were replica plated to a YPD plate containing a lawn of the tester strain, incubated overnight at 30°C, and replica plated to selective medium for diploids. Mating efficiencies (in parentheses) were calculated as for panel A.
On the other hand, disruption of KlGpa2 did not affect viability or mating (Fig. 2A). As we showed, cells lacking KlGpa2p have a slight increase in doubling time and reach stationary phase much earlier than wild-type cells (25), indicating that they have a defect in monitoring nutrient conditions of the medium.
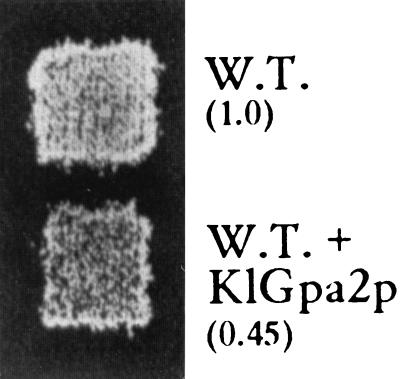
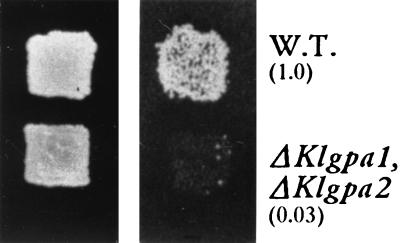
One possible explanation for the apparent sterile phenotype of ΔKlGpa1 is that an excess of KlGpa2p is able to bind free βγ produced either by pheromone activation or by lack of KlGpa1p. If this assumption is true, then overproduction of KlGpa2p should block the mating of wild-type cells and the double mutant (ΔKlgpa1 ΔKlgpa2) should be lethal and transiently able to mate. To test this hypothesis, we transfected wild-type cells with a multicopy plasmid carrying KlGpa2 under the control of its own promoter. As shown in Fig. 3, mating was slightly reduced, indicating that KlGpa2p has the potentiality to interfere with the pheromone response pathway. As mentioned earlier, KlGpa2p does not participate in the mating pathway, since cells devoid of this subunit are viable and able to mate at wild-type levels. In addition we constructed the ΔKlgpa1 ΔKlgpa2 mutant. Disruption of both Gα alleles was done in a homozygous ura3/ura3 hisA/hisA diploid with the Klgpa1::Ura3 and Klgpa2::His3 cassettes. Disruption was further confirmed by Southern blot analysis, and the lack of their messengers was confirmed by Northern blotting. The double-mutant diploid was then transferred to sporulation medium, and 10 tetrads were dissected. A first observation was that sporulation of the double mutant was deficient compared with that of wild-type cells and with that of the ΔKlgpa2 strain, i.e., wild-type cells and the mutant lacking KlGpa2p yielded four viable spores. Sporulation of the double mutant was deficient, and in most cases only three spores were able to grow. The inviable spores had either a Δgpa1 or Δgpa1 Δgpa2 genotype. This result may suggest that the product of KlGpa1 is necessary for proper sporulation of diploid cells. Auxotrophic markers were tested in haploid segregants, and surprisingly clones having both Ura+ and His+ alleles were obtained. This striking result indicates that the ΔKlgpa1 ΔKlgpa2 double mutant is viable (Fig. 4). In this double mutant the sterility impairment observed in the ΔKlgpa1 strain persisted (Fig. 4). Double-mutant cells carrying the KlGpa1 gene under the control of its own promoter grew normally and were able to mate (not shown), which indicates that KlGpa1p is required to trigger the pheromone response pathway in K. lactis.
FIG. 3.
Strain 155, transfected with either YEpKDHis or YEpKDHis-KlGpa2, was grown and mated as indicated for Fig. 2B. Patches of diploid cells were selected on SD supplemented with uracil. Mating efficiencies (in parentheses) were calculated as indicated for Fig. 2A. W.T., wild type.
FIG. 4.
The wild type (W.T.) and the ΔKlgpa1 ΔKlgpa2 double mutant were grown and mated as indicated for Fig. 2. (Right) Growth on YPD medium. (Left) Diploids grown on SD selective medium. Mating efficiencies (in parentheses) were calculated as indicated for Fig. 2A.
All these observations suggest that KlGpa1p has a positive role in mating and that its activation by a pheromone via a receptor (if this is the case) is a required step for conjugation. In S. cerevisiae cells, constitutive activation of Gpa1p, achieved by replacing asparagine 388 with lysine, induces growth arrest and morphology defects in the absence of a pheromone (15). We tested the effect that the equivalent constitutively active KlGpa1 allele, i.e., KlGpa1p(K364) (Fig. 1), has in the mating properties of K. lactis by transfecting both wild-type and ΔKlgpa1 cells. Figure 5 shows that, rather than bypassing the pheromone requirement for mating, KlGpa1p(K364) reduces mating in wild-type cells and is unable to restore the mating competence of the ΔKlgpa1 mutant. Cells expressing the constitutively active KlGpa1 allele are still viable and do not have a significantly different doubling time.
FIG. 5.
Wild-type (W.T.) (A) and ΔKlgpa1 (B) strains transfected with vector alone, vector plus KlGpa1, or vector plus KlGpa1(K364) were grown and mated as indicated for Fig. 2. Diploids were selected on SD medium. Mating efficiencies (in parentheses) were calculated as indicated for Fig. 2A.
Finally we tested the effects that expression of both KlGpa1p and KlGpa2p have on the mating response pathway of S. cerevisiae. While no significant effect was observed in a qualitative experiment of diploid formation, i.e., S. cerevisiae cells carrying either KlGpa1 or KlGpa2 mate normally, we observed a weak effect of KlGpa1p on the expression of Fus1 measured with the Fus1-LacZ fusion (21). Wild-type cells expressing KlGpa1 in a multicopy plasmid show 1.5-times-lower β-galactosidase activity than cells carrying the vector alone. Although in vivo KlGpa1p does not seem to rescue S. cerevisiae Δgpa1 cells from lethality, we were able to observe the association between KlGpa1p and the Ste4p (Gβ) subunit using the two-hybrid assay (data not shown). Indeed, association experiments indicated that KlGpa1p has an interaction that is almost 20% that of the cognate Gpa1p-Ste4p association. KlGpa2p fails to show any association with Ste4p in these assays.
DISCUSSION
We have isolated the K. lactis Gpa1 gene, which encodes a heterotrimeric Gα subunit homologue. The deduced primary structure of the protein predicts similarities with Gα subunits from different species of yeast. In fact it shows, depending on the exact alignment, 72% similarity and 64% identity with its S. cerevisiae counterpart.
The functional role of KlGpa1p was inferred from genetic studies of a Klgpa1-disrupted strain. Disruption of the KlGpa1 allele resulted in haploid viable cells with a strong deficiency in mating. In contrast, Gpa1p from S. cerevisiae has a negative role in the pheromone response pathway, i.e., disruption of the associated gene causes arrested cells in G1 phase with a transient ability to mate, while its overexpression reduces the pheromone response and mating. Therefore, K. lactis seems to be the first example of budding yeast where Gpa1p is a positive element in the mating pathway. Gα subunits playing positive roles in mating have been described in other yeasts: Gpa1p and Gpa3p in the basidiomycetes Cryptococcus neoformans (1, 27) and Ustilago maydis (23), respectively, and Gpa1p in the fission yeast Schizosaccharomyces pombe (20).
In S. cerevisiae cells, the Gβγ dimer transmits a pheromone signal to downstream elements to facilitate mating (5, 29). If parallelism can be drawn between the K. lactis and S. cerevisiae mating pathways, the mating inhibition effect of KlGpa2p could come from its interaction with the Gβγ dimer. In fact ΔKlgpa1 cells, although sterile, show a weak ability to mate (Fig. 2), which indicates that other gene products (e.g., Gβγ) are participating in the mating pathway. In this model of cooperative KlGpa1p and Gβγ signaling, activation of KlGpa1p by artificial means [i.e., KlGpa1p(K364)] should bypass the pheromone requirement and cells should increase mating. In fact the opposite is true, i.e., the activated form of KlGpa1p (GTP bound) does not rescue ΔKlgap1 cells from sterility and also reduces the mating of wild-type cells (Fig. 5). This indicates that the real situation is more complex and suggests that mating depends on a delicate balance of active and inactive forms of Gα. The hypothesis that points to a cooperative role for Gβγ in mating remains to be proved, especially since the identified Gβ subunit in S. pombe may play an inhibitory role in mating as described previously (13) and/or could be involved in the regulation of adenylate cyclase in response to glucose detection (16). In fact, we have identified the putative KlSte4 gene (unpublished results), which encodes a protein with 48% identity to ScSte4p. Unlike the Gβ subunit of S. pombe (13, 16), KlSte4p contains the amino-terminal coiled-coil structure found in ScSte4p, which indicates that this protein has the potentiality to associate to a Gγ subunit. Therefore, it becomes essential to investigate the putative role that the Gβ subunit may play in the pheromone response pathway of K. lactis. Failure of cells expressing the KlGpa1p(K364) subunit to enter into growth arrest and induce mating may serve as a useful screen to identify the putative positive effector of KlGpa1p.
K. lactis has at least two G-protein α subunits that are involved in two different pathways. While KlGpa1p participates in the mating pathway, KlGpa2p is implicated in regulation of the adenylyl cyclase (25) and, as we showed here, is able to partially interfere with mating. These data suggest cross talk between these two proteins and, although we have not investigated a possible role of KlGpa1p in cAMP regulation, a relationship between cAMP and mating signaling pathways may exist. In fact, it has been shown that, in S. cerevisiae cells, the α pheromone suppresses glucose-stimulated cAMP formation (2). This effect is dependent on the Ste2 receptor and the Ste4p subunit and may involve the Gα subunit encoded by the Gpa2 gene (22).
Although KlGpa1p from K. lactis is structurally colinear and shows high similarity with its S. cerevisiae counterpart, it plays a divergent role. In fact, it is unable to functionally replace Gpa1p, which, in S. cerevisiae, connects the pheromone stimulus with the mating response. It may be that, although KlGpa1p may interact with Ste4p, as seen in the association experiments, it cannot be activated by the pheromone-bound receptor.
The results here described point to the model in which, in K. lactis, Gpa1p functions as a positive factor that transmits the signal from a pheromone receptor to a downstream effector(s). In this respect the KlGpa1p subunit is equivalent to Gα subunits from other yeast species and even to mammalian subunits, which places the Gpa1p of S. cerevisiae in a special context within the metazoan Gα subunits.
Besides the evolutionary relationship between these budding yeasts and the similarities that they share in sexual reproduction, it seems that they have developed divergent mechanisms for pursuing the same goal, i.e., mating of haploid cells.
ACKNOWLEDGMENTS
This work was supported in part by grants 2259PN and 28015N from the Consejo Nacional de Ciencia y Tecnología, México, and grant 102369 from PAEP, UNAM, to A.L.S.-T.
We are grateful to Gerardo Coello and Ana M. Escalante for sequence analysis of KlGpa1. We acknowledge the technical assistance of Soledad Guevara. We are grateful to Marcela Sosa and Guadalupe Codiz (staff of the Molecular Biology Facilities at the IFC) for assistance.
REFERENCES
- 1.Alspaugh J A, Perfect J R, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit Gpa1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkinstall S J, Papasavvas S G, Payton M A. Yeast α-mating factor receptor-linked G-protein signal transduction suppresses Ras-dependent activity. FEBS Lett. 1991;284:123–128. doi: 10.1016/0014-5793(91)80777-z. [DOI] [PubMed] [Google Scholar]
- 3.Brake A, Irvine B, Masiarz F, Shultz K. Structure of genes encoding precursors of two Kluyveromyces lactis transported proteins. Yeast. 1988;4:S436. [Google Scholar]
- 4.Cheol-Won Y, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. G-protein coupled receptor from Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1997;240:287–292. doi: 10.1006/bbrc.1997.7649. [DOI] [PubMed] [Google Scholar]
- 5.Cole G M, Stone D E, Reed S I. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol Cell Biol. 1990;10:510–517. doi: 10.1128/mcb.10.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzel C, Kurjan J. The yeast SCG1 gene: a Gα-like protein implicated in the a- and α-factor response pathway. Cell. 1987;50:1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- 7.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 8.Herman A, Roman H. Allele specific determinants of homothalism in Saccharomyces lactis. Genetics. 1966;53:727–740. doi: 10.1093/genetics/53.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 10.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 11.Johnson D R, Bhatnagar R S, Knoll L J, Gordon J I. Genetic and biochemical studies of protein N-myristoylation. Annu Rev Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 12.Johnson G L, Kaslow H R, Bourne H R. Genetic evidence that cholera toxin substrates are regulatory components of adenylyl cyclase. J Biol Chem. 1978;253:7120–7123. [PubMed] [Google Scholar]
- 13.Kim D-U, Park S-K, Chung K-Y, Choi M-U, Yoo H-S. The G protein β subunit Gpb1 of Schizosaccharomyces pombe is a negative regulator of sexual development. Mol Gen Genet. 1996;252:20–32. doi: 10.1007/BF02173201. [DOI] [PubMed] [Google Scholar]
- 14.Kübler E, Mösch H-U, Rupp S, Lisanti M P. Gpa2p, a G-protein α-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 15.Kurjan J, Hirsch J P, Dietzel C. Mutations in the guanine nucleotide-binding domains of a yeast Gα protein confer a constitutive or uninducible state to the pheromone response pathway. Genes Dev. 1991;5:475–483. doi: 10.1101/gad.5.3.475. [DOI] [PubMed] [Google Scholar]
- 16.Landry S, Pettit M T, Apolinaro E, Hoffman C S. The fission yeast git5 gene encodes a Gβ subunit required for glucose-triggered adenylate cyclase activation. Genetics. 2000;154:1463–1471. doi: 10.1093/genetics/154.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 18.Miranda M, Ongay-Larios L, Guevara S, Ramírez J, Peña A, Coria R. Nucleotide sequence and chromosomal localization of the gene encoding the old yellow enzyme from Kluyveromyces lactis. Yeast. 1995;11:459–465. doi: 10.1002/yea.320110509. [DOI] [PubMed] [Google Scholar]
- 19.Nakafuku M, Obara T, Kaibuchi K, Miyajima I, Miyajima A, Itoh H, Nakamura S, Arai K I, Matsumoto K, Kaziro Y. Isolation of a second yeast Saccharomyces cerevisiae gene (Gpa2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc Natl Acad Sci USA. 1988;85:1374–1378. doi: 10.1073/pnas.85.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obara T, Nakafuku M, Yamamoto M, Kaziro Y. Isolation and characterization of a gene encoding a G-protein α subunit from Schizosaccharomyces pombe: involvement in mating and sporulation pathways. Proc Natl Acad Sci USA. 1991;88:5877–5881. doi: 10.1073/pnas.88.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ongay-Larios L, Saviñón-Tejeda A, Williamson M J, Jr, Durán-Avelar M J, Coria R. The Leu-132 of the Ste4(Gβ) subunit is essential for proper coupling of the G protein with the Ste2 α factor receptor during the mating pheromone response in yeast. FEBS Lett. 2000;467:22–26. doi: 10.1016/s0014-5793(00)01106-6. [DOI] [PubMed] [Google Scholar]
- 22.Papasavvas S G, Arkinstall S, Reid J, Payton M. Yeast α-mating factor receptor and G-protein-linked adenylyl cyclase inhibition requires RAS2 and GPA2 activities. Biochem Biophys Res Commun. 1992;184:1378–1385. doi: 10.1016/s0006-291x(05)80035-x. [DOI] [PubMed] [Google Scholar]
- 23.Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bölker M, Kahmann R. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 1996;16:1934–1942. doi: 10.1093/emboj/16.8.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Saviñón-Tejeda A, Ongay-Larios L, Ramírez J, Coria R. Isolation of a gene encoding a G protein α subunit involved in the regulation of cAMP levels in the yeast Kluyveromyces lactis. Yeast. 1996;12:1125–1133. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1125::AID-YEA7%3E3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Sherman F, Fink G R, Hicks J. Methods in yeast genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 27.Tolkacheva T, McNamara P, Piekarz E, Courchesne W. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein α-subunit homolog. Infect Immun. 1994;62:2849–2856. doi: 10.1128/iai.62.7.2849-2856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wésolowski-Louvel M, Breunig K D, Fukuhara H. Kluyveromyces lactis: genetics, biochemistry, and molecular biology of non-conventional yeast. Berlin, Germany: Springer-Verlag KG; 1996. [Google Scholar]
- 29.Whiteway M, Hougan L, Dignard D, Thomas D Y, Bell L, Saari G C, Grant F J, O'Hara P, MacKay V L. The Ste4 and Ste18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 30.Xue Y, Batlle M, Hirsch J P. Gpr1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan Y O, Stroke I L, Fields S. Coupling of cell identity to signal response in yeast: interaction between the α1 and Ste12 proteins. Genes Dev. 1993;7:1584–1597. doi: 10.1101/gad.7.8.1584. [DOI] [PubMed] [Google Scholar]