Abstract
A transposon Tn5-mob insertional mutant of Paracoccus pantotrophus GB17, strain TP43, was unable to oxidize thiosulfate aerobically or to reduce nitrite anaerobically, and the cellular yields were generally decreased by 11 to 20%. Strain TP43 was unable to form functional c-type cytochromes, as determined by difference spectroscopy and heme staining. However, formation of apocytochromes and their transport to the periplasm were not affected, as seen with SoxD, a c-type cytochrome associated with the periplasmic sulfite dehydrogenase homologue. The Tn5-mob-containing DNA region of strain TP43 was cloned into pSUP205 to produce pE18TP43. With the aid of pE18TP43 the corresponding wild-type gene region of 15 kb was isolated from a heterogenote recombinant to produce pEF15. Sequence analysis of 2.8 kb of the relevant region uncovered three open reading frames, designated ORFA, ccdA, and ORFB, with the latter being oriented divergently. ORFA and ccdA were constitutively cotranscribed as determined by primer extension analysis. In strain TP43 Tn5-mob was inserted into ccdA. The deduced ORFA product showed no similarity to any protein in databases. However, the ccdA gene product exhibited similarities to proteins assigned to different functions in bacteria, such as cytochrome c biogenesis. For these proteins at least six transmembrane helices are predicted with the potential to form a channel with two conserved cysteines. This structural identity suggests that these proteins transfer reducing equivalents from the cytoplasm to the periplasm and that the cysteines bring about this transfer to enable the various specific functions via specific redox mediators such as thioredoxins. CcdA of P. pantotrophus is 42% identical to a protein predicted by ORF2, and its location within the sox gene cluster coding for lithotrophic sulfur oxidation suggested a different function.
The neutrophilic facultatively lithoautotrophic gram-negative bacterium Paracoccus pantotrophus grows aerobically with a large variety of carbon sources and with molecular hydrogen or thiosulfate as energy source, and nitrate serves as electron acceptor under anaerobic conditions (41). P. pantotrophus (formerly Paracoccus denitrificans and Thiosphaera pantotropha [31, 38, 41]) has a branched electron transfer network. The electron flux is regulated by the reduction of the Q pool (33). While the transfer of electrons by the terminal cytochrome oxidases aa3 and cbb3 results in a translocation of six protons (6H+/2e−) (36, 54), the reduction of a terminal electron acceptor by the quinol oxidase o results in a translocation of only four protons (4H+/2e−) (39).
To identify the components involved in energy transformation by oxidation of reduced inorganic sulfur compounds (Sox) of P. pantotrophus GB17, genetic studies were initiated by the isolation of mutants. Transpositional mutagenesis with Tn5-mob coding for kanamycin resistance yielded mutants defective in Sox. One mutant, TP43, is unable to grow with thiosulfate (Sox−) or reduce nitrite under anaerobic conditions (Nitd−), while reduction of nitrate to nitrite is not affected (11).
Cytochromes of the c type are distinguished from cytochromes of other classes by the covalent attachment of their prosthetic heme group to the conserved CXXCH motif of the apoprotein. The biogenesis of c-type cytochromes consists of several steps performed in the cytoplasm and in the periplasm (reviewed in reference 53). Two different systems with respect to the late steps of cytochrome c biogenesis have been described for bacteria (reviewed in reference 28). Most gram-positive bacteria possess the class II system, consisting of at least 4 proteins (ResA, CcsA, CcsI, and CcdA), while most gram-negative bacteria harbor the class I system, consisting of at least 11 proteins (DsbABD and CcmABCDEFGH). In gram-negative bacteria the addition of the heme group to the apoprotein takes place in the oxidative environment of the periplasm, where disulfide bonds between pairs of protein cysteine thiols are generated by the DsbA/B system (4). Thus, cytochrome c apoproteins have to be rereduced prior to heme binding. Recently three proteins, CcmH, CcmG, and DsbD, were proposed to be responsible for this step in cytochrome c maturation in Escherichia coli (15). According to this proposal, the oxidized cytochrome c apoprotein is reduced by CcmH and CcmG and the latter is rereduced by DsbD. In Rhodobacter capsulatus, CcdA has been described as homologous to DsbD of E. coli, having similar predicted structural characteristics and essential cysteine residues (14). In P. pantotrophus within the sox gene cluster coding for lithotrophic sulfur oxidation, ORF2 has been identified, with a predicted product that is about 35% identical to CcdA of Bacillus subtilis and about 45% identical to CcdA of R. capsulatus. Eight genes required for cytochrome c biogenesis including ccmG have been identified so far in P. denitrificans. CcmG of P. denitrificans was proposed to reduce disulfide bonds in vivo (34) and may have the same function in cytochrome c biogenesis as that proposed for CcmG of E. coli.
To determine the nature of the mutation in the Sox− mutant TP43, we have reexamined its pleiotropic character and have identified the complete absence of c-type cytochromes in this strain. We have cloned the Tn5-mob-containing DNA region of strain TP43 (11) and have isolated and sequenced the corresponding wild-type gene region. Below we describe a new gene, ccdA, of P. pantotrophus GB17 which is essential for cytochrome c biogenesis and which is suggested to be involved in the transfer of reducing equivalents from the cytoplasm to the periplasm.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used and constructed in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant geno- or phenotypea | Reference or source |
|---|---|---|
| E. coli | ||
| S17-1 | recA pro thi hsdS, RP4 tra functions, supE44 | 48 |
| XL1-Blue | hsdR17 recA1 endA1 gyrA46 thi relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)] | 9, Stratagene |
| P. pantotrophus | ||
| GB17 | Sox+ Hox + | 31, 38, 41; L. A. Robertson |
| TP43 | Sox− Nitd− cyt c− | 11; this study |
| TPX18 | Sox+ Nitd+ cyt c+ Kmr Tcr, heterogenote of GB17 | This study |
| TP43(pVK1) | Sox+ Nitd+ cyt c+ Tcr | This study |
| Plasmids | ||
| pBluescript SK− | AprlacZ, f1 ori, T7 Phil 10 promoter | Stratagene |
| pSUP205 | Cmr Tcr Tra− Mob+ | 48 |
| pVK101 | Kmr Tcr Tra− Mob+ | 27 |
| pE18TP43 | 18-kb EcoRI fragment containing Tn5-mob from TP43 in pSUP205 | This study |
| pEF15 | 15.2-kb EcoRI wild-type fragment extending the corresponding insert of pE18TP43 by 1.5 kb | This study |
| pBSK−XP6.8 | 6.8-kb XhoI-PstI fragment from pEF15 in pBSK− | This study |
| pBSK−B3.8 | 3.8-kb BamHI fragment from pEF15 in pBSK− | |
| pBSK−ccdA | 744-bp PCR amplification product homologous to ccdA in pBSK− | This study |
| pVK1 | 744-bp PCR amplification product homologous to ccdA in pVK101 | This study |
Sox, lithotrophic growth with thiosulfate; Hox, lithotrophic growth with molecular hydrogen; Nitd, dissimilatory nitrite reduction; Tra, transfer of mobilizable plasmids; Mob, mobilizability; cyt, cytochrome.
Media and growth conditions.
Mineral media were identical for heterotrophic and lithotrophic growth of P. pantotrophus and were described previously (11). P. pantotrophus was cultivated at 30°C. For mixotrophic growth with thiosulfate, mineral media contained 20 mM sodium succinate and 20 mM sodium thiosulfate at an initial pH of 8. For anaerobic growth, bacteria were cultivated in Luria-Bertani broth containing 0.1% (wt/vol) sodium nitrite or 0.1% (wt/vol) potassium nitrate. The following antibiotics were used when appropriate for P. pantotrophus GB17: kanamycin (KM) at 300 μg/ml, tetracycline (TC) at 5 μg/ml, and chloramphenicol (CM) at 5 μg/ml. For E. coli the antibiotics were KM at 50 μg/ml, ampicillin (AP) at 50 μg/ml, TC at 12.5 μg/ml, and CM at 30 μg/ml. Antimycin A was used at a concentration of 50 μg/ml. Cellular yields of P. pantotrophus were determined as described for Ralstonia eutropha (19).
Enzyme assays.
Whole cells were used for enzyme assays. Thiosulfate oxidizing activity was determined with an oxygen electrode as described elsewhere (56). N, N,N′,N′-Tetramethyl-p-phenylenediamine (TMPD) oxidase activity was measured in an oxygen electrode in the presence of 1 mM TMPD. One unit of enzyme activity was defined as 1 μmol of substrate converted per min at 30°C.
Immunoblot analysis.
The level of Sox-specific c-type cytochrome SoxD and the purity of periplasmic extracts were determined by immunoblot analysis as described elsewhere (56). Antibodies against SoxD antigens were obtained against the oligopeptide FYPDDRDQTEYPLF, deduced from the soxD nucleotide sequence. Antibodies were raised in rabbits at the facilities of Eurogentec (Seraing, Belgium). Antibodies against ribulose-bisphosphate carboxylase of R. eutropha were obtained from B. Bowien, Goettingen, Germany (7).
Cytochrome analysis.
Cytochromes of P. pantotrophus GB17 and its derivative strain TP43 were analyzed by using cell extracts. Cells were cultivated as specified below, washed twice at 0°C, and resuspended in 10 mM HEPES buffer, pH 7.4. Cells were disrupted by French press, and cell extracts were obtained from the 30,000 × g supernatant (16). For analysis of c-type cytochromes from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), samples were prepared without boiling. Cytochromes were stained as described by Francis and Becker (18). For identification of the cytochromes, cell extracts (10 mg of protein/ml) were subjected to redox difference spectroscopy. Cell extracts reduced by addition of solid sodium dithionite were measured at 20°C against air-oxidized extracts with a Shimadzu UV 160 A spectrophotometer. Cytochromes were quantified from the 30,000 × g supernatant by the pyridine extraction procedure (5).
Periplasmic proteins were selectively extracted from whole cells by the chloroform procedure described in reference 3. Periplasmic extracts were examined for contamination by cytoplasmic proteins by immunoblot analysis of ribulose-bisphosphate carboxylase antigens.
Analytical procedures.
Formate, thiosulfate, and nitrite were quantified by colorimetric methods. Formate was measured by the procedure described by Lang and Lang (30), thiosulfate was measured by the procedure described by Sørbø (49), and nitrite was measured by the sulfanilamide method (23). Protein from cell extracts was quantified by the procedure described by Bradford (8).
DNA and RNA techniques.
Standard DNA techniques (43) were used. Plasmid DNA was isolated by the procedure described by Birnboim and Doly (6) or by using the high pure-plasmid isolation kit (Boehringer, Mannheim, Germany) according to the manufacturer's protocol. Total RNA was isolated as described elsewhere (24) or by using the high pure-RNA isolation kit of Boehringer. DNA sequencing was performed by primer walking with the thermostable DNA polymerase of Thermus aquaticus and 7-deaza-dGTP (Amersham-Buchler, Braunschweig, Germany) by the dideoxy-chain termination method (44) with an automated DNA sequencing system (Li-Cor; MWG-Biotech, Munich, Germany). The plasmids pBSK−XP6.8 and pBSK−B3.8, used for sequencing, are described in Table 1.
For primer extension experiments P. pantotrophus strains were grown heterotrophically with glucose in mineral media as described above. RNA was isolated from cells in the late exponential growth phase and quantified at 260 nm. Primers were designated PE1 (5′-CGGTCATATAGGCCAGATAGGGG-3′), complementary to bases 1204 to 1227; PE2 (5′-GTGTCCAGGAAGGGGATGCGGAT-3′), complementary to bases 1436 to 1452; PE3 (5′-CCTCGGCGGGTTCCTCGTCATCG-3′), complementary to bases 261 to 284; and PE4 (5′-CGGCGCGGTCTTCCTCCCCGGCG-3′), complementary to bases 300 to 322. The oligonucleotides were labeled at the 5′ end with the fluorescent dye IRD-800 (MWG Biotech). One picomole of primer was annealed to 1 μg of total RNA and extended for 1 h at 42°C by using reverse transcriptase of a variant of Moloney murine leukemia virus (Boehringer) according to the manufacturer's protocol. The primer extension products were analyzed with a sequencing gel as described above.
Sequence analysis was done with the DNASIS (Hitachi Software Engineering, San Bruno, Calif.) and PC/GENE (IntelliGenetics Inc., Mountain View, Calif.) software packages. Nucleotide and amino acid homology searches were done with the BLAST algorithm (2). Higher-order structure analysis of amino acid sequences was done with the PROSIS (Hitachi Software Engineering) and TMpred (25) software packages.
Construction of pEF15.
The Tn5-mob-containing EcoRI fragment of strain TP43 was cloned in pSUP205 at the facilities of Biodelta (Bad Oeynhausen, Germany). Total DNA of strain TP43 was digested with EcoRI. Fragments of 18 to 6 kb were separated by agarose gel electrophoresis and purified with the extraction kit Jetsorb (Genomed, Bad Oeynhausen, Germany). Fragments were ligated with pSUP205, and the resulting plasmids were transformed into E. coli XL1-Blue. Transformants exhibiting the Tcr, Kmr, Apr, and CmS phenotypes were examined for hybrid plasmids. One clone contained the 18-kb hybrid plasmid pE18TP43. The insertion of Tn5-mob was verified by Southern hybridization, and its location was determined by physical mapping as well as by DNA sequencing (Fig. 1).
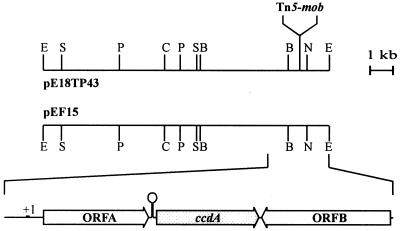
FIG. 1.
Physical map of the insert DNAs of the plasmids pE18TP43 and pEF15 and ORFs within the 2.8-kb ccdA gene region. E, EcoRI; S, SmaI; P, PstI; C, ClaI; B, BamHI; N, NotI.
For isolation of the respective wild-type gene region, plasmid pE18TP43 was transformed into E. coli S17–1 and conjugated therefrom into P. pantotrophus GB17. Heterogenote recombinants such as strain TPX18 exhibiting the Kmr Tcr phenotype were isolated. Total DNA of strain TPX18 was partially digested with EcoRI, religated, and transformed into E. coli S17–1. Transformants exhibiting the KmS Tcr phenotype were examined for hybrid plasmids by colony hybridization using the 1,560-bp ClaI-BamHI fragment of pE18TP43. Positive clones were screened for insert sizes, and plasmid pEF15 was selected (Table 1).
Construction of pVK1 to complement ccdA::Tn5 in TP43.
Primer C1 (5′-ATGTTGGGAATCGAGCTTGCA-3′) and primer C2 (5′-CTAACCCAGCGTGGCCAGCCA-3′) were used to amplify the ccdA gene region by PCR (32). The resulting PCR product was cloned into the EcoRV site of pBSK− to produce pBSK−ccdA. The insert DNA was then isolated by EcoRI and HindIII and cloned into the plasmid pVK101. The resulting plasmid pVK1 was transformed into E. coli S17–1 and conjugated therefrom into the mutant strain TP43 to produce P. pantotrophus TP43(pVK1).
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the EMBL and GenBank databases under accession number AF308446.
RESULTS
Physiological characterization of strain TP43.
Strain TP43 is unable to grow lithotrophically with thiosulfate and unable to reduce nitrite anaerobically (11). The pleiotropic character may have resulted from a mutation in a regulatory mechanism or may have been related to the energy metabolism. To distinguish between these possibilities, the cellular yields were determined. When cultivated with glucose, glyoxylate, or formate, respectively, strain TP43 exhibited generally lower cellular yields of 11.3 to 20.7% compared to the wild type (Table 2). The reduced yield was evidence that the mutation (i) was not restricted to the phenotypes described initially (11) but was a general characteristic of strain TP43 and (ii) was linked to the energy metabolism. Therefore, the effect of antimycin A, an inhibitor of the electron transfer to the bc1 complex of the respiratory chain in various bacteria (reviewed in reference 40), was examined under anaerobic growth conditions with nitrate as electron acceptor. These conditions allowed the determination of whether c-type or other types of cytochromes were affected. Nitrate reduction does not require c-type cytochromes in P. denitrificans, whereas nitrite reduction involves different c-type cytochromes linked to the bc1 complex, nitrite reductase, nitric oxide, and nitrous oxide reductase (10, 52). Under anaerobic conditions in the presence of antimycin A the yield of the wild type was reduced by 55% and the specific growth rate was reduced from 0.38 to 0.25 per h. The extent and the rate of growth of strain TP43 without antimycin A were identical to those of the wild type cultivated with antimycin A, and addition of the drug did not affect the growth characteristics of TP43 (data not shown). Under anaerobic growth conditions addition of antimycin A to the wild type allowed quantitative reduction of nitrate; however, nitrite was not metabolized further (data not shown). These results pointed to the inability of strain TP43 to form a component essential for the cytochrome bc1 complex or the electron flow via this complex. This conclusion was supported by the complete absence of TMPD oxidase activity in strain TP43 (Table 2). For oxidation of TMPD, c-type cytochromes are essential (26).
TABLE 2.
Growth yields of P. pantotrophus GB17 and strain TP43 with different carbon sourcesa and specific TMPD oxidase activity
| Strain |
Ysb
|
TMPD oxidasec | ||
|---|---|---|---|---|
| Glucose | Glyoxylate | Formate | ||
| GB17 | 0.627 | 0.118 | 0.080 | 1.0 |
| TP43 | 0.546 | 0.105 | 0.064 | 0 |
| TP43(PVK1) | 0.620 | 0.120 | 0.078 | 0.95 |
Media and growth conditions are given in the text.
In grams (dry weight) per gram of substrate.
O2 consumption (in micromoles per minute per milligram of protein). Cells were grown with glucose as carbon source.
Addition of hemin (45 μM) to glucose mineral medium led to an increase in the concentration of b-type cytochromes but did not enable strain TP43 to form c-type cytochromes or to grow anaerobically with nitrite (data not shown). This suggested that hemin was taken up by the cell and that heme deficiency was not responsible for the phenotype of TP43.
Cytochrome c analysis.
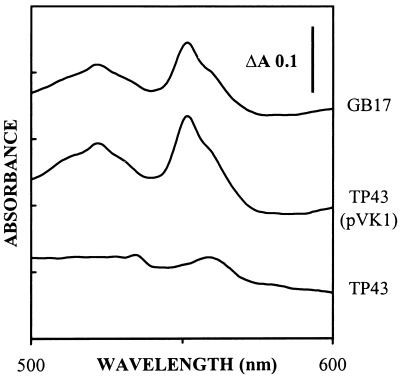
To analyze components involved in electron transport, cytochromes were analyzed for P. pantotrophus GB17 and TP43. Redox difference spectroscopy of extracts from glucose-grown cells of P. pantotrophus GB17 showed an absorption maximum at 551 nm, diagnostic of c-type cytochromes, while extracts from glucose-grown cells of TP43 showed an absorption maximum at 560 nm, typical of b-type cytochromes (Fig. 2). Quantitative analysis of pyrimidine hemochromes (5) from the wild type and strain TP43 revealed a maximum concentration of 1.83 μmol of cytochrome c per g of protein for the wild type, while only traces (0.09 μmol of cytochrome c per g of protein) were detected from strain TP43. Using this method, the concentrations of b-type cytochromes (5) from the wild type and strain TP43 were identical (0.3 μmol per g of protein; data not shown). The c-type cytochromes observed for strain TP43 were less than 5% of those for the wild type. This amount was attributed to the method of detection and its interference with the spectrometric determination of b- and a-type cytochromes as previously discussed (5).
FIG. 2.
Visible absorption spectra of total soluble cell extracts from P. pantotrophus GB17, strain TP43, and strain TP43(pVK1). The cuvettes contained 10 mg of protein per ml. The samples were reduced by addition of solid sodium dithionite, and spectra were measured against air-oxidized references.
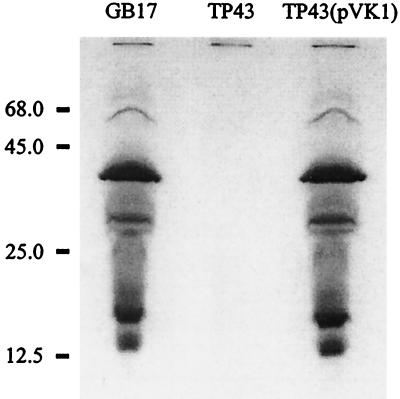
Using heme staining (18), major c-type cytochromes were detected for crude extracts of the wild-type GB17 after separation of proteins by SDS-PAGE while no c-type cytochromes were detected for extracts of strain TP43 (Fig. 3). Therefore, c-type cytochromes were absent in strain TP43.
FIG. 3.
Heme staining of total soluble extracts from P. pantotrophus GB17, strain TP43, and TP43(pVK1). Samples of cell extract of each strain (0.3 mg of protein) were subjected to SDS-PAGE without boiling.
Cloning and physical map of the ccdA gene region.
The wild-type gene region of 15 kb was cloned as described in Materials and Methods and was physically mapped. The map was compared with the Tn5-mob-containing insert cloned in pE18TP43. Identical restriction sites were obtained within the 12.5-kb EcoRI fragments of the two plasmids. The position of Tn5-mob mapped between the EcoRI and BamHI sites of pE18TP43. The precise position of the Tn5-mob inverted-repeat chromosome junction was determined by sequencing to be between nucleotides 1697 and 1698 (Fig. 1).
Sequence analysis
The nucleotide sequence of 2.8 kb was determined for both strands starting from the EcoRI restriction site. The sequence was consistent with the restriction enzyme cleavage map of pEF15. Analysis of this sequence revealed three open reading frames (ORFs) designated ORFA, ccdA, and ORFB. These ORFs revealed coding characteristics according to codon preference analysis (50). ORFA was separated from ccdA by 48 nucleotides. Within this stretch was located an inverted repeat of 23 nucleotides with the potential to form a hairpin structure with a free energy of formation of −100.4 kJ/mol. ORFB was separated from ccdA by 9 bp and transcribed in the opposite direction.
ORFA predicted a protein of 251 amino acids (27,995 Da). A well-conserved ribosome binding site was located eight nucleotides before the start codon. The putative ORFA gene product was hydrophilic, with a hydropathy index of −1.55, and exhibited a very alkaline pI of 12.95 (29). The predicted protein had a high content of 54 proline residues distributed over the whole sequence and accumulating at the C terminus (data not shown). Amino acid sequence analysis suggested only a weak possibility for a signal peptide of 20 amino acids, consistent with the −3/−1 rule (55). Since the stretch of hydrophobic amino acids was missing, it was questionable whether the ORFA gene product was located in the periplasm. Amino acid sequence comparison of the deduced ORFA gene product revealed no significant similarity to other proteins.
The ccdA gene predicted a protein of 247 amino acids (25,838 Da). A putative ribosome binding site was located six nucleotides 5′ of the start codon. The ccdA gene product was highly hydrophobic, with an overall hydropathy index of +1.05 (29). The pI was 9.25. The deduced CcdA exhibited an identity of 55% to proteins involved in cytochrome c biogenesis of R. capsulatus (14) or 32% to a protein involved in sporulation of B. subtilis (46) or in heavy metal resistance of different gram-negative bacteria (22, 47), while an identity of 42% was observed to the ORF2 gene product of the sox gene cluster of P. pantotrophus (data not shown). Therefore, the CcdA protein was analyzed in detail. Amino acid sequence analysis suggested six transmembrane helices of 19 to 26 amino acids each. These helices were separated by segments of 7 to 25 amino acids. According to the algorithm used (25) the amino- and carboxy-terminal ends faced the periplasm. Helix 1 and helix 4 each contained one highly conserved cysteine residue flanked by conserved proline residues previously described for a putative protein disulfide reductase of Pseudomonas aeruginosa (35). The essential function of these cysteines of CcdA of R. capsulatus related to DipZ of P. aeruginosa with respect to transport of reductant from the cytoplasm to the periplasm has recently been demonstrated (14).
ORFB predicted a protein of 273 amino acids (29,015 Da) with a well-conserved putative ribosome binding site six nucleotides 5′ of the start codon. The predicted protein was slightly hydrophobic, with a hydropathy index of +0.06 and a pI of 4.87 (29). Amino acid sequence comparison of the deduced ORFB gene product revealed an identity of 30% to a putative enoyl coenzyme A hydratase of E. coli (1).
Complementation of the mutation in strain TP43.
To analyze if the insertion of the transposon Tn5-mob in ccdA caused any polar effects which could have been responsible for the pleiotropy of TP43, plasmid pVK1 was constructed. The ccdA gene region was cloned into the vector pVK101 and used for complementation. Strain TP43 harboring plasmid pVK1 in trans was able to grow aerobically with thiosulfate and anaerobically with nitrite (data not shown). When cultivated heterotrophically with glucose, glyoxylate, or formate, the cellular yields of TP43(pVK1) were similar to those of the wild type (Table 2). Spectrophotometric analysis demonstrated that the ability to form holo- c-type cytochromes was fully restored (Fig. 2), as was also evident from heme staining (Fig. 3). These data were convincing evidence that the pleiotropy of strain TP43 resulted from the inactivation of ccdA.
Transcription of ORFA and ccdA.
Transcription of ORFA started with an adenine 114 nucleotides before the translation start codon as determined by primer extension analysis using the oligonucleotides PE3 and PE4 (data not shown). Using the oligonucleotides PE1 and PE2, complementary to the beginning of the subsequent ccdA gene, the signal appeared at sizes of about 1,000 and 1,200 nucleotides, respectively (data not shown). This result was evidence that ORFA and ccdA were cotranscribed. Constitutive expression of ORFA and ccdA was deduced from primer extension analysis using oligonucleotide PE3. Signals were equally intense in cells grown aerobically with glucose or succinate and cells grown anaerobically with nitrate as electron acceptor (data not shown).
Biochemical analysis of the ccdA function.
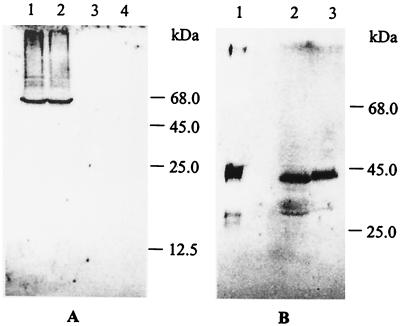
To determine the function of ccdA expression of SoxD, a c-type cytochrome was monitored. The c-type cytochrome SoxD is associated with a sulfite dehydrogenase homologue, located in the periplasm and beneficial for thiosulfate oxidation (37, 56). SoxD antigens were formed by strain TP43 when cultivated in the presence of thiosulfate, demonstrating that expression of the sox genes was not affected by the mutation. Moreover, SoxD antigens were detected in the periplasm using the chloroform procedure which specifically extracts proteins therefrom (Fig. 4). This result demonstrated that the ccdA gene was not involved in the transport of the apoprotein SoxD to the periplasm but was involved in a late step of cytochrome c maturation.
FIG. 4.
Western blot analysis after SDS-PAGE of cell extracts of P. pantotrophus GB17 and strain TP43 for ribulose-bisphosphate carboxylase (A) and cytochrome SoxD antigens (B). (A) Proteins (15 μg) of the cytoplasmic and periplasmic fractions of strain GB17 (lanes 1 and 3, respectively) and strain TP43 (lanes 2 and 4, respectively). (B) homogenous SoxD (2 μg; lane 1), periplasmic fraction of strain GB17 (10 μg of protein; lane 2), and periplasmic fraction of strain TP43 (20 μg of protein; lane 3).
A mutation in the dipZ gene of E. coli inactivates DipZ, and this inactivation has been overcome by adding compounds containing thiol groups (42). In P. pantotrophus addition of cysteine, cystine, or other thiols did not compensate the mutation described for strain TP43.
DISCUSSION
Three new ORFs, ORFA, ccdA, and ORFB, were identified for the genome of P. pantotrophus GB17. ORFA and ccdA were constitutively expressed and cotranscribed. Disruption of ccdA by transposon Tn5-mob caused inability to form functional c-type cytochromes. Convincing evidence for the requirement of ccdA in cytochrome c biogenesis was obtained from (i) physiological studies, (ii) complementation analysis, (iii) biochemical and immunochemical analysis, and (iv) sequence analysis of the deduced ccdA gene product.
Strain TP43 not only was impaired in aerobic thiosulfate oxidation or dissimilatory nitrite reduction but also exhibited significantly reduced cellular yields compared to the wild type. The reduction in cellular yields was evidence of a general defect in energy metabolism. This evidence was supported by the inability of strain TP43 to oxidize TMPD and the absence of any effect of antimycin A, an inhibitor of electron transfer to the bc1 complex, which caused reduction in the cellular yield of the wild type but not in that of strain TP43. Without antimycin A the yield of strain TP43 was identical to that of the wild type grown in the presence of antimycin A. This was evidence that electron transfer was abolished from ubiquinone to the bc1 complex and further to cytochrome c551 (40).
Analysis of cytochromes revealed the complete absence of holocytochromes c, whereas apoproteins of c-type cytochromes were still formed in strain TP43, as demonstrated by the immunochemical detection of SoxD, the periplasmic c-type cytochrome which is part of the sulfur-oxidizing enzyme system in P. pantotrophus GB17 (37, 56). Moreover, the SoxD apoprotein was transported to the periplasm as demonstrated by its specific extraction and by immunochemical analysis. Therefore, disruption of ccdA eliminated an essential late step in cytochrome c biogenesis. This in turn affected energy conservation and caused reduced cellular yields.
To exclude a polar effect of the Tn5-mob which could have been responsible for the pleiotropy of strain TP43, the ccdA gene region was cloned into pVK101. TP43 harboring pVK1 was fully restored in its ability to form c-type cytochromes. This demonstrated that the inactivation of only ccdA caused the inability to form holo- c-type cytochromes, and thiol compounds could not compensate this defect as described for E. coli (42).
CcdA of P. pantotrophus is homologous to proteins of other bacteria involved in copper tolerance (17, 22), mercury resistance (47), spore synthesis (46), and cytochrome c biogenesis (13, 35, 45). CcdA was not cotranscribed with a thioredoxin in P. pantotrophus, and it missed the thioredoxin stretch at the C terminus as described for DipZ homologous proteins found in E. coli (13) or P. aeruginosa (35). CcdA homologues have been proposed to participate in class II cytochrome c biogenesis systems, whereas the class I cytochrome c biogenesis systems present in most gram-negative bacteria are proposed to involve a DipZ homologue (28). The participation of a CcdA homologue in the biogenesis of a class I cytochrome c has been described only for R. capsulatus (14) and CcdA of P. pantotrophus, closely related to R. capsulatus (31).
In analogy to previous findings we suggest that CcdA of P. pantotrophus transports electrons from the cytoplasm to the periplasm via the two cysteine residues, although its electron donor is still unknown. Such involvement has been suggested for CcdA of R. capsulatus (14) and for DsbD of E. coli (12, 21, 51).
The disruption of ccmG of P. denitrificans caused a partial deficiency in holo- c-type cytochromes, and it was proposed that CcmG participates in cytochrome c biogenesis by reducing disulfide bonds in the periplasm (34). However, it is still unknown in which way CcmG derives the reducing equivalents. On the basis of the data presented we suggest that CcdA may act in reducing CcmG directly or via an unknown mediator in P. pantotrophus GB17 which seems to play an important role in the rereduction of cytochrome c apoproteins in the periplasm prior to heme binding.
The primary structure of CcdA is 42% identical and 80% similar to that of the predicted ORF2 gene product of P. pantotrophus located within the sox gene cluster (20). ORF2, however, cannot complement the disrupted ccdA, as shown with strain TP43. Therefore, the possible ORF2 function is proposed to be different from that of ccdA; whether it is specific for or linked to lithotrophic sulfur oxidation remains to be analyzed.
REFERENCES
- 1.Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kasai H, Kishimoto K, Kimura S, Kitakawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motumura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Horiuchi T, et al. A 570-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 28.0 - 40.1 min region on the linkage map. DNA Res. 1996;3:363–377. doi: 10.1093/dnares/3.6.363. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ames G F-L, Prody C, Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol. 1984;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardwell J C, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 5.Berry E A, Trumpower B L. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowien B, Mayer F, Codd G A, Schlegel H G. Purification, some properties, and quaternary structure of the d-ribulose 1,5-diphosphate carboxylase of Alcaligenes eutrophus. Arch Microbiol. 1976;110:157–167. doi: 10.1007/BF00690223. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strains with beta-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- 10.Carr G J, Ferguson S J. The nitric oxide reductase of Paracoccus denitrificans. Biochem J. 1990;269:423–429. doi: 10.1042/bj2690423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandra T S, Friedrich C G. Tn5-induced mutations affecting sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha. J Bacteriol. 1986;166:446–452. doi: 10.1128/jb.166.2.446-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung J, Chen T, Missiakas D. Transfer of electrons across the cytoplasmic membrane by DsbD, a membrane protein involved in thiol-disulphide exchange and protein folding in the bacterial periplasm. Mol Microbiol. 2000;35:1099–1109. doi: 10.1046/j.1365-2958.2000.01778.x. [DOI] [PubMed] [Google Scholar]
- 13.Crooke H, Cole J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol Microbiol. 1995;15:1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 14.Deshmukh M, Brasseur G, Daldal F. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol Microbiol. 2000;35:123–138. doi: 10.1046/j.1365-2958.2000.01683.x. [DOI] [PubMed] [Google Scholar]
- 15.Fabianek R A, Hofer T, Thöny-Meyer L. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch Microbiol. 1999;171:92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- 16.Fischer J, Quentmeier A, Kostka S, Kraft R, Friedrich C G. Purification and characterization of the hydrogenase from Thiobacillus ferrooxidans. Arch Microbiol. 1996;165:289–296. doi: 10.1007/s002030050329. [DOI] [PubMed] [Google Scholar]
- 17.Fong S-T, Camakaris J, Lee B T O. Molecular genetics of a chromosomal locus involved in copper tolerance in Escherichia coli K-12. Mol Microbiol. 1995;15:1127–1137. doi: 10.1111/j.1365-2958.1995.tb02286.x. [DOI] [PubMed] [Google Scholar]
- 18.Francis R T, Becker R R. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal Biochem. 1984;136:509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- 19.Friedrich C G, Bowien B, Friedrich B. Formate and oxalate metabolism in Alcaligenes eutrophus. J Gen Microbiol. 1979;115:185–192. [Google Scholar]
- 20.Friedrich C G, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, Prinz H. Novel genes coding for lithotrophic sulfur oxidation in Paracoccus pantotrophus GB17. J Bacteriol. 2000;182:4677–4687. doi: 10.1128/jb.182.17.4677-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon E H J, Page M D, Willis A C, Ferguson S J. Escherichia coli DipZ: anatomy of a transmembrane protein disulphide reductase in which three pairs of cysteine residues, one in each of three domains, contribute differentially to function. Mol Microbiol. 2000;35:1360–1374. doi: 10.1046/j.1365-2958.2000.01796.x. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S D, Wu H C, Rick P D. A Salmonella typhimurium genetic locus which confers copper tolerance on copper-sensitive mutants of Escherichia coli. J Bacteriol. 1997;179:4977–4984. doi: 10.1128/jb.179.16.4977-4984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson R S, Phillips J A. Chemical composition. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general microbiology. Washington, D.C.: American Society for Microbiology; 1981. p. 355. [Google Scholar]
- 24.Heilmann H-J. Isolierung von RNA aus E. coli. In: Bertram S, Gassen H G, editors. Gentechnische Methoden. Stuttgart, Germany: Gustav Fischer; 1991. pp. 199–200. [Google Scholar]
- 25.Hoffmann K, Stoffel W. TMBASE—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 26.Keilin D, editor. The history of cell respiration and cytochrome. Cambridge, United Kingdom: Cambridge University Press; 1966. [Google Scholar]
- 27.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmic clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 28.Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Lang E, Lang H. Spezifische Farbreaktion zum direkten Nachweis von Ameisensäure. Z Anal Chem. 1972;260:8–10. [Google Scholar]
- 31.Ludwig W, Mittenhuber G, Friedrich C G. Transfer of Thiosphaera pantotropha to Paracoccus denitrificans. Int J Syst Bacteriol. 1993;43:363–367. doi: 10.1099/00207713-43-2-363. [DOI] [PubMed] [Google Scholar]
- 32.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp Quant Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 33.Otten F M, Reijnders W N M, Bedaux J J M, Westerhoff H V, Krab K, Van Spanning R J M. The reduction state of the Q-pool regulates the electron flux through the branched respiratory network of Paracoccus denitrificans. Eur J Biochem. 1999;261:767–774. doi: 10.1046/j.1432-1327.1999.00334.x. [DOI] [PubMed] [Google Scholar]
- 34.Page D M, Ferguson S J. Paracoccus denitrificans CcmG is a periplasmic protein-disulphide oxidoreductase required for c- and aa3-type cytochrome biogenesis; evidence for a reductase role in vivo. Mol Microbiol. 1997;24:977–990. doi: 10.1046/j.1365-2958.1997.4061775.x. [DOI] [PubMed] [Google Scholar]
- 35.Page D M, Saunders N F, Ferguson S J. Disruption of the Pseudomonas aeruginosa dipZ gene, encoding a putative protein-disulfide reductase, leads to partial pleiotropic deficiency in c-type cytochrome biogenesis. Microbiology. 1997;143:3111–3122. doi: 10.1099/00221287-143-10-3111. [DOI] [PubMed] [Google Scholar]
- 36.Puustinen A, Finel M, Virkki M, Wikström M. Cytochrome o (bo) is a proton pump in Paracoccus denitrificans and Escherichia coli. FEBS Lett. 1989;249:163–167. doi: 10.1016/0014-5793(89)80616-7. [DOI] [PubMed] [Google Scholar]
- 37.Quentmeier A, Kraft R, Kostka S, Klockenkämper R, Friedrich C G. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch Microbiol. 2000;173:117–125. doi: 10.1007/s002039900118. [DOI] [PubMed] [Google Scholar]
- 38.Rainey F A, Kelly D P, Stackebrandt E, Burhardt J, Hiraishi A, Katayama Y, Wood A P. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int J Syst Bacteriol. 1999;49:645–651. doi: 10.1099/00207713-49-2-645. [DOI] [PubMed] [Google Scholar]
- 39.Raitio M, Wikström M. An alternative cytochrome oxidase of Paracoccus denitrificans functions as a proton pump. Biochim Biophys Acta. 1994;1186:100–106. [Google Scholar]
- 40.Rieske J S. Antimycin A. In: Gottlieb D, editor. Antibiotics I. Mechanism of action. Berlin, Germany: Springer; 1967. pp. 542–580. [Google Scholar]
- 41.Robertson L A, Kuenen J G. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultative autotrophic sulphur bacterium. J Gen Microbiol. 1983;129:2847–2855. [Google Scholar]
- 42.Sambongi Y, Ferguson S J. Specific thiol compounds complement deficiency in c-type cytochrome biogenesis in Escherichia coli carrying a mutation in a membrane bound disulphide isomerase-like protein. FEBS Lett. 1994;353:235–238. doi: 10.1016/0014-5793(94)01053-6. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiött T, von Wachenfeldt C, Hederstedt L. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J Bacteriol. 1997;179:1962–1973. doi: 10.1128/jb.179.6.1962-1973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiött T, Hederstedt L. Efficient spore synthesis in Bacillus subtilis depends on the CcdA protein. J Bacteriol. 2000;182:2845–2854. doi: 10.1128/jb.182.10.2845-2854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sedlmeier R, Altenbuchner J. Cloning and DNA sequence analysis of the mercury resistance genes of Streptomyces lividans. Mol Gen Genet. 1992;236:76–85. doi: 10.1007/BF00279645. [DOI] [PubMed] [Google Scholar]
- 48.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 49.Sørbø B. A colorimetric method for the determination of thiosulfate. Biochim Biophys Acta. 1957;23:412–416. doi: 10.1016/0006-3002(57)90346-3. [DOI] [PubMed] [Google Scholar]
- 50.Steinrücke P, Ludwig B. Genetics of Paracoccus denitrificans. FEMS Microbiol Rev. 1993;104:83–118. doi: 10.1016/0378-1097(93)90505-v. [DOI] [PubMed] [Google Scholar]
- 51.Stewart E J, Katzen F, Beckwith J. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 1999;18:5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stouthamer A H. Metabolic regulation including anaerobic metabolism in Paracoccus denitrificans. J Bioenerg Biomembr. 1991;23:163–185. doi: 10.1007/BF00762216. [DOI] [PubMed] [Google Scholar]
- 53.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trumpower B L. The three subunit cytochrome bc1 complex of Paracoccus denitrificans. It's physiological function, structure, and mechanism of electron transfer and energy transduction. J Bioenerg Biomembr. 1991;23:241–255. doi: 10.1007/BF00762220. [DOI] [PubMed] [Google Scholar]
- 55.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 56.Wodara C, Bardischewsky F, Friedrich C G. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J Bacteriol. 1997;179:5014–5023. doi: 10.1128/jb.179.16.5014-5023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]