Abstract
The advent of PCR has improved the identification of viruses in patients with community-acquired pneumonia (CAP). Several studies have used PCR to establish the importance of viruses in the aetiology of CAP.
We performed a systematic review and meta-analysis of the studies that reported the proportion of viral infection detected via PCR in patients with CAP. We excluded studies with paediatric populations. The primary outcome was the proportion of patients with viral infection. The secondary outcome was short-term mortality.
Our review included 31 studies. Most obtained PCR via nasopharyngeal or oropharyngeal swab. The pooled proportion of patients with viral infection was 24.5% (95% CI 21.5–27.5%). In studies that obtained lower respiratory samples in >50% of patients, the proportion was 44.2% (95% CI 35.1–53.3%). The odds of death were higher in patients with dual bacterial and viral infection (OR 2.1, 95% CI 1.32–3.31).
Viral infection is present in a high proportion of patients with CAP. The true proportion of viral infection is probably underestimated because of negative test results from nasopharyngeal or oropharyngeal swab PCR. There is increased mortality in patients with dual bacterial and viral infection.
Short abstract
Viral infection is present in a high proportion of patients with community-acquired pneumonia http://ow.ly/X03pK
Introduction
Establishing the aetiological diagnosis of community-acquired pneumonia (CAP) is important for different reasons. From a public health perspective, it allows the monitoring of trends in CAP aetiology and the development of guidelines for treatment. From a patient perspective, it allows the choice of correct and narrowed antimicrobial therapy. New diagnostic techniques have shed a light on the role of viruses as a cause of CAP. The advent of PCR has greatly improved the identification of viruses in patients with CAP [1]. Over the last decade, several studies have used PCR to establish the importance of viruses in the aetiology of CAP [1–12]. An updated and objective literature review that quantitatively summarises and critically appraises the results of these studies is warranted.
The aim of this study was to synthesise the results of studies that evaluated viral infection in patients with CAP. More specifically, we aimed to establish the most common viral pathogens, the proportion of viral infection and risk of death with viral infection in patients with CAP. For that, we carried out a systematic review of the literature and meta-analysis.
Methods
Eligibility criteria
Studies that included patients with diagnosis of CAP, performed PCR for the investigation of viral pathogens in a systematic way, and reported the proportion of viral infection in this patient population were eligible to be included in this review.
Exclusion criteria
We excluded studies that included a paediatric population, systematically selected an immunosuppressed population, were not restricted to pneumonia (e.g. included lower respiratory tract infection) or deliberately selected a patient population whose microbiological results would not be representative of all patients with CAP (e.g. patients with bilateral pneumonia). We also excluded studies that investigated only one virus, as the focus of these studies was, in general, the assessment of outbreaks or pandemic.
Measurements
The primary outcome was the proportion of patients with viral infection. The secondary outcome was the short-term (in-hospital or <30-day) mortality. We also provide meta-analysis of the proportion of the most common viruses. We evaluated the odds of death in those with viral infection compared with the odds in those without it.
For the primary outcome, we planned the following subgroup analyses a priori: inpatient versus outpatient, intensive care unit (ICU; most patients) versus non-ICU, and lower respiratory tract sample (most patients) versus no lower respiratory tract sample. Sensitivity analyses were carried out based on the reporting quality of studies.
Search strategy
Two investigators (M. Burk and R. Cavallazzi) independently searched PubMed and Embase up to August 2015. The search strategy in PubMed included the following: “pneumonia” (Medical Subject Headings (MeSH)) AND (virus OR viral) AND (polymerase chain reaction* OR pcr). In Embase, we used the following terms: “community-acquired pneumonia” AND pcr.
Risk of bias
Risk of bias was assessed by the use of items listed in the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement [13]. We evaluated whether each individual study reported the following items: objectives, study design, setting, eligibility criteria, participants (description of numbers eligible and enrolled) and descriptive data (description of number of patients with PCR performed).
Data extraction
Using a structured data collection form, two authors (M. Burk and R. Cavallazzi) independently collected data from the primary studies. Discrepancies were resolved by discussion amongst all authors. Study characteristics extracted included country where the study was conducted, date of enrolment, study design, study setting, patient population features, tests employed to detect virus, quality items and aim of the study. Numerical data extracted included number of patients in each study, number of patients with viral infection, mortality in the study population, mortality in patients with viral infection, mortality in patients without viral infection and number of infections by each specific virus.
Statistical analysis
For meta-analysis of proportion, we computed a summary proportion with corresponding 95% confidence interval (CI) for each individual study. Subsequently, we performed meta-analysis by means of a random-effects (DerSimonian and Laird) method, which assumes some degree of heterogeneity among the studies, and present a pooled estimate with 95% CI [14]. For meta-analysis of mortality, we computed a summary odds ratio (OR) and corresponding 95% CI for each individual study. The odds of death was placed on the numerator for patients with viral infection, and on the denominator for those without viral infection. Again, we performed meta-analysis by means of a random-effects (DerSimonian and Laird) method and present a pooled OR with 95% CI. We assessed heterogeneity with a Chi-squared test and the I2. Evidence of heterogeneity included I2 >30%. For all analyses, a p-value <0.05 indicated statistical significance. The meta-analyses were carried out with the statistical software package Stata 10 (StataCorp LP, College Station, TX, USA). We used the commands “metaprop” for meta-analysis of proportion, and “metan” for meta-analysis of mortality [15, 16].
Results
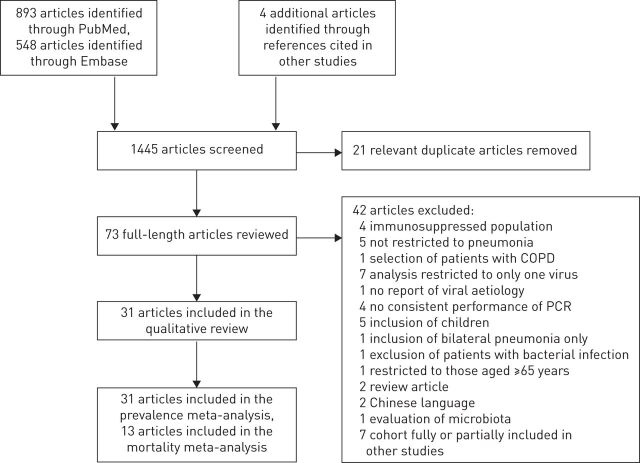
Our searches retrieved a total of 1441 citations (PubMed n=893, Embase n=548) up to August 9, 2015. After reading the title and abstract (when available) of these citations, we selected 94 articles for full text review. After removing 21 duplicate articles, a total of 73 articles were fully reviewed. Of these, 42 were excluded for a number of reasons (see online supplementary material). Figure 1 shows the flow diagram of our literature search.
FIGURE 1.
Flow diagram of our literature search for studies. COPD: chronic obstructive pulmonary disease.
We were left with 31 studies [1–3, 5–12, 17–36], which included 10 762 patients (table 1). Of these, 29 studies were prospective. They were observational cohort studies except for one case–control study [34]. They were published between 2005 and 2015 and carried out in Europe (n=14) [1, 3, 5, 18–24, 26, 28, 33, 35], Asia (n=9) [2, 6, 10, 29–32, 34, 36], North America (n=4) [11, 12, 25, 27], Oceania (n=3) [7, 8, 17] and South America (n=1) [9]. These studies enrolled a predominantly adult population although two of them used a cut-off of age >14 years for inclusion [3, 31]. Our original protocol anticipated the exclusion of studies with paediatric populations. We felt that our systematic review would still be representative of an adult population with the inclusion of the latter two studies, given the mean/median age of their sample. The patient population consisted of inpatients in 25 studies [1–3, 5–12, 17, 18, 20, 21, 23–28, 30, 33–35], outpatients in one study [36], and both inpatients and outpatients in five studies [19, 22, 29, 31, 32].
TABLE 1.
Characteristics of the studies
| First author [ref.] | Study setting | Study design | Patient population | Patients n | Viral tests | Age years | Male % | Aims | Mortality % |
| Angeles Marcos [3] | Spain, 2003–2004, inpatient | Prospective cohort | Patients aged >14 years admitted to the hospital with CAP | 198 | PCR, IFA, culture, serology | 70 (median) | 58 | Incidence and characteristics of viral CAP, and yield of tests for viral infection | 2.5 |
| van Gageldonk-Lafeber [5] | The Netherlands, 2007–2010, inpatient | Prospective cohort | Patients aged >18 years with CAP in the ED | 339 | PCR | 66 (median) | 62.5 | Aetiology of CAP | Not reported |
| Hara [6] | Thailand, 2006–2008, inpatient | Prospective cohort | Patients aged >16 years admitted to the hospital with CAP | 119 | PCR | 55 (mean) | 71.4 | Aetiology of CAP | 18.5 |
| Holm [22] | Denmark, 2002–2003, inpatient and outpatient | Prospective cohort | Patients aged >18 years with suspected CAP seen in outpatient clinic | 48 | PCR | 50 (median) | 49 | Aetiology of CAP and outcomes | 0 |
| Mermond [7] | New Caledonia, 2006–2007, inpatient | Prospective cohort | Patients aged >16 years admitted to the hospital with CAP | 137 | PCR, IFA | 58 (median) | 58.4 | Aetiology of CAP, performance of tests, and assessment of vaccination-preventable proportion of causative organisms | 5.8 |
| Jennings [8] | New Zealand, 1999–2000, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 304 | PCR, IFA, culture, serology | 70 (median) | 52 | Aetiology of CAP and characteristics of viral pneumonia | 6.8 (only reported for viral subset) |
| Luchsinger [9] | Chile, 2005–2007, inpatient and outpatient | Prospective cohort | Patients aged >18 years presenting with CAP | 356 | PCR, IFA, culture, serology | 61 (median) | 53.6 | Aetiology of CAP and relationship between severity and pathogen | 7.8 |
| Takahashi [10] | Vietnam, 2009–2010, inpatient | Prospective cohort | Patients aged >15 years admitted to the hospital with CAP | 174 | PCR | 42 (median) | 52 | Incidence, aetiology and clinical features of CAP | 9.8 |
| Wiemken [11] | USA, 2008–2011, inpatient ICU | Prospective cohort | Patients aged >18 years with CAP admitted to the ICU | 393 | PCR | 52 (mean)# | 56# | Incidence of respiratory viruses in CAP patients in ICU | Not reported |
| Choi [2] | Republic of Korea, 2010–2011, inpatient | Retrospective analysis of a prospective cohort | Patients aged >18 years admitted to the hospital with severe CAP | 64 | PCR, culture | 49 (mean) | 76.6 | Role of viral infection in patients with severe CAP | 32.8 |
| Templeton [1] | The Netherlands, 2000–2002, inpatient and outpatient | Prospective cohort | Patients aged >18 years with CAP | 105 | PCR, culture, IFA, complement fixation |
>60 (50% of patients) | 71 | Aetiology of CAP | 3.8 |
| Musher [12] | USA, 2011–2012, inpatient | Prospective cohort | Patients admitted to a Veteran Affairs hospital | 259 | PCR | 70 (mean) | 95.7 | Yield of traditional and new diagnostic tests | Not reported |
| Shibli [34] | Israel, 2006–2007, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 126 | PCR, DNA/RNA genomic extraction, serology | 58.3 (mean) | 57.9 | Identify common pathogens causing CAP in hospitalised patients and evaluate correlation of aetiology and disease severity | 3.96 |
| Sangil [33] | Spain, 2009–2010, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 131 | Multiplex PCR, serology | 64.4 (mean) | 68.7 | Determine aetiology of CAP by adding PCR to conventional methods and describe clinical and laboratory features between patients with bacterial and viral CAP | 5.3 |
| Johansson [26] | Sweden, 2004–2005, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 184 | PCR, serology, viral culture | 61.3 (mean) | 51 | Determine aetiology of CAP among adults using new PCR platform and conventional methods | 3.8 |
| Huijskens [24] | The Netherlands, 2008–2009, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 408 | PCR, serology | 68 (median) | 61.3 | Evaluate potential for targeted diagnostics and directed treatment | 39 |
| Saito [32] | Japan, 1999–2000, outpatient and inpatient | Prospective cohort | Patients aged >17 years admitted to the hospital with CAP | 232 | PCR, antigen immunoassay, serology | 60 (mean) | 57.8 | Identify aetiology of CAP | 1.7 |
| Yin [36] | China, 2008–2009, infectious disease outpatient clinic | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 215 | PCR | 36.1 (mean of those reported) | 47.1 (of those with aetiologies) | Evaluate Japanese Respiratory Society guidelines for identification of Mycoplasma pneumoniae | 0 |
| Diederen [20] | The Netherlands, 1998–2000, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 242 | PCR, immunoassay/ELISA, serology | Not reported | Not reported | Detection of respiratory viruses and Legionella by PCR in CAP patients | 9.5 |
| Hohenthal [21] | Finland, 1999–2004, inpatient | Prospective cohort | Patients aged >16 years admitted to the hospital with CAP | 384 | PCR, immunoassay/IFA, serology | 49.8 (mean) | 52.3 | Assess utility of rapid micro tests in different disease severities of CAP | Not reported |
| Lieberman [30] | Israel, Nov 2004–March 2005 and Nov 2005–April 2006, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 183 | PCR | 59.5 (mean) | 57 | Identify viruses in CAP versus control versus lower respiratory infections | Not reported |
| Kim [29] | Republic of Korea, 2010–2013, inpatient and outpatient | Retrospective cohort | Patients aged >18 years admitted to the hospital with CAP | 327 | PCR, immunoassay for influenza antigen | 67 (mean) in viral pneumonia; 70 (mean) in nonviral pneumonia |
72% in viral CAP; 78% in nonviral CAP |
Investigate predictors of viral CAP | 18.9 |
| Karhu [28] | Finland, 2008–2013, inpatient ICU | Prospective cohort | Patients aged >18 years admitted to the hospital with severe CAP | 49 | PCR | 54 (median) | 42.9 | Evaluate presence and course of viral infections in ventilated patients using PCR | 12.2 |
| Holter [23] | Norway, 2008–2011, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 264 | PCR, serology | 66 (mean) | 52 | Describe aetiology and evaluate diagnostic results | 3.7 |
| Qu [31] | China, 2010–2012, inpatient and outpatient | Prospective cohort | Patients aged >14 years admitted to the hospital with CAP | 954 | PCR | 45.2 (mean) | 56.6 | Investigate respiratory virus aetiology in adolescents and adults with CAP, by age and PSI | 0.4 |
| Charles [17] | Australia, 2004–2006, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 865 | PCR, serology for influenza | 65.1 (mean) | 60.7 | Evaluate aetiology, severity assessment tools and outcomes of CAP |
5.6 |
| Viasus [35] | Spain, 2010–2011, inpatient | Prospective cohort | Adults admitted to the hospital with CAP | 747 | PCR | 65 (median) | 56.8 | Evaluate aetiology, clinical features and prognosis of CAP | 8.2 |
| Das [19] | France, 2011–1012, ED | Prospective cohort | Patients aged >18 years presenting to the ED with CAP | 125 | PCR | 65 (mean)# | 49# | Evaluate aetiology of CAP | Not reported |
| Jain [25] | USA, 2010–2012, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 2259 | PCR, serology | 57 (median) | Not reported | Population-based surveillance study of CAP aetiology | 2 |
| Johnstone [27] | Canada, 2004–2006, inpatient | Prospective cohort | Patients aged >18 years admitted to the hospital with CAP | 193 | NAT, IFA | 71 (median) | 51 | Describe the pathogens, clinical presentation and outcomes of patients with CAP |
3 |
| Cillóniz [18] | Spain, 2003–2010, inpatient ICU | Prospective cohort | Patients aged >18 years admitted to the ICU with CAP | 362 | PCR, IFA | 63 (mean) | 64 | Study the prevalence, clinical characteristics and outcomes of severe CAP in the ICU |
10 |
CAP: community-acquired pneumonia; IFA: immunofluorescence assay; ED: emergency department; ICU: intensive care unit; PSI: pneumonia severity index; NAT: nucleic acid amplification tests. #: for all patients, including some not included in our analysis.
Six studies reported that bronchoalveolar lavage was one of the diagnostic tools used to obtain a microbiological diagnosis [2, 7, 18, 25, 28, 35]. However, only four of them reported the number of patients who had lower respiratory tract sampling [2, 18, 25, 28]. In two of them, >50% of the patients had lower respiratory tract sampling [2, 28]. Table 1 gives details of these studies.
The risk of bias assessment revealed reporting of the following items by the studies: objectives (100%), design (94%), setting (100%), eligibility (87%), description of numbers eligible and enrolled (45%), and description of number of patients who had PCR performed (81%).
Primary outcome
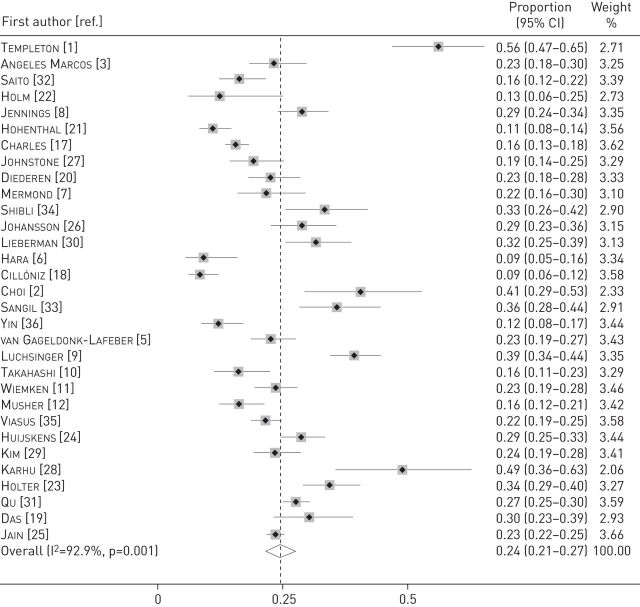
The pooled proportion of patients with viral infection was 24.5% (95% CI 21.5–27.5%; I2=92.9%) (figure 2). Among the individual studies, it ranged from 8.6% to 56.2%. The pooled proportion of viral infection was 25.5% (95% CI 22–29%; I2=93.6%) in studies with an inpatient population [1–3, 5–12, 17, 18, 20, 21, 23–28, 30, 33–35], 12.1% (95% CI 7.7–16.5%; I2=0.0%) in a study with an outpatient population [36] and 22.4% (95% CI 16.8–28.1%; I2=83.2%) in studies with mixed inpatients and outpatients [19, 22, 29, 31, 32]. In the two studies that obtained lower respiratory samples in >50% of the patients [2, 28], it was 44.2% (95% CI 35.1–53.3%; I2=0%), compared with 23.5% (95% CI 20.5–26.6%; I2=93%) in the other studies. The proportion was 29% (95% CI 14.5–43.4%; I2=96%) in studies that included only patients requiring ICU admission [2, 11, 18, 28], compared with 24.1% (95% CI 21.1–27.1%; I2=91.9%) in other studies.
FIGURE 2.
Forest plot displaying meta-analysis of the proportion of viral infection in patients with community-acquired pneumonia. Weights are from random-effects analysis.
Data from 25 studies revealed a pooled proportion of dual bacterial and viral infection of 10% (95% CI 8–11%; I2=93.1%) [2, 3, 5–9, 12, 17, 19–21, 23–29, 31–36].
Individual viruses
The number of viruses tested across all studies ranged from four to 11 (median=7). The most commonly identified viruses were influenza in 18 studies [3, 6, 7, 10, 11, 17–19, 21, 23, 26, 27, 29, 31, 32, 34–36], rhinovirus in eight studies [1, 5, 8, 12, 22, 24, 25, 28], respiratory syncytial virus in three studies [2, 9, 33] and coronavirus in two studies [20, 30]. Meta-analyses revealed that the proportion of infection by influenza (31 studies) was 8% (95% CI 6.3–9.6%; I2=91.9%), by rhinovirus (25 studies) was 5.7% (95% CI 4.3–7.1%; I2=90.3%), by respiratory syncytial virus (29 studies) was 2.2% (95% CI 1.6–2.8%; I2=80%) and by coronavirus (18 studies) was 3.3% (95% CI 2.3–4.2%; I2=80.2%).
Short-term mortality
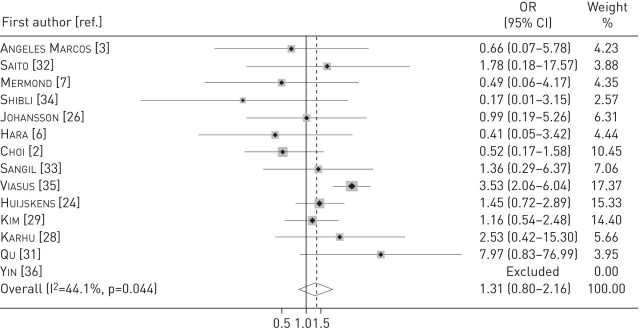
For the outcome short-term mortality, we pooled data from 13 studies [2, 3, 6, 7, 24, 26, 28, 29, 31–35]. The odds of death were higher in patients with viral infection, but there was no statistical significance (OR 1.3, 95% CI 0.8–2.16; p=0.283; I2=44.1%) (figure 3). In patients with dual bacterial and viral infection (10 studies), the odds of death were significantly higher compared with patients without dual infection (OR 2.1, 95% CI 1.32–3.31; p=0.002; I2=0%) [2, 3, 24, 26, 28, 31–35].
FIGURE 3.
Forest plot displaying the odds ratio (OR) of death in the presence of viral infection in patients with community-acquired pneumonia. Weights are from random-effects analysis.
Sensitivity analyses
After exclusion of the study with largest sample [25], the pooled proportion of viral infection remained similar (24.6%, 95% CI 21.2–27.9%; I2=93%). Studies that met all the reporting criteria set in our risk of bias assessment were considered of low risk of bias. Analysis of these studies showed a proportion of viral infection of 22.6% (95% CI 18.7–26.5%; I2=92.6%) [2, 3, 7, 10, 12, 17, 18, 22, 23, 25, 28, 29, 31, 35].
Discussion
Our analysis showed a high proportion of viral infection in patients with CAP. The proportion was higher in studies of inpatients compared with studies of outpatients. It was not significantly increased in studies performed in the ICU setting. However, the proportion was significantly increased in studies that obtained lower respiratory tract samples in most patients. There was no significant increase in mortality for patients with viral infection. However, patients with dual bacterial and viral infection had twice the risk of death compared with patients without dual infection.
While it is well established that early diagnosis of influenza in CAP has substantial therapeutic implications, this is less so with non-influenza viruses. Historically, antiviral therapy for non-influenza pneumonia had been evaluated mostly in immunosuppressed patients and infants. More recently, there has been interest in the use of antivirals or immunomodulating agents for other populations [37]. An example is a recent case series demonstrating good outcomes with early administration of cidofovir for non-immunocompromised patients with severe CAP [38]. Additionally, there are a number of new antivirals being developed [39–41]. The prospect of new therapeutic options for viral infection in CAP highlights the increasing role that diagnostic tests for viral infection are likely to have in the near future.
An important and controversial consideration is whether the viruses detected in the nasopharynx represent only colonisation in some patients. In support of this notion, approximately one-third of subjects who are experimentally infected with wild-type influenza virus remain asymptomatic [42]. Furthermore, even when viruses are detected by lower respiratory tract sampling, contamination from the upper airways may have occurred. Conversely, a study using PCR found that asymptomatic nasopharyngeal carriage in adults was common for Staphylococcus aureus (9.1%) and Streptococcus pneumoniae (6.8%), but absent for the four viruses tested: influenza virus A, influenza virus B, respiratory syncytial virus A and respiratory syncytial virus B [43].
Perhaps a more misleading factor is that nasopharyngeal swab PCR may provide negative test results in cases of lower respiratory tract viral infection. This is highlighted by two studies that obtained both nasopharyngeal swab and lower respiratory tract samples in some patients [2, 28]. In one of these studies, of the 21 viruses detected by lower respiratory tract sample, only seven were detected by nasopharyngeal swab [28]. In the other study, of the 23 patients that had both a nasopharyngeal swab and bronchoalveolar lavage PCR test, five had positive virus in the bronchoalveolar lavage but negative in the nasopharyngeal swab [2]. In fact, a subgroup analysis including these two studies revealed a pooled proportion of viral infection that was almost twice as high as that of other studies.
Analysis of the individual viruses showed that influenza and rhinovirus were the most commonly detected viruses. Respiratory syncytial virus, coronavirus, parainfluenza, adenovirus and human metapneumovirus have a lesser yet prominent role in CAP, being detected in 1–4% of this patient population. Viruses that have been detected in <1% of the CAP population include enterovirus, polyomavirus, varicella-zoster virus and herpes simplex virus. As expected, studies did not consistently examine the same set of viruses.
We did not find a significant association between viral infection and increased mortality. However, because of the relatively small sample size (most studies did not contribute to the mortality meta-analysis) and wide confidence interval, we cannot conclusively exclude that the presence of viral infection confers a higher risk of death. Indirect evidence that viral infection confers higher morbidity would be that inpatients had a higher proportion of viral infection compared with outpatients. However, studies with ICU populations did not demonstrate a significantly higher proportion of viral infection compared with other studies.
In the presence of bacterial and viral co-infection, the data are less equivocal and point to an interaction leading to worse outcomes. For instance, some (but not all) studies included in our meta-analysis showed higher severity of pneumonia with bacterial and viral co-infection [1, 20]. The interaction seems particularly relevant with rhinovirus [1, 8] and influenza [44]. In a comprehensive analysis of both literature and tissue specimens from the 1918 influenza pandemic, Morens et al. [44] showed the facilitating effect that influenza has upon upper respiratory tract bacteria, which were ultimately the probable cause of most deaths related to that pandemic. While our analysis does not allow us to evaluate the prognostic impact of individual viruses, it clearly shows an increased mortality in the presence of dual viral and bacterial infection.
Another common occurrence, which was not fully scrutinised by our analysis, is the presence of secondary bacterial infection after a viral infection. This is distinct from concomitant viral/bacterial infection and is usually caused by S. pneumoniae infection preceded by influenza [45]. There is typically a lag time of 2–4 weeks between the two infections, and a study indicates that 12–20% of seasonal cases of invasive pneumococcal disease can be attributed to prior influenza infection [46]. Data from the 2009 influenza A(H1N1) pandemic indicate that other bacterial pathogens associated with influenza A infection include S. aureus, Gram-negative rods and group A streptococcus [47].
Two meta-analyses evaluating the proportion of viral infection in CAP have been published [48, 49]. The study by Wang et al. [48] addressed childhood CAP and found a pooled proportion of viral infection of 57.4% in that patient population. The study by Wu et al. [49] included adults with CAP and found a pooled proportion of viral infection of 22.4%. However, the latter analysis included fewer studies and did not evaluate mortality [49].
Our study has a number of limitations. We did not restrict our search to any language; however, we were unable to include some articles in languages other than English. We deliberately excluded abstracts because often they represent preliminary findings and do not provide enough information for data extraction. This focus on quality comes at the expense of excluding some studies from our review. In some instances, we were unable to obtain data from authors of the original studies despite attempting contact via e-mail. There was significant heterogeneity among the studies, which could not be fully explained by the subgroup analyses.
Future research should focus on practical applications of viral PCR tests. A potential area for research is the evaluation of whether the use of PCR tests in a clinical pathway would allow a faster de-escalation of antimicrobials. Further research on the importance of viral colonisation of the nasopharynx and the impact of false-negative nasopharyngeal tests is warranted. The relevance of viruses that have not been tested in most studies should be clarified. Finally, the pathogenic role of the viral infections is not clear from the included studies. In what proportion do these viral infections represent only upper respiratory tract infection? In the presence of mixed viral and bacterial infection, what is the mechanistic role underlying the worse outcomes? Does viral infection act as a facilitator to the entry and invasiveness of bacteria, or does it have a direct pathogenic effect? Could some of the cases of dual bacterial and viral infection actually represent secondary bacterial infection? It is known that PCR may remain positive for 2–5 weeks after an acute infection by some of the respiratory viruses [50]. Thus, in the group classified as having dual bacterial and viral infection, active viral replication may not always occur at the time of the diagnosis. These are points that need to be further explored in future investigations.
We conclude that viral infection is present in a high proportion of patients with CAP. While upper airway colonisation by viruses could lead to overestimation of their role in CAP, we believe that it is far more likely that the true proportion of viral infection in CAP is underestimated because of negative test results from nasopharyngeal swab PCR in some cases of lower respiratory infection, failure to evaluate potentially relevant viruses, and inability to test all patients for PCR in some studies. Furthermore, the analysis of the evidence shows a significant increase in mortality in CAP patients with dual bacterial and viral infection.
Supplementary Material
Footnotes
This article has supplementary material available from err.ersjournals.com
Conflict of interest: Disclosures can be found alongside the online version of this article at err.ersjournals.com
Provenance: Submitted article, peer reviewed.
References
- 1.Templeton KE, Scheltinga SA, van den Eeden WC, et al. . Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis 2005; 41: 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi SH, Hong SB, Ko GB, et al. . Viral infection in patients with severe pneumonia requiring intensive care unit admission. Am J Respir Crit Care Med 2012; 186: 325–332. [DOI] [PubMed] [Google Scholar]
- 3.Angeles Marcos M, Camps M, Pumarola T, et al. . The role of viruses in the aetiology of community-acquired pneumonia in adults. Antivir Ther 2006; 11: 351–359. [PubMed] [Google Scholar]
- 4.Cao B, Ren LL, Zhao F, et al. . Viral and Mycoplasma pneumoniae community-acquired pneumonia and novel clinical outcome evaluation in ambulatory adult patients in China. Eur J Clin Microbiol Infect Dis 2010; 29: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Gageldonk-Lafeber AB, Wever PC, van der Lubben IM, et al. . The aetiology of community-acquired pneumonia and implications for patient management. Neth J Med 2013; 71: 418–425. [PubMed] [Google Scholar]
- 6.Hara K, Yahara K, Gotoh K, et al. . Clinical study concerning the relationship between community-acquired pneumonia and viral infection in northern Thailand. Intern Med 2011; 50: 991–998. [DOI] [PubMed] [Google Scholar]
- 7.Mermond S, Berlioz-Arthaud A, Estivals M, et al. . Aetiology of community-acquired pneumonia in hospitalized adult patients in New Caledonia. Trop Med Int Health 2010; 15: 1517–1524. [DOI] [PubMed] [Google Scholar]
- 8.Jennings LC, Anderson TP, Beynon KA, et al. . Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax 2008; 63: 42–48. [DOI] [PubMed] [Google Scholar]
- 9.Luchsinger V, Ruiz M, Zunino E, et al. . Community-acquired pneumonia in Chile: the clinical relevance in the detection of viruses and atypical bacteria. Thorax 2013; 68: 1000–1006. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Suzuki M, Minh le N, et al. . The incidence and aetiology of hospitalised community-acquired pneumonia among Vietnamese adults: a prospective surveillance in Central Vietnam. BMC Infect Dis 2013; 13: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiemken T, Peyrani P, Bryant K, et al. . Incidence of respiratory viruses in patients with community-acquired pneumonia admitted to the intensive care unit: results from the Severe Influenza Pneumonia Surveillance (SIPS) project. Eur J Clin Microbiol Infect Dis 2013; 32: 705–710. [DOI] [PubMed] [Google Scholar]
- 12.Musher DM, Roig IL, Cazares G, et al. . Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: results of a one-year study. J Infect 2013; 67: 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Elm E, Altman DG, Egger M, et al. . The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med 2007; 147: 573–577. [DOI] [PubMed] [Google Scholar]
- 14.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 15.Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Health 2014; 72: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris RJ, Bradburn MJ, Deeks JJ, et al. . Metan: fixed- and random-effects meta-analysis. Stata J 2008; 8: 3–28. [Google Scholar]
- 17.Charles PG, Whitby M, Fuller AJ, et al. . The etiology of community-acquired pneumonia in Australia: why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin Infect Dis 2008; 46: 1513–1521. [DOI] [PubMed] [Google Scholar]
- 18.Cillóniz C, Ewig S, Ferrer M, et al. . Community-acquired polymicrobial pneumonia in the intensive care unit: aetiology and prognosis. Crit Care 2011; 15: R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das D, Le Floch H, Houhou N, et al. . Viruses detected by systematic multiplex polymerase chain reaction in adults with suspected community-acquired pneumonia attending emergency departments in France. Clin Microbiol Infect 2015; 21: 608.e1–608.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diederen BM, Van Der Eerden MM, Vlaspolder F, et al. . Detection of respiratory viruses and Legionella spp. by real-time polymerase chain reaction in patients with community acquired pneumonia. Scand J Infect Dis 2009; 41: 45–50. [DOI] [PubMed] [Google Scholar]
- 21.Hohenthal U, Vainionpää R, Meurman O, et al. . Aetiological diagnosis of community acquired pneumonia: utility of rapid microbiological methods with respect to disease severity. Scand J Infect Dis 2008; 40: 131–138. [DOI] [PubMed] [Google Scholar]
- 22.Holm A, Nexoe J, Bistrup LA, et al. . Aetiology and prediction of pneumonia in lower respiratory tract infection in primary care. Br J Gen Pract 2007; 57: 547–554. [PMC free article] [PubMed] [Google Scholar]
- 23.Holter JC, Müller F, Bjørang O, et al. . Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: a 3-year prospective study in Norway. BMC Infect Dis 2015; 15: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huijskens EG, Koopmans M, Palmen FM, et al. . The value of signs and symptoms in differentiating between bacterial, viral and mixed aetiology in patients with community-acquired pneumonia. J Med Microbiol 2014; 63: 441–452. [DOI] [PubMed] [Google Scholar]
- 25.Jain S, Self WH, Wunderink RG, et al. . Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 2015; 373: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson N, Kalin M, Tiveljung-Lindell A, et al. . Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis 2010; 50: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnstone J, Majumdar SR, Fox JD, et al. . Viral infection in adults hospitalized with community-acquired pneumonia: prevalence, pathogens, and presentation. Chest 2008; 134: 1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karhu J, Ala-Kokko TI, Vuorinen T, et al. . Lower respiratory tract virus findings in mechanically ventilated patients with severe community-acquired pneumonia. Clin Infect Dis 2014; 59: 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JE, Kim UJ, Kim HK, et al. . Predictors of viral pneumonia in patients with community-acquired pneumonia. PLoS One 2014; 9: e114710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman D, Shimoni A, Shemer-Avni Y, et al. . Respiratory viruses in adults with community-acquired pneumonia. Chest 2010; 138: 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu JX, Gu L, Pu ZH, et al. . Viral etiology of community-acquired pneumonia among adolescents and adults with mild or moderate severity and its relation to age and severity. BMC Infect Dis 2015; 15: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito A, Kohno S, Matsushima T, et al. . Prospective multicenter study of the causative organisms of community-acquired pneumonia in adults in Japan. J Infect Chemother 2006; 12: 63–69. [DOI] [PubMed] [Google Scholar]
- 33.Sangil A, Calbo E, Robles A, et al. . Aetiology of community-acquired pneumonia among adults in an H1N1 pandemic year: the role of respiratory viruses. Eur J Clin Microbiol Infect Dis 2012; 31: 2765–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibli F, Chazan B, Nitzan O, et al. . Etiology of community-acquired pneumonia in hospitalized patients in northern Israel. Isr Med Assoc J 2010; 12: 477–482. [PubMed] [Google Scholar]
- 35.Viasus D, Marinescu C, Villoslada A, et al. . Community-acquired pneumonia during the first post-pandemic influenza season: a prospective, multicentre cohort study. J Infect 2013; 67: 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin YD, Zhao F, Ren LL, et al. . Evaluation of the Japanese Respiratory Society guidelines for the identification of Mycoplasma pneumoniae pneumonia. Respirology 2012; 17: 1131–1136. [DOI] [PubMed] [Google Scholar]
- 37.Hayden FG. Advances in antivirals for non-influenza respiratory virus infections. Influenza Other Respir Viruses 2013; 7: Suppl. 3, 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SJ, Kim K, Park SB, et al. . Outcomes of early administration of cidofovir in non-immunocompromised patients with severe adenovirus pneumonia. PLoS One 2015; 10: e0122642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li BJ, Tang Q, Cheng D, et al. . Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med 2005; 11: 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raj VS, Mou H, Smits SL, et al. . Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013; 495: 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez R, Elbashir S, Borland T, et al. . RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob Agents Chemother 2009; 53: 3952–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrat F, Vergu E, Ferguson NM, et al. . Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008; 167: 775–785. [DOI] [PubMed] [Google Scholar]
- 43.Kumar S, Wang L, Fan J, et al. . Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J Clin Microbiol 2008; 46: 3063–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008; 198: 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Sluijs KF, van der Poll T, Lutter R, et al. . Bench-to-bedside review: bacterial pneumonia with influenza – pathogenesis and clinical implications. Crit Care 2010; 14: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grabowska K, Högberg L, Penttinen P, et al. . Occurrence of invasive pneumococcal disease and number of excess cases due to influenza. BMC Infect Dis 2006; 6: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louie JK, Acosta M, Winter K, et al. . Factors associated with death or hospitalization due to pandemic 2009 influenza A(H1N1) infection in California. JAMA 2009; 302: 1896–1902. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Cai F, Wu X, et al. . Incidence of viral infection detected by PCR and real-time PCR in childhood community-acquired pneumonia: a meta-analysis. Respirology 2015; 20: 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X, Wang Q, Wang M, et al. . Incidence of respiratory viral infections detected by PCR and real-time PCR in adult patients with community-acquired pneumonia: a meta-analysis. Respiration 2015; 89: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jartti T, Lehtinen P, Vuorinen T, et al. . Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol 2004; 72: 695–699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.