Abstract
Impaired baseline lung function is associated with mortality after pediatric allogeneic hematopoietic cell transplantation (HCT), yet limited knowledge of the molecular pathways that characterize pre-transplant lung function has hindered the development of lung-targeted interventions. In this study, we quantified the association between bronchoalveolar lavage (BAL) metatranscriptomes and paired pulmonary function tests performed a median of 1 to 2 weeks prior to allogeneic HCT in 104 children in the Netherlands. Abnormal pulmonary function was recorded in more than half the cohort, consisted most commonly of restriction and impaired diffusion, and was associated with both all-cause and lung-injury related mortality after HCT. Depletion of commensal supraglottic taxa, such as Haemophilus, and enrichment of nasal and skin taxa, such as Staphylococcus, in the BAL microbiome were associated with worse measures of lung capacity and gas diffusion. In addition, BAL gene expression signatures of alveolar epithelial activation, epithelial-mesenchymal transition, and downregulated immunity were associated with impaired lung capacity and diffusion, suggesting a post-injury pro-fibrotic response. Detection of microbial depletion and abnormal epithelial gene expression in BAL enhanced the prognostic utility of pre-HCT pulmonary function tests for the outcome of post-HCT mortality. These findings suggest a potentially actionable connection between microbiome depletion, alveolar injury, and pulmonary fibrosis in the pathogenesis of pre-HCT lung dysfunction.
One Sentence Summary:
Poor lung function in children preparing to undergo stem cell transplantation is linked to microbial dysbiosis, alveolar injury, and fibrosis.
INTRODUCTION
Allogeneic hematopoietic cell transplantation (HCT) is a life-saving cellular therapy used to cure underlying hematopoietic defects, such as hematologic malignancies, primary immunodeficiencies, bone marrow failure syndromes, and hemoglobinopathies. Although more than 7,500 children receive this treatment annually in the United States and Europe, the success of this therapy is limited by a broad range of post-HCT pulmonary injuries that develop in 10 to 40% of pediatric recipients in response to chemotherapeutic toxicity, infection, and impaired or dysregulated immunity (1–3). Comprehensive assessment of pulmonary health prior to HCT conditioning is critical in order to identify high-risk patients who may benefit from modified conditioning chemotherapy regimens, closer surveillance, and risk-stratified treatments aimed at mitigating the development of irreversible pulmonary toxicity (4–6).
In children, evaluation of pulmonary health prior to HCT is not standardized, but may include symptom screening and physical exam, nasopharyngeal swab for common respiratory viruses, pulmonary function testing (PFT) if developmentally able (typically age greater than or equal to 6 years), and targeted high-resolution chest tomography (HR-CT), followed by bronchoalveolar lavage (BAL) in patients with identified or suspected abnormalities (7, 8). Despite the challenges of completing effort-dependent testing in children, when performed with adequate quality, abnormal PFTs are detected in up to 60% of pediatric HCT candidates, predominantly due to restriction and impaired diffusion (9–11), and are associated with the development of post-HCT complications. These include interstitial pneumonitis, pulmonary graft-versus-host disease (GVHD), and transplant-related mortality (TRM) (2, 3, 12–16).
To improve outcomes, it remains crucial to identify risk factors for pre-HCT pulmonary dysfunction and elucidate pathobiologic correlates. To date, known risk factors include exposure to alkylating and other pulmotoxic chemotherapy (such as busulfan, bleomycin, cyclophosphamide, carmustine, fludarabine, lomustine), thoracic radiation (particularly in concert with radiomimetics), thoracic surgery, chronic endothelial injury due to hemolysis or transfusion dependence, chronic inflammation, and recurrent pulmonary infections (17–19). Most patients with substantially impaired pre-HCT pulmonary function experience a number of these risk factors, suggesting a multimodal pathogenesis. Despite the need for deeper understanding of the pathogenesis of pre-HCT pulmonary dysfunction, translational studies are limited by scarcity of investigable pulmonary samples from children.
In this study, we aimed to evaluate associations between pre-HCT pulmonary function and measures of pulmonary biology as assessed by metatranscriptomic RNA sequencing of paired BALs. We recently analyzed pre-HCT BAL samples from a cohort of asymptomatic pediatric HCT candidates who all underwent routine pre-HCT BAL in the absence of lower respiratory symptoms as part of an aggressive standard-of-care screening program. We found that pre-HCT pulmonary bacterial depletion, viral enrichment, inflammation, and epithelial activation were associated with the development of post-HCT pulmonary toxicity (20). A subset of these patients were developmentally able to perform contemporaneous PFTs, presenting an opportunity to link the domains of organ function, biology, and microbiology within a cohort of high-risk patients. We hypothesized that the combination of pulmonary microbiome data paired with gene expression data could be associated with abnormal PFTs prior to HCT, thus providing further evidence for the pathobiologic role of the pulmonary microenvironment in pediatric HCT candidates.
RESULTS
Patient Characteristics
Contemporaneous pre-HCT PFTs and BAL fluid were available for 104 patients. PFTs and BAL were typically performed 1 to 2 days apart and 1 to 2 weeks prior to HCT conditioning. Most patients were between 8 and 19 years of age, of self-reported Northern European ancestry, and preparing to undergo first allogeneic HCT for underlying hematologic malignancy (Table 1). Most patients received myeloablative conditioning chemotherapy, followed by an umbilical cord blood allograft. At 2-year post-HCT follow-up, 33 of 104 patients (31.7%) had died from relapsed disease or transplant toxicity.
Table 1. Pediatric Allogeneic HCT Candidate Patient Characteristics.
Includes n=104 total patients.
| Pre-HCT Characteristics | ||
|---|---|---|
| 4≥ age <8 | 24 (23.1) | |
| 8≥ age <13 | 32 (30.8) | |
| 13≥ age <20 | 48 (46.2) | |
| Male | 61 (58.3) | |
| Female | 43 (41.4) | |
| Northern European | 76 (73.1) | |
| Multiracial/Other | 10 (9.6) | |
| Middle Eastern/Persian/Turkish | 7 (6.7) | |
| African/North-African | 6 (5.8) | |
| Asian/Southeast Asian | 3 (2.9) | |
| Eastern European/Russian | 2 (1.9) | |
| 2005 to 2009 | 23 (22.1) | |
| 2011 to 2013 | 40 (38.5) | |
| 2014 to 2016 | 41 (39.4) | |
| Hematologic Malignancy | 64 (61.5) | |
| Bone Marrow Failure Syndrome | 17 (16.4) | |
| Primary Immunodeficiency | 16 (15.4) | |
| Metabolic/Inborn Errors | 4 (3.9) | |
| Autoimmune Disease | 3 (2.9) | |
| Positive | 56 (53.9) | |
| Negative | 33 (31.7) | |
| N/A | 15 (14.4) | |
| Any abnormality | 42 (40.4) | |
| – Virus Detected on PCR | − 23 (22.1) | |
| – Bacterial Culture Growth | − 10 (9.6) | |
| – Fungal Culture Growth | − 12 (11.5) | |
| – Galactomannan Positivity | − 11 (11.2) | |
| Days from PFT to HCT (median, IQR) | 11 (8 to 17) | |
| Days from BAL to HCT (median, IQR) | 10.5 (8 to 14) | |
| Days from PFT to BAL (median, IQR) | 1 (−1 to 2) | |
| HCT Characteristics and Outcomes | ||
| Mismatched Umbilical Cord Blood | 32 (30.8) | |
| Matched Umbilical Cord Blood | 23 (22.1) | |
| Matched Sibling Donor | 29 (27.9) | |
| Matched Unrelated Donor | 20 (19.2) | |
| Myeloablative | 97 (93.3) | |
| Reduced Intensity | 7 (6.7) | |
| ATG or alemtuzumab | 64 (61.5) | |
| All-Cause Mortality | 33 (31.7) | |
| Non-Relapse Mortality | 21 (20.2) | |
| – Due to Lung Injury | − 15 (14.4) | |
| – Not Due to Lung Injury | − 6 (5.7) | |
| Relapse Mortality | 12 (11.5) | |
Race is self-reported. Multiracial/other includes mixed race (n=8), Hispanic (n=1), Surinamese (n=1).
Malignancy includes acute lymphoid leukemia (ALL, n=23), acute myeloid leukemia (AML, n=17), myelodysplastic syndrome (MDS, n=9), chronic myeloid leukemia (CML, n=4), and other (n=11). Bone marrow failure syndromes include Fanconi anemia (n=11), severe aplastic anemia (SAA, n=4), and other (n=2). Primary immunodeficiency includes combined immunodeficiency (CID, n=9), chronic granulomatous disease (CGD, n=3), hemophagocytic lymphohistiocytosis (HLH, n=2), and other (n=2). Metabolic/Inborn Errors includes metachromatic leukodystrophy (MLD, n=2), adrenoleukodystrophy (ALD, n=1), β-thalassemia (n=1). Autoimmune disease includes systemic lupus erythematosus (SLE, n=1), refractory multiple sclerosis (MS, n=1), and systemic juvenile idiopathic arthritis (sJIA, n=1).
Cytomegalovirus (CMV) serostatus unknown or assumed positive in many recipients of umbilical cord blood (UCB) allografts.
No organisms were detected with other diagnostic tests such as cytology for pneumocystis jirovecii pneumonia (PJP) or PCR for mycoplasma or legionella. Some patients had more than one abnormality.
Matched sibling donor includes one 8/10 human leukocyte antigen (HLA) match. There were no HLA-mismatched or haploidentical allograft donors.
Myeloablative conditioning regimens were busulfan and fludarabine (Bu/Flu, n=35); busulfan, fludarabine, and clofarabine (Bu/Flu/Clo, n=37); total body irradiation with etoposide (TBI/etoposide, n=11); fludarabine and cytarabine (Flu/Cy, n=10); and other (n=4). All reduced intensity conditioning (RIC) regimens were fludarabine-based.
Causes of post-HCT death due to lung injury included invasive fungal infection (n=3), viral infection (n=1), other infection (n=1), alloimmune lung injury including idiopathic pneumonia syndrome (n=4), idiopathic multi-organ failure (n=4), or other (n=2). IQR, interquartile range.
Impaired pulmonary function prior to HCT is common.
All patients performed spirometry, and a subset of typically older children performed plethysmography (n=78) and diffusion testing (n=63, Table 2). To determine the inter-relatedness of the different PFT measurements, we constructed a correlation matrix which showed broad grouping of PFTs into 4 clades pertaining to lung capacity, obstruction, diffusion, and air trapping (fig. S1). Using conventional cutoffs of absolute ratios and percentage deviations from the population mean (% predicted), PFT abnormalities were detected in 80 of 104 patients due to restriction (n=35 of 104), obstruction (n=12 of 104), mixed restriction and obstruction (n=1 of 104), impaired diffusion (n=21 of 63), and air trapping (n=44 of 78). Using Global Lung Index (GLI) cutoffs based on standard deviations from the population mean (z-scores), PFT abnormalities were detected in 54 of 104 patients due to restriction (n=29 of 104), obstruction (n=7 of 104), impaired diffusion (n=14 of 63), and air trapping (n=16 of 78, table S1). As GLI z-scores are not calculable for all measured PFTs, we used PFTs expressed as percentage of predicted for subsequent analyses.
Table 2. Pulmonary Function Tests in Pediatric Allogeneic HCT Candidates.
Pulmonary Function Tests in Pediatric Allogeneic HCT Candidates.
| Spirometrya (n=104) | Actual | % of Predicted | Z-score |
|---|---|---|---|
| Forced Vital Capacity (FVC, liters) | 2.21 (1.50–3.21) | 0.87 (0.74–0.96) | −1.07 (−2.28 to −0.35) |
| Forced Expiratory Volume in 1 second (FEV1, liters) | 1.94 (1.36–2.77) | 0.86 (0.75–0.96) | −1.14 (−2.08 to −0.40) |
| Forced Expiratory Volume in 1 second, adjusted (FEV1/FVC) | 0.88 (0.84–0.94) | 1.00 (0.95–1.06) | −0.17 (−0.83 to 0.92) |
| Peak Expiratory Flow (PEF, liters /second) | 4.44 (3.12–6.02) | 0.85 (0.76–0.97) | -- |
| Forced Expiratory Flow at 25% (FEF25, liters/second) | 3.85 (2.83–5.47) | 0.87 (0.72–0.99) | -- |
| Forced Expiratory Flow at 50% (FEF50, liters /second) | 2.58 (1.96–3.59) | 0.91 (0.69–1.05) | -- |
| Forced Expiratory Flow at 75% (FEF75, liters /second) | 1.23 (0.86–1.76) | 0.86 (0.62–1.06) | −0.40 (−1.28 to 0.32) |
| Forced Expiratory Flow 25%−75% (FEF25–75, liters/second) | 2.68 (1.73–3.81) | 0.98 (0.79–1.15) | −0.10 (−1.00 to 0.55) |
| Plethysmographyb (n=78) | |||
| Total Lung Capacity (TLC, liters) | 3.75 (2.81–4.73) | 0.85 (0.74–0.95) | −0.76 (−1.45 to 0.09) |
| Vital Capacity (VC, liters) | 2.74 (1.94–3.33) | 0.90 (0.75–0.99) | −1.32 (−2.62 to −0.42) |
| Alveolar Volume (Va, liters) | 3.56 (2.69–4.63) | 0.91 (0.80–1.00) | −0.77 (−1.83 to −0.01) |
| Alveolar Volume/Total Lung Capacity (Va/TLC) | 0.95 (0.89–1.01) | 0.85 (0.93–0.97) | -- |
| Residual Volume (RV, liters) | 0.89 (0.71–1.35) | 0.99 (0.77–1.30) | 0.11 (−0.54 to 0.69) |
| Residual Volume/Total Lung Capacity (RV/TLC) | 0.26 (0.21–0.35) | 1.06 (0.83–1.44) | 0.51 (−0.11 to 1.47) |
| Diffusionc (n=63) | |||
| Diffusion Capacity for Carbon Monoxide (DLCO, mmol/min/kPa) | 5.78 (4.35–6.92) | 0.78 (0.68–0.95) | −1.65 (−2.53 to −0.34) |
| Diffusion Capacity for Carbon Monoxide/Va (DLCO/Va) | 1.50 (1.37–1.73) | 0.88 (0.76–1.02) | −0.69 (−1.58 to 0.12) |
n=104 for spirometry,
n=78 for plethysmography,
n=63 for diffusion factor testing.
Abnormal pulmonary function varied according to age and sex and was associated with HCT outcomes.
We tested for clinical characteristics associated with PFT abnormalities and identified that older age was associated with restrictive lung disease, as well as with worse measures of obstruction such as forced expiratory flow (FEF25, FEF50, FEF75) and peak expiratory flow (PEF) (Fig. 1A). Female sex was associated with worse measures of both lung capacity (including forced vital capacity [FVC] and alveolar volume [Va]) and diffusion (measured as diffusing capacity for carbon monoxide accounting for Va [DLCO/Va]) but better measures of obstruction (such as forced expiratory volume in one second adjusted for FVC [FEV1/FVC], Fig. 1B). We tested for, but did not identify, an association between HCT indication and pre-HCT PFTs (data file S1).
Figure 1. Association between Pre-HCT PFTs, Clinical Characteristics, and Post-HCT Outcomes.
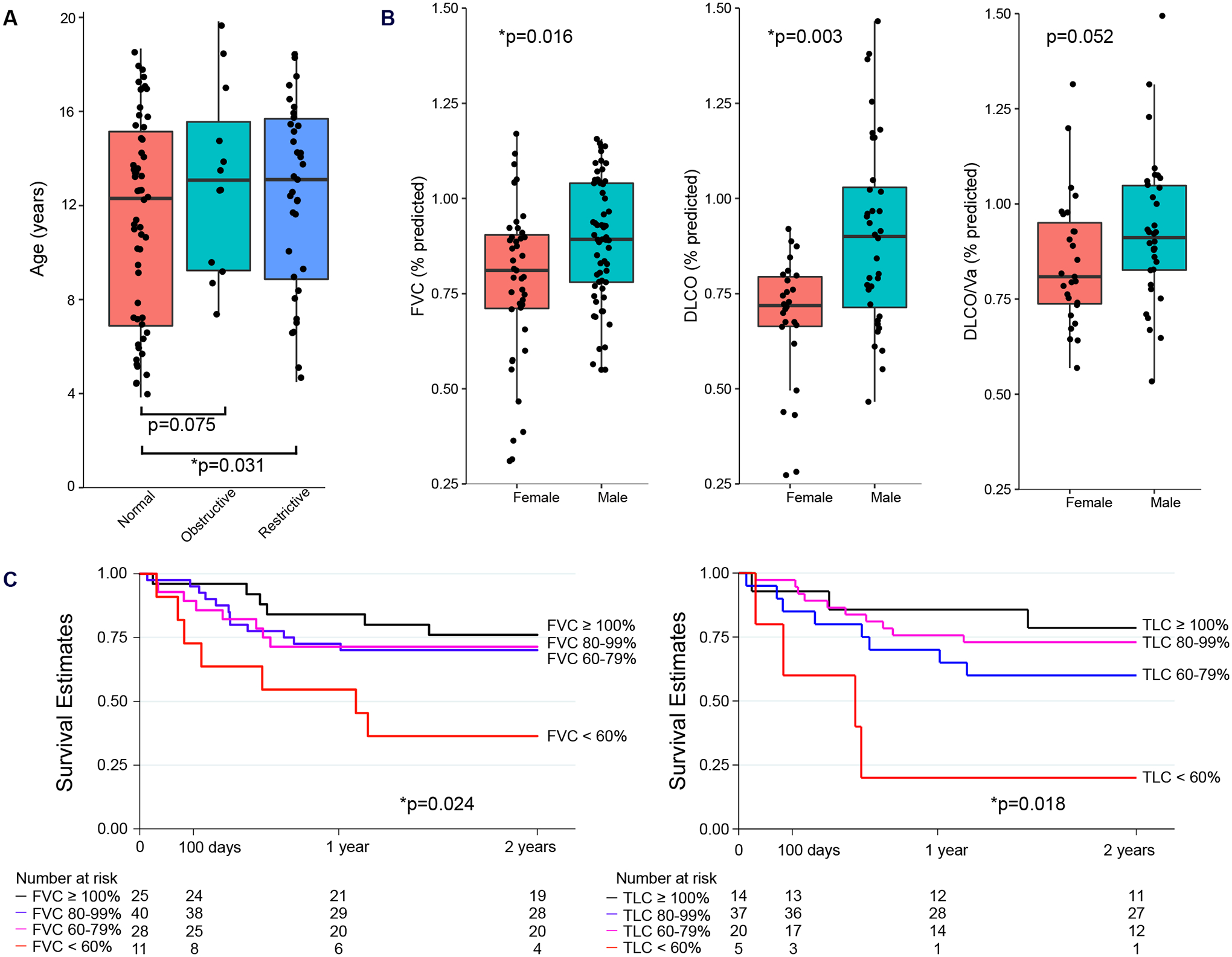
(A) Box-and-whisker plot of age distribution are shown for patients with restriction (n=35), obstruction (n=12), or neither (n=57) using conventional PFT categorization. Patients with either restriction or obstruction were older than patients with normal PFTs. Data were analyzed using Dunn’s Test. (B) Box-and-whisker plot of PFT distributions are shown according to patient sex (females, n=43; males, n=61). Data were analyzed using Wilcoxon rank-sum tests. Box-and-whisker plots in (A) and (B) depict the median and interquartile range (box) with 1.5 times the interquartile range (whiskers). (C) Kaplan-Meier post-HCT survival estimates are shown according to pre-HCT FVC%pred (left) and TLC%pred (right). Both FVC%pred and TLC%pred were associated with post-HCT survival. Data were analyzed by Cox regression.
Previous studies have shown impaired PFTs to be associated with post-HCT mortality (2, 3, 12–16). We confirmed these findings by demonstrating an association between reduced pre-HCT lung capacity and the development of all-cause mortality after HCT (hazard ratio [HR] 1.21 per 10% decline in FVC, 95% CI 1.03–1.43, p=0.024, Fig. 1C). To test whether these poor outcomes were due to acute lung injury specifically, we calculated cumulative incidence with competing risks regression and found that FVC%pred was associated with the development of fatal lung injury in the first 180 days post-HCT (p<0.001, fig. S2).
BAL microbiome composition varied with pulmonary function.
Our prior studies have associated alterations in the pre-HCT pulmonary microbiome composition with the later development of adverse post-HCT outcomes (20). To determine whether the composition of the pre-HCT pulmonary microbiome is associated with lung function at the time of BAL measurement, we compared microbiome findings to contemporaneously performed PFTs. Microbe RNA sequencing counts are relative in nature and thus were converted to RNA mass abundances using the reference spike-in (21). We first surveyed the composition of BAL microbiomes and found two oppositely correlated clusters of microbes: one containing common supraglottic taxa (such as Haemophilus, Streptococcus, Neisseria) and a second inversely correlated cluster containing common nasal, skin, and environmental taxa (such as Staphylococcus, Corynebacterium, fig. S3). We represented these clusters mathematically using principal component analysis (PCA) and found that lower coordinates along principal component 1 (with top contributions from oropharyngeal taxa Prevotella, Veillonella, Rothia, and Streptococcus) were associated with worse measures of lung capacity (including FVC, fig. S4, data file S2).
Next, we used negative binomial generalized linear models (NB-GLM) with adjustment for false discovery to identify specific taxa associated with each PFT. In models adjusted for patient age and sex, we found that lower BAL quantities of supraglottic taxa (such as Haemophilus, Neisseria) and greater BAL quantities of skin or environmental taxa (including Staphylococcus, Saccharomyces, Corynebacterium) were associated with worse measures of lung capacity (FVC), obstruction (FEF25), diffusion impairment (DLCO), and air trapping (RV/TLC) (Fig. 2A, fig. S5, data file S3). For example, progressively lower FVC was associated with progressively less Haemophilus RNA and progressively more Staphylococcus RNA (Fig. 2, B and C). In addition, greater pulmonary fungal RNA mass was associated with worse measures of lung capacity (FVC) and diffusion (DLCO), and greater RNA mass of the Actinobacteria phylum, which includes the skin-associated genera Corynebacterium, Micrococcus, and Cutibacterium, was associated with worse measures of lung capacity (FVC) and obstruction (FEF25, FEF75). In contrast, lower RNA mass of the Fusobacterium phylum was associated with worse measures of lung capacity (Va) and diffusion (DLCO). We did not find an association between PFTs and total viral RNA mass (data file S2). Some microbiome studies have suggested that aggregate microbiome traits such as alpha diversity, dominance, and richness are associated with severity of diseases such as chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF) (22–26). In this cohort, neither alpha diversity, richness, nor dominance were associated with PFTs.
Figure 2. Association between BAL Microbiome and FVC%pred.
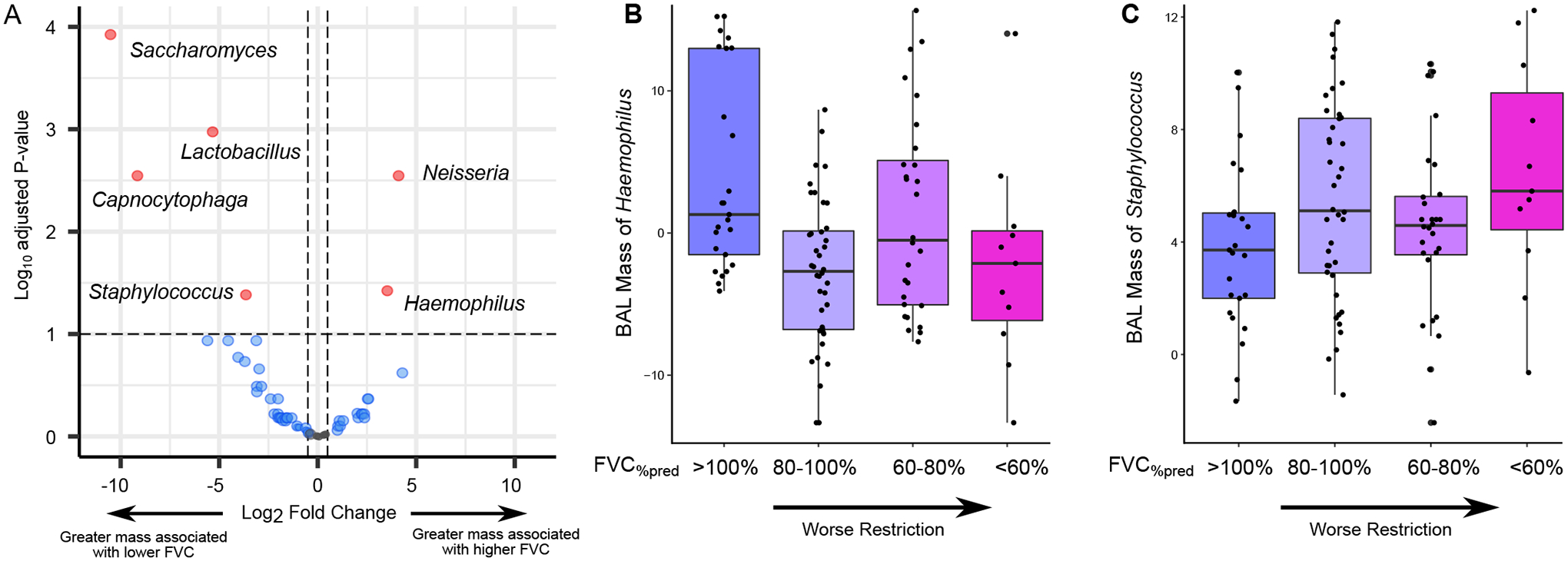
(A) The volcano plot shows the association between BAL microbe RNA mass and FVC%pred. Associations were tested using negative binomial generalized linear models accounting for patient age and sex covariates. Positive log2 fold change (x-axis) indicates that greater microbial RNA mass is associated with greater FVC%pred; negative log2 fold change indicates that greater microbial RNA mass is associated with lower FVC%pred. (B and C) Box-and-whisker plot of variance stabilizing transformed (vst) RNA masses of Haemophilus (B) and Staphylococcus (C) are shown according to category of FVC%pred (>100%, n=25; 80–100%, n=40; 60–80%, n=28; <60%, n=11). Worse restriction is associated with lower Haemophilus and greater Staphylococcus RNA mass in BAL fluid.Box-and-whisker plots in (B) and (C) depict the median and interquartile range (box) with 1.5 times the interquartile range (whiskers).
BAL gene expression was correlated with pulmonary function.
To determine whether impaired pulmonary function was associated with unique gene expression signatures, we first utilized an overview approach and calculated BAL gene expression enrichment scores for each of the n=50 Hallmark Gene Sets in the Molecular Signatures Database. We found that measures of lung capacity and diffusion were positively associated with enrichment of pathways relating to immune activation, metabolism, and cell division and were negatively associated with pathways relating to epithelial mesenchymal transition and cell fate or differentiation (Fig. 3).
Figure 3. Association between BAL Hallmark Gene Expression and Pulmonary Function.
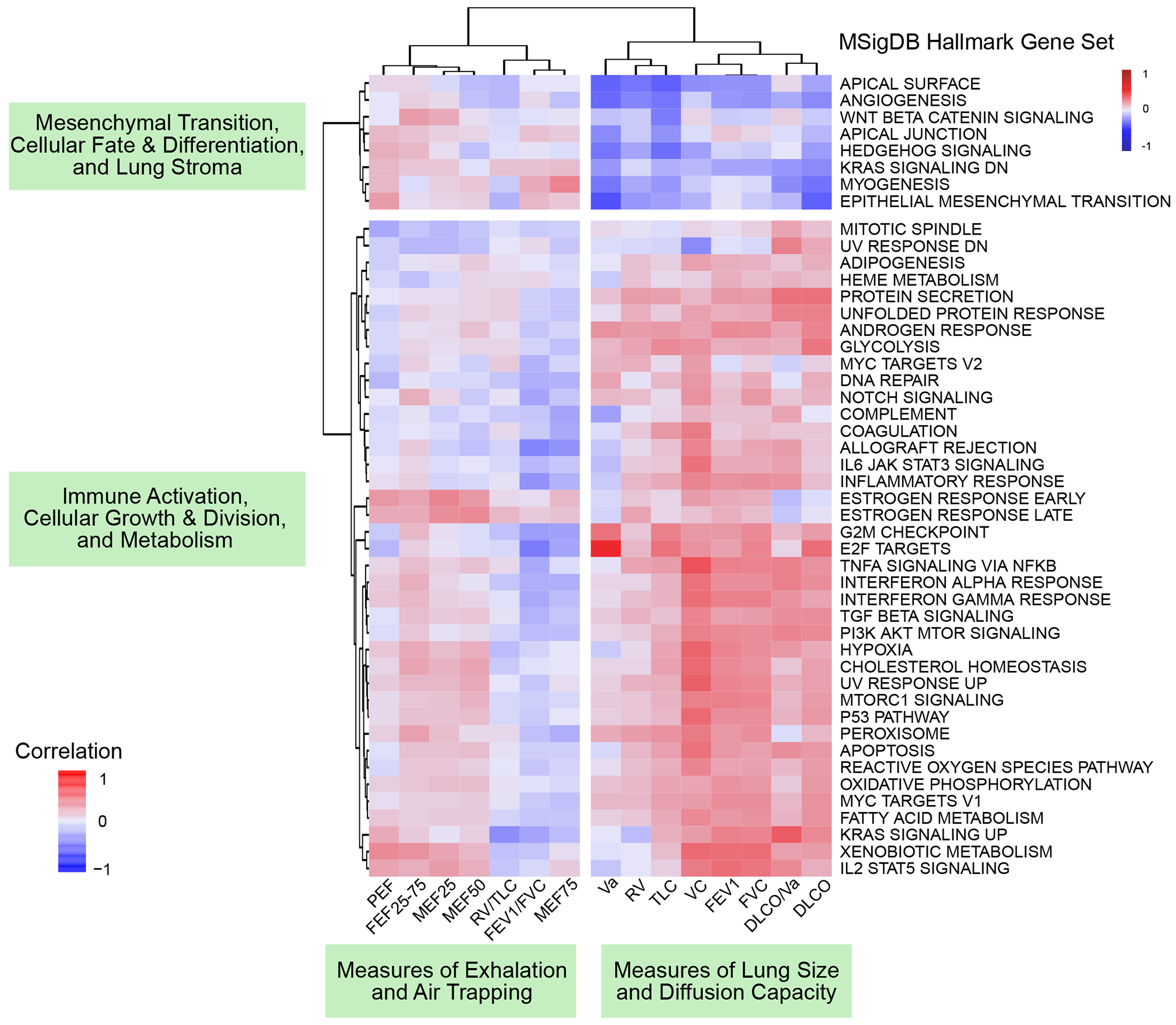
Hallmark gene set variance analysis using Poisson gene count distributions was used to calculate gene set enrichment scores for each patient. Spearman rank-based correlations between gene set enrichment scores (rows) and PFTs (columns) were calculated and plotted (cells) with hierarchical clustering of columns and rows according to Euclidean distance. Dendrogram-based clustering shows that PFT measures of lung size and diffusion were positively associated with a cluster of hallmark gene sets corresponding to immune activation, cellular growth and division, and metabolism and were negatively associated with a cluster of hallmark gene sets corresponding to mesenchymal transition, cellular fate and differentiation, and the lung stroma.
We next tested for specific genes whose expression was associated with each PFT. Pathway analysis showed that lower values of both FVC and DLCO/Va were associated with greater expression of pulmonary epithelial genes, including surfactant genes (such as SFTPA1/2, SFTPC, and SFTPD), keratinization genes (such as KRTAP3.2 and KRTAP5.6), and epithelial sensory and stimulus response genes (such as OR2A1 and OR4S2), as well as lower expression of innate immunity genes (such as CXCL8, HLA-DRA, IL1B, and LYZ; Fig. 4). In contrast, lower FEV1/FVC was associated with greater expression of innate immunity genes and lower expression of alveolar epithelial genes. These findings persisted after adjusting models for age and sex. Few genes (n=9) were associated with a metric of air trapping (RV/TLC, data file S4).
Figure 4. Association between BAL Gene Expression and Pulmonary Function Testing.
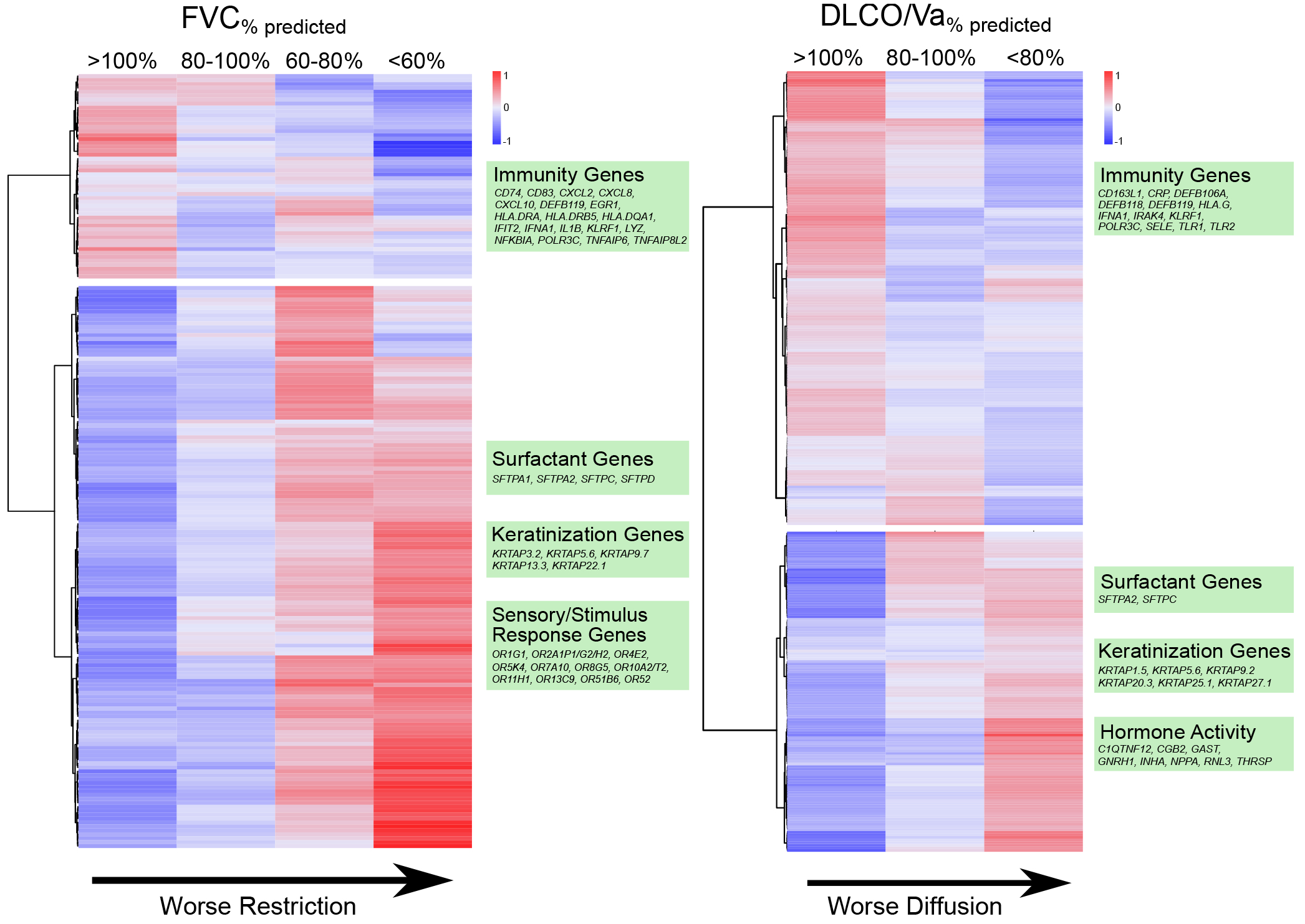
Heatmaps of differential gene expression are plotted according to pulmonary restriction (left) and impaired diffusion (right). Negative binomial models were used to identify differentially expressed genes (rows) across the continuum of FVC%pred and DLCO/Va%pred (columns). Cell shading indicates mean gene expression for each PFT subgroup after variance stabilizing transformation, centering, and scaling. Dendrogram-based clustering shows that declining FVC%pred was associated with a decrease in expression of immune-related genes and an increase in expression of airway epithelial genes relating to surfactant, keratinization, and olfactory responses. Declining DLCO/Va%pred was also associated with a decrease in expression of immune-related genes and an increase in expression of airway epithelial genes as well as hormone response genes.
The BAL cell fraction was associated with pulmonary function.
Since BAL samples are heterogeneous cell mixtures, we posited that these results could be due to either different proportions of cell types within samples, different expression patterns of each cell type within samples, or both. Therefore, we used in silico cell deconvolution techniques based on the Human Lung Cell Atlas to impute lung cell proportions in each sample. BAL cell types clustered into 3 groups consisting of innate immune cells, upper airway cells, and lower airway and adaptive immune cells (Fig. 5A). Patients with restriction had greater BAL enrichment scores for type 2 alveolar epithelial cells (AECs) (Fig. 5B). In addition, patients with impaired diffusion had greater BAL enrichment scores for type 2 AECs (p=0.011). Next, using the CiberSortX algorithm (27), we estimated the average gene expression profile for each BAL cell type. In patients with restriction, type 1 AECs showed increased transcription of genes related to epithelial-mesenchymal transition, hedgehog signaling, and KRAS signaling. Additionally, T- and B-lymphocytes showed an overall decrease in transcription of genes relating to cell proliferation and inflammation (data file S5). We did not detect differences in innate immune cell or upper airway epithelial cell-specific gene expression among patients with versus without restriction.
Figure 5. BAL Cell Type Differential.
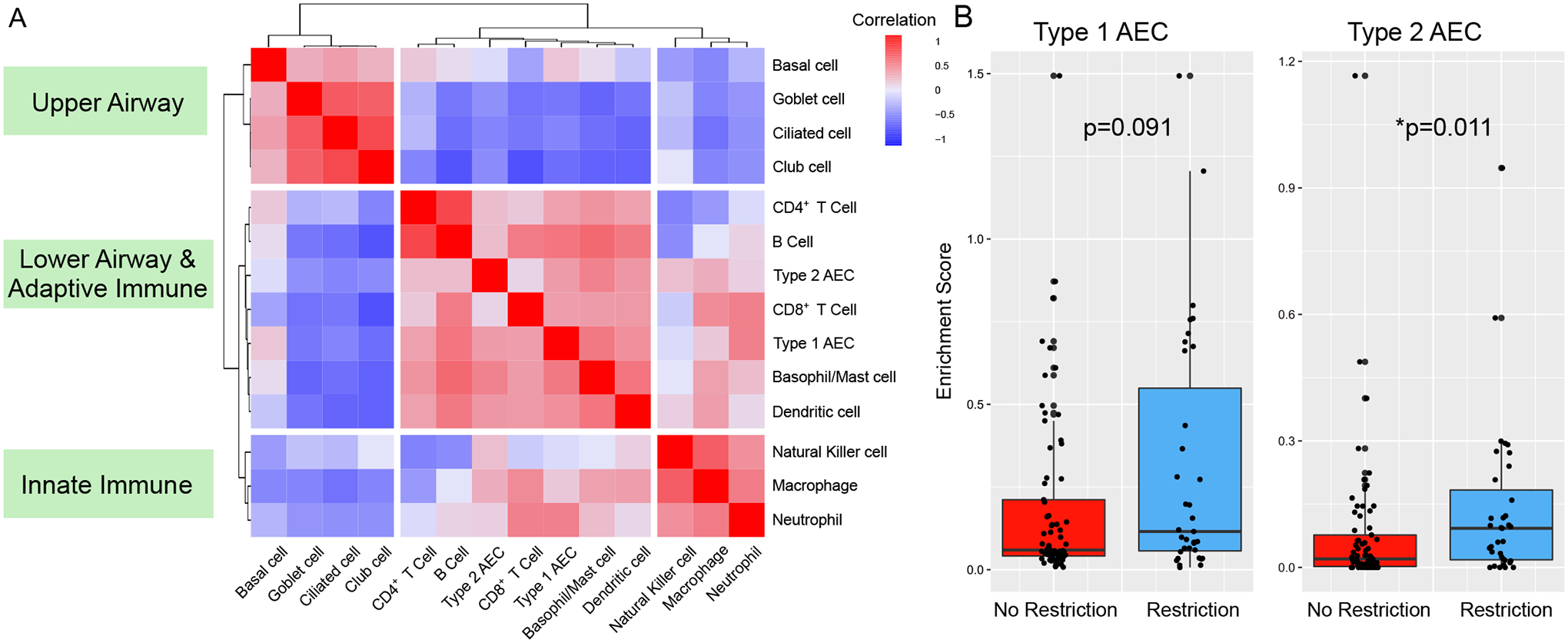
Single cell RNA sequencing data from The Human Lung Cell Atlas was used to calculate cell-type specific marker genes and impute cell-type fractions and enrichment scores (CiberSortX). (A) Highly correlated cell types were identified by calculating Spearman rank-based correlation of cell type fractions followed by hierarchical clustering of correlation distances. (B) Cell type enrichment scores were tested for association with pulmonary restriction (No restriction, n=69; restriction, n=35) using Kruskal-Wallis testing.
PFTs and the BAL metatranscriptome are jointly associated with risk of post-HCT mortality.
Given prior findings that pre-HCT BAL metatranscriptomes are associated with post-HCT mortality (20), we next sought to determine whether these associations were independent of measured pulmonary function. Using multivariable modeling, we found that lower BAL RNA mass of oropharyngeal taxa (such as Haemophilus, Neisseria) was associated with post-HCT all-cause mortality independent of FVC, age, and sex. Further, progressively lower BAL RNA mass of oropharyngeal taxa strengthened the association between poor pre-HCT FVC and post-HCT mortality (interaction p<0.05). In addition, lower BAL expression of genes related to airway epithelial cell integrity, polarity, and junction formation (Hallmark Apical Surface) were also associated with post-HCT mortality independent of FVC, age, and sex (table S2). The addition of BAL Haemophilus RNA mass and BAL Hallmark Apical Surface gene set enrichment to a Cox survival model of FVC, age, and sex improved model performance across strata of lung function (likelihood ratio test p=0.002, Fig. 6).
Figure 6. Survival Estimates by Pre-HCT BAL Metatranscriptome and Pre-HCT FVC.
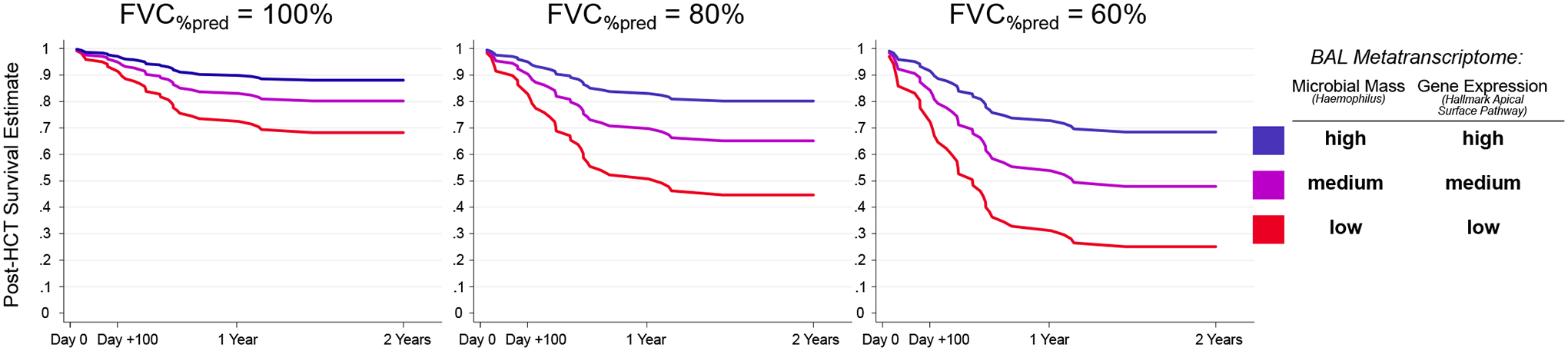
A multivariable Cox regression model for the outcome of post-HCT all-cause mortality was fit for all patients (n=104) using pre-HCT FVC%pred, pre-HCT BAL RNA mass of Haemophilus (vst transformed), and pre-HCT BAL enrichment score of MSigDB Hallmark Apical Surface Gene Set with covariate adjustment for patient age and sex. Model-based mortality estimates are plotted according to pre-HCT FVC (100% versus 80% versus 60%) and high, medium, and low Haemophilus RNA mass and Apical Surface gene expression. High, medium, and low refer to 75th, 50th, and 25th percentile within the cohort.
DISCUSSION
The primary finding of this study is that, in pediatric HCT candidates, pulmonary microbiome and gene expression signatures were associated with pulmonary function and may augment the utility of PFTs in the stratification of post-HCT mortality risk. Specifically, we found that the pulmonary microbiomes of patients with restriction and diffusion impairment were depleted of common supraglottic taxa and instead enriched in skin, nasal, and environmental taxa. In addition, the pulmonary transcriptomes of patients with restriction and diffusion impairment showed increased activation of alveolar epithelial cells and a deficit in immune cell signaling consistent with post-injury fibrotic transformation. Further, BAL signatures of commensal microbial depletion and diminished epithelial homeostasis were associated with post-HCT mortality independent of pre-HCT pulmonary function. Together these findings suggest a multidimensional connection between lung function, biology, and microbiology in pediatric HCT candidates and suggest possible biologic targets to optimize lung function in children preparing to undergo HCT.
First, our finding that pulmonary microbiome depletion is associated with simultaneously impaired lung function in pediatric HCT candidates supports our previous finding that pre-HCT pulmonary microbiome depletion is associated with post-HCT lung injury (20). Although studies of the pulmonary microbiome in children are rare, investigations of the more readily-studied intestinal microbiome have similarly shown that dysbiosis is associated with intestinal disease as well as poor overall post-HCT outcomes (28). Such landmark studies lend credence to contextualizing an organ-specific microbiome with the harboring organ’s function and molecular biology. However, such observations do not prove causality, and therefore whether pulmonary microbial dysbiosis can directly contribute to lung disease or is simply a biomarker of other exposures remains unclear. On the one hand, microbial dysbiosis is linked to antimicrobial exposure, which is more common in patients with underlying diseases such as malignancy; such patients also have confounding reasons for poor lung function, such as greater exposure to pulmotoxic chemotherapy. However, emerging studies suggest that microbial dysbiosis disrupts the control of pathogen overgrowth and contributes to a loss of immune tolerance at the epithelial-parenchymal boundary; therefore reciprocal causality between lung dysfunction and microbiome disturbances seems likely (22, 29). To that end, we found that the pulmonary RNA mass of taxa that are typically supraglottic commensals was inversely related to the RNA mass of taxa that typically reside in the nose and skin, supporting the theory that maintenance of the respiratory microbiomes may exclude invasion by outside organisms. Future studies directly measuring nasal, supraglottic, and pulmonary microbiomes in each patient may better test whether and how the upper and lower respiratory microbiomes interact and whether the microbiome structures at these upper respiratory sites are indeed microbiologically similar when compared to other populations (30–32). For example, detection of Staphyloccocus overgrowth in the nose and lungs and Haemophilus depletion in the oropharynx and lungs would provide more direct evidence for the assertion that the supraglottic microbiome protects the lungs from nasal, skin, and environmental microbes. In addition, strategies to characterize gut-lung cross-talk may further elucidate factors influencing the pulmonary microbiome (33, 34). Ultimately, strategies to modulate the lung microbiome in order to improve pulmonary health remain in development and include investigation of both ingested and inhaled probiotics and other microbial metabolites (35, 36).
A second finding of this work is the association between altered alveolar epithelial gene expression, restriction, and impaired diffusion. By combining differential gene expression with cell type imputation, we identified a greater proportion of type 2 AECs in patients with restriction and diffusion; further, cell-type specific gene expression suggested an increased transcriptional state of these cells, associated with growth factors and increased kinase expression. The increased expression of surfactant genes and overall AEC activity is consistent with response to alveolar injury and has been demonstrated in numerous cohorts of lung-injured patients (37, 38). By combining hallmark gene set enrichment with these findings, we found that pathways related to the lung stroma were upregulated, including angiogenesis, myogenesis, epithelial mesenchymal transition, and cell fate (Wnt/β-catenin signaling and hedgehog signaling). Activation of stromal pathways in the lung has been associated with a pro-fibrotic signaling state and has been identified in cohorts of patients with IPF, thus providing a plausible mechanistic link between these gene expression findings and the observed impairment in PFTs (39, 40). As lung-resident mesenchymal stem cells undergoing myofibroblastic differentiation strongly express genes in the Wnt/β-catenin and hedgehog signaling pathways, anti-fibrotic agents targeting this pathway, such as pirfenidone, remain of utmost interest (41, 42).
A third finding of this work is that patients with pulmonary restriction and impaired diffusion displayed an overall deficit in lower airway immune signaling. In addition to lower overall expression of immune genes among patents with restriction, cell type deconvolution inferred a less active transcriptional state for lymphocytes specifically. It seems counterintuitive that patients with poor PFTs would have a relative deficit in pulmonary immune activity, especially given our prior findings that pre-HCT pulmonary inflammation was associated with post-HCT lung injury. There are three potential explanations. First, the latter finding of pulmonary inflammation was tied to pre-HCT viral infection, which was more prevalent in children younger than 4 years old. As younger children are not typically able to perform PFTs, they were excluded from this present study. Second, although pulmonary inflammation is a hallmark of acute lung injury including chemoradiation-induced lung injury, it typically subsides after the initial insult and is followed by a profound anti-inflammatory and pro-fibrotic phase (43), which more likely represents the phase of injury for the patients in this study. Third, patients with impaired lower airway immunity may be at greater risk for infections and associated sequelae such as post-infectious fibrosis. Ultimately, the relevance of impaired pulmonary immune environment in pediatric HCT candidates with poor lung function needs to be explored with additional approaches in the future.
A fourth and unanticipated finding of this work was the identification of substantial sex differences in pulmonary function. Although both male and female study participants had worse lung function than age- and sex-matched controls, females had profoundly worse measures of both restriction and impaired diffusion, and males had slightly worse measures of obstruction. Long-term follow-up studies of pediatric oncology and HCT patients have also demonstrated associations between female sex, restriction, and impaired diffusion as well as between male sex and obstruction; this study adds to the field by identifying these disparities prior to HCT. A likely culprit for these toxicities are underlying differences in chemotherapy pharmacokinetics and pharmacodynamics, which merit exploration in future studies (44–46).
Our study has several strengths. First, the analysis of contemporaneously measured PFTs and BAL is unique among pediatric cohorts and allows multidimensional insights into lung function, biology, and microbiology. Second, we provide PFT measurements using both percent of predicted and GLI Z-scores according to updated ATS/ERS recommendations. Third, we analyzed BAL using a paired assessment of microbiome and gene expression.
Several limitations merit comment. First, as with all observational human studies, we are unable to prove mechanism or causation among the factors measured and described herein. Second, we were unable to ascertain all possible risk factors that may have influenced the pulmonary microenvironment, including history of pulmonary infections, prior antimicrobial exposure, and detailed chemotherapeutic regimens. Third, BAL samples were not paired with supraglottic and nasopharyngeal samples, and therefore assumptions about the possible origin of BAL taxa described as typically supraglottic or typically nasal or environmental are based on published literature rather than direct observation. Fourth, the identification of microbial RNA does not directly imply viable, intact organisms in the lower airways; future studies leveraging functional microbial genomics and metabolomics may provide deeper insight into the activity states of detected organisms (47, 48). Fifth, we did not have documentation of the fractional return of each patient’s lavage. Low fractional return is associated with both restrictive and obstructive lung disease and yield below 30% can indicate undersample of the distal airways. Of note, undersampling of the distal airways would be associated with greater microbial recovery and lower AEC recovery, which is the opposite of what we found in our patients with pulmonary restriction (49–54). Finally, younger children were necessarily excluded due to inability to perform PFTs. However, given our findings associating PFTs with lower respiratory biology, this study raises the possibility that BAL RNA sequencing could be a useful surrogate to detect pro-fibrotic states in those unable to perform PFTs. Greater correlation between biologic measures, standard spirometry, and newer PFT approaches such as forced impulse oscillometry and multiple breath washout may yield additional insights in children unable to perform standard PFTs (55–58).
In conclusion, in this study we found that among children preparing to undergo allogeneic HCT, PFT abnormalities were common and consisted mostly of restriction and impaired diffusion. Abnormalities were associated with pulmonary microbiome depletion, pro-fibrotic signaling, and post-HCT mortality. These findings suggest a potentially actionable connection between microbiome depletion, alveolar injury, and pulmonary fibrosis in the pathogenesis of HCT-related lung dysfunction.
MATERIALS AND METHODS
Study Design
As previously reported, pediatric allogeneic HCT candidates up to 19 years of age with malignant and non-malignant diseases underwent protocolized evaluation of pre-HCT pulmonary health between 2005 and 2016 in Utrecht, the Netherlands (11). Patients were not randomized and investigators were not blinded. This study included the subset who completed both pre-HCT PFTs and pre-HCT BAL prior to the start of conditioning chemotherapy (n=104). Informed consent was obtained with Institutional Review Board approval (#05/143 and #11/063). Details of approach to peri-HCT clinical care are described in the Supplementary Methods. Due to the exploratory nature of this study, a priori sample size estimates were not performed for the analyses described herein; individual level data for analyses with groups consisting of fewer than twenty patients are provided in data file S6.
PFT Measurements
PFTs included spirometry, whole body plethysmography, and carbon monoxide diffusion capacity and were performed in developmentally capable children ages 4 and older according to established criteria (59, 60). Pre-bronchodilator PFT values of adequate performance quality as assessed by each patient’s clinical pulmonologist were normalized for age, height, sex, and race and expressed as (1) a percentage deviation from the mean (% predicted) according to healthy Dutch children and (2) a standardized deviation from the mean (z-score) according to the GLI (61, 62). Restriction, obstruction, mixed restriction and obstruction, diffusion impairment, and air trapping were identified first using conventional PFT cutoffs and were compared to GLI-recommended cutoffs wherein PFTs below the 5th percentile (z-score <−1.64) were considered below the lower limit of normal (Supplementary Methods) (59, 62).
BAL Measurements
BAL was performed in all pediatric HCT candidates as standard-of-care routine clinical practice in order to identify potential pulmonary pathogens and reduce the risk of post-HCT lung injury. BAL was performed in the absence of lower respiratory tract symptoms and at the time of another clinically-indicated procedure (such as bone marrow aspirate, central line placement) (19, 63). Cryopreserved aliquots of BAL underwent metatranscriptomic RNA sequencing as previously described (20). Briefly, samples underwent mechanical homogenization followed by RNA extraction and purification (64, 65). Sample RNA was combined with control spike-in RNA and underwent reverse transcription, library preparation, and 125nt paired-end sequencing on a NovaSeq 6000 instrument to a target depth of 40 million read-pairs per sample (Supplementary Methods). Resultant .fastq files underwent quality filtration followed by parallel alignment to both human and microbial reference genomes using the IDseq pipeline (66). Microbe count data were adjusted for laboratory contamination and estimated microbe RNA mass was calculated using the reference ERCC spike-in (Supplementary Methods) (21).
Statistical Analysis
Associations between PFTs and patient characteristics were tested using Kruskal-Wallis tests and Spearman correlations for categorical and continuous traits, respectively. Associations between pre-HCT PFTs and post-HCT outcomes were tested with Kaplan-Meier estimates and Cox proportional hazards models for all-cause mortality and cumulative incidence function with competing risks regression for fatal lung injury. The pulmonary microbiome was described using taxa correlation matrices and PCA. Associations between PFTs and microbial RNA masses were tested using NB-GLM with adjustment for false discovery in the R statistical environment (edgeR) (67). Gene expression was characterized by calculating enrichment scores for the Hallmark Gene Sets in the Molecular Signatures Database (MSigDB) (68). Associations between PFTs and differentially expressed genes were tested using NB-GLM and characterized using pathway analysis (enrichR) (69). BAL cell type proportions and cell type-specific gene expression profiles were imputed using CiberSortX and the Human Lung Cell Atlas (Supplementary Methods, fig. S6) (27, 70).
Supplementary Material
Acknowledgments:
We thank Dr. Dean Sheppard, MD for his critical review of this manuscript.
Funding:
This study was funded by the National Marrow Donor Program Amy Strelzer Manasevit Grant (to MSZ), NICHD K12HD000850 (to MSZ), NHLBI K23HL146936 (to MSZ), An American Thoracic Society Foundation Grant (to MSZ), NIAID F31AI150007 (to SS), NHLBI R01HL134828 (to MAM), NHLBI R35HL140026 (to MAM), and by the Chan Zuckerberg Biohub (to JLD).
Footnotes
This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at www.sciencetranslationalmedicine.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior written permission of AAAS.
Supplementary Materials
Supplementary Materials and Methods
MDAR Reproducibility Checklist
Data File S1 to S6
Competing interests:
CAL participated in a data safety and monitoring board for Excellthera and advisory boards for Bluebird Bio, Orchard Therapeutics, Sobi. EKF participated in an advisory board for Boehringer Ingelheim. MAM participated in advisory boards for Johnson & Johnson and Citius Pharmaceuticals. CCD participated in advisory boards for Jazz Pharma, Alexion Inc, and Omeros Corp. JJB participated in advisory boards for Race Oncology, Omeros, BlueRock, Advanced Clinical, Sanofi, and Avrobio. JLD received consulting fees from Allen & Co and PHC, Inc. All other authors (MSZ, ABV, MC, MYM, GR, SS, SK, MC, SP, MS) declare that they have no competing interests.
Data and materials availability:
All data are available in the main text or the supplementary materials. Sequencing files are available through dbGaP study accession phs002208.v1.p1. Requests for data sharing should be addressed to matt.zinter@ucsf.edu.
References and Notes
- 1.D’Souza A, Fretham C, Lee SJ, Aurora M, Brunner J, Chhabra S, Devine S, Eapen M, Hamadani M, Hari P, Pasquini MC, Phelan RA, Riches ML, Rizzo JD, Saber W, Shaw BE, Spellman SR, Steinert P, Weisdorf DJ, Horowitz MM, Current Use and Trends in Hematopoietic Cell Transplantation in the United States. Biol.Blood Marrow Transplant 26, e177–182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broglie L, Fretham C, Al-Seraihy A, George B, Kurtzberg J, Loren A, MacMillan M, Martinez C, Davies SM, Pasquini MC, Pulmonary Complications in Pediatric and Adolescent Patients Following Allogeneic Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant 25, 2024–2030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaya Z, Weiner DJ, Yilmaz D, Rowan J, Goyal RK, Lung function, pulmonary complications, and mortality after allogeneic blood and marrow transplantation in children. Biol Blood Marrow Transplant 15, 817–826 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Tamburro RF, Cooke KR, Davies SM, Goldfarb S, Hagood JS, Srinivasan A, Steiner ME, Stokes D, DiFronzo N, El-Kassar N, Shelburne N, Natarajan A, Pulmonary Complications of Pediatric Hematopoietic Stem Cell Transplantation Workshop Participants, Pulmonary Complications of Pediatric Hematopoietic Stem Cell Transplantation (HCT): An NIH Workshop Summary. Ann Am Thorac Soc (2020), doi: 10.1513/AnnalsATS.202001-006OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duque-Afonso J, Ihorst G, Waterhouse M, Zeiser R, Wäsch R, Bertz H, Yücel M, Köhler T, Müller-Quernheim J, Marks R, Finke J, Comparison of reduced-toxicity conditioning protocols using fludarabine, melphalan combined with thiotepa or carmustine in allogeneic hematopoietic cell transplantation. Bone Marrow Transplant 56, 110–120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schechter T, Naqvi A, Weitzman S, Risk for complications in patients with hemophagocytic lymphohistiocytosis who undergo hematopoietic stem cell transplantation: myeloablative versus reduced-intensity conditioning regimens. Expert Rev Clin Immunol 10, 1101–1106 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Cheng G-S, Pulmonary Function and Pretransplant Evaluation of the Hematopoietic Cell Transplant Candidate. Clinics in Chest Medicine 38, 307–316 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Chien JW, Madtes DK, Clark JG, Pulmonary function testing prior to hematopoietic stem cell transplantation. Bone Marrow Transplant 35, 429–435 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Wieringa J, van Kralingen KW, Sont JK, Bresters D, Pulmonary function impairment in children following hematopoietic stem cell transplantation. Pediatr Blood Cancer 45, 318–323 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Smith AR, Majhail NS, MacMillan ML, DeFor TE, Jodele S, Lehmann LE, Krance R, Davies SM, Hematopoietic cell transplantation comorbidity index predicts transplantation outcomes in pediatric patients. Blood 117, 2728–2734 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Versluys AB, van der Ent K, Boelens JJ, Wolfs T, de Jong P, Bierings MB, High Diagnostic Yield of Dedicated Pulmonary Screening before Hematopoietic Cell Transplantation in Children. Biol Blood Marrow Transplant 21, 1622–1626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan A, Srinivasan S, Sunthankar S, Sunkara A, Kang G, Stokes DC, Leung W, Pre-hematopoietic stem cell transplant lung function and pulmonary complications in children. Ann Am Thorac Soc 11, 1576–1585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsberg JP, Aplenc R, McDonough J, Bethel J, Doyle J, Weiner DJ, Pre-transplant lung function is predictive of survival following pediatric bone marrow transplantation. Pediatr Blood Cancer 54, 454–460 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Quigg TC, Kim Y-J, Goebel WS, Haut PR, Lung function before and after pediatric allogeneic hematopoietic stem cell transplantation: a predictive role for DLCOa/VA. J Pediatr Hematol Oncol 34, 304–309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herasevich S, Frank RD, Bo H, Alkhateeb H, Hogan WJ, Gajic O, Yadav H, Pretransplant Risk Factors Can Predict Development of Acute Respiratory Distress Syndrome after Hematopoietic Stem Cell Transplantation. Ann Am Thorac Soc 18, 1004–1012 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Lee HJ, Kim K, Kim SK, Lee JW, Yoon J-S, Chung N-G, Bin C, Hb-adjusted DLCO with GLI reference predicts long-term survival after HSCT in children. Bone Marrow Transplant 56, 1929–1936 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Huang T-T, Hudson MM, Stokes DC, Krasin MJ, Spunt SL, Ness KK, Pulmonary outcomes in survivors of childhood cancer: a systematic review. Chest 140, 881–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kharbanda S, Panoskaltsis-Mortari A, Haddad IY, Blazar BR, Orchard PJ, Cornfield DN, Grewal SS, Peters C, Regelmann WE, Milla CE, Baker KS, Inflammatory cytokines and the development of pulmonary complications after allogeneic hematopoietic cell transplantation in patients with inherited metabolic storage disorders. Biol Blood Marrow Transplant 12, 430–437 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Versluys B, Bierings M, Murk JL, Wolfs T, Lindemans C, Vd Ent K, Boelens JJ, Infection with a respiratory virus before hematopoietic cell transplantation is associated with alloimmune-mediated lung syndromes. J Allergy Clin Immunol 141, 697–703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinter MS, Lindemans CA, Versluys BA, Mayday MY, Sunshine S, Reyes G, Sirota M, Sapru A, Matthay MA, Kharbanda S, Dvorak CC, Boelens JJ, DeRisi JL, The pulmonary metatranscriptome prior to pediatric HCT identifies post-HCT lung injury. Blood 137, 1679–1689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinter MS, Mayday MY, Ryckman KK, Jelliffe-Pawlowski LL, DeRisi JL, Towards precision quantification of contamination in metagenomic sequencing experiments. Microbiome 7, 62–6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Dwyer DN, Zhou X, Wilke CA, Xia M, Falkowski NR, Norman KC, Arnold KB, Huffnagle GB, Murray S, Erb-Downward JR, Yanik GA, Moore BB, Dickson RP, Lung Dysbiosis, Inflammation, and Injury in Hematopoietic Cell Transplantation. Am.J.Respir.Crit.Care Med 198, 1312–1321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Dwyer DN, Ashley SL, Gurczynski SJ, Xia M, Wilke C, Falkowski NR, Norman KC, Arnold KB, Huffnagle GB, Salisbury ML, Han MK, Flaherty KR, White ES, Martinez FJ, Erb-Downward JR, Murray S, Moore BB, Dickson RP, Lung Microbiota Contribute to Pulmonary Inflammation and Disease Progression in Pulmonary Fibrosis. Am J Respir Crit Care Med 199, 1127–1138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Einarsson GG, Comer DM, McIlreavey L, Parkhill J, Ennis M, Tunney MM, Elborn JS, Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 71, 795–803 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE, The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One 7, e47305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S, Shariff M, Chaturvedi G, Sharma A, Goel N, Yadav M, Mortensen MS, Sørensen SJ, Mukerji M, Chauhan NS, Comparative analysis of the alveolar microbiome in COPD, ECOPD, Sarcoidosis, and ILD patients to identify respiratory illnesses specific microbial signatures. Sci Rep 11, 3963 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, Diehn M, Alizadeh AA, Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37, 773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, Weber D, Hashimoto D, Slingerland AE, Slingerland JB, Maloy M, Clurman AG, Stein-Thoeringer CK, Markey KA, Docampo MD, Burgos da Silva M, Khan N, Gessner A, Messina JA, Romero K, Lew MV, Bush A, Bohannon L, Brereton DG, Fontana E, Amoretti LA, Wright RJ, Armijo GK, Shono Y, Sanchez-Escamilla M, Castillo Flores N, Alarcon Tomas A, Lin RJ, Yanez San Segundo L, Shah GL, Cho C, Scordo M, Politikos I, Hayasaka K, Hasegawa Y, Gyurkocza B, Ponce DM, Barker JN, Perales MA, Giralt SA, Jenq RR, Teshima T, Chao NJ, Holler E, Xavier JB, Pamer EG, van den Brink MRM, Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N.Engl.J.Med 382, 822–834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, Lynch SV, Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci.Transl.Med 4, 151ra124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, Weiden MD, Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1, 19–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, Curtis JL, Bacterial Topography of the Healthy Human Lower Respiratory Tract. MBio 8, 10.1128/mBio.02287-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB, The Microbiome and the Respiratory Tract. Annu.Rev.Physiol 78, 481–504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trivedi R, Barve K, Gut microbiome a promising target for management of respiratory diseases. Biochem J 477, 2679–2696 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJTH, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, de Vos WM, van der Poll T, Wiersinga WJ, The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65, 575–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H-S, Arellano K, Park H, Todorov SD, Kim B, Kang H, Park YJ, Suh DH, Jung ES, Ji Y, Holzapfel WH, Assessment of the safety and anti-inflammatory effects of three Bacillus strains in the respiratory tract. Environ Microbiol (2021), doi: 10.1111/1462-2920.15530. [DOI] [PubMed] [Google Scholar]
- 36.Nieto A, Mazón A, Nieto M, Calderón R, Calaforra S, Selva B, Uixera S, Palao MJ, Brandi P, Conejero L, Saz-Leal P, Fernández-Pérez C, Sancho D, Subiza JL, Casanovas M, Bacterial Mucosal Immunotherapy with MV130 Prevents Recurrent Wheezing in Children: A Randomized, Double-Blind, Placebo-controlled Clinical Trial. Am J Respir Crit Care Med 204, 462–472 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia O, Hiatt MJ, Lundin A, Lee J, Reddy R, Navarro S, Kikuchi A, Driscoll B, Targeted Type 2 Alveolar Cell Depletion. A Dynamic Functional Model for Lung Injury Repair. Am J Respir Cell Mol Biol 54, 319–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahmer MK, Flori H, Sapru A, Kohne J, Weeks HM, Curley MAQ, Matthay MA, Quasney MW, BALI and RESTORE Study Investigators and Pediatric Acute Lung Injury and Sepsis Investigators (PALISI) Network, Surfactant Protein D Is Associated With Severe Pediatric ARDS, Prolonged Ventilation, and Death in Children With Acute Respiratory Failure. Chest 158, 1027–1035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Distler JHW, Györfi A-H, Ramanujam M, Whitfield ML, Königshoff M, Lafyatis R, Shared and distinct mechanisms of fibrosis. Nat Rev Rheumatol 15, 705–730 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Crosby LM, Waters CM, Epithelial repair mechanisms in the lung. Am J Physiol Lung Cell Mol Physiol 298, L715–731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu B, Liu J, Wu Z, Liu T, Ullenbruch MR, Ding L, Henke CA, Bitterman PB, Phan SH, Reemergence of hedgehog mediates epithelial-mesenchymal crosstalk in pulmonary fibrosis. Am J Respir Cell Mol Biol 52, 418–428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Didiasova M, Singh R, Wilhelm J, Kwapiszewska G, Wujak L, Zakrzewicz D, Schaefer L, Markart P, Seeger W, Lauth M, Wygrecka M, Pirfenidone exerts antifibrotic effects through inhibition of GLI transcription factors. FASEB J 31, 1916–1928 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Malaviya R, Kipen HM, Businaro R, Laskin JD, Laskin DL, Pulmonary toxicants and fibrosis: innate and adaptive immune mechanisms. Toxicol Appl Pharmacol 409, 115272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haupt S, Caramia F, Klein SL, Rubin JB, Haupt Y, Sex disparities matter in cancer development and therapy. Nat Rev Cancer (2021), doi: 10.1038/s41568-021-00348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner AD, Grothey A, Andre T, Dixon JG, Wolmark N, Haller DG, Allegra CJ, de Gramont A, VanCutsem E, Alberts SR, George TJ, O’Connell MJ, Twelves C, Taieb J, Saltz LB, Blanke CD, Francini E, Kerr R, Yothers G, Seitz JF, Marsoni S, Goldberg RM, Shi Q, Sex and Adverse Events of Adjuvant Chemotherapy in Colon Cancer: An Analysis of 34 640 Patients in the ACCENT Database. J Natl Cancer Inst 113, 400–407 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diefenhardt M, Ludmir EB, Hofheinz R-D, Ghadimi M, Minsky BD, Rödel C, Fokas E, Association of Sex With Toxic Effects, Treatment Adherence, and Oncologic Outcomes in the CAO/ARO/AIO-94 and CAO/ARO/AIO-04 Phase 3 Randomized Clinical Trials of Rectal Cancer. JAMA Oncol 6, 294–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sulaiman I, Wu BG, Li Y, Tsay J-C, Sauthoff M, Scott AS, Ji K, Koralov SB, Weiden M, Clemente JC, Jones D, Huang YJ, Stringer KA, Zhang L, Geber A, Banakis S, Tipton L, Ghedin E, Segal LN, Functional lower airways genomic profiling of the microbiome to capture active microbial metabolism. Eur Respir J 58, 2003434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Yang Y, Yan Z, Liu H, Chen B, Liang Z, Wang F, Miller BE, Tal-Singer R, Yi X, Li J, Stampfli MR, Zhou H, Brightling CE, Brown JR, Wu M, Chen R, Shu W, Multi-omic meta-analysis identifies functional signatures of airway microbiome in chronic obstructive pulmonary disease. ISME J 14, 2748–2765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer KC, Raghu G, Baughman RP, Brown KK, Costabel U, du Bois RM, Drent M, Haslam PL, Kim DS, Nagai S, Rottoli P, Saltini C, Selman M, Strange C, Wood B, American Thoracic Society Committee on BAL in Interstitial Lung Disease, An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med 185, 1004–1014 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Heron M, Grutters JC, ten Dam-Molenkamp KM, Hijdra D, van Heugten-Roeling A, Claessen AME, Ruven HJT, van den Bosch JMM, van Velzen-Blad H, Bronchoalveolar lavage cell pattern from healthy human lung. Clin Exp Immunol 167, 523–531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shikano K, Abe M, Shiko Y, Tsushima K, Yoshioka K, Ishiwata T, Kawasaki T, Ikari J, Terada J, Kawasaki Y, Tatsumi K, What are the factors affecting the recovery rate of bronchoalveolar lavage fluid? Clin Respir J (2021), doi: 10.1111/crj.13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schildge J, Nagel C, Grun C, Bronchoalveolar lavage in interstitial lung diseases: does the recovery rate affect the results? Respiration 74, 553–557 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Koda K, Hozumi H, Yasui H, Suzuki Y, Karayama M, Furuhashi K, Enomoto N, Fujisawa T, Inui N, Nakamura Y, Suda T, Predictors for bronchoalveolar lavage recovery failure in diffuse parenchymal lung disease. Sci Rep 11, 1682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Löfdahl JM, Cederlund K, Nathell L, Eklund A, Sköld CM, Bronchoalveolar lavage in COPD: fluid recovery correlates with the degree of emphysema. Eur Respir J 25, 275–281 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Uhlving HH, Mathiesen S, Buchvald F, Green K, Heilmann C, Gustafsson P, Müller K, Nielsen KG, Small airways dysfunction in long-term survivors of pediatric stem cell transplantation. Pediatr Pulmonol 50, 704–712 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Nyilas S, Baumeler L, Tamm M, Halter JP, Savic S, Korten I, Meyer A, Singer F, Passweg JR, Latzin P, Stolz D, Inert Gas Washout in Bronchiolitis Obliterans Following Hematopoietic Cell Transplantation. Chest 154, 157–168 (2018). [DOI] [PubMed] [Google Scholar]
- 57.King GG, Bates J, Berger KI, Calverley P, de Melo PL, Dellacà RL, Farré R, Hall GL, Ioan I, Irvin CG, Kaczka DW, Kaminsky DA, Kurosawa H, Lombardi E, Maksym GN, Marchal F, Oppenheimer BW, Simpson SJ, Thamrin C, van den Berge M, Oostveen E, Technical standards for respiratory oscillometry. Eur Respir J 55, 1900753 (2020). [DOI] [PubMed] [Google Scholar]
- 58.De Assumpcao MS, da Silva Goncalves E, Oliveira MS, Ribeiro JD, Dalbo Contrera Toro AA, de Azevedo Barros-Filho A, de Monteiro MARG, Santos Schivinski CI, Impulse Oscillometry System and Anthropometric Variables of Preschoolers, Children and Adolescents Systematic Review. Curr Pediatr Rev 13, 126–135 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Stanojevic S, Graham BL, Cooper BG, Thompson BR, Carter KW, Francis RW, Hall GL, Global Lung Function Initiative TLCO working group, Global Lung Function Initiative (GLI) TLCO, Official ERS technical standards: Global Lung Function Initiative reference values for the carbon monoxide transfer factor for Caucasians. Eur Respir J 50 (2017), doi: 10.1183/13993003.00010-2017. [DOI] [PubMed] [Google Scholar]
- 60.Culver BH, Graham BL, Coates AL, Wanger J, Berry CE, Clarke PK, Hallstrand TS, Hankinson JL, Kaminsky DA, MacIntyre NR, McCormack MC, Rosenfeld M, Stanojevic S, Weiner DJ, ATS Committee on Proficiency Standards for Pulmonary Function Laboratories, Recommendations for a Standardized Pulmonary Function Report. An Official American Thoracic Society Technical Statement. Am J Respir Crit Care Med 196, 1463–1472 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Koopman M, Zanen P, Kruitwagen CLJJ, van der Ent CK, Arets HGM, Reference values for paediatric pulmonary function testing: The Utrecht dataset. Respir Med 105, 15–23 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Quanjer PH, Stanojevic S, Cole TJ, Baur X, Hall GL, Culver BH, Enright PL, Hankinson JL, Ip MSM, Zheng J, Stocks J, ERS Global Lung Function Initiative, Multi-ethnic reference values for spirometry for the 3–95-yr age range: the global lung function 2012 equations. Eur Respir J 40, 1324–1343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Versluys AB, Rossen JW, van Ewijk B, Schuurman R, Bierings MB, Boelens JJ, Strong association between respiratory viral infection early after hematopoietic stem cell transplantation and the development of life-threatening acute and chronic alloimmune lung syndromes. Biol Blood Marrow Transplant 16, 782–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zinter MS, Dvorak CC, Mayday MY, Iwanaga K, Ly NP, McGarry ME, Church GD, Faricy LE, Rowan CM, Hume JR, Steiner ME, Crawford ED, Langelier C, Kalantar K, Chow ED, Miller S, Shimano K, Melton A, Yanik GA, Sapru A, DeRisi JL, Pulmonary Metagenomic Sequencing Suggests Missed Infections in Immunocompromised Children. Clin Infect Dis 68, 1847–1855 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mayday MY, Khan LM, Chow ED, Zinter MS, DeRisi JL, Miniaturization and optimization of 384-well compatible RNA sequencing library preparation. PLoS One 14, e0206194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalantar KL, Carvalho T, de Bourcy CFA, Dimitrov B, Dingle G, Egger R, Han J, Holmes OB, Juan Y-F, King R, Kislyuk A, Lin MF, Mariano M, Morse T, Reynoso LV, Cruz DR, Sheu J, Tang J, Wang J, Zhang MA, Zhong E, Ahyong V, Lay S, Chea S, Bohl JA, Manning JE, Tato CM, DeRisi JL, IDseq-An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascience 9, giaa111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD, Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat.Protoc 8, 1765–1786 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128–128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, Berry GJ, Shrager JB, Metzger RJ, Kuo CS, Neff N, Weissman IL, Quake SR, Krasnow MA, A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panoskaltsis-Mortari A, Griese M, Madtes DK, Belperio JA, Haddad IY, Folz RJ, Cooke KR, American Thoracic Society Committee on Idiopathic Pneumonia Syndrome, An official American Thoracic Society research statement: noninfectious lung injury after hematopoietic stem cell transplantation: idiopathic pneumonia syndrome. Am.J.Respir.Crit.Care Med 183, 1262–1279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jagasia MH, Greinix HT, Arora M, Williams KM, Wolff D, Cowen EW, Palmer J, Weisdorf D, Treister NS, Cheng GS, Kerr H, Stratton P, Duarte RF, McDonald GB, Inamoto Y, Vigorito A, Arai S, Datiles MB, Jacobsohn D, Heller T, Kitko CL, Mitchell SA, Martin PJ, Shulman H, Wu RS, Cutler CS, Vogelsang GB, Lee SJ, Pavletic SZ, Flowers ME, National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group report. Biol.Blood Marrow Transplant 21, 389–401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ, Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226–2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. Sequencing files are available through dbGaP study accession phs002208.v1.p1. Requests for data sharing should be addressed to matt.zinter@ucsf.edu.


