Abstract
We employed two separate genetic approaches to examine the roles of various OmpF residues in assembly. In one approach, intragenic suppressors of a temperature-sensitive OmpF assembly mutant carrying a W214E substitution were sought at 42°C, or at 37°C in a genetic background lacking the periplasmic folding factor SurA. In the majority of cases (58 out of 61 revertants), the suppressors mapped either at the original site (position 214) or two residues downstream from it. In the remaining three revertants that were obtained in a surA background, an alteration of N230Y was located 16 residues away from the original site. The N230Y suppressor also corrected OmpF315 assembly at 42°C in a surA+ background, indicating that the two different physiological environments imposed similar assembly constraints. The specificity of N230Y was tested against five different residues at position 214 of mature OmpF. Clear specificity was displayed, with maximum suppression observed for the original substitution at position 214 (E214) against which the N230Y suppressor was isolated, and no negative effect on OmpF assembly was noted when the wild-type W214 residue was present. The mechanism of suppression may involve compensation for a specific conformational defect. The second approach involved the application of informational suppressors (Su-tRNA) in combination with ompF amber mutations to generate variant OmpF proteins. In this approach we targeted the Y40, Q66, W214, and Y231 residues of mature OmpF and replaced them with S, Q, L, and Y through the action of Su-tRNAs. Thus, a total of 16 variant OmpF proteins were generated, of which three were identical to the parental protein, and two variants carrying W214Q and Y231Q substitutions were similar to assembly-defective proteins isolated previously (R. Misra, J. Bacteriol. 175:5049–5056, 1993). The results obtained from these analyses provided useful information regarding the compatibility of various alterations in OmpF assembly.
The assembly and targeting of outer membrane proteins (OMPs) in gram-negative bacteria are complex biological processes (for reviews, see references 1 and 5). Part of this complexity originates from the fact that during their biogenesis, OMPs have to interact transiently or permanently with the biochemically distinct environments of the inner membrane, periplasm, and outer membrane. Naturally, compositional changes in any of these three compartments can potentially influence the biogenesis of OMPs. Accordingly, inner membrane components influence protein translocation (for a review, see reference 21), periplasmic components influence the early stages of OMP assembly (2, 14, 19, 23), and outer membrane components such as lipopolysaccharide (LPS) can affect the later stages of OMP assembly (6, 8, 11, 12, 13, 22, 24).
The structural features of assembling molecules, which are governed by their primary sequences, also influence their ability to be correctly targeted and assembled. Thus, alterations in the signal sequence can cause defects in translocation across the inner membrane (1, 21), whereas changes in the mature portion can affect folding and assembly (15, 16, 18, 26, 27). An impressive collection of signal sequence mutants have been obtained, permitting examination of the role of almost every single residue of the signal sequence during protein translocation (9). Such an analysis has not been attempted for the mature region of OMPs because there is no distinct signal sequence-like linear stretch of residues that constitute an outer membrane-sorting signal. It has been suggested that the conserved carboxy-terminal phenylalanine residue of many OMPs may be a part of the outer membrane-sorting signal (4, 16, 26). Indeed, alterations of this residue affect assembly, but this is likely due to misfolding of early assembly intermediates, which are then diverted to a degradation pathway (16). However, it is unlikely that the terminal phenylalanine residue of OMPs or even the last β-strand contains all the necessary information for targeting and assembly.
If the critical information for outer membrane targeting and assembly lies within the folded structure, then we must identify residues or regions of mature OMPs that influence folding. Since targeting and assembly are dynamic cellular processes, knowing the atomic structure of a fully assembled protein may not be immediately useful in allowing us to see the role of a residue during these processes. This has been the case with OmpF, for which structural resolution (3) failed to provide meaningful clues as to what residues are critical for targeting and assembly.
Influenced by an elegant piece of work carried out in Jon King's laboratory on the assembly of the P22 tail spike protein (10), we devised a novel genetic scheme which led to the isolation of OmpF mutants with a conditional (temperature-sensitive) assembly defect (15). Reversion analysis of these temperature-sensitive assembly mutants resulted in extragenic suppressor mutations in asm genes, whose products influence the outer membrane lipid environment (6, 11, 12, 17). In this study, we focused our attention on intragenic suppressor mutations in hopes of identifying additional residues within OmpF that play a role during assembly. In addition, we utilized informational suppressors in combination with various ompF amber mutations to effectively carry out in vivo site-directed mutagenesis. A combination of these two genetic approaches allowed us to investigate the role of various OmpF residues in assembly.
MATERIALS AND METHODS
Bacterial strains, genetic manipulations, media, and biochemicals.
All strains were of MC4100 descent [F− araD139 Δ(argF-lac)U139 rpsL150 flbB5301 ptsF25 deoC1 thi-1 rbsR relA]. RAM472 (MC4100 ΔlamB106 ompF205) was used as the maltodextrin-positive (Dex+) parental strain from which ompF amber mutations were isolated as described previously (15). RAM496 (MC4100 ΔlamB106 ΦompC′-lacZ+ ompF315) expressed a variant OmpF that shows a temperature-sensitive assembly defect (15). Thus, RAM496 displayed Dex+ and Dex− phenotypes at 30 and 42°C, respectively. RAM496 was utilized in the isolation of intragenic ompF revertants. Strains containing various suppressor tRNA (Su-tRNA) alleles were obtained from Promega Biochemicals. P1 transductions were performed by the method of Silhavy et al. (25). Minimal medium (M63 salts based) and Luria broth were prepared as described by Silhavy et al. (25). [35S]methionine-cysteine was purchased from Du Pont-New England Nuclear. Heat-killed Staphylococcus aureus cells (Pansorbin) were purchased from Calbiochem. The DNA mutagen 1-methyl-3-nitro-1-nitrosoguanidine (NTG) was purchased from Sigma. Other chemicals were of analytical grade.
Site-directed mutagenesis and cloning of recombinant ompF genes.
Site-directed mutagenesis of ompF to alter codon 214 was carried out using GeneEditor in vitro site-directed mutagenesis system from Promega Biochemicals. The mutagenic primer (5′-GCTGAACAGNNNGCTACTGGTCTGAAG-3′) was used to randomize codon 214 (NNN) of ompF.
Various recombinant ompF genes with or without the suppressor mutation affecting residue 230 of OmpF were constructed in two steps. In the first step, the 3′ end of the gene corresponding to residues 224 to 330 of mature OmpF was cloned into pTrc99A (Pharmacia). To do this, ompF DNA from strains with or without the suppressor alteration was amplified by PCR using primers complementary to the middle (5′-GTACGACGCGAATTCCATCTACCTGG-3′) and end (5′-GACGAGGATCCATTATGGTTACAGAAGG-3′) of the gene. Restriction sites for EcoRI (middle of the gene in italics) and BamHI (end of the gene in italics) were incorporated for cloning into a pTrc99A vector plasmid. The resulting clones contained either a wild-type residue (N230) or a suppressor alteration (N230Y). Subsequently, the 5′ ends of ompF genes, corresponding to residues 1 to 223 of mature OmpF from parental (W214) strains or mutant strains carrying a W214A, W214D, W214E, or W214T alteration, were amplified by PCR using primers complementary to the start (5′-GAGGGTAATAAACCATGGTGAAGCGCAATATTCTGGCAGTG-3′) or middle (5′-CCAGGTAGATGGAATTCGCGTCGTAC-3′) of the ompF gene. Restriction sites for NcoI (start of the gene in italics) and EcoRI (middle of the gene in italics) were used for cloning into the 3′-end ompF recombinants generated in the first step. The introduction of an EcoRI site in the middle of the ompF gene created a neutral N224S alteration in the periplasmic loop of OmpF that had no effect on the protein's biogenesis. An isopropyl β-d-thiogalactopyranoside (IPTG)-inducible promoter controlled the expression of the recombinant ompF genes.
Protein methods.
Radioactive labeling of proteins, immunoprecipitation reactions, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis were carried out as described previously (15). Whole-cell envelopes were isolated by the French press lysis procedure (15) and were treated with sodium sarcosyl (1% final concentration) at room temperature for 30 min. A sarcosyl-insoluble pellet was obtained by centrifuging samples at 110,000 × g for 30 min at 4°C in a tabletop ultracentrifuge. Western blot analysis of OmpF was carried out as described by Misra et al. (16).
RESULTS
Intragenic suppressors of an OmpF assembly mutant.
The isolation of OmpF assembly mutants in which a conditional (temperature sensitive) folding defect within an assembly intermediate prevented the formation of stable OmpF trimers has been described previously (15). Cells that failed to accumulate sufficient amounts of stable OmpF trimers in the outer membrane were unable to grow on a medium with maltodextrins as the sole carbon source and hence expressed a Dex− phenotype. In this genetic background, due to a mutation in the lamB gene, an OmpF variant is the sole means of maltodextrin entry (15). By utilizing a severely defective assembly mutant, OmpF315, suppressor mutations conferring a Dex+ phenotype were isolated. Further examination revealed that all the suppressor mutations mapped outside the ompF gene (11, 12, 15, 17). A factor contributing to our inability to isolate intragenic suppressor mutations could have been the availability of a large number of potential chromosomal targets other than ompF that can be mutated to yield a Dex+ phenotype. In this study, we sought suppressor mutations offsetting the original assembly defect that map within the ompF gene. To enrich the desired class of intragenic suppressor mutations, localized NTG-induced mutagenesis was carried out.
Nine independent cultures of cells expressing an assembly-defective OmpF315 protein were mutagenized with NTG, plated on maltodextrin minimal medium, and incubated at the nonpermissive temperature of 42°C for 48 h. To enrich Dex+ revertants over Dex− background cells, Dex+ colonies were replica plated onto maltodextrin minimal medium and incubated for an additional 8 h at 42°C. After the second incubation period, no background Dex− lawn was visible on the plates. Dex+ revertants from each plate were then pooled by rinsing colonies with 2 ml of M63 salts and diluted into Luria broth from which P1 lysates were prepared. This exercise created P1 lysates on nine independently mutagenized cultures that were specifically enriched for Dex+ revertants at 42°C.
ompF alleles from mutagenized cultures were moved into a mapping strain by P1 transduction utilizing a linked genetic marker, pyrD+ (approximately 50% cotransducible with ompF). The pyrD+ allele was transduced from the Dex+ mutant pools into a pyrD ΦompF′-lacZ+ (Dex−) recipient strain (RAM473) (15) by selecting for growth on glucose minimal medium. The presence or absence of ΦompF′-lacZ+ among the PyrD+ transductants was monitored by spreading a chromogenic indicator, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) on the plates. PyrD+ transductants that had lost the fusion (white colonies) due to its replacement by an ompF allele from the donor were purified and tested for their Dex+ phenotype at 30 and 42°C. Colonies that displayed a temperature-resistant Dex+ phenotype were expected to contain a mutant ompF allele in which a second (NTG-induced) mutation within ompF corrected the temperature-sensitive defect of the ompF315 allele. The results showed that all nine independently mutagenized pools provided the desired intragenic ompF mutations. Four isolates from each of the nine pooled cultures were further analyzed.
To determine the genetic alterations within ompF, nucleotide sequence analysis of PCR-amplified ompF DNA fragments was carried out. This revealed either an E214K (12 isolates) or a T216I (24 isolates) substitution within OmpF (Table 1). These two types of substitutions were found in each of the nine independently mutagenized cultures.
TABLE 1.
Suppressor alterations within OmpF315
| Genetic background | Reversion temperature (°C) | No. of isolates | Suppressor alterationa |
|---|---|---|---|
| ompF315 | 42 | 12 | E214K |
| ompF315 | 42 | 24 | T216I (W214E) |
| ompF315 surA | 37 | 6 | E214K |
| ompF315 surA | 37 | 3 | E214V |
| ompF315 surA | 37 | 13 | T216I (W214E) |
| ompF315 surA | 37 | 3 | N230Y (W214E) |
The original alteration (W214E) was maintained in some suppressors.
Isolation of suppressors in a surA background.
The above analysis showed that all 36 revertants contained a suppressor alteration either at the mutant codon 214 or two codons downstream from it. This suggested that the initial assembly defect imposed by W214E at 42°C could be corrected only by a limited number of suppressor alterations within the protein. This constraint was highlighted by the fact that all nine independently mutagenized cultures repeatedly produced the same mutations.
We reasoned that, by varying the starting genetic background, different intragenic ompF suppressor mutations could be obtained. Disruption of surA has been shown to have a profound effect on OMP assembly (14, 23). This is because SurA, which has a cis-trans peptidyl prolyl isomerase activity in the periplasm, influences the folding of OMP assembly intermediates (14, 23). Recently it has been shown that SurA can also act as a chaperone independent of its isomerase activity (Hans de Cock, personal communication). Thus, we sought revertants in a surA background. The introduction of a surA null allele in a strain producing the parental OmpF205 protein had no effect on OmpF205-mediated Dex+ phenotype. However, its introduction into a strain background expressing the mutant OmpF315 conferred a Dex− phenotype at 37°C; normally at this temperature, an OmpF315 strain displays a Dex+ phenotype in a surA+ background. Several independent Dex+ revertants were obtained as described above except that the cells were incubated at 37°C.
DNA sequence analysis of ompF from 25 mutants obtained from six independent cultures revealed four different alterations among them (Table 1). Interestingly, two alterations were identical to those obtained at 42°C. Although the third alteration also affected codon 214, it produced an E214V substitution not obtained at 42°C. The last alteration turned out to be quite unusual, as it affected residue 230 of the mature OmpF protein. Thus, exploitation of a surA null allele did indeed produce a unique suppressor.
Characterization of OmpF revertants.
Examination of envelope protein profiles showed that the revertants bearing a E214V, T216I, or N230Y suppressor substitution had OmpF levels similar to that of a strain expressing the assembly-proficient OmpF205; OmpF levels in revertants with an E214K substitution were lower than levels of OmpF205. Interestingly, N230Y, which was obtained in a surA background, also elevated OmpF315 levels at 42°C in a surA+ background.
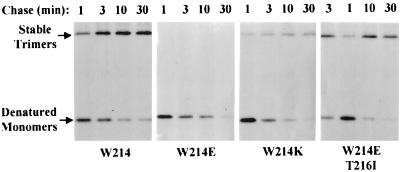
We then examined the kinetics of stable trimer formation. Pulse-chase labeling experiments were followed by immunoprecipitation of OmpF trimers with OmpF trimer-specific polyclonal antibodies. These antibodies recognize both stable and metastable trimers. The results showed that in contrast to the assembly-defective OmpF315, the revertant OmpF proteins proceeded to form stable trimers (Fig. 1). However, while revertants bearing a T216I substitution formed stable OmpF trimers with kinetics similar to that observed for the assembly- proficient Dex+ protein (OmpF205), assembly of the revertant bearing an E214K substitution was still defective, albeit better than that observed for OmpF315 (W214E). Revertants bearing an E214V or N230Y suppressor showed trimerization kinetics similar to those obtained with OmpF205 (data not shown).
FIG. 1.
OmpF trimer assays. Cells expressing various OmpF proteins were grown to mid-log phase at 42oC in glycerol minimal medium, labeled for 20 s with [35S]methionine-cysteine, and chased with an excess of nonradioactive methionine. Chase samples were removed after 1, 3, 10, and 30 min. Proteins were extracted, and OmpF was immunoprecipitated using trimer-specific antibodies. Immunoprecipitates were heated at 60°C for 15 min prior to analysis by SDS-PAGE. The gel was dried and subjected to autoradiography at −70°C. Positions of OmpF trimers (T) and denatured monomers (M) are shown. Relevant alterations in various OmpF proteins are shown below the gel. The loading of 1- and 3-min chase samples carrying OmpF with a T216I alteration was inadvertently reversed.
Suppression specificity of N230Y.
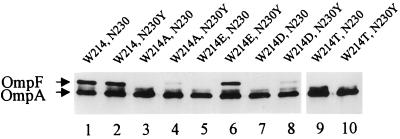
The N230Y mutation suppressed an assembly defect caused by W214E. We tested N230Y's ability to suppress other alterations at position 214 and whether its presence is detrimental for assembly when the wild-type tryptophan residue is present at position 214. Through site-directed mutagenesis, position 214 was altered to introduce A, D, and T substitutions, all of which conferred a severe assembly defect (Fig. 2). Recombinant ompF genes were constructed (see Material and Methods) so as to express 10 different OmpF proteins carrying one of five substitutions at position 214 (A, D, E, T, or W) and either N (wild type) or Y (suppressor) at position 230. The results showed that the N230Y suppressor had no effect when combined with the wild-type W214 residue (Fig. 2). Furthermore, although N230Y was able to suppress to various degrees the assembly defect caused by the replacement of W214 by A, D, or E, it had the greatest effect on the E214 mutation against which it was isolated and was unable to suppress the T214-mediated assembly defect.
FIG. 2.
SDS-PAGE analysis of sarcosyl-extracted cell envelopes obtained from strains producing either the parental OmpF205 protein (lane 1) or various mutant OmpF proteins with indicated alterations at position 214 (lanes 3 to 10). The suppressor alteration N230Y was present in proteins analyzed in even lanes.
Informational suppression of ompF amber mutations.
In the original mutant isolation strategy, various ompF(Dex) amber mutations were genetically reverted in a conditional manner, and only two of the four amber sites resulted in assembly mutants (15). In this scheme, the reversion to Dex+ at 30°C required that OmpF(Dex) be properly synthesized, translocated across the inner membrane, and inserted and assembled into functional trimers in the outer membrane. It is conceivable that this demand for functional trimers may have precluded us from examining other OmpF residues important for assembly but also involved in channel function or β-barrel stability. Thus, an approach involving informational suppression of amber mutations was exploited to overcome this limitation. In this scheme, ompF amber mutations were suppressed without demanding functionality of the suppressed protein. To achieve this, we introduced various amber suppressor tRNA (Su-tRNA) alleles into genetic backgrounds harboring different ompF(Dex) amber mutations. In all, four different ompF(Dex) amber mutations, affecting residue Y40, Q66, W214, or Y231 of mature OmpF, were suppressed with four different Su-tRNAs having the capacity to insert an S, Y, Q, or L residue at the respective amber sites, thus generating 16 different variant OmpF(Dex) proteins. It may be noted that three such combinations produced the parental protein, while two variant OmpF(Dex) proteins, bearing W214Q and Y231Q substitutions, were identical to those synthesized when amber mutations at these sites were genetically reverted (15). The Su-tRNAs chosen in this study produced efficient suppression of OmpF amber codons.
The various Su-tRNA alleles were introduced by utilizing a separate amber mutation in the argE gene. The mutant argE gene was transduced, using a linked thiA::Tn10 marker (50% cotransducible with argE), into strains harboring various ompF(Dex) amber alleles. The Su-tRNA alleles were then transduced into this background by selecting for Arg+ transductants on glucose minimal medium supplemented with thiamine. Thus, the presence of the argE amber mutation in the background permitted an unbiased introduction of the various Su-tRNAs and provided an easy means of ensuring their proper functionality.
Characteristics of tRNA-suppressed OmpFs.
Strains carrying different combinations of ompF(Dex) amber mutations and Su-tRNAs were tested for sensitivity to an OmpF-specific bacteriophage, K20, and growth on maltodextrin medium. Phage plaquing efficiencies of all 16 mutants were similar to that of a strain expressing the parental OmpF protein, showing that (i) the phage receptor activity of the mutant OmpF proteins was unaffected and (ii) sufficient levels of the variant proteins were assembled in the outer membrane under steady-state growth conditions. In contrast to the phage sensitivity phenotype, mutants showed different growth patterns on maltodextrin minimal medium (data not shown), which could be due either to alterations in structure or to the level of OmpF trimers. On glucose or glycerol minimal medium, all mutants formed similar-sized colonies, showing that the argE amber mutation was efficiently suppressed by the Su-tRNAs.
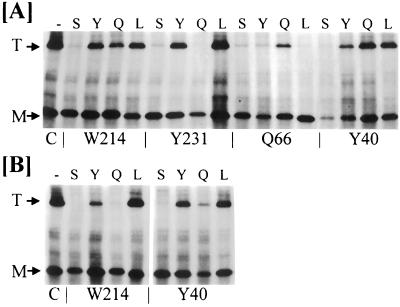
Next, we examined the biogenesis of all 16 mutant OmpF proteins (Fig. 3). By performing pulse-chase experiments, it was determined that parental OmpF stable trimers and assembly intermediates reached a roughly 1:1 ratio by 3 min postchase. The extent of mutant trimerizations at this time point was then compared with that of the parental protein. Since it was conceivable that mutant trimers dissociated into monomers at temperatures lower than that required to dissociate parental trimers (62.5°C), the temperature required to examine mutant stable trimers was determined prior to the above experiment. The optimal conversion temperature for the 16 mutants ranged from 47.5 to 62.5°C.
FIG. 3.
Trimer assays of various OmpF proteins generated through the informational suppression of amber codons located at various positions in ompF (shown at the bottom). Various amino acids introduced through the suppressor tRNAs are shown on top. Strains were grown to mid-log phase at 37°C (A) or 40°C (B) in glycerol minimal medium. Labeling and immunoprecipitation conditions were similar to those described for Fig. 1 except that only 3-min chase samples were analyzed. C, control (assembly-proficient OmpF205 protein); T, OmpF trimers; M, denatured monomers.
Suppression of various ompF(Dex) amber mutations by Su-tRNASer resulted in the poorest recovery of trimerization from all four amber sites. In contrast, leucine was found to be quite acceptable at three of the four amber sites; its insertion at position 66, replacing the wild-type glutamine residue, severely affected the accumulation of stable trimers. The insertion of tyrosine at various positions gave results that were similar to those obtained with the leucine insertion. Insertion of glutamine at all sites except Y231 was acceptable for assembly. As expected, a glutamine inserted at position 66, which was originally occupied by glutamine, was the most effective replacement. Interestingly, assembly of OmpFW214Q and OmpFY40Q, which appeared normal at 37oC, became severely defective when pulse-chase experiments were carried out at 40oC (Fig. 3B). OmpFW214Q was also obtained through an amber reversion analysis and shown to be temperature sensitive for assembly (15). Thus, informational suppressors allowed us to examine the role of various residues within OmpF in assembly when it was not possible through the Dex+ reversion analysis.
DISCUSSION
Suppression with or without requiring porin function.
We took two fundamentally different genetic approaches to examine the roles of various residues within OmpF in trimer assembly. In the first approach, we asked for the correction of an assembly defect, imposed by a W214E substitution, with a second alteration within OmpF. Because the starting strain produces an assembly-defective OmpF, all revertants are expected to possess an alteration that compensates for the assembly defect. However, since revertants were selected for their abilities to form functional OmpF channels, isolates in which a substitution simultaneously reduced channel activity were lost. In the second approach, amber codons located at various positions within ompF were suppressed by utilizing various Su-tRNAs and the resulting proteins were analyzed for assembly proficiencies. So, unlike with the previous strategy, active channels were not sought, and thus various substitutions were introduced without a functional bias.
Reversion at 42°C.
The strategy for isolating revertants at high growth temperatures revealed that the negative effect of the W214E substitution on OmpF assembly was correctable by only two alterations located at or near the original substitution. One such alteration affected residue E214 itself, changing it to a K. While in the second group of revertants, the W214E substitution was maintained but a change of T216 to I was introduced. A T216I substitution may contribute positively by increasing regional hydrophobicity within the β-strand, thus offsetting the effect of W214E. The results showed that although W214 is not absolutely required for OmpF assembly, its replacement by lysine is preferred over polar glutamate. It is worth pointing out that revertants bearing a true reversion substitution of E214W were not obtained, as this would have required two simultaneous base pair changes.
OmpF's crystal structure shows that W214 is present in the upper girdle of an aromatic ring that surrounds the β-barrel (3). Side chains of these aromatic residues are exposed to the lipid environment of the outer membrane, where they are proposed to stabilize LPS-protein interactions. It is conceivable that the side chain of W214 is important in establishing critical interactions between the porin and LPS core during metastable-to-stable trimer conversion. The W214E substitution in OmpF315 would be expected to alter this interaction. Intragenic suppressors may overcome this defect by restoring normal interactions of OmpF315 with LPS during assembly. Curiously, while both lysine and glutamate have highly polar and charged side chains, lysine is acceptable but glutamate is not. One explanation for this may be that phosphate groups in the LPS inner core electrostatically repel the negatively charged side chain of glutamate. In contrast, the positively charged lysine residue would have a stabilizing effect. Indeed, interaction between several positively charged residues of the FhuA barrel and phosphate groups of the LPS core have recently been reported (7).
Overlapping assembly constraints of temperature and surA on OmpF315 assembly.
SurA is a major folding factor whose activity in the periplasm is needed for proper OMP assembly (14, 23). The phenotypic defect of OmpF315 at 37°C in a surA background is identical to that observed at 42°C in a surA+ background. Thus, SurA's removal from the cell exaggerates the assembly defect of OmpF315 and hence permits the isolation of suppressors at 37°C. Moreover, these two physiologically different environments must produce overlapping assembly constraints because identical suppressors of OmpF315 were obtained under these conditions. It is noteworthy that the N230Y suppressor, which was obtained only at 37°C in a surA background, also corrected the assembly defect of OmpF315 at 42oC in a surA+ background. As none of the suppressors obtained affected OmpF315's proline residues, it is conceivable that it is the absence of the general chaperone function rather than the loss of the peptidyl prolyl isomerase activity of SurA that conferred an assembly defect.
Suppression specificity.
All intragenic suppressors acted by fully or partially restoring the assembly defect. The N230Y suppressor showed specificity towards alterations at position 214. In the folded OmpF molecule, residues 214 and 230 are present in two consecutive β-strands of the monomer. The side chain of W214 projects outward in the exterior lipid environment of the membrane, but N230's side chain is oriented inward in the barrel. Thus, if suppression involves a specific side chain interaction, it can occur only transiently and must occur within partially folded structures in which the two side chains have not yet assumed the opposite orientation with respect to each other. Alternatively, suppression may involve compensation for a conformational defect. As the conformation may vary depending on the nature of the side chain, the ability of N230Y to suppress will change. Regardless of the mechanism of suppression, our data showed that the residues or regions encompassing 214 and 230 of OmpF have a role in assembly. This is further substantiated by our earlier finding that changes at position 231 of OmpF can also confer an assembly defect (15).
Informational suppression.
Amber suppressor analysis provided useful information regarding the acceptability of various substitutions at particular sites. Of the four amber sites analyzed, two were located at residues 214 and 231, where assembly mutants have previously been isolated (15; also, see above). In the wild-type OmpF trimer molecule, the side chain of Y40 extends inwards within the channel, and substitutions at this position would therefore be expected to alter the interior of the channel in the folded molecule. The data presented here showed that alterations at Y40 can also influence OmpF trimerization, indicating an additional role of Y40 during OmpF biogenesis. Two quite different amino acids, Q and L, can replace Y40 at 37°C, but the Y40Q substitution was temperature sensitive for trimerization, indicating a preference for a hydrophobic or an aromatic residue at this position.
In the folded OmpF trimer, Q66 is present at the end of a surface loop, L2, with its side chain oriented inward (3). As several residues of L2 from one subunit interact with the neighboring subunit, certain substitutions in this loop are expected to affect trimer assembly and or stability (20). Consistent with this notion, we observed a severe defect in the assembly of stable trimer formation when Q66 was replaced with S, Y, or L. With the exception of S, substitutions involving Y, Q, and L at position W214 were well accommodated at 37°C, and as noted before (15), its replacement by Q at 40°C resulted in inefficient trimerization. At Y231, Y and L replacements were well tolerated but Q and S replacements affected OmpF trimerization negatively. Again, the finding that the Y231Q substitution is unacceptable for OmpF assembly substantiated our previous genetic data (15). The aromatic side chains of W214 and Y231 in wild-type OmpF trimers protrude outward towards the membrane environment. Our data clearly show that it is the hydrophobic nature of these residues rather than the aromatic rings that is crucial during assembly.
The amber suppression approach described above shows potential for use in the study of the role of a relatively large number of OmpF residues in assembly or function. A limitation may rest with our ability to isolate all the possible amber mutations within ompF. There are 42 sense codons scattered throughout the coding sequence corresponding to the mature portion of OmpF that can be converted into amber codons by single base substitutions. Of these, 11 codons represent hydrophilic residues, including Q, D, and K, whereas the remaining 30 codons specify the aromatic residues Y and W. The genetic selection for isolating amber mutations among phage-resistant mutants is relatively simple, and in our estimates, anywhere from 1 to 10% of null mutations are amber alleles. Moreover, aside from the four Su-tRNA alleles used here, additional amber suppressor tRNA alleles are available, and their combination with additional ompF amber mutations should yield a greater number of variant OmpF proteins.
ACKNOWLEDGMENTS
We are grateful to Leanne Misra for her constructive criticisms.
This work was supported by a grant (GM48167 to R.M.) from the National Institutes of Health.
REFERENCES
- 1.Baker K, Mackman N, Holland I B. Genetics and biochemistry of the assembly of proteins into the outer membrane of E. coli. Prog Biophys Mol Biol. 1987;49:89–115. doi: 10.1016/0079-6107(87)90010-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Henning U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol. 1996;19:1287–1294. doi: 10.1111/j.1365-2958.1996.tb02473.x. [DOI] [PubMed] [Google Scholar]
- 3.Cowen S W, Schirmer T, Rummel G, Steiert M, Ghose R, Pauptit A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature (London) 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 4.de Cock H, Struyvé M, Kleerebezem M, van der Krift T, Tommassen J. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein of PhoE of Escherichia coli K-12. J Mol Biol. 1997;269:473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- 5.Denese P N, Silhavy T. Targeting and assembly of periplasmic and outer membrane proteins in Escherichia coli. Annu Rev Genet. 1999;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 6.Deng M, Misra R. Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins. Mol Microbiol. 1996;21:605–612. doi: 10.1111/j.1365-2958.1996.tb02568.x. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson A D, Welte W, Hofmann E, Lindner B, Holst O, Coulton J W, Diederichs K. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Struct Fold Design. 2000;8:585–592. doi: 10.1016/s0969-2126(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 8.Fourel D, Mizushima S, Pages J-M. Dynamics of the exposure of epitopes on OmpF, an outer membrane protein of Escherichia coli. Eur J Biochem. 1992;206:109–114. doi: 10.1111/j.1432-1033.1992.tb16907.x. [DOI] [PubMed] [Google Scholar]
- 9.Gennity J, Goldstein J, Inouye M. Signal peptide mutants of Escherichia coli. J Bioenerg Biomembr. 1990;22:233–269. doi: 10.1007/BF00763167. [DOI] [PubMed] [Google Scholar]
- 10.King J, Fane B, Haase-Pattingell C, Mitraki A, Villafane R, Yu M-H. Identification of amino acid sequences influencing intracellular folding pathways using temperature sensitive folding mutations. In: Gierasch L, King J, editors. Protein folding. Washington, D.C.: American Association for the Advancement of Science; 1989. pp. 225–240. [Google Scholar]
- 11.Kloser A W, Laird M W, Misra R. asmB, a suppressor locus for assembly-defective OmpF mutants of Escherichia coli, is allelic to envA (lpxC) J Bacteriol. 1996;178:5138–5143. doi: 10.1128/jb.178.17.5138-5143.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloser A W, Laird M W, Deng M, Misra R. Modulation in lipid A and phospholipid biosynthesis pathways influence outer membrane protein assembly in Escherichia coli K-12. Mol Microbiol. 1998;27:1003–1008. doi: 10.1046/j.1365-2958.1998.00746.x. [DOI] [PubMed] [Google Scholar]
- 13.Laird W M, Kloser A W, Misra R. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. J Bacteriol. 1994;176:2259–2264. doi: 10.1128/jb.176.8.2259-2264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazar S W, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra R. OmpF assembly mutants of Escherichia coli K-12: isolation, characterization, and suppressor analysis. J Bacteriol. 1993;175:5049–5056. doi: 10.1128/jb.175.16.5049-5056.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misra R, CastilloKeller M, Deng M. Overexpression of protease deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J Bacteriol. 2000;182:4882–4888. doi: 10.1128/jb.182.17.4882-4888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misra R, Miao Y. Molecular analysis of asmA, a locus identified as the suppressor of OmpF assembly mutants of Escherichia coli K-12. Mol Microbiol. 1995;16:779–788. doi: 10.1111/j.1365-2958.1995.tb02439.x. [DOI] [PubMed] [Google Scholar]
- 18.Misra R, Peterson A, Ferenci T, Silhavy T J. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J Biol Chem. 1991;266:13592–13597. [PubMed] [Google Scholar]
- 19.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phale P S, Philippsen A, Kiefhaber T, Koebnik R, Phale V P, Schirmer T, Rosenbusch J P. Stability of trimeric OmpF porin: the contributions of the latching loop L2. Biochemistry. 1998;37:15663–15670. doi: 10.1021/bi981215c. [DOI] [PubMed] [Google Scholar]
- 21.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ried G, Hindenach I, Henning U. Role of lipopolysaccharide in assembly of Escherichia coli outer membrane proteins OmpA, OmpC, and OmpF. J Bacteriol. 1990;172:6048–6053. doi: 10.1128/jb.172.10.6048-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouvière P E, Gross C A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 24.Sen K, Nikaido H. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J Bacteriol. 1991;173:926–928. doi: 10.1128/jb.173.2.926-928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 26.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 27.Xiong X, Deeter J N, Misra R. Assembly-defective OmpC mutants of Escherichia coli K-12. J Bacteriol. 1996;178:1213–1215. doi: 10.1128/jb.178.4.1213-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]