Abstract
Dinitrogenase reductase is posttranslationally regulated by dinitrogenase reductase ADP-ribosyltransferase (DRAT) via ADP-ribosylation of the arginine 101 residue in some bacteria. Rhodospirillum rubrum strains in which the arginine 101 of dinitrogenase reductase was replaced by tyrosine, phenylalanine, or leucine were constructed by site-directed mutagenesis of the nifH gene. The strain containing the R101F form of dinitrogenase reductase retains 91%, the strain containing the R101Y form retains 72%, and the strain containing the R101L form retains only 28% of in vivo nitrogenase activity of the strain containing the dinitrogenase reductase with arginine at position 101. In vivo acetylene reduction assays, immunoblotting with anti-dinitrogenase reductase antibody, and [adenylate-32P]NAD labeling experiments showed that no switch-off of nitrogenase activity occurred in any of the three mutants and no ADP-ribosylation of altered dinitrogenase reductases occurred either in vivo or in vitro. Altered dinitrogenase reductases from strains UR629 (R101Y) and UR630 (R101F) were purified to homogeneity. The R101F and R101Y forms of dinitrogenase reductase were able to form a complex with DRAT that could be chemically cross-linked by 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide. The R101F form of dinitrogenase reductase and DRAT together were not able to cleave NAD. This suggests that arginine 101 is not critical for the binding of DRAT to dinitrogenase reductase but that the availability of arginine 101 is important for NAD cleavage. Both DRAT and dinitrogenase reductase can be labeled by [carbonyl-14C]NAD individually upon UV irradiation, but most 14C label is incorporated into DRAT when both proteins are present. The ability of R101F dinitrogenase reductase to be labeled by [carbonyl-14C]NAD suggested that Arg 101 is not absolutely required for NAD binding.
Various procaryotic microorganisms are capable of fixing nitrogen. The essential enzyme in this reaction is the nitrogenase complex, which catalyzes the reduction of nitrogen to ammonium (4). This complex is composed of two separable metalloenzymes designated dinitrogenase and dinitrogenase reductase. Dinitrogenase reductase transfers electrons from an electron donor to dinitrogenase, which then can catalyze the reduction of nitrogen to ammonium. Because nitrogen fixation is an energy-demanding process, it is tightly regulated in response to a number of environmental factors. In addition to transcriptional regulation, a posttranslational nitrogenase regulatory mechanism also exists in certain organisms, such as Rhodospirillum rubrum (33, 35). R. rubrum is a free-living, purple, nonsulfur photosynthetic bacterium that fixes nitrogen (20). The activity of its nitrogenase is regulated by the reversible ADP-ribosylation of the arginine 101 of the dinitrogenase reductase (31, 42). Dinitrogenase reductase ADP-ribosyltransferase (DRAT) can transfer ADP-ribose from NAD to dinitrogenase reductase in response to darkness or the presence of ammonium. This ADP-ribosylation prevents the productive association of dinitrogenase reductase with dinitrogenase, such that neither electron transfer nor ATP hydrolysis can occur. In these circumstances, the nitrogenase activity is said to be switched off (38). Only one of the two arginine 101 residues present within a homodimeric dinitrogenase reductase is modified (42). Dinitrogenase reductase-activating glycohydrolase can remove ADP-ribose from inactivated dinitrogenase reductase in vivo when illumination resumes or upon exhaustion of ammonium in the medium and activate the nitrogenase system (43).
Dinitrogenase reductases with substitutions at arginine 101 or its equivalent residues have been studied for their ability to transfer electrons and to serve as a substitute for DRAT. No substitution of arginine at position 101 results in a dinitrogenase reductase capable of being ADP-ribosylated by DRAT. However, several dinitrogenase reductases with substitutions at arginine 101 accumulate as native proteins with a redox-active Fe4S4 center and have substantial electron transfer ability. These include R100Y from Azotobacter vinelandii (44) as well as R101Y and R101F from Azospirillum brasilense (46). The R100H forms of dinitrogenase reductase from both A. vinelandii and Klebsiella pneumoniae accumulate as native proteins but have only a trace of electron transfer activity (30, 44). The electron transfer activity of R100H is very sensitive to ionic strength (44). In the Rhodobacter capsulatus background, R102F and R102Y again show electron transfer activity (41).
Although the role of Arg 101 in the interaction between dinitrogenase reductase and dinitrogenase has been studied, the role of this residue in the interaction between dinitrogenase reductase and DRAT has not been explored. A chemical cross-linking method has been used to study the interaction between these two proteins (17). The cross-linking reagent 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) can cross-link dinitrogenase reductase and DRAT in the presence of NAD. The crystal structure of A. vinelandii dinitrogenase reductase shows that the two arginine 100 side chains are located at the same surface as the metal cluster (15), which is the region speculated to interact with DRAT (17). The effects of substitutions at several other residues have been reported (18). The ability of dinitrogenase reductase with substitution at arginine 101 to participate in a complex with DRAT is reported here.
Most ADP-ribosyltransferases, such as diphtheria toxin, exotoxin A, cholera toxin, and pertussis toxin, can bind and glycohydrolyze NAD without the presence of target proteins (5, 6, 14, 29). Unlike these other ADP-ribosyltransferases, however, the ability of DRAT alone to glycohydrolyze NAD is undetectable in vitro (31) and an affinity of DRAT for NAD has not so far been detected. To study the role of arginine 101 in NAD binding and NAD glycohydrolysis, as well as in DRAT binding, dinitrogenase reductases altered at this residue have been constructed.
MATERIALS AND METHODS
Strains, growth, and whole-cell nitrogenase activity assay.
The strains used in this study are listed in Table 1. Throughout this report, R101 (Arg-101, which is the wild type), R101Y (the Tyr-101 substitution), R101F (the Phe-101 substitution), and R101L (the Leu-101 substitution) refer to R. rubrum dinitrogenase reductase with the specified amino acid at position 101. Growth conditions and concentrations of antibiotics used for Escherichia coli and R. rubrum cells were the same as those used previously (28). Derepression and the measurement of nitrogenase activity in R. rubrum were done as before (27). For NH4+ treatment, 50 μl of 1 M NH4Cl was added into 10 ml of derepressed R. rubrum cultures. For dark treatment, vials containing derepressed R. rubrum cells were wrapped with aluminum foil.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| UR2 | Smrnif+ | 25 |
| UR206 | SmrnifH::kan derivative of UR156 lackiing pPH1JI | 25 |
| UR628 | Wild-type (Arg 101) nifHD′ genes were integrated into chromosome of UR206 | This study |
| UR629 | Tyr 101 nifHD′ genes were integrated into chromosome of UR206 | This study |
| UR630 | Phe 101 nifHD′ genes were integrated into chromosome of UR206 | This study |
| UR631 | Leu 101 nifHD′ genes were integrated into chromosome of UR206 | This study |
| Plasmids | ||
| pLL102 | pUC19 containing R. rubrum nifHDK | 25 |
| pLL106 | pUC19 containing R. rubrum nifH′ | 25 |
| pSUP202 | Ampr Cmr mob+ | 26 |
| pYM1 | pUC19 containing R. rubrum nifH′ | This study |
| pYM2 | pSUP202 containing R. rubrum nifHD′ | This study |
Site-directed mutagenesis of nifH
A unique site elimination method (11) was modified to generate mutations (46).
Mobilization of the cloned nifHD′ region into R. rubrum cells.
To mobilize mutated R. rubrum nifH genes into the R. rubrum chromosome, plasmid vector pYM2 was constructed by blunt-end ligation of a 2.5-kb AflIII-BglII fragment from pLL102 and a 7.2-kb EcoRI-PstI fragment of pSUP202 (25); then the 0.6-kb EcoRI-PstI fragment containing nifH encoding either the R101Y, R101F, or R101L form was subcloned into pYM2 to replace the wild-type fragment. The desired plasmids were transformed into E. coli strain UQ324 and mated with UR206 under the conditions described previously (16). A single crossover event was desired, so that Smr Kmr Tcr colonies were selected. The constructed strains were named UR628, UR629, UR630, and UR631 (Table 1).
Purification of dinitrogenase reductase and DRAT.
DRAT was purified from R. rubrum strain UR356 as described previously (17, 31). DRAT protein was monitored throughout the purification by immunoblotting with anti-DRAT antibody. The wild type, R101Y, and R101F dinitrogenase reductases were purified from R. rubrum strains UR472, UR629, and UR630, respectively, as described previously (34). Dinitrogenase reductase activity was monitored throughout the purification by the in vitro acetylene reduction nitrogenase assay (3).
DRAT-dinitrogenase reductase cross-linking.
Cross-linking reactions were performed in Bonam vials as described previously (17, 32) with modification. The cross-linking reaction in a 40-μl mixture contained 8 μg of purified dinitrogenase reductase, 0.1 μg of purified DRAT, 0.1 mM sodium dithionite, and other nucleotide components as indicated in 25 mM HEPES buffer, pH 7.6. The reaction was started by adding EDC (Pierce Chemicals) to the reaction mixture to a final concentration of 8 mM, the reaction mixture was incubated at 30°C for 5 min, and the reaction was stopped by addition of 40 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (130 mM Tris [pH 6.8], 4.2% [wt/vol] SDS, 20% [vol/vol] glycerol, 0.003% [wt/vol] bromphenol blue, and 10% [vol/vol] 2-mercaptoethanol). After boiling for 1 min, 5 μl of each reaction sample was loaded onto a low cross-linker gel (22). Immunoblots with anti-DRAT antibody were visualized by either alkaline phosphatase color development (2) or chemiluminescence (Amersham) (17).
DRAT assay, in vitro ADP-ribosylation of dinitrogenase reductase, and [14C]nicotinamide release experiment.
DRAT activity was assayed as described previously (31) except as noted. For in vitro ADP-ribosylation of dinitrogenase reductase, assay mixtures contained 16 μg of dinitrogenase reductase, 0.4 μg of DRAT, 1 mM ADP, 5 mM MgCl2, and 0.2 mM [α-32P]NAD (15 mCi/mmol), which was prepared from [α-32P]ATP (3,000 Ci/mmol; Amersham) (9), in 100 mM morpholinepropanesulfonic acid (MOPS) buffer, pH 7, to a total volume of 50 μl. After a 1-h incubation at 30°C, proteins were precipitated by 5% trichloroacetic acid (TCA), washed two times with 5% TCA to remove free [32P]NAD, and then resuspended in SDS-PAGE sample buffer and loaded on a low cross-linker gel. After drying, the gel was exposed to a storage phosphor screen (Molecular Dynamics, Inc) to detect the [32P]ADP-ribosylated modified dinitrogenase reductase. In order to test the ability of DRAT to hydrolyze NAD in the presence of the R101F dinitrogenase reductase (which cannot be ADP-ribosylated), 250 μg of dinitrogenase reductase was incubated with 3 μg of DRAT in the presence of 1 mM ADP, 5 mM MgCl2, and 0.15 μCi of [carbonyl-14C]NAD (35 mCi/mmol; Amersham) in 100 mM MOPS buffer, pH 7, in a total volume of 55 μl. After incubation at 30°C for 3 h, 1 μl of reaction mixture was spotted on a Polygram CEL 300 PEI thin-layer plate with a 254-nm fluorescence indicator (Macherey-Nagel) and was developed in isobutyrate-NH4OH-H2O (66:1:33). After drying, the plate was exposed to a Molecular Dynamics storage phosphor screen and quantitated by using ImageQuaNT software on a PhosphorImager (Molecular Dynamics, Inc.).
UV photoaffinity labeling.
A solution containing 30 μg of DRAT, 1 mM ADP, and 5 mM MgCl2 in 100 mM MOPS buffer, pH 7, was placed in a quartz cuvette (Starna Cells, Inc.) with 71.5 μM [carbonyl-14C]NAD (35 mCi/mmol; Amersham) and then made anaerobic. Then 32 μg of dinitrogenase reductase was added to the anaerobic cuvette with a Hamilton syringe to bring the final volume to 100 μl. The cuvette was placed on ice with the optical transmission side 4 cm away from a 25-W germicidal lamp (General Electric) for 1 h. Following UV irradiation, the proteins in the reaction mixture were precipitated by 5% TCA and were washed two times with 5% TCA to remove free [carbonyl-14C]NAD; the pellet was then resuspended in 80 μl of SDS sample buffer. Then 20 μl was loaded on a standard SDS-PAGE gel (24). After Coomassie blue staining and drying, the gel was exposed to a Molecular Dynamics storage phosphor screen.
Preparation of R. rubrum crude extract samples, SDS-PAGE, and immunoblotting.
For samples to be analyzed for the ADP-ribosylation status of dinitrogenase reductase, TCA precipitation was used to extract proteins quickly from R. rubrum cells as described before (45). Low cross-linker gels were used to obtain separation of the dinitrogenase reductase subunits (22). Proteins separated on an SDS-PAGE gel were transferred to a nitrocellulose membrane, which was incubated with a polyclonal antibody against dinitrogenase reductase or DRAT, and then were visualized by either alkaline phosphatase color development (2) or chemiluminescence (Amersham).
RESULTS
Nitrogenase activity and immunoblot of arginine substitution mutants of R. rubrum in derepression medium.
To study the effects of substitutions at the Arg 101 position of dinitrogenase reductase in R. rubrum, the R101Y, R101F, and R101L mutants were constructed as described in Materials and Methods. In order to compare the nitrogenase activities of mutants with a strain containing the wild-type nifH gene in the same genetic background, UR628, which encodes the wild-type dinitrogenase reductase, was constructed in the UR206 genetic background (Table 1). Cultures were grown in malate-glutamate derepression medium, and their in vivo nitrogenase activities were measured before and after dark or ammonium treatment. As shown in Table 2, UR628 had 73% of the activity of the wild-type R. rubrum strain, UR2, which may be due to the less efficient dinitrogenase reductase expression in UR628. Compared to UR628, UR629 (R101Y) had 91%, UR630 (R101F) had 72%, and UR631 (R101L) had 28% of the nitrogenase activity of UR628. These results differ from those for R. capsulatus and A. brasilense, in which a Tyr substitution of dinitrogenase reductase Arg 101 gave a higher nitrogenase activity than a Phe substitution (41, 46). After a 60-min dark treatment or a 90-min 5 mM ammonium treatment, both UR2 and UR628 were switched off, while none of the mutants with substitutions at Arg 101 lost activity. UR206, the nifH mutant host strain for Arg substitution, itself had a very low nitrogenase activity which showed dark- and ammonium-induced switch-off; this was due to the expression of an alternative nitrogenase (27).
TABLE 2.
Responses of in vivo acetylene reduction activities of R. rubrum strains to inactivating conditionsa
| Strain | Residue at position 101 | Initial activity before dark treatment | Activity after 60-min dark treatment | Initial activity before ammonium treatment | Activity after 90-min ammonium treatment (5 mM) |
|---|---|---|---|---|---|
| UR2 | Arg (wild type) | 1,334 ± 132 | 184 ± 23 | 1,080 ± 187 | 248 ± 16 |
| UR206 | Noneb | 84 ± 6 | 16 ± 5 | 77 ± 3 | 42 ± 17 |
| UR628 | Arg | 957 ± 86 | 136 ± 37 | 804 ± 32 | 170 ± 56 |
| UR629 | Tyr | 688 ± 113 | 708 ± 23 | 567 ± 77 | 556 ± 145 |
| UR630 | Phe | 881 ± 35 | 666 ± 88 | 719 ± 91 | 600 ± 133 |
| UR631 | Leu | 283 ± 49 | 255 ± 44 | 203 ± 43 | 180 ± 28 |
The activities were calculated as nanomoles of C2H4 per hour · optical density unit. The values shown are the averages of triplicates ± standard deviations. Strains were derepressed in glutamate-malate medium, and the in vivo acetylene reduction activities were measured before and after dark or ammonium treatment at the indicated time.
The nifH gene is deleted in strain UR206.
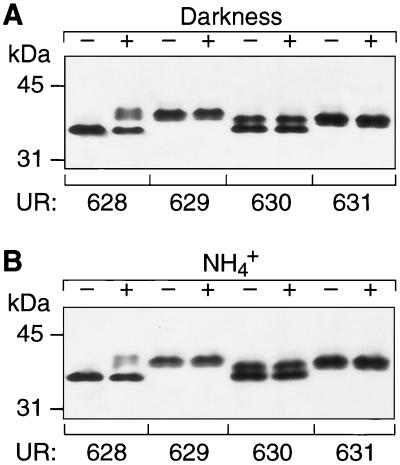
Extracts of strains expressing R101R, R101Y, R101F, and R101L dinitrogenase reductase were analyzed by immunoblotting. Immunoblots of SDS-PAGE gels developed with anti-dinitrogenase reductase antibody showed that each mutant synthesized dinitrogenase reductase with electrophoretic properties slightly different than those of the wild-type enzyme (Fig. 1). This is consistent with the observation that dinitrogenase reductases substituted at Arg 101 often run differently than the wild type on SDS-PAGE gels (41). R101F dinitrogenase reductase ran as two equally intense bands, which has also been observed for A. vinelandii dinitrogenase reductase (13) and Azotobacter chroococcum dinitrogenase reductase (37). The reason for this is unknown, and no relation between this phenomenon and the activity of the dinitrogenase reductase was found. No shifted bands for any of the altered dinitrogenase reductases were detected on immunoblots, in contrast to the band shift caused by ADP-ribosylation of wild-type dinitrogenase reductase from strain UR628 subjected to dark or ammonium treatment (Fig. 1). Immunoblots of two-dimensional gels of extracts of strains producing R101Y, R101F, and R101L failed to show any anf-encoded alternative nitrogenase (data not shown).
FIG. 1.
Immunoblot of crude extracts of strains UR628, UR629, UR630, and UR631 developed with anti-dinitrogenase reductase antibody. Samples were taken before and after a 60-min dark (A) or 90-min ammonium (B) treatment.
To further characterize these altered proteins, wild-type, R101Y, and R101F dinitrogenase reductases were purified to homogeneity. DRAT-catalyzed ADP-ribosylation was then attempted with the different forms of dinitrogenase reductases in the presence of MgADP and [32P]NAD. Neither R101Y nor R101F dinitrogenase reductase was able to incorporate the 32P label, while two positive controls, wild-type A. vinelandii and R. rubrum dinitrogenase reductase, incorporated the 32P label, indicating ADP-ribosylation (data not shown).
Cross-linking of DRAT to dinitrogenase reductase.
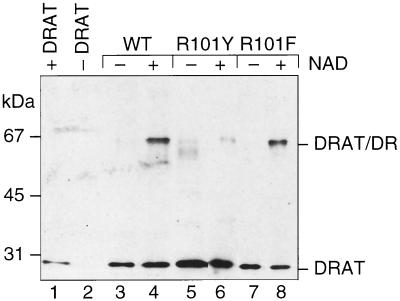
The above data demonstrated that dinitrogenase reductase altered at the Arg 101 site cannot be ADP-ribosylated by DRAT either in vivo or in vitro. The role of Arg 101 in the association of dinitrogenase reductase with DRAT is unclear, however. The crystal structure of A. vinelandii dinitrogenase reductase (15) suggested that the Arg 101 region might interact with DRAT (17), and in order to explore the role of Arg 101 in the interaction of dinitrogenase reductase and DRAT, a chemical cross-linking method was used. The ability of R101Y and R101F dinitrogenase reductases to be chemically cross-linked with DRAT in the presence of NAD was tested using EDC, and the results are presented in Fig. 2. To our surprise, both altered dinitrogenase reductases were able to be cross-linked with DRAT in the presence of NAD or some of its analogs. Previous studies (17) showed that the protein band labeled DRAT/DR in Fig. 2 contained both DRAT and dinitrogenase reductase. R101F dinitrogenase reductase showed a very strong cross-linked band with DRAT, while R101Y dinitrogenase reductase showed a less intense cross-linked band on gels. The degree of cross-linking of the altered dinitrogenase reductases corresponds to the level of their abilities to transfer electrons to dinitrogenase. NADP and nicotinamide mononucleotide are also able to promote cross-linking of DRAT to R101Y and R101F dinitrogenase reductase, as they are with the wild-type protein.
FIG. 2.
Immunoblot with anti-DRAT antibody, showing the nucleotide dependence of DRAT-dinitrogenase reductase EDC-dependent cross-linking. Cross-linking reactions were performed as described in Materials and Methods except that the following proteins and nucleotides (1.25 mM MgADP, 2 mM NAD) were added to the reaction: lane 1, only DRAT; lane 2, no DRAT; lanes 3 and 4, DRAT and wild-type dinitrogenase reductase; lanes 5 and 6, DRAT and R101Y dinitrogenase reductase; lanes 7 and 8, DRAT and R101F dinitrogenase reductase. Lanes 3, 5, and 7, no NAD; lanes 4, 6, and 8, NAD. All reactions contained 1.25 mM MgADP.
Having shown by EDC-dependent chemical cross-inking that altered dinitrogenase reductases are able to interact with DRAT, we then wanted to determine if the interaction is similar to that observed with wild-type protein. Two lines of evidence suggested that it is. First, cross-linking was supported by the same set of nucleotides in both altered and wild-type dinitrogenase reductases, as indicated in Fig. 2. The second line of evidence involved salt inhibition of the cross-linking reaction. It is known that salt can inhibit the wild-type dinitrogenase reductase cross-linking to DRAT, and it is assumed from this that ionic interactions are important in the cross-linking of these two proteins (17). Similar salt inhibition was observed in these altered proteins' cross-linking with DRAT (data not shown). The tolerance of R101F dinitrogenase reductase cross-linking to salt is almost the same as that of the wild-type protein, while the R101Y substitution is much more sensitive to salt inhibition.
This surprising result—that dinitrogenase reductase altered in its arginine 101 site still supports apparently normal cross-linking—suggests that the arginine 101 site is probably not essential for dinitrogenase reductase in the interaction with DRAT, even though it is the residue to which the ADP-ribose is attached.
[14C]nicotinamide release experiment.
Because DRAT shows no ability to cleave NAD in the absence of dinitrogenase reductase (31), it appears that NAD cleavage occurs only when NAD is recruited into the DRAT-dinitrogenase reductase protein complex. The EDC cross-linking experiment shows that altered dinitrogenase reductases can still associate with DRAT. The next question is whether NAD cleavage is dependent upon arginine at position 101. There are two possible fates proposed for the NAD buried in the protein complex of DRAT and dinitrogenase reductase R101F. One possibility is that NAD is cleaved into ADP-ribose and nicotinamide; the other is that no NAD cleavage occurs.
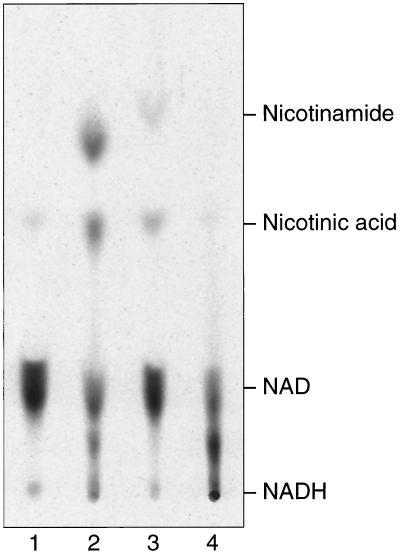
The ability of the DRAT-R101F dinitrogenase reductase complex to cleave NAD was tested by mixing the two proteins with [carbonyl-14C]NAD. The production of nicotinamide was tested by chromatography of the reaction mixture on thin-layer plates; various standards were cochromatographed to establish the chromatographic position of products. As positive controls, the wild-type dinitrogenase reductases from A. vinelandii and R. rubrum were mixed with DRAT and the production of nicotinamide resulting from ADP-ribosylation of the dinitrogenase reductase was observed (Fig. 3, lanes 2 and 3, respectively) (31). Interestingly, in these positive controls, a spot corresponding to nicotinic acid was observed in addition to the nicotinamide spot. A very small amount of the putative nicotinic acid spot was observed in the negative control (DRAT only), but the intensity of the spot clearly increased when DRAT and [14C]NAD were incubated with either A. vinelandii or R. rubrum dinitrogenase reductase. Perhaps the NAD contained some nicotinic acid adenine dinucleotide or perhaps nicotinamide is deamidated in the ADP-ribosylation reaction. Nicotinic acid adenine dinucleotide is not effective in the ADP-ribosylation reaction, so it is more likely that a deamidation occurs.
FIG. 3.
Phosphorimage of a thin-layer chromatogram showing [14C]nicotinamide release from [carbonyl-14C]NAD. Reactions of different dinitrogenase reductases with DRAT were performed as described in Materials and Methods except that different proteins were added to the reaction as follows: lane 1, DRAT only; lane 2, DRAT plus A. vinelandii dinitrogenase reductase; lane 3, DRAT plus wild-type R. rubrum dinitrogenase reductase; lane 4, DRAT plus R101F dinitrogenase reductase.
When DRAT and R101F dinitrogenase reductase are incubated with [14C]NAD, no increase in nicotinamide above background is observed (Fig. 3, lane 4). It is not possible to absolutely rule out the cleavage of NAD by this complex, but if it occurs, it does so at a rate less than 1% of that of the ADP-ribosylation reaction. No cleavage of NAD is observed when NAD is incubated with the dinitrogenase reductase protein in the absence of DRAT (data not shown).
UV photoaffinity labeling.
Wild-type dinitrogenase reductase is ADP-ribosylated by DRAT during cross-linking reactions, and ADP-ribosylated dinitrogenase reductase is no longer able to cross-link with DRAT. This suggests that ADP-ribosylation disrupts the complex (17). No detectable ADP-ribosylation occurs in the protein complex formed between R101F dinitrogenase reductase and DRAT, however, nor is there any detectable NAD cleavage. This suggests that the DRAT-R101F dinitrogenase reductase might form a stable protein complex. EDC-dependent cross-linking experiments suggest that the cross-linkable protein complex is NAD dependent but do not show whether NAD exists within this complex. Because of its presumed greater stability, this unique altered dinitrogenase reductase may serve as a good model in which to look for the presence of NAD in the DRAT-R101F dinitrogenase reductase protein complex.
Collier and coworkers discovered that UV irradiation of diphtheria toxin in the presence of [carbonyl-14C]NAD results in labeling of Glu 148 of the protein (5, 8). Bacterial toxins usually have a strong binding affinity for NAD. The NAD binding Kd is 27 μM for pertussis toxin (29) and 8 μM for diphtheria toxin (21). In diphtheria toxin, UV irradiation results in the decarboxylation of glutamic acid 148 of the toxin and the formation of a new bond between the γ-methylene carbon of glutamic acid and carbon 6 of the nicotinamide ring, with release of the ADP-ribose moiety of NAD (8).
In order to test if NAD is present within the dinitrogenase reductase-DRAT protein complex, similar UV labeling experiments were done. After UV irradiation of the proposed protein complex, SDS-PAGE was performed to separate the proteins.
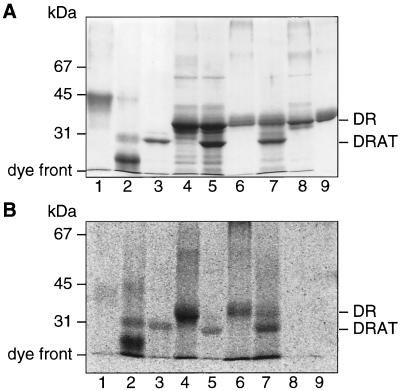
Surprisingly, DRAT, wild-type dinitrogenase reductase and R101F dinitrogenase reductase were each labeled by [14C]NAD when irradiated by UV separately. When dinitrogenase reductase (or altered dinitrogenase reductase) and DRAT were present together with [14C]NAD, most of the 14C label appeared in the DRAT band (Fig. 4). The negative control in lane 1 shows that chicken egg albumin did not incorporate the 14C label under the same conditions, while the positive control in lane 2 shows that cholera toxin A1 can incorporate the 14C label. The 14C label incorporation efficiency of cholera toxin, DRAT, dinitrogenase reductase, and altered dinitrogenase reductase were all in the same range, around 0.01 mol of NAD/mol of protein. The oxygen-denatured wild-type or R101F dinitrogenase reductase did not incorporate 14C label (Fig. 4, lanes 8 and 9). A small amount of protein-protein cross-linking may have occurred in all the UV-irradiated samples, but most proteins were still intact after 1 h under UV light (Fig. 4A). The acetylene reduction activity of dinitrogenase reductase was decreased to almost 0 after 1 h of UV irradiation, and the ADP-ribosyltransferase activity of DRAT still retained 20% of original activity after 1 h of UV irradiation (data not shown). A similar set of experiments was done with [adenylate-32P]NAD, and the 32P label incorporation efficiency was less than 0.001 mol of NAD/mol of protein.
FIG. 4.
Coomassie blue-stained SDS-PAGE gel (A) and its phosphorimage (B) showing proteins labeled upon UV irradiation in the presence of [carbonyl-14C]NAD. They include chicken egg albumin (lanes 1), cholera toxin A (lanes 2), DRAT (lanes 3), dinitrogenase reductase (DR) (lanes 4), DRAT and dinitrogenase reductase (lanes 5), R101F dinitrogenase reductase (lanes 6), DRAT and R101F dinitrogenase reductase (lanes 7), oxygen-denatured dinitrogenase reductase (lanes 8), and oxygen-denatured R101F dinitrogenase reductase (lanes 9). After reactions were performed as described in Materials and Methods, except with the above-noted proteins added, the SDS-PAGE gel was Coomassie blue stained and exposed to a Molecular Dynamics storage phosphor screen.
Most ADP-ribosyltransferases exhibit very strong affinity for NAD alone, while their substrates have not been reported to have affinity for NAD. The results of these UV experiments, in which it is shown that both DRAT and dinitrogenase reductase have affinity for NAD, indicate that the DRAT-dinitrogenase reductase ADP-ribosylation system is very unusual.
DISCUSSION
R. rubrum strains UR629, UR630, and UR631, which have the arginine 101 site of dinitrogenase reductase changed to tyrosine, phenylalanine, or leucine, respectively, were constructed. In vivo acetylene reduction assay before and after dark or ammonium treatment showed that no switch-off occurred in any of the three mutants. This further demonstrates both that arginine 101 of dinitrogenase reductase is the site of ADP-ribosylation by DRAT in R. rubrum and that there is no other posttranslational regulatory system in R. rubrum (such as those found in R. capsulatus and A. brasilense, in which ammonium treatment of Arg 101 substitution mutants caused switch-off while dark treatment did not) (40, 46). The UR630 mutant (R101F) retains 91%, the UR629 mutant (R101Y) retains 72%, and the UR631 mutant (R101L) retains 28% of nitrogenase activity of the corresponding wild-type strain, UR628 (Table 2). Immunoblotting (Fig. 1) and [32P]NAD labeling experiments (results not shown) reveal that no ADP-ribosylation occurs either in vivo or in vitro. EDC-dependent chemical cross-linking experiments show that although both R101Y and R101F dinitrogenase reductases cannot be modified by DRAT, they can form an EDC-dependent cross-linkable complex with DRAT in the presence of NAD and some NAD analogs. The complex formation between R101F dinitrogenase reductase and DRAT is as efficient as with wild-type proteins, and its sensitivity to salt inhibition is also similar to that of the wild type. The complex formation between R101Y dinitrogenase reductase and DRAT is much less efficient and is more sensitive to salt inhibition. The pattern of nucleotide dependence of the cross-linkable complex is the same as that of the wild-type protein. The fact that these chemical cross-linking features are similar between altered and wild-type proteins suggests that the complexes formed are also similar. The [carbonyl-14C]nicotinamide-releasing experiment revealed that R101F dinitrogenase reductase and DRAT together cannot cleave NAD, although they can form a strong cross-linkable protein complex.
UV photoaffinity labeling experiments demonstrated that both DRAT and dinitrogenase reductase can be labeled independently by [carbonyl-14C]NAD under UV treatment, although most 14C label is incorporated into DRAT when both proteins are present simultaneously. The oxygen-denatured dinitrogenase reductase cannot be labeled by [14C]NAD. The efficiency of labeling by [adenylate-32P]NAD is at least 10-fold less than by [carbonyl-14C]NAD. The R101F substitution of dinitrogenase reductase does not obviously affect the efficiency of UV-induced labeling of either dinitrogenase reductase or DRAT. These results support several hypotheses: (i) DRAT alone can bind NAD; (ii) dinitrogenase reductase alone can bind NAD, and this binding is dependent on the protein conformation of dinitrogenase reductase; (iii) most NAD is bound by DRAT when DRAT and dinitrogenase reductase are present together (the reactive species generated by UV irradiation are more reactive towards DRAT); (iv) only the carbonyl-14C-labeled portion, but not the adenylate-32P-labeled portion, of NAD is transferred to proteins under UV treatment; and (v) since UV-dependent labeling of R101F dinitrogenase reductase also occurs, the arginine 101 residue must not be essential for this process.
The following conclusions can be drawn about the R. rubrum mutants that produce altered dinitrogenase reductases. (i) Although dinitrogenase reductase with arginine at position 101 gave the best activity, other amino acids at the 101 position can also support electron transfer to a lesser degree. This is consistent with the results obtained with dinitrogenase reductases from other organisms. (ii) The arginine 101 residue is required for ADP-ribosylation of dinitrogenase reductase by DRAT in R. rubrum. (iii) The formation of a chemical cross-linkable protein complex is not dependent on arginine 101, although some arginine substitutions (e.g., R101Y) supported only weak cross-linking. (iv) Arginine 101 is important for NAD cleavage. (v) Arginine 101 is not critical for UV-dependent labeling by [14C]NAD either in dinitrogenase reductase alone or in the dinitrogenase reductase-DRAT system.
UV photolabeling was used to examine NAD binding in the DRAT-dinitrogenase reductase system. It suggests that the mechanism used in this system is different from the ones used in the diphtheria toxin and exotoxin A systems. With diphtheria toxin and exotoxin A, NAD binds the bacterial toxins to form a binary complex which then binds to their substrate—elongation factor 2 (1, 10, 23). For the DRAT-dinitrogenase reductase system, NAD can bind both DRAT and dinitrogenase reductase individually, and the nicotinamide portion of NAD is oriented toward DRAT in the DRAT-dinitrogenase reductase complex.
Although DRAT had not previously been observed to bind NAD, it had been assumed that this binding could occur because of the sequence similarity of DRAT to NAD binding bacterial toxin ADP-ribosyltransferases (39). UV-induced photolabeling experiments have been used to identify amino acids in contact with NAD in several bacterial toxins. Such amino acids are presumed to be within the active site (7). An XXE motif that is the site of NAD binding has been identified in ADP-ribosyltransferases. When the first X is a glutamyl residue (i.e., EXE), the transferase is always arginine specific. DRAT sequences from four species, R. rubrum, R. capsulatus, A. brasilense, and Azospirillum lipoferum, are known, and they all have this EXE motif (12, 19, 36, 47). Glu 259, which is the first glutamic acid in this region in R. rubrum DRAT, is the potential amino acid to which a part of NAD is attached by UV irradiation, although further experiments are needed to test this hypothesis.
UV photolabeling experiments also showed that dinitrogenase reductase alone can bind NAD. But at present there is no available information allowing us to predict the region of dinitrogenase reductase involved in NAD binding.
ACKNOWLEDGMENTS
This work was supported by NIGMS grant 54910 to P.W.L.
We thank Sandra Grunwald, Yaoping Zhang, Robert Kerby, and Gary Roberts for assistance and useful discussions. We also thank R. John Collier for advice on the UV-labeling experiments.
REFERENCES
- 1.Bell C E, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 2.Blake S M, Johnston K H, Russell-Jones G J, Gotschlich E C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 3.Burris R H. Nitrogen fixation-assay methods and techniques. Methods Enzymol. 1972;24:415–431. doi: 10.1016/0076-6879(72)24088-5. [DOI] [PubMed] [Google Scholar]
- 4.Burris R H. Nitrogenases. J Biol Chem. 1991;266:9339–9342. [PubMed] [Google Scholar]
- 5.Carroll S F, Collier R J. NAD binding site of diphtheria toxin: identification of a residue within the nicotinamide subsite by photochemical modification with NAD. Proc Natl Acad Sci USA. 1984;81:3307–3311. doi: 10.1073/pnas.81.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll S F, Collier R J. Active site of Pseudomonas aeruginosa exotoxin A. Glutamic acid 553 is photolabeled by NAD and shows functional homology with glutamic acid 148 of diphtheria toxin. J Biol Chem. 1987;262:8707–8711. [PubMed] [Google Scholar]
- 7.Carroll S F, Collier R J. Photoaffinity labeling of active site residues in ADP-ribosylating toxins. Methods Enzymol. 1994;235:631–639. doi: 10.1016/0076-6879(94)35176-7. [DOI] [PubMed] [Google Scholar]
- 8.Carroll S F, McCloskey J A, Crain P F, Oppenheimer N J, Marschner T M, Collier R J. Photoaffinity labeling of diphtheria toxin fragment A with NAD: structure of the photoproduct at position 148. Proc Natl Acad Sci USA. 1985;82:7237–7241. doi: 10.1073/pnas.82.21.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassel D, Pfeuffer T. Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc Natl Acad Sci USA. 1978;75:2669–2673. doi: 10.1073/pnas.75.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung D W, Collier R J. The mechanism of ADP-ribosylation of elongation factor 2 catalyzed by fragment A from diphtheria toxin. Biochim Biophys Acta. 1977;483:248–257. doi: 10.1016/0005-2744(77)90053-5. [DOI] [PubMed] [Google Scholar]
- 11.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 12.Fitzmaurice W P, Saari L L, Lowery R G, Ludden P W, Roberts G P. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol Gen Genet. 1989;218:340–347. doi: 10.1007/BF00331287. [DOI] [PubMed] [Google Scholar]
- 13.Fu H-A, Hartmann A, Lowery R G, Fitzmaurice W P, Roberts G P, Burris R H. Posttranslational regulatory system for nitrogenase activity in Azospirillum spp. J Bacteriol. 1989;171:4679–4685. doi: 10.1128/jb.171.9.4679-4685.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galloway T S, van Heyningen S. Binding of NAD+ by cholera toxin. Biochem J. 1987;244:225–230. doi: 10.1042/bj2440225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgiadis M M, Komiya H, Chakrabarti P, Woo D, Kornuc J J, Rees D C. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 16.Grunwald S K, Lies D P, Roberts G P, Ludden P W. Posttranslational regulation of nitrogenase in Rhodospirillum rubrum strains overexpressing the regulatory enzymes dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase activating glycohydrolase. J Bacteriol. 1995;177:628–635. doi: 10.1128/jb.177.3.628-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunwald S K, Ludden P W. NAD-dependent cross-linking of dinitrogenase reductase and dinitrogenase reductase ADP-ribosyltransferse from Rhodospirillum rubrum. J Bacteriol. 1997;179:3277–3283. doi: 10.1128/jb.179.10.3277-3283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grunwald S K, Ryle M J, Lanzilotta W N, Ludden P W. ADP-ribosylation of variants of Azotobacter vinelandii dinitrogenase reductase by Rhodospirillum rubrum dinitrogenase reductase ADP-ribosyltransferase. J Bacteriol. 2000;182:2597–2603. doi: 10.1128/jb.182.9.2597-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue A, Shigematsu T, Hidaka M, Masaki H, Uozumi T. Cloning, sequencing and transcriptional regulation of the draT and draG genes of Azospirillum lipoferum FS. Gene. 1996;170:101–106. doi: 10.1016/0378-1119(95)00852-7. [DOI] [PubMed] [Google Scholar]
- 20.Kamen D M, Gest H. Evidence for a nitrogenase system in the photosynthetic bacterium Rhodospirillum rubrum. Science. 1949;109:560. doi: 10.1126/science.109.2840.560. [DOI] [PubMed] [Google Scholar]
- 21.Kandel J, Collier R J, Chung D W. Interaction of fragment A from diphtheria toxin with nicotinamide adenine dinucleotide. J Biol Chem. 1974;249:2088–2097. [PubMed] [Google Scholar]
- 22.Kanemoto R H, Ludden P W. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J Bacteriol. 1984;158:713–720. doi: 10.1128/jb.158.2.713-720.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler S P, Galloway D R. Pseudomonas aeruginosa exotoxin A interaction with eucaryotic elongation factor 2. Role of the His426 residue. J Biol Chem. 1992;267:19107–19111. [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lehman L J, Fitzmaurice W P, Roberts G P. The cloning and functional characterization of the nifH gene of Rhodospirillum rubrum. Gene. 1990;95:143–147. doi: 10.1016/0378-1119(90)90426-r. [DOI] [PubMed] [Google Scholar]
- 26.Lehman L J, Roberts G P. Glycine 100 in the dinitrogenase reductase of Rhodospirillum rubrum is required for nitrogen fixation but not for ADP-ribosylation. J Bacteriol. 1991;173:6159–6161. doi: 10.1128/jb.173.19.6159-6161.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehman L J, Roberts G P. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J Bacteriol. 1991;173:5705–5711. doi: 10.1128/jb.173.18.5705-5711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang J H, Nielsen G M, Lies D P, Burris R H, Roberts G P, Ludden P W. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobban D M, Irons L I, van Heyningen S. Binding of NAD+ to pertussis toxin. Biochim Biophys Acta. 1991;1078:155–160. doi: 10.1016/0167-4838(91)99004-c. [DOI] [PubMed] [Google Scholar]
- 30.Lowery R G, Chang C L, Davis L C, McKenna M C, Stephens P J, Ludden P W. Substitution of histidine for arginine-101 of dinitrogenase reductase disrupts electron transfer to dinitrogenase. Biochemistry. 1989;28:1206–1212. doi: 10.1021/bi00429a038. [DOI] [PubMed] [Google Scholar]
- 31.Lowery R G, Ludden P W. Purification and properties of dinitrogenase reductase ADP-ribosyltransferase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1988;263:16714–16719. [PubMed] [Google Scholar]
- 32.Lowery R G, Saari L L, Ludden P W. Reversible regulation of the nitrogenase iron protein from Rhodospirillum rubrum by ADP-ribosylation in vitro. J Bacteriol. 1986;166:513–518. doi: 10.1128/jb.166.2.513-518.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludden P W. Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes. Mol Cell Biochem. 1994;138:123–129. doi: 10.1007/BF00928453. [DOI] [PubMed] [Google Scholar]
- 34.Ludden P W, Burris R H. Purification and properties of nitrogenase from Rhodospirillum rubrum, and evidence for phosphate, ribose and an adenine-like unit covalently bound to the iron protein. Biochem J. 1978;175:251–259. doi: 10.1042/bj1750251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludden P W, Roberts G P. The biochemistry and genetics of nitrogen fixation by photosynthetic bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 929–947. [Google Scholar]
- 36.Masepohl B, Krey R, Klipp W. The draTG gene region of Rhodobacter capsulatus is required for post-translational regulation of both the molybdenum and the alternative nitrogenase. J Gen Microbiol. 1993;139:2667–2675. doi: 10.1099/00221287-139-11-2667. [DOI] [PubMed] [Google Scholar]
- 37.Munoz-Centeno M C, Ruiz M T, Paneque A, Cejudo F J. Posttranslational regulation of nitrogenase activity by fixed nitrogen in Azotobacter chroococcum. Biochim Biophys Acta. 1996;1291:67–74. doi: 10.1016/0304-4165(96)00045-1. [DOI] [PubMed] [Google Scholar]
- 38.Murrell S A, Lowery R G, Ludden P W. ADP-ribosylation of dinitrogenase reductase from Clostridium pasteurianum prevents its inhibition of nitrogenase from Azotobacter vinelandii. Biochem J. 1988;251:609–612. doi: 10.1042/bj2510609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okazaki I J, Moss J. Common structure of the catalytic sites of mammalian and bacterial toxin ADP-ribosyltransferases. Mol Cell Biochem. 1994;138:177–181. doi: 10.1007/BF00928460. [DOI] [PubMed] [Google Scholar]
- 40.Pierrard J, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase in Rhodobacter capsulatus: existence of two independent regulatory effects of ammonium. J Bacteriol. 1993;175:1358–1366. doi: 10.1128/jb.175.5.1358-1366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierrard J, Willison J C, Vignais P M, Gaspar J L, Ludden P W, Roberts G P. Site-directed mutagenesis of the target arginine for ADP-ribosylation of nitrogenase component II in Rhodobacter capsulatus. Biochem Biophys Res Commun. 1993;192:1223–1229. doi: 10.1006/bbrc.1993.1547. [DOI] [PubMed] [Google Scholar]
- 42.Pope R M, Murrell S A, Ludden P W. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenosine diphosphoribosylation of a specific arginine residue. Proc Natl Acad Sci USA. 1985;82:3173–3177. doi: 10.1073/pnas.82.10.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saari L L, Triplett E W, Ludden P W. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1984;259:15502–15508. [PubMed] [Google Scholar]
- 44.Wolle D, Kim C, Dean D, Howard J B. Ionic interactions in the nitrogenase complex. Properties of Fe-protein containing substitutions for Arg-100. J Biol Chem. 1992;267:3667–3673. [PubMed] [Google Scholar]
- 45.Zhang Y, Burris R H, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J Bacteriol. 1993;175:6781–6788. doi: 10.1128/jb.175.21.6781-6788.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Burris R H, Ludden P W, Roberts G P. Presence of a second mechanism for the posttranslational regulation of nitrogenase activity in Azospirillum brasilense in response to ammonium. J Bacteriol. 1996;178:2948–2953. doi: 10.1128/jb.178.10.2948-2953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Burris R H, Roberts G P. Cloning, sequencing, mutagenesis, and functional characterization of draT and draG genes from Azospirillum brasilense. J Bacteriol. 1992;174:3364–3369. doi: 10.1128/jb.174.10.3364-3369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]