ABSTRACT
Aspergillus fumigatus is the main etiological agent of aspergillosis. The antifungal drug caspofungin (CSP) can be used against A. fumigatus, and CSP tolerance is observed. We have previously shown that the transcription factor FhdA is important for mitochondrial activity. Here, we show that FhdA regulates genes transcribed by RNA polymerase II and III. FhdA influences the expression of tRNAs that are important for mitochondrial function upon CSP. Our results show a completely novel mechanism that is impacted by CSP.
KEYWORDS: Aspergillus fumigatus, caspofungin paradoxical effect, transcription factor, gene regulation, codon usage
TEXT
Aspergillus fumigatus is a filamentous, saprophytic fungus and an opportunistic pathogen (1). It is the main pathogen responsible for invasive pulmonary aspergillosis (IPA), one of the most severe infections in immunosuppressed patients in terms of morbidity and mortality (2). The echinocandin caspofungin (CSP) is a fungistatic drug against filamentous fungi and can be used as salvage therapy for IPA (3). It acts by noncompetitively inhibiting the fungal β-1,3-glucan synthase (Fks1), which is required for the biosynthesis of the primary fungal cell wall carbohydrate, β-1,3-glucan (4). However, in a certain range of higher concentrations, there is a reduction of CSP activity. This phenomenon, which is known as the “caspofungin paradoxical effect” (CPE), results from a tolerance cellular response that alters both the cell wall content and fungal growth (5).
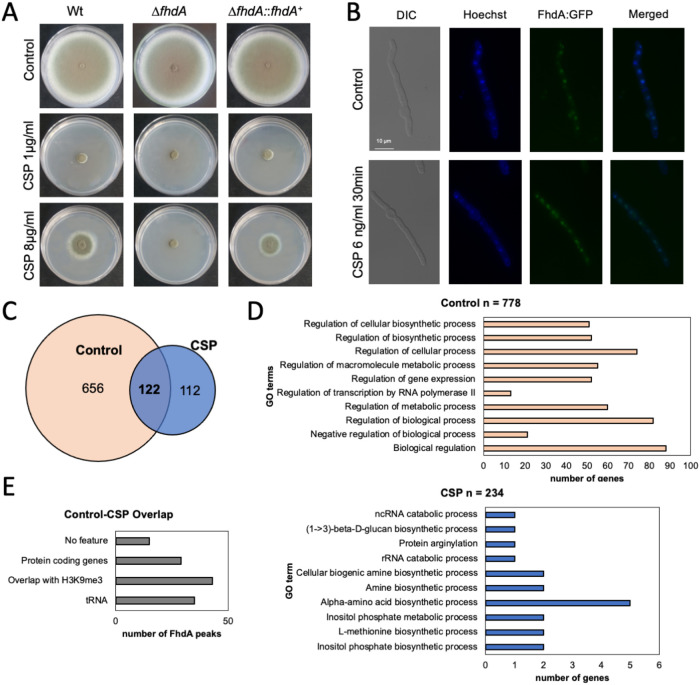
A. fumigatus is a highly successful opportunistic pathogen, mostly due to its ability to rapidly adapt to diverse environments. To this end, changes in physicochemical conditions and nutrient availability generate signals at the cell surface that are conveyed by a system of signaling pathways to the nucleus and converge at transcription factors (TFs) (1). TFs regulate the transcription of gene sets that drive metabolic reprogramming to enable adaptation to the new conditions and promote proliferation inside the host (6). Recently, by screening a library of 484 TF null mutants (7), we identified FhdA (AFUB_091020), a novel TF that plays a role in the CSP response (8). The ΔfhdA strain is more sensitive to CSP and lacks the CPE (Fig. 1A), and a functional FhdA:GFP (Fig. S1; 10.6084/m9.figshare.20254623) strain shows that FhdA is constitutively located inside the nucleus (Fig. 1B).
FIG 1.
FhdA is essential to the caspofungin paradoxical effect (CPE) and binds to several targets before and after caspofungin (CSP) exposure. (A) Radial growth of the wild-type strain (Wt, CEA17), ΔfhdA, and ΔfhdA:fhdA+ incubated for 5 days at 37°C in minimal medium containing 0, 1, and 8 μg/mL of CSP. (B) Fluorescence microscopy of a FdhA:GFP functional strain before and after exposure to 6 ng/mL of CSP for 30 min. Hoechst was used to dye the nuclei. (C) A Venn diagram showing the number of FhdA targets before and after exposure to 2 μg/mL of CSP for 1 h. (D) Top 10 GO terms categorizing the genes closest to the FhdA targets before and after CSP exposure. (E) Functional classification of the 122 targets constitutively bound by FhdA. All of the methods are described in the Supplementary Text S1 (10.6084/m9.figshare.20254623).
In order to explore the pathways affected by FhdA, we investigated its direct targets by determining the FhdA:3xHA (Fig. S1; 10.6084/m9.figshare.20254623) binding sites using genome-wide ChIP-seq, chromatin immunoprecipitation coupled to a DNA sequencing analysis (BioProject ID PRJNA855589). We detected a total of 890 regions bound by FhdA at their promoters (Fig. 1C). Most of the promoters (n = 778) were bound by FhdA before CSP exposure. 122 of them were constitutively bound by the TF both before and after exposure to 2 μg/mL of CSP (Fig. 1C). In contrast, 112 genes were bound by FhdA exclusively after CSP exposure (Fig. 1C).
An enrichment analysis of the 778 genes bound by FhdA in control conditions suggests that this TF is involved in the regulation of processes related to gene expression and transcriptional control (Fig. 1D, upper panel), while the 234 genes bound after CSP exposure are mostly involved in amino acid metabolism (Fig. 1D, bottom panel). Of the 122 constitutive targets of FhdA, 15 have no feature, 29 encode proteins, 43 overlap intergenic regions bound by the modified histone H3K9me3 previously described by our group (9), and 35 encode tRNAs (Fig. 1E). Since tRNAs are transcribed by RNA polymerase III (PolIII), these results suggest that FhdA modulates the expression of not only genes transcribed by RNA polymerase II (PolII) but also genes transcribed by PolIII.
FhdA is important for mitochondrial respiratory function (8). We analyzed the 29 protein coding genes bound by TF using MitoProt (https://openebench.bsc.es/tool/mitoprot_ii), which revealed 8 genes that had a mitochondrial targeting signature. These are a putative 30S ribosomal subunit S4 (AFUB_007360), the TF LeuB (AFUB_020530), the putative ER Hsp70 chaperone BipA (AFUB_021670), the putative ABC transporter Adp1 (AFUB_073240), a ketol-acid reductoisomerase (AFUB_034740), a putative glucosamine-6-phosphate deaminase (AFUB_083490), a protein predicted to bind chromatin (AFUB_096570), and a hypothetical protein (AFUB_077560). It remains to be determined whether these proteins localize at the mitochondria and how they affect CSP tolerance.
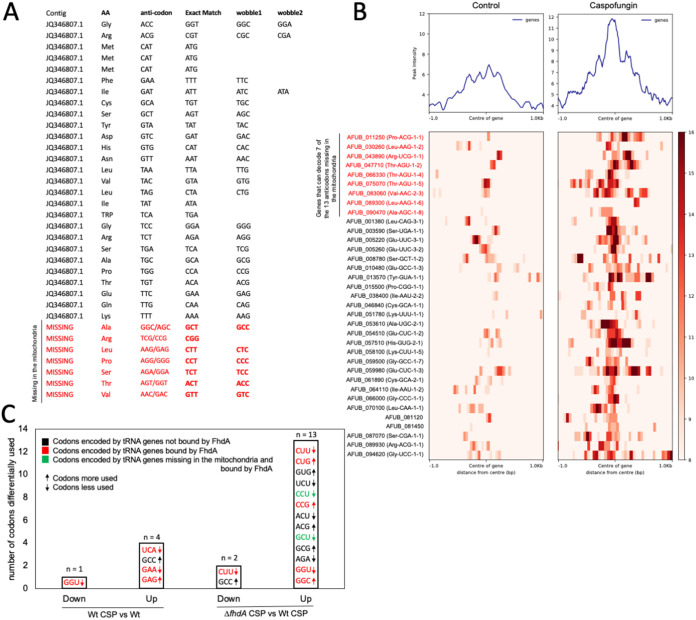
We then hypothesized that the tRNA genes controlled by FhdA could influence the mitochondrial tRNA pool. A. fumigatus has 27 predicted mitochondrially encoded tRNA anti-codons, with 47 codons that can be decoded from wobble or exact match pairing. That leaves 13 codons that cannot be decoded by the mitochondrially encoded tRNAs without tRNA modifications (Fig. 2A). 7 of these 13 codons without a tRNA encoded in the mitochondria can be decoded by one of the nine tRNAs identified in the ChIP-seq experiment (Fig. 2B).
FIG 2.
FhdA binds tRNA promoters, including promoters of several tRNAs that are not encoded by the mitochondrial genome. (A) The tRNAs encoded by the mitochondrial genome and the ones that are absent from it. (B) A heat map representing the FhdA binding intensity to the 35 tRNA genes detected in our ChIP-seq. (C) Relative synonymous codon usage (RSCU) analysis showing the number of codons differentially used for the transcription of the 5-fold upregulated and downregulated genes detected in the RNA-seq experiment. All of the methods are described in the Supplementary Text S1 (10.6084/m9.figshare.20254623).
We then asked if the fhdA deletion effect could be related to an altered tRNA preference. To test for codon specific effects on nuclear gene expression, we compared the Relative-Synonymous-Codon-Usage (RSCU), defined as the ratio of the observed frequency of codons to the expected frequency, given that all of the synonymous codons for the same amino acids are used equally (10), using the RNAseq data set, in which genes regulated by FhdA in the absence or presence of CSP are 5-fold upregulated or downregulated (8). When we compared the wild-type exposed to CSP with the corresponding control, genes that were 5-fold upregulated or downregulated during exposure to CSP had lower usage of the codons, UCA, GAA, and GGU, and higher usage of the codons, GAG and GCC. Four of these five codons are decoded by tRNAs identified in the ChIP-seq experiment (Fig. 2C). Genes overexpressed or underexpressed in the ΔfhdA mutant compared to the wild-type, both treated with CSP, were enriched or depleted for 15 different codons in upexpressed or downexpressed genes; 8 of these codons are decoded by tRNAs identified in the ChIP-seq, and two of these codons are not present in the mitochondria (Fig. 2C). These results suggest that there are differences in codon usage during the CSP response; there are about 3-fold more differences in codon usage in ΔfdhA during CSP, suggesting that the combination of ΔfhdA and CSP affects gene expression in a codon-specific manner that may be related to changes in tRNA expression.
In summary, our results suggest that FhdA can bind to the tRNA promoter regions and most likely collaborates with PolIII in the regulation of their expression levels. The binding of FhdA to the tRNA promoter regions is affected by CSP, indicating that CSP can modulate the tRNA pool utilization and that several side effects related to CSP activity, such as a decrease in the mitochondrial function, could be caused by the depletion of essential tRNAs. FhdA seems to play an important roles in the control of the expression of protein-encoding genes and in tRNA expression, suggesting that FhdA can influence RNA PolII and RNA PolIII targets. Transcriptional regulation of tRNAs may represent a novel route of translation regulation in filamentous fungi.
ACKNOWLEDGMENTS
We thank the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grants numbers 2017/07536-4 (A.C.C.), 2018/00715-3 (C.V.), and 2016/07870-9 (G.H.G.), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grants numbers 301058/2019-9 and 404735/2018-5 (G.H.G.), both from Brazil, and the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01AI153356) from the USA (A.R. and G.H.G.).
REFERENCES
- 1.Dagenais TR, Keller NP. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev 22:447–465. 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latgé JP, Chamilos G. 2019. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev 33:e00140-18. 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenks JD, Hoenigl M. 2018. Treatment of aspergillosis. JoF 4:98. 10.3390/jof4030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perlin DS. 2015. Echinocandin resistance in Candida. Clin Infect Dis 61 Suppl:S612–S617. 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aruanno M, Bachmann D, Sanglard D, Lamoth F. 2019. Link between heat shock protein 90 and the mitochondrial respiratory chain in the caspofungin stress response of Aspergillus fumigatus. Antimicrob Agents Chemother 63:e00208-19. 10.1128/AAC.00208-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bultman KM, Kowalski CH, Cramer RA. 2017. Aspergillus fumigatus virulence through the lens of transcription factors. Med Mycol 55:24–38. 10.1093/mmy/myw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furukawa T, van Rhijn N, Fraczek M, Gsaller F, Davies E, Carr P, Gago S, Fortune-Grant R, Rahman S, Gilsenan JM, Houlder E, Kowalski CH, Raj S, Paul S, Cook P, Parker JE, Kelly S, Cramer RA, Latgé J-P, Moye-Rowley S, Bignell E, Bowyer P, Bromley MJ. 2020. The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nat Commun 11:1–16. 10.1038/s41467-019-14191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valero C, Colabardini AC, Chiaratto J, Pardeshi L, de Castro PA, Ferreira Filho JA, Silva LP, Rocha MC, Malavazi I, Costa JH, Fill T, Barros MH, Wong SSW, Aimanianda V, Wong KH, Goldman GH. 2020. Aspergillus fumigatus transcription factors involved in the caspofungin paradoxical effect. mBio 11:e00816-20. 10.1128/mBio.00816-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colabardini AC, Wang F, Miao Z, Pardeshi L, Valero C, de Castro PA, Akiyama DY, Tan K, Nora LC, Silva-Rocha R, Marcet-Houben M, Gabaldón T, Fill T, Wong KH, Goldman GH. 2022. Chromatin profiling reveals heterogeneity in clinical isolates of the human pathogen Aspergillus fumigatus. PLoS Genet 18:e1010001. 10.1371/journal.pgen.1010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grantham R, Gautier C, Gouy M. 1980. Codon frequencies in 119 individual genes confirm consistent choices of degenerate base according to genome type. Nucleic Acids Res 8:1892–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]