Abstract
The discovery of epidermal growth factor receptor (EGFR) mutations in nonsmall cell lung cancer (NSCLC) has allowed the identification of a subset of patients whose tumours are exquisitely sensitive to EGFR tyrosine kinase inhibitors (TKIs). Despite the efficacy and superiority of EGFR TKIs over chemotherapy as first-line therapy, all patients will ultimately develop progressive disease, with a median of 9–13 months progression-free survival. A better understanding of the molecular mechanisms underlying resistance to EGFR TKIs can help design new drugs and therapeutic strategies to overcome resistance. This has been illustrated by the new generation TKIs that are effective on the T790M mutation, which is the most frequent mechanism of acquired resistance to EGFR TKIs. In this article, we will address the main molecular mechanisms of primary and acquired resistance to EGFR TKIs in EGFR-mutant NSCLC.
Introduction
Recent insights in the molecular mechanisms of nonsmall cell lung cancer (NSCLC) have led to the development of targeted treatments associated with substantial clinical benefit. These include epidermal growth factor receptor (EGFR) inhibitors such as gefitinib, erlotinib and afatinib, which have shown superiority over chemotherapy as a first-line treatment for EGFR-mutant NSCLC.‘‘Topics in Thoracic Oncology’’Edited by G. Zalcman and N. GirardNumber 5 in the Series
EGFR (HER1) is a transmembrane tyrosine kinase receptor that belongs to the HER (or ErbB) family that includes four members. Upon ligand binding, EGFR homo- or hetero-dimerises with other HER family receptors, which results in phosphorylation of tyrosine residues that will serve as docking sites for recruiting and activating downstream pathways. The main downstream signalling involves the phosphatidylinositol 3-kinase (PI3K)/Akt and mitogen-activated protein kinase (MAPK) pathways, which promote cell survival and proliferation. EGFR is deregulated in a broad range of cancers including NSCLC, pancreatic cancer, breast cancer, colorectal cancer, and head and neck cancer. Most NSCLCs overexpress EGFR and a high EGFR gene copy number is observed in 10–30% of NSCLCs. Mutations of EGFR occurs in 10% of NSCLC patients of Caucasian ethnicity. They are more frequent in never-smokers, females, patients of Asian ethnicity and adenocarcinomas. EGFR mutations affect the EGFR tyrosine kinase domain, within exons 18–21. The L858R substitution in exon 21 and in-frame deletions in exon 19 account for 90% of all EGFR mutations. Apart from these two hotspot mutations, rare mutations include insertions in exon 20 and substitutions in exon 18 or 20 [1]. Mutations in EGFR exons 19 and 21 force EGFR into an active conformation, resulting in constitutive activation independent of ligand binding. EGFR mutant cells are highly dependent on the EGFR pathway, a phenomenon often referred to as “oncogene addiction” [2]. They display permanent activation of the PI3K/Akt and MAPK pathways. EGFR can be targeted by monoclonal antibodies directed against its extracellular domain, such as cetuximab or panitumumab, or by tyrosine kinase inhibitors (TKIs) such as gefitinib or erlotinib, which are small molecules that act as reversible competitive inhibitors of ATP. EGFR mutant cells are exquisitely sensitive to inhibition by TKIs. In these cells, EGFR TKIs decrease activation of EGFR, PI3K/Akt and MAPK pathways, and inhibit cell proliferation and induce apoptosis [3, 4].
The first trial comparing EGFR TKIs to chemotherapy in EGFR-mutant NSCLC was the IRESSA Pan-Asia Study (IPASS), which enrolled previously untreated patients from East Asia with advanced NSCLC who were light or nonsmokers. Patients were randomly assigned to treatment with carboplatin and paclitaxel or gefitinib. Among the 261 patients harbouring an EGFR mutation, objective response rate (71.2 versus 47.3%) and progression-free survival (PFS) (9.5 versus 6.3 months, hazard ratio 0.48; p<0.001) were significantly improved in patients receiving gefitinib compared with those receiving chemotherapy [5]. Further studies confirmed the superiority of EGFR TKIs over chemotherapy for gefitinib, erlotinib and afatinib, an irreversible EGFR TKI, with median PFS ranging from 9 to 13 months (table 1) [6–11]. Treatment with EGFR TKIs was usually well tolerated, with most adverse events involving skin rash and diarrhoea, and quality of life was better in patients treated with TKIs than in those treated with standard chemotherapy [12]. Given these results, treatment with an EGFR TKI has been recommended as the standard of care for first-line treatment of EGFR-mutant NSCLC [13–15]. However, all patients with EGFR-mutant NSCLC treated with EGFR TKIs will ultimately develop resistance. In this article, we will address the molecular mechanisms that underlie primary and acquired resistance to EGFR TKIs in EGFR-mutant NSCLC.
Table 1. Primary resistance and PFS in EGFR-mutant nonsmall cell lung cancer patients treated with EGFR TKIs as first-line therapy in major phase III trials.
| First author [ref.] | EGFR mutation type | Patients n | TKI | Progressive disease as best overall response % | Progressive or stable disease as best overall response % | Median PFS months |
| Mok [5] | Common/rare | 132 | Gefitinib | 8 | 28 | 9.5 |
| Mistudomi [6] | Common | 86 | Gefitinib | 7 | 38 | 9.2 |
| Maemondo [7] | Common/rare | 228 | Gefitinib | 10 | 25 | 10.8 |
| Rosell [8] | Common | 86 | Erlotinib | 7 | 28 | 9.7 |
| Zhou [9] | Common | 83 | Erlotinib | 4# | 20¶ | 13.1 |
| Sequist [10] | Common/rare | 230 | Afatinib | 10# | 44¶ | 11.1 |
| Wu [11] | Common/rare | 242 | Afatinib | 4 | 25 | 11.0 |
PFS: progression-free survival; EGFR: epidermal growth factor receptor; TKI: tyrosine kinase inhibitor. #: calculated as percentage of patients who did not achieve disease control; ¶: calculated as percentage of patients who did not achieve an objective response.
Definition of resistance
The study of resistance to EGFR TKIs in EGFR-mutant NSCLC patients can be divided into primary and acquired resistances, which have different origins. Primary resistance refers to patients who had progressive disease or stable disease as the best response to EGFR TKI, whereas acquired, or secondary, resistance refers to patients who had progressive disease following an initial objective response or prolonged stable disease. Jackman et al. [16] have proposed a detailed definition of acquired resistance to EGFR TKIs, which relies on: 1) the presence of a known activating EGFR mutation associated with sensitivity to EGFR TKI or a prolonged response or stable disease to EGFR TKI (>6 months); 2) treatment with an EGFR TKI as a monotherapy; 3) disease progression upon uninterrupted exposure to EGFR TKI; and 4) no additional systemic therapy since discontinuation of EGFR TKI. The occurrence of osteoblastic reactions during treatment with TKIs, while the primary tumour and metastases are stable or in response, should not be considered as disease progression [17].
Primary resistance to EGFR TKIs
The definition of primary resistance to EGFR TKI varies among authors, from “a lack of objective response” to “progressive disease as the best response”. Using the latter definition, primary resistance usually occurs in 4–10% of EGFR-mutant NSCLC patients (table 1) [5–11, 18]. Although mechanisms of primary resistance are not fully understood, they can be divided into host-dependent and tumour-dependent factors.
Host-dependent factors
Cigarette smoke
Cigarette smoke is known to induce activation of cytochrome CYP 1A1, which is one of the cytochromes involved in the metabolism of EGFR TKIs [19]. Cigarette smoke also induces release of reactive oxidative stress species, and promotes autophosphorylation leading to impaired receptor ubiquitination/degradation of wild-type and mutant EGFR even in the presence of EGFR TKI [20, 21]. Recently, Filosto et al. [22] found that activation of EGFR through cigarette smoke involves activation of Src, and that Src inhibitors could reverse cigarette-induced resistance to EGFR TKI. Several studies have found that current smokers with EGFR-mutant NSCLC experience a lower benefit from EGFR TKI [23]. However, when comparing characteristics of patients with or without primary resistance to EGFR TKI, Lee et al. [24] have not found smoking status to be significantly associated with primary resistance.
BIM polymorphism
BIM is a pro-apoptotic member of the BCL-2 family of proteins, which is required for apoptosis induced by several TKIs, including EGFR TKIs [25–27]. In vitro, inhibition of BIM expression has been found to induce intrinsic resistance to EGFR TKI. In the EURTAC study (Erlotinib versus Chemotherapy as First-line Treatment for EGFR-mutant NSCLC Patients), patients treated with erlotinib who had a low or intermediate levels of expression of BIM mRNA had a worse median PFS than those with high levels of BIM mRNA (7.2 versus 12.9 months; p=0.0003), whereas BIM expression levels had no impact on PFS in chemotherapy-treated patients [28]. Moreover, Ng et al. [29] have reported a polymorphism of BIM, that has been exclusively described in Asian populations, with no cases in series of African–American or Caucasian patients, in which alternative splicing has resulted in BIM isoforms lacking the pro-apoptotic BH3 domain. Expression of these isoforms was associated with lack of sensitivity to EGFR TKIs in EGFR-mutant cell lines. Importantly, the authors found that patients with BIM polymorphism had a lower median PFS than those without polymorphism (6.6 versus 11.9 months; p=0.0027) [29].
Tumour-dependent factors
EGFR mutations associated with resistance to EGFR TKI
Not all EGFR mutations have the same consequences on sensitivity of EGFR to EGFR TKIs. Thus, although the two “hotspot” mutations of EGFR, L858R substitution and exon 19 deletions, confer sensitivity by increasing affinity for EGFR TKI, other rarer EGFR mutations confer resistance. Exon 20 insertions account for 5–10% of all EGFR mutations and are highly variable, ranging from 3 to 12 bp [30–32]. In vitro, exon 20 insertions have been found to activate EGFR by promoting the active conformation of the receptor [33]. Unlike EGFR L858R mutants, EGFR harbouring an exon 20 insertion does not show increased affinity for EGFR TKI, thus, resulting in intrinsic resistance. However, this may not be true for all exon 20 insertions, since one insertion (EGFR-A763_Y764insFQEA) was found to be highly sensitive to EGFR TKI in vitro [33]. In the French ERMETIC-IFCT study, Beau-Faller et al. [30] identified 102 (10%) rare EGFR mutations among 1047 EGFR-mutated NSCLC samples. Rare mutations were divided into exon 18 mutations, exon 20 mutations or complex mutations (a double mutation involving at least one mutation in exon 18 or exon 20). The authors found an objective response rate to EGFR TKIs in patients harbouring a mutation in exon 18 or exon 20 as low as 8%, whereas the response rate was 57% for patients with complex mutations (table 2) [30]. Overall survival for patients with exon 20 mutations ranges from 9.5 to 16 months [30, 31].
Table 2. Frequency of rare EGFR mutations and efficacy of EGFR TKIs.
| Rare mutations | Frequency among EGFR rare mutations % | ORR % | DCR % | PFS ≥6 months on EGFR TKI % |
| Exon 18 mutations | 38 | 7 | 34 | 21 |
| Exon 20 mutations | 51 | 8 | 44 | 20 |
| Complex exon 18 and exon 20 mutations | 11 | 57 | 86 | 43 |
EGFR: epidermal growth factor receptor; TKI: tyrosine kinase inhibitors; ORR: objective response rate; DCR: disease control rate; PFS: progression-free survival. Data from [30].
T790M mutation is the main mechanism of acquired resistance to EGFR TKIs, found in up to 60% of post-TKI tumour samples (see later). However, it can be rarely found in TKI naïve NSCLC patients. Pre-treatment T790M mutations are usually associated in cis to another activating EGFR mutation (i.e. on the same allele) such as L858R substitution or a deletion in exon 19. Some germline T790M mutations have been observed at a low frequency, sometimes in a context of familial cancer syndrome [34, 35]. The rate of pre-treatment T790M mutation is highly dependent on the sensitivity of the method of detection and the true frequency associated with drug resistance remains unknown. Costa et al. [28] recently found a 65% T790M mutation rate using a technique based on laser microdissection and peptide–nucleic acid clamping PCR, Rosell et al. [36] found a 35% rate of T790M mutations using a TaqMan assay (Applied Biosystems, Carlsbad, CA, USA), and Yu et al. [37] found a 2% T790M mutation rate using a mass spectrometry-based assay. Among 107 EGFR-mutant NSCLC patients, Su et al. [38] detected a T790M mutation in 27 (25.2%) patients using the highly sensitive method of matrix-assisted laser desorption ionisation time-of-flight mass spectrometry, and in only three (2.8%) patients using direct sequencing. Impact of pre-treatment T790M mutation in a minority clone is controversial. The presence of a pre-treatment T790M mutation seems to be associated with a lower rate of objective response to EGFR TKI when it is detected by routine molecular genotyping [37]. It is also associated with shorter PFS compared with patients with activation EGFR mutations but without any detected T790M mutation. However, PFS is still significantly longer than for patients with wild-type EGFR [36, 37].
Molecular alterations associated with EGFR mutations
Alterations occurring in genes other than the EGFR gene may affect sensitivity to EGFR TKIs in EGFR-mutant NSCLC. As mentioned earlier, alterations in apoptotic pathways such as low expression of BIM may impact response to TKI. Mutations occurring within the EGFR pathway may also induce resistance to EGFR TKI by promoting activation of downstream pathways, independent of EGFR activation. Loss of expression of the phosphatase and tensin homolog, which is a tumour suppressor gene controlling the PI3K/Akt pathway, has been found to induce resistance to EGFR TKI in EGFR-mutant models in vitro [39–41]. Somatic mutations of PIK3CA, which encodes the catalytic domain of PI3K, have been detected in 1–3% of all NSCLC, sometimes in association with EGFR mutations [42]. These mutations are also thought to be involved in acquired resistance to EGFR TKIs [43]. In vitro, PIK3CA mutations are responsible for continued activation of PI3K and abrogate gefitinib-induced apoptosis in EGFR-mutant cell lines [44]. PIK3CA mutations have been found to be associated with poor prognosis in EGFR/KRAS wild-type NSCLC patients [45]. However, it remains to be determined whether PIK3CA mutations actually decrease sensitivity to EGFR TKI in EGFR-mutant NSCLC patients. Other pathways have been shown to play a role in resistance to EGFR TKI in pre-clinical EGFR-mutant models, such as insulin-like growth factor 1 receptor (IGF1R) or nuclear factor-κB pathways, although translational validation of these alterations is still awaited [46, 47].
Acquired resistance to EGFR TKI
Patients whose tumours harbour sensitising mutations in the EGFR kinase domain typically initially respond to EGFR TKIs such as gefitinib, erlotinib or afatinib, but invariably develop progressive disease after a progression-free period of approximately 9–13 months. Recent studies have allowed a better understanding of the molecular mechanisms that are responsible for acquired resistance.
Tools to study acquired resistance
The study of mechanisms of acquired resistance is usually based on two strategies [48, 49]. The first strategy is to compare the molecular alterations between samples obtained before and after the patient has relapsed following TKI treatment. This strategy is limited by the difficulty of obtaining post-relapse samples, which sometimes requires invasive investigations, and if so, by the quality and amount of cancer cells. However, recent studies have demonstrated the feasibility of such rebiopsies, and it is expected that the prospect of a biology-driven treatment will facilitate patients’ adherence [43, 50]. The second strategy is based on in vitro experiments, which involve making TKI-sensitive cell lines resistant by progressively increasing the concentration of the drug. This approach offers a useful tool that has allowed, for example, modelling of various mechanisms of resistance to gefitinib or erlotinib in EGFR TKI-sensitive cell lines such as the T790M mutation or the MET amplification [51, 52]. The study of mechanisms of resistance to kinase inhibitors also requires the most sensitive and accurate methods for detection of mutations. Indeed, detection of resistance mutations can be challenging when only a small subset of cells is implicated [44]. These technical limitations may explain in part the variations observed in the incidence of resistance mutations among various studies.
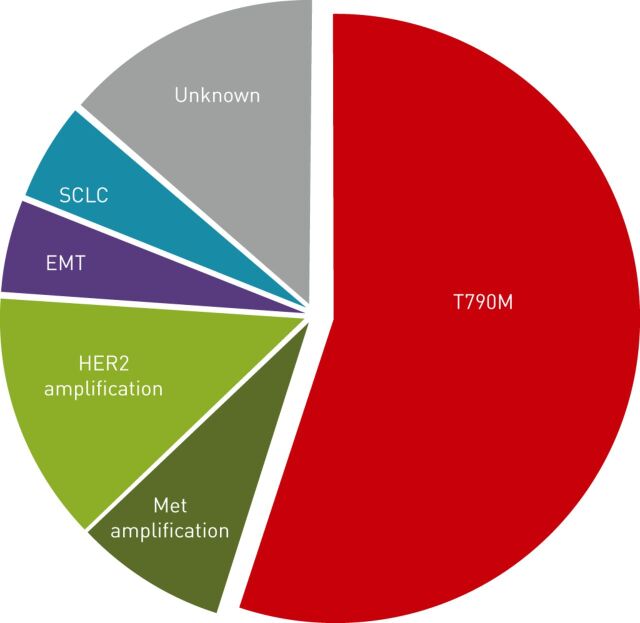
Sequist et al. [43] reported a series of EGFR-mutant NSCLC patients who had undergone a rebiopsy following progression on EGFR TKI. Among 37 patients, repeat tissue biopsies were obtained by percutaneous biopsy in most cases (68%) or by bronchoscopy, mediastinoscopy or surgery [43]. Arcila et al. [53] obtained 153 tumour samples from 121 patients, of which 82% could be analysed. Biopsies, fine-needle aspirations and pleural fluids had the highest success rates [53]. More recently, Yu et al. [50] reported the results of rebiopsy in 155 patients after the development of acquired resistance to gefitinib or erlotinib. Post-treatment tumour samples were mainly lung biopsies (core biopsies or fine-needle aspiration), pleural effusions, liver biopsies (core biopsies or fine-needle aspiration) and brain metastases resections. Only one serious adverse event occurred (pneumothorax requiring chest tube placement) [50]. In these three large series, the initial EGFR mutation was found in nearly all samples. The main mechanisms of acquired resistance to EGFR TKI that were identified could be classified as follows: 1) the T790M mutation; 2) alternative pathway activation; and 3) phenotypic transformation (fig. 1).
Figure 1.
Main mechanisms of acquired resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors in EGFR-mutant nonsmall cell lung cancer. SCLC: small cell lung cancer; EMT: epithelial–mesenchymal transition.
The T790M mutation
In analogy with the T315I gatekeeper mutation of Bcr-Abl in chronic myeloid leukaemia, researchers investigated whether the same C to T single nucleotide change, resulting in a T790M substitution at the gatekeeper residue of EGFR, could confer resistance to an EGFR TKI, namely gefitinib, and showed that this was the case [54]. Two groups of investigators further confirmed that the T790M mutation is detected in patients who develop acquired resistance to EGFR TKI treatment [55, 56]. As already mentioned, the EGFR T790M mutation occurs in cis with the primary activating mutation, and is found in 50–63% of patients with acquired resistance to EGFR TKI treatment [43, 50]. Introduction of a T790M mutation in a gefitinib-sensitive cell line confers resistance to gefitinib, which is mediated by maintenance of PI3K/Akt activation in the presence of gefitinib [57]. The T790M mutation accounts for >90% of secondary EGFR mutations. So far, three other resistance mutations have been described, D761Y, L747S and T854A, all of which have a very low prevalence and their clinical relevance remains to be determined [25, 58, 59].
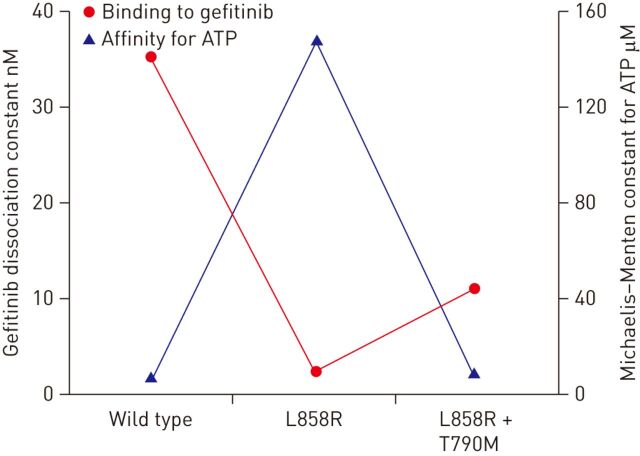
The substitution of a bulky methionine residue for threonine at the gatekeeper residue in position 790 in the kinase domain of EGFR was initially thought to cause resistance by steric interference with binding of TKIs, as seen with the T315I mutation in Bcr-Abl, since the gatekeeper residue plays a crucial role in controlling binding to the ATP pocket. However, contrary to the T315I mutant, the T790M mutant kinase remains sensitive to “irreversible” inhibitors, which does not support the hypothesis of steric hindrance as the mechanism of resistance, as if this were the case it should affect binding of both reversible and irreversible anilinoquinazoline inhibitors [60]. Indeed, Yun et al. [60] used a direct binding assay to show that the T790M mutation only modestly affects binding of gefitinib in the context of the L858R mutant, which cannot explain the clinically observed drug resistance. Subsequently, the T790M mutation was found to confer a marked decrease in the Michaelis–Menten constant (Km) for ATP (which is inversely proportional to ATP affinity) (fig. 2). The L858R mutant is known to activate EGFR, but it also reduces the apparent affinity for ATP. Strikingly, the T790M mutation restores the ATP affinity to near wild-type levels in the L858R/T790M double mutant (Km for ATP of 8.4 μM, as compared with 148 μM for the L858R mutant) (fig. 2). Thus, as highlighted by Yun et al. [60], the L858R mutation opens a therapeutic window by decreasing the affinity of the kinase for ATP, and the addition of the T790M mutation merely closes this therapeutic window by restoring ATP affinity to its baseline level.
Figure 2.
Gefitinib dissociation constants and Michaelis–Menten constants for the wild type and mutant epidermal growth factor receptor kinases. Data from [60].
Although it was initially reported that the kinase activity of the EGFR T790M mutant was indistinguishable from wild-type EGFR, Vikis et al. [61] further showed that the T790M mutation alone is responsible for increased phosphorylation levels of EGFR and a growth advantage with respect to wild-type EGFR. However, TKI-resistant cells, harbouring a double mutation including T790M, show slower growth than TKI-sensitive cells that don’t have a T790M mutation [62]. Moreover, Oxnard et al. [31] showed that among patients with acquired resistance to EGFR TKI, the presence of T790M mutation has been found to be associated with a more favourable prognosis, with a median post-progression survival of 19 months and 12 months for patients with and without T790M, respectively (p=0.036). These findings were further confirmed in several independent series [63, 64].
Activation of alternative pathways
Another frequent mechanism of resistance to EGFR TKI is activation of growth and survival pathways. The most studied example of this kind of resistance is c-Met amplification. c-Met is a transmembrane tyrosine kinase receptor, as is EGFR, which induces activation of PI3K/Akt and MAPK pathways following binding of its ligand, hepatocyte growth factor (HGF). In vitro, c-Met amplification confers resistance to EGFR TKIs in EGFR-mutant NSCLC cell lines through phosphorylation of ErbB3, resulting in activation of the PI3K/Akt pathway even in the presence of EGFR TKIs [51]. Inhibition of proliferation cannot be achieved with Met inhibitors alone, but requires inhibition of both EGFR and Met. Whether c-Met amplification induces other properties in these cells, aside from maintaining proliferation in the presence of EGFR TKI, remains undetermined. c-Met amplification was detected in 5–22% of tumour samples from patients who had progressive disease following EGFR TKI [43, 50, 51]. Variations in the frequency of c-Met amplification may be due to different definitions and assays used for the detection of c-Met amplification. The bona fide method for detection of true c-Met amplification is fluorescence in situ hybridisation (FISH), with a c-Met/CEP7 fluorescent signal ratio of five or more, rather than only the gene copy number. HGF has also been found to promote resistance to various targeted therapies in oncogene-addicted cell lines, especially EGFR TKIs [65]. In vitro, HGF induces resistance through Gab-1 mediated activation of the PI3K/Akt pathway without recruitment of ErbB3 [66, 67]. HGF can also favour c-Met amplification [67]. High levels of HGF in the tumour have been detected in 61% of EGFR-mutant Japanese patients with progressive disease following EGFR TKI [68]. The concentrations of HGF were found to be higher in the serum of patients with progressive disease following EGFR TKI, than among those without progressive disease, and high HGF levels were significantly associated with poor PFS and overall survival in patients treated with EGFR TKI [69].
Activation of other tyrosine kinase receptors can mediate resistance to EGFR TKIs. HER2 seems to play a key role in EGFR-mutant cells. Takezawa et al. [70] recently showed that overexpression of HER2 could confer resistance to afatinib, an EGFR TKI that inhibits both EGFR and HER2 kinases, in all EGFR-mutant cell lines tested. Conversely, knockdown of HER2 enhanced sensitivity to afatinib [70]. HER2 was shown to heterodimerise with mutant EGFR harbouring both L858R and T790M mutations. The authors further observed that HER2 amplification, assessed by FISH analysis, was detected in three (12%) out of 26 tumour samples from patients with EGFR-mutant NSCLC and acquired resistance to gefitinib or erlotinib. None of these samples had a concurrent T790M mutation. Moreover, HER2 amplification was found in only 1% of pre-treatment tumour samples, suggesting that this molecular alteration plays a role in acquisition of resistance to TKIs in a subset of patients [70]. Yu et al. [50] found a similar HER2 amplification frequency of 13% in patients with progressive disease following EGFR TKI treatment, with a case of HER2 amplification and T790M mutation found in the same patient. Interestingly, it has been shown that a combination of afatinib and cetuximab, a monoclonal antibody against EGFR, can overcome acquired resistance to EGFR TKI in murine models and human patients, independent of the presence of a T790M mutation [71].
Rare mutations occurring in signalling pathways downstream of EGFR have also been reported at a very low frequency, including PIK3CA and BRAF mutations [43, 72].
Phenotypic transformation
Phenotypic transformation encompasses two mechanisms of resistance to EGFR TKI: transformation to small cell lung cancer (SCLC) and epithelial–mesenchymal transition (EMT).
Transformation of EGFR-mutant NSCLC, mostly adenocarcinoma, to SCLC has been reported in 2–14% of patients following acquired resistance to EGFR TKI [43, 50, 53]. In the reported cases, no evidence for pre-existing SCLC contingent in the pre-treatment tumour was found, although all pre-treatment samples were core biopsy which cannot give a complete view of tumour heterogeneity and, therefore, could not exclude the clonal selection of the SCLC component of a composite tumour, rather than a phenotypic “transformation” at the individual cell level. Interestingly, these new SCLC still harboured the initial EGFR mutation and treatment with SCLC chemotherapy was effective. The mechanisms leading to SCLC transformation are unknown.
EMT is a process in which cells undergo a switch from an epithelial phenotype to a mesenchymal phenotype, characterised by loss of tight junctions leading to high motility and increased invasion capacity. Detection of EMT usually relies on immunohistochemistry staining, with the typical phenotype showing loss of E-cadherin and β-catenin, and gain of vimentin and N-cadherin. EMT induction can be triggered by various signals, including release of cytokines such as transforming growth factor (TGF)-β or activation of growth factor receptors. The mechanisms leading to EMT in TKI treated patients are not fully understood and may involve NOTCH-1 upregulation, TGF-β overexpression and/or Met activation [73–75]. Activation of the tyrosine kinase receptor AXL, through its overexpression or upregulation of its ligand GAS6, has also been proposed as a mechanism leading to EMT. In pre-clinical models, inhibition of AXL restored sensitivity to EGFR TKI. Interestingly, AXL overexpression has been detected in 20% of tumour samples from patients with progressive disease upon treatment with EGFR TKI [76].
Origins of acquired resistance
Resistance mutations may arise via two different mechanisms: either the mutation is acquired through selective pressure during treatment, or it exists before treatment and grows in prevalence owing to the selective pressure of treatment.
In NSCLC, it remains unclear how tumour cells harbouring the T790M mutation arise in patients receiving EGFR TKI. As mentioned previously, the T790M mutation can be found in pre-treatment samples with highly variable frequencies, ranging from <1–65%, depending on the techniques used to detect the resistance mutation. Using the SARMS (scorpion amplification refractory mutation system) technology (DxS, Manchester, UK), Maheswaran et al. [77] were among the first to detect low levels of T790M in pre-treatment samples from 10 (38%) out of 26 patients, in addition to the known activating EGFR mutation. The high number of amplification cycles needed to detect T790M suggests that the mutation was present in a small subset of cells. Interestingly, although the presence of the T790M mutation didn’t affect the response rate, PFS was significantly shorter in these patients compared with those without the T790M mutation (7.7 versus 16.5 months, hazard ratio for progression for the T790M allele 11.5, 95% CI 2.94–45.1; p<0.001) [77]. Thus, T790M may initially arise by virtue of its oncogenicity and may rapidly emerge as a dominant allele after treatment. c-Met amplification has also been observed in pre-TKI treatment samples from EGFR-mutant NSCLC patients. Turke et al. [67] found that resistance through c-Met amplification could be triggered by transient exposure of cells to HGF.
Heterogeneity of mechanisms of resistance
Whereas EGFR mutations are homogeneous among tumour sites and over time, mechanisms of resistance are more heterogeneous. Multiple mechanisms of resistance can be detected in a single tumour sample, and multiple mechanisms of resistance can arise from different tumour sites within the same patient, suggesting the polyclonal nature of resistance. As an example, analysis of various progressive lesions in an autopsy case from an individual with acquired resistance to EGFR TKI revealed that the seven different sites all harboured the exon 19 deletion, whereas the T790M mutation was found in six out of the seven sites, but was not detected in the brain metastasis, despite the use of highly sensitive methods [58]. As mentioned previously, Yu et al. [50] reported the case of a patient with a T790M mutation in one site and a HER2 amplification in another site. Moreover, c-Met amplification has been found to occur frequently with T790M mutations [59]. These results suggest that the selection pressure for resistant tumour cells that grow in the presence of EGFR TKI may be different among the different tumour sites. Furthermore, this may explain the cause of the “flare-up” effect observed when EGFR TKIs are withdrawn. Mechanisms of resistance can also vary over time: following chemotherapy, the T790M mutation has become undetectable in some patients [43]. Whether the same phenomenon will be observed with T790M-specific inhibitors will be of particular interest for monitoring efficacy of these drugs.
Resistance to new generation EGFR inhibitors
Second-generation EGFR TKIs have been developed in order to overcome acquired resistance to first-generation TKIs, such as gefitinib and erlotinib. Irreversible EGFR TKIs such as neratinib, afatinib and dacomitinib bind covalently to EGFR. In vitro, irreversible TKIs are able to decrease EGFR and PI3K/Akt activation, as well as cell proliferation in EGFR-mutant cell lines harbouring a T790M mutation [78, 79]. However, clinical efficacy of these irreversible TKIs has been disappointing, with an objective response rate of <10% in patients who had progressive disease following gefitinib or erlotinib treatment [80]. This may be due to a narrow therapeutic index, resulting in low TKI concentrations in patients’ blood. At lower concentrations, i.e. those achievable in patients, irreversible TKIs are not able to inhibit EGFR-T790M anymore, which subsequently mediates resistance [81]. At higher concentrations, T790M is still implicated in resistance through an amplification of the T790M allele [82]. Indeed, patients treated with irreversible TKIs develop resistance through acquisition of a T790M mutation [71]. Resistance to the combination of afatinib and cetuximab involves activation of mTORC1, which can be reversed using mTOR inhibitors such as rapamycin [83].
A new third-generation of EGFR inhibitors effective against T790M has recently emerged. These inhibitors, for example WZ4002, AZD9291 or CO-1686, have been specifically designed to inhibit EGFR-T790M and are different to all previous inhibitors from the biochemical point of view. In vitro, they are able to inhibit EGFR harbouring an activating mutation with or without T790M [84, 85]. Interestingly, they don’t inhibit wild-type EGFR, which should result into decreased toxicity compared with previous TKIs. AZD9291 and CO-1686 are currently in clinical development. It is remarkable that pre-clinical models have shown that first-line use of such third-generation TKIs, in contrast with first-generation TKIs, totally avoids the occurrence of T790M resistant clones, warranting studies in the first-line setting for confirmation [86]. Despite these promising preliminary results, it is anticipated that most, if not all, patients would ultimately develop progressive disease even with these drugs. The molecular mechanisms that may arise in tumours exposed to T790M-effective TKIs have only been studied in pre-clinical models so far. The mechanisms include bypass signalling, especially through activation of the IGF1R pathway, activation of the MAPK pathway and may also involve EMT [85–87]. Whether these mechanisms of resistance will be identified in tumour samples from patients treated with these new generation TKIs remains to be studied.
Conclusion
Kinase inhibitors are effective clinical therapies for several human malignancies and represent a new paradigm of treatment for EGFR-mutated NSCLC. However, their clinical efficacy is ultimately limited by the development of drug resistance. Recent studies have identified resistance mechanisms to kinase inhibitors and used these findings to develop more effective therapeutic approaches. By inhibiting a specific kinase that plays a crucial role in the cancer cell, kinase inhibitors induce a strong selective pressure on their target for emergence of resistance mechanisms. Recent studies have offered new insights into the precise mechanisms implicated in resistance, such as the finding that the T790M mutation in EGFR confers resistance through a change in ATP affinity rather than because of steric hindrance. Such findings allow development of a new generation of inhibitors. However, it is expected that resistance will also occur with these inhibitors. As new kinase inhibitors enter the clinical area, it will be necessary to prospectively study drug resistance to these agents. The continued interaction between drug development, basic biology and translational studies will make new therapeutic approaches emerge, such as first-line combinations of specific inhibitors targeting both EGFR activation mutations and oncoproteins involved in cell resistance (c-Met, IGF1R, HER2, etc.), defining “highly active antitumour therapy” [88] that will further improve clinical outcomes for these patients.
Footnotes
Previous articles in this series: No. 1: Girard N. Thymic epithelial tumours: from basic principles to individualised treatment strategies. Eur Respir Rev 2013; 22: 75–87. No. 2: Dooms C, Muylle I, Yserbyt J, et al. Endobronchial ultrasound in the management of nonsmall cell lung cancer. Eur Respir Rev 2013; 22: 169–177. No. 3: Lang-Lazdunski L. Surgery for nonsmall cell lung cancer. Eur Respir Rev 2013; 22: 382–404. No. 4: Bergot E, Levallet G, Campbell K, et al. Predictive biomarkers in patients with resected non-small cell lung cancer treated with perioperative chemotherapy. Eur Respir Rev 2013; 22: 565–576.
Provenance: Submitted article, peer reviewed.
Conflict of interest: Disclosures can be found alongside the online version of this article at err.ersjournals.com
References
- 1.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer 2006; 118: 257–262. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein IB. Cancer. Addiction to oncogenes – the Achilles heal of cancer. Science 2002; 297: 63–64. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947–957. [DOI] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010; 11: 121–128. [DOI] [PubMed] [Google Scholar]
- 7.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–2388. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012; 13: 239–246. [DOI] [PubMed] [Google Scholar]
- 9.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–742. [DOI] [PubMed] [Google Scholar]
- 10.Sequist LV, Yang JC-H, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013; 31: 3327–3334. [DOI] [PubMed] [Google Scholar]
- 11.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014; 15: 213–222. [DOI] [PubMed] [Google Scholar]
- 12.Yang JC-H, Hirsh V, Schuler M, et al. Symptom control and quality of life in LUX-Lung 3: a phase III study of afatinib or cisplatin/pemetrexed in patients with advanced lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013; 31: 3342–3350. [DOI] [PubMed] [Google Scholar]
- 13.Besse B, Adjei A, Baas P, et al. 2nd ESMO Consensus Conference on Lung Cancer: non-small-cell lung cancer first-line/second and further lines of treatment in advanced disease. Ann Oncol 2014 [In press DOI: 10.1093/annonc/mdu123]. [DOI] [PubMed] [Google Scholar]
- 14.Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw 2013; 11: 645–653. [DOI] [PubMed] [Google Scholar]
- 15.Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol 2011; 29: 2121–2127. [DOI] [PubMed] [Google Scholar]
- 16.Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010; 28: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pluquet E, Cadranel J, Legendre A, et al. Osteoblastic reaction in non-small cell lung carcinoma and its association to epidermal growth factor receptor tyrosine kinase inhibitors response and prolonged survival. J Thorac Oncol 2010; 5: 491–496. [DOI] [PubMed] [Google Scholar]
- 18.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361: 958–967. [DOI] [PubMed] [Google Scholar]
- 19.Alfieri RR, Galetti M, Tramonti S, et al. Metabolism of the EGFR tyrosin kinase inhibitor gefitinib by cytochrome P450 1A1 enzyme in EGFR-wild type non small cell lung cancer cell lines. Mol Cancer 2011; 10: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filosto S, Khan EM, Tognon E, et al. EGF receptor exposed to oxidative stress acquires abnormal phosphorylation and aberrant activated conformation that impairs canonical dimerization. PLoS One 2011; 6: e23240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filosto S, Becker CR, Goldkorn T. Cigarette smoke induces aberrant EGF receptor activation that mediates lung cancer development and resistance to tyrosine kinase inhibitors. Mol Cancer Ther 2012; 11: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filosto S, Baston DS, Chung S, et al. Src mediates cigarette smoke-induced resistance to tyrosine kinase inhibitors in NSCLC cells. Mol Cancer Ther 2013; 12: 1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006; 12: 839–844. [DOI] [PubMed] [Google Scholar]
- 24.Lee JK, Shin JY, Kim S, et al. Primary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: an exploratory study. Ann Oncol 2013; 24: 2080–2087. [DOI] [PubMed] [Google Scholar]
- 25.Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 2007; 4: 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cragg MS, Kuroda J, Puthalakath H, et al. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med 2007; 4: 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong Y, Somwar R, Politi K, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med 2007; 4: e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa C, Molina MA, Drozdowskyj A, et al. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin Cancer Res 2014; 20: 2001–2010. [DOI] [PubMed] [Google Scholar]
- 29.Ng KP, Hillmer AM, Chuah CTH, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med 2012; 18: 521–528. [DOI] [PubMed] [Google Scholar]
- 30.Beau-Faller M, Prim N, Ruppert AM, et al. Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Ann Oncol 2014; 25: 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oxnard GR, Lo PC, Nishino M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 2013; 8: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 2013; 12: 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 2013; 5: 216ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girard N, Lou E, Azzoli CG, et al. Analysis of genetic variants in never-smokers with lung cancer facilitated by an internet-based blood collection protocol: a preliminary report. Clin Cancer Res 2010; 16: 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005; 37: 1315–1316. [DOI] [PubMed] [Google Scholar]
- 36.Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 2011; 17: 1160–1168. [DOI] [PubMed] [Google Scholar]
- 37.Yu HA, Arcila ME, Hellmann MD, et al. Poor response to erlotinib in patients with tumours containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol 2014; 25: 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su K-Y, Chen H-Y, Li K-C, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol 2012; 30: 433–440. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto C, Basaki Y, Kawahara A, et al. Loss of PTEN expression by blocking nuclear translocation of EGR1 in gefitinib-resistant lung cancer cells harboring epidermal growth factor receptor-activating mutations. Cancer Res 2010; 70: 8715–8725. [DOI] [PubMed] [Google Scholar]
- 40.Sos ML, Koker M, Weir BA, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res 2009; 69: 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vivanco I, Rohle D, Versele M, et al. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci USA 2010; 107: 6459–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer 2006; 54: 209–215. [DOI] [PubMed] [Google Scholar]
- 43.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 2006; 116: 2695–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Hu H, Pan Y, et al. PIK3CA mutations frequently coexist with EGFR/KRAS mutations in non-small cell lung cancer and suggest poor prognosis in EGFR/KRAS wildtype subgroup. PLoS One 2014; 9: e88291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010; 141: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bivona TG, Hieronymus H, Parker J, et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature 2011; 471: 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev 2008; 18: 73–79. [DOI] [PubMed] [Google Scholar]
- 49.Jänne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov 2009; 8: 709–723. [DOI] [PubMed] [Google Scholar]
- 50.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumour specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013; 19: 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: 1039–1043. [DOI] [PubMed] [Google Scholar]
- 52.Ogino A, Kitao H, Hirano S, et al. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res 2007; 67: 7807–7814. [DOI] [PubMed] [Google Scholar]
- 53.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011; 17: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blencke S, Ullrich A, Daub H. Mutation of threonine 766 in the epidermal growth factor receptor reveals a hotspot for resistance formation against selective tyrosine kinase inhibitors. J Biol Chem 2003; 278: 15435–15440. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 352: 786–792. [DOI] [PubMed] [Google Scholar]
- 56.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005; 2: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosaka T, Yatabe Y, Endoh H, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res 2006; 12: 5764–5769. [DOI] [PubMed] [Google Scholar]
- 58.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res 2006; 12: 6494–6501. [DOI] [PubMed] [Google Scholar]
- 59.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumours with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA 2007; 104: 20932–20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yun C-H, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA 2008; 105: 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vikis H, Sato M, James M, et al. EGFR-T790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Res 2007; 67: 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med 2011; 3: 90ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuiper JL, Heideman DA, Thunnissen E, et al. Incidence of T790M mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients. Lung Cancer 2014; 85: 19–24. [DOI] [PubMed] [Google Scholar]
- 64.Li W, Ren S, Li J, et al. T790M mutation is associated with better efficacy of treatment beyond progression with EGFR-TKI in advanced NSCLC patients. Lung Cancer 2014; 84: 295–300. [DOI] [PubMed] [Google Scholar]
- 65.Wilson TR, Fridlyand J, Yan Y, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012; 487: 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res 2008; 68: 9479–9487. [DOI] [PubMed] [Google Scholar]
- 67.Turke AB, Zejnullahu K, Wu Y-L, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010; 17: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yano S, Yamada T, Takeuchi S, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol 2011; 6: 2011–2017. [DOI] [PubMed] [Google Scholar]
- 69.Kasahara K, Arao T, Sakai K, et al. Impact of serum hepatocyte growth factor on treatment response to epidermal growth factor receptor tyrosine kinase inhibitors in patients with non-small cell lung adenocarcinoma. Clin Cancer Res 2010; 16: 4616–4624. [DOI] [PubMed] [Google Scholar]
- 70.Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012; 2: 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Regales L, Gong Y, Shen R, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest 2009; 119: 3000–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci USA 2012; 109: E2127–E2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suda K, Tomizawa K, Fujii M, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol 2011; 6: 1152–1161. [DOI] [PubMed] [Google Scholar]
- 74.Xie M, Zhang L, He CS, et al. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem 2012; 113: 1501–1513. [DOI] [PubMed] [Google Scholar]
- 75.La Monica S, Caffarra C, Saccani F, et al. Gefitinib inhibits invasive phenotype and epithelial-mesenchymal transition in drug-resistant NSCLC cells with MET amplification. PLoS One 2013; 8: e78656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012; 44: 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008; 359: 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Engelman JA, Zejnullahu K, Gale CM, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007; 67: 11924–11932. [DOI] [PubMed] [Google Scholar]
- 79.Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 2008; 27: 4702–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reckamp KL, Giaccone G, Camidge DR, et al. A phase 2 trial of dacomitinib (PF-00299804), an oral, irreversible pan-HER (human epidermal growth factor receptor) inhibitor, in patients with advanced non-small cell lung cancer after failure of prior chemotherapy and erlotinib. Cancer 2014; 120: 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Godin-Heymann N, Ulkus L, Brannigan BW, et al. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther 2008; 7: 874–879. [DOI] [PubMed] [Google Scholar]
- 82.Ercan D, Zejnullahu K, Yonesaka K, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene 2010; 29: 2346–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pirazzoli V, Nebhan C, Song X, et al. Acquired resistance of EGFR-mutant lung adenocarcinomas to afatinib plus cetuximab is associated with activation of mTORC1. Cell Rep 2014; 7: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009; 462: 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walter AO, Sjin RTT, Haringsma HJ, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov 2013; 3: 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cortot AB, Repellin CE, Shimamura T, et al. Resistance to irreversible EGF receptor tyrosine kinase inhibitors through a multistep mechanism involving the IGF1R pathway. Cancer Res 2013; 73: 834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ercan D, Xu C, Yanagita M, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov 2012; 2: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chmielecki J, Pao W. Highly active antitumor therapy (HAATT) for epidermal growth factor-mutant lung cancer. Clin Cancer Res 2010; 16: 5371–5373. [DOI] [PubMed] [Google Scholar]