ABSTRACT
Patients with burn injuries are at high risk for infectious complications, and infections are the most common cause of death after the first 72 h of hospitalization. Hospital-acquired infections caused by multidrug resistant (MDR) Gram-negative bacteria (GNB) in this population are concerning. Here, we evaluated carriage with MDR GNB in patients in a large tertiary-care burn intensive care unit. Twenty-nine patients in the burn unit were screened for intestinal carriage. Samples were cultured on selective media. Median time from admission to the burn unit to first sample collection was 9 days (IQR 5 – 17 days). In 21 (72%) patients, MDR GNB were recovered; the most common bacterial species isolated was Pseudomonas aeruginosa, which was found in 11/29 (38%) of patients. Two of these patients later developed bloodstream infections with P. aeruginosa. Transmission of KPC-31-producing ST22 Citrobacter freundii was detected. Samples from two patients grew genetically similar C. freundii isolates that were resistant to ceftazidime-avibactam. On analysis of whole-genome sequencing, blaKPC-31 was part of a Tn4401b transposon that was present on two different plasmids in each C. freundii isolate. Plasmid curing experiments showed that removal of both copies of blaKPC-31 was required to restore susceptibility to ceftazidime-avibactam. In summary, MDR GNB colonization is common in burn patients and patient-to-patient transmission of highly resistant GNB occurs. These results emphasize the ongoing need for infection prevention and antimicrobial stewardship efforts in this highly vulnerable population.
KEYWORDS: Citrobacter, Pseudomonas aeruginosa, burn, carbapenemase, ceftazidime-avibactam, hospital infections, plasmid-mediated resistance
INTRODUCTION
Patients with burn injuries are at high risk for infectious complications, and infections are the primary cause of death in these patients after the first 72 h of hospitalization (1). In particular, patients with large burns and/or inhalational injury often have prolonged hospitalizations during which they are frequently treated with broad-spectrum antibiotics. This exposure, combined with loss of skin barrier function and the need for invasive medical devices leads to high antimicrobial resistance rates in burn units (1, 2). Unsurprisingly, the risk of infection caused by multidrug-resistant organisms (MDRO) increases with longer hospitalization durations, and colonization with MDRO generally precedes infection (2, 3). Clinically important MDRO include expanded-spectrum cephalosporin-resistant Enterobacterales, carbapenem-resistant Enterobacterales (CRE), and MDR non-lactose-fermenting Gram-negative bacteria, such as Pseudomonas spp., Stenotrophomonas maltophilia, and Acinetobacter baumannii (4–7). Common underlying enzymatic mechanisms for β-lactam resistance in Gram-negative bacteria include extended-spectrum β-lactamases (ESBL), AmpC enzymes, and carbapenemases. In recent years, novel β-lactam antibiotics have become available for the treatment of MDR Gram-negative bacteria. These include ceftolozane-tazobactam, ceftazidime-avibactam, meropenem-vaborbactam, imipenem-relebactam, and cefiderocol (8, 9). In the treatment of CRE infections, novel agents were shown to result in better outcomes compared to polymyxin-based regimens (10–12). However, treatment-emergent resistance is a concern, especially for ceftazidime-avibactam (13, 14).
Here, we evaluated MDR Gram-negative intestinal colonization and transmission in a large tertiary-care burn intensive care unit.
RESULTS
Patients.
Twenty-nine patients in the burn unit were screened for MDR Gram-negative bacterial colonization during the study period (Table 1). Median age was 58 years (interquartile range, IQR 46 to 66 years), and 22 (76%) were male. Burn injury was the reason for admission in 21 (72%) patients; and flame burns were the most common. Sixteen (55%) patients were transferred from other hospitals, primarily (15/16) from other hospitals in North Carolina. In patients with burn injuries, the median total body surface area (TBSA) involved was 12% (IQR 9% to 28%). Three patients had inhalational injury. The remaining 8 (28%) patients were admitted to the burn unit for other reasons, including nonburn skin disorders (n = 4). Median time from admission to the burn unit to first sample collection was 9 days (IQR 5 to 17 days).
TABLE 1.
Clinical characteristicsa
| Characteristic | All | MDR GNB colonized | Not MDR GNB colonized |
|---|---|---|---|
| n | 29 | 21 | 8 |
| Age, yrs, median (range) | 58 (22–88) | 58 (22–76) | 56 (43–88) |
| Sex, male | 22 (76) | 16 (76) | 6 (75) |
| Time from admission to sample, days, median (range) | 9 (0–271) | 9 (0–133) | 12 (0–271) |
| Total length of stay, median (range) | 37 (2–564) | 37 (2–259) | 47 (4–564) |
| Antibiotic exposuresb | |||
| Cefazolin | 11 (38) | 9 (43) | 2 (25) |
| Cefepime | 12 (41) | 8 (38) | 4 (50) |
| Vancomycin | 15 (52) | 11 (52) | 4 (50) |
| Comorbidities | |||
| Hypertension | 19 (66) | 14 (67) | 5 (63) |
| Alcohol use | 17 (59) | 15 (71) | 2 (25) |
| Lung disease | 8 (28) | 7 (33) | 1 (13) |
| Heart disease | 10 (34) | 6 (29) | 4 (50) |
| Diabetes | 10 (34) | 8 (38) | 2 (25) |
| Kidney disease | 4 (14) | 1 (5) | 3 (38) |
| Mechanical ventilationc | 14 (48) | 10 (48) | 4 (50) |
| Burn characteristics | |||
| Revised Baux score | 69 (31–126) | 69 (31–110) | 92 (53–126) |
| TBSA, %, median (range) | 12 (0–87) | 12 (0–87) | 14.25 (10–56) |
| Inhalational injury | 3 (10) | 2 (10) | 1 (13) |
| Mechanism | |||
| Flame | 9 (31) | 5 (24) | 4 (50) |
| Scald | 5 (17) | 5 (24) | 0 |
| Other burnd | 7 (24) | 5 (24) | 2 (25) |
| Nonburn injury | 8 (28) | 6 (29) | 2 (25) |
| Hospital mortality | 4 (14) | 3 (14) | 1 (13) |
All data in n(%), unless otherwise indicated.
Antibiotic exposures preceded collection of the isolates.
Mechanical ventilation on the day of sample collection.
One person with an unknown mechanism of burn was included in “Other.”
Intestinal colonization.
Thirty-four MDR GNB were recovered from 21 (72%) patients. Most clinical characteristics, including burn size were similar between patients with and without MDR Gram-negative bacterial carriage (Table 1). Numerically, more patients with a history of alcohol use were colonized with MDR GNB. The median number of species was one per patient (range 1 to 5). Carbapenemase genes were only found in Citrobacter freundii isolates. The most common bacterial species isolated was Pseudomonas aeruginosa, which was found in 10/29 (34%) of patients (Table 2). Eight of these isolates were nonsusceptible to carbapenems in vitro. Three P. aeruginosa isolates were ST167 by MLST, and one additional isolate was very similar on MLST (6 identical alleles and a single-nucleotide variant for the nuoD allele). The other 6 isolates belonged to 6 different strain types by MLST. Two patients with ST167 P. aeruginosa were admitted to adjacent rooms in the unit.
TABLE 2.
Bacterial species
| Species | Alla | Carbapenem nonsusceptiblec |
|---|---|---|
| Pseudomonas aeruginosa | 10 (34) | 8 (80) |
| Enterobacter cloacae complex | 5 (17) | 2 (40) |
| Other Pseudomonas speciesb | 5 (17) | 1 (20) |
| Klebsiella pneumoniae | 4 (14) | 2 (50) |
| Escherichia coli | 2 (7) | 0 |
| Citrobacter freundii | 2 (7) | 2 (100) |
| Stenotrophomonas maltophilia | 2 (7) | 2 (100) |
| Enterobacter bugandensis | 1 (3) | 1 (100) |
| Hafnia alvei | 1 (3) | 0 |
| Klebsiella aerogenes | 1 (3) | 0 |
| Achromobacter dentrificans | 1 (3) | 0 |
Numbers are shown as n (% of 29 total patients). Totals add to more than 100% as some patients were colonized by more than one species.
Other Pseudomonas species included P. luteola (1), P. putida (1), P. otitidis (1), P. guariconensis (1).
Defined as nonsusceptibility to imipenem and/or meropenem. Numbers are shown as n (% of patients colonized with that species).
In one patient colonized with Enterobacter cloacae complex, the same species was cultured from a respiratory sample collected for clinical reasons prior to obtaining the study sample. Two patients colonized with P. aeruginosa developed subsequent bacteremia with P. aeruginosa after research sample collection. In one of these patients, a similar antimicrobial susceptibility pattern was observed in the infecting strain compared to the colonizing strain. No other patients had pre- or postsample clinical cultures with growth of the colonizing species.
Transmission of KPC-producing C. freundii.
Two patients were colonized with carbapenem nonsusceptible C. freundii; blaKPC-31 was detected in both isolates. These isolates displayed in vitro resistance to ceftazidime-avibactam (MIC >64 μg/mL). The index patient was admitted from another hospital 44 days prior to sample collection and stayed in the ICU 60 days after sample collection. The second patient’s sample was collected on the seventh day of hospitalization, 9 days after the first patient’s sample collection date. The second patient stayed in the burn ICU for 10 days after sample collection. Neither patient had a clinical culture positive for carbapenem-resistant C. freundii. C. freundii was also not recovered from any other clinical cultures during the study period in the burn ICU.
Genomics of C. freundii.
The two C. freundii isolates from two unique burn patients were sequenced. The two genomes both belonged to ST22 and differed by 5 SNPs in the core genome. They carried the same resistance genes. In addition to blaKPC-31, they contained the following notable resistance genes: ant(2′')-Ia, aadA1, sat2 (encoding aminoglycoside resistance), blaFOX-5, blaTEM-1, blaCMY-48 (β-lactam resistance), catA2, catB3 (phenicol resistance), qacEdelta1 (quaternary ammonium resistance), sul1 (sulfonamide resistance), tet(D) (tetracycline resistance) and dfrA1 (trimethoprim resistance). Hybrid assembly of the first isolate (59174) showed that this strain harbors four plasmids of 43.6 (p59174-43.6, pKPC-CAV1193-like), 57.6 (p59174-57.6, IncX8), 80.5 (p59174-80.5, IncFIB), and 171.4 (p59174-171.4, IncC) kb in length. Interestingly, blaKPC-31 was present on two plasmids (p59174-43.6 and p59174-80.5, Fig. 1). On both plasmids, blaKPC-31 was part of a Tn4401b transposon. p59174-43.6 was nearly identical to plasmid pKPC_UVA01, with 100% blast query coverage and >99.99% nucleotide identities, recovered in a Virginia hospital in 2007 (15). In p59174-80.5, the Tn4401b element was inserted an IS5 element, generating 5-bp target duplicate sequences of GTTTT. The result suggested that the origin of blaKPC-31 in p59174-80.5 was likely the result of duplication and transposition of Tn4401b from p59174-43.6. Further comparison of the sequences of the index-2 isolate with the close genome of the first isolate revealed that the index-2 isolate contained the same four plasmids as those of 59174, but harbored an additional IncFIB(pHCM2) type plasmid; ~106 kb in length). Antimicrobial resistance genes were not found in this IncFIB(pHCM2) plasmid. To determine the impact of two copies of blaKPC-31 on ceftazidime-avibactam susceptibility, we used a CRISPR-Cas-mediated plasmid curing approach to remove p59174-43.6, p59174-80.5, respectively and together. The results showed the curing on both plasmids restored susceptibility to ceftazidime-avibactam, but deletion of only a single copy did not (Table 3).
FIG 1.
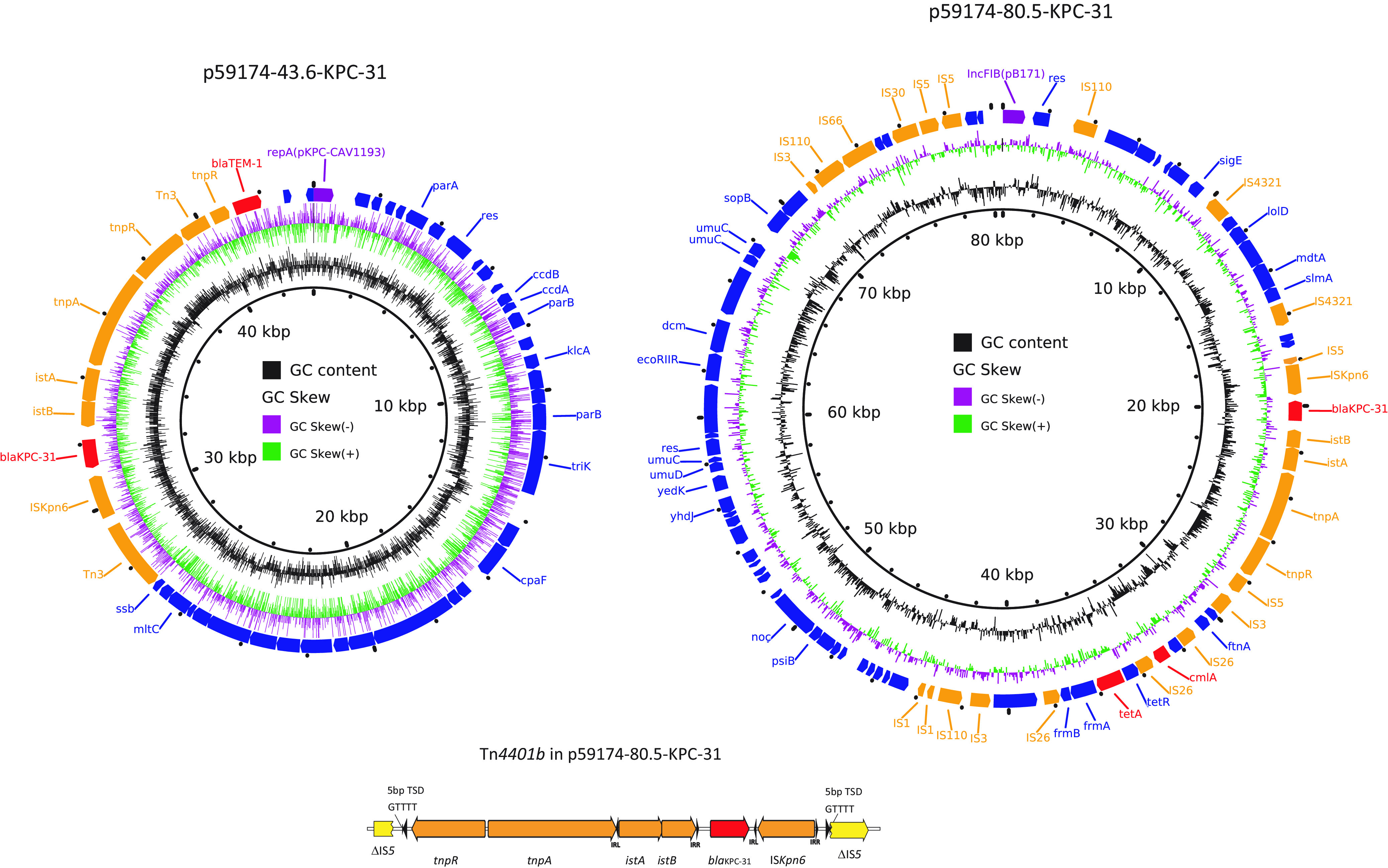
The plasmid structures of the two blaKPC-31 harboring plasmids, p59174-43.6 (left) and p59174-80.5 (right). Colored arrows indicate open reading frames, with purple, orange, green, red, and blue arrows representing replication genes, mobile elements, plasmid transfer genes, the antimicrobial and heavy metal resistance gene, and plasmid backbone genes, respectively. The bottom panel shows the Tn4401 insertion in the IS5 in p59174-80.5.
TABLE 3.
Antimicrobial susceptibility of C. freundiia
| Isolate | CAZ | CZA | IPM | ETP | MEM | I-R |
|---|---|---|---|---|---|---|
| Index-1 | >64 | >64 | 4 | 1 | <0.5 | 1 |
| Index-2 | >64 | >64 | 8 | 4 | 4 | 1 |
| KPC-31 on p59174-43.6 plasmid cured | >64 | 32 | 4 | 2 | 4 | 1 |
| KPC-31 on p59174-80.5 plasmid cured | >64 | 64 | 8 | 2 | 2 | 1 |
| Both copies cured | >64 | 2 | 8 | 1 | 2 | 1 |
CAZ, ceftazidime; CZA, ceftazidime-avibactam; ETP, ertapenem; IPM, imipenem; I-R, imipenem-relebactam; MEM, meropenem.
DISCUSSION
Burn injuries are a common cause of morbidity and mortality in the United States and across the world (16). Infection is the most frequent cause of mortality in patients who survive the first 72 h after the initial burn injury (1, 2). In this cohort of patients admitted to a large burn ICU, intestinal carriage of multidrug resistant Gram-negative bacteria was very common. Hospitalizations for burn injuries are associated with several risk factors for MDRO infections. The local immune barrier function of the skin is decreased, and burn injuries—especially those involving greater total body surface areas—are also associated with a central immunomodulatory impact (17). Furthermore, antibiotic use and long-term need for invasive medical devices is common in this population (1). Given the high risk for infection and the potential for poor outcomes after infection, empirical antibiotic use is common in burn patients. These antibiotics are often directed against MDR bacteria frequently seen in the burn ICU, and this increased antibiotic pressure thus creates a vicious cycle.
P. aeruginosa was the most frequently encountered species. P. aeruginosa is a frequent cause of invasive infections in burn patients which are associated with high rates of morbidity and mortality (1, 2, 18, 19). Infections with P. aeruginosa after burn injury generally occur during extended hospitalizations; the reported median time from admission to first positive P. aeruginosa culture in other burn centers is between 10 days to 32 days (2, 20–23). This prolonged period between admission and first positive culture suggests a nosocomial acquisition. The finding of genetically related ST167 and ST167-like P. aeruginosa strains in four patients—including two patients who were in neighboring rooms—supports transmission within the unit. Our data further support intestinal colonization as an intermediary step to infection. In fact, two patients in our cohort had subsequent pseudomonal bacteremia.
The spread of highly resistant bacteria in hospital settings is alarming. We report here the spread of blaKPC-31 carrying C. freundii between two patients. As clinical infections were not documented to result from these bacteria, investigations by infection preventionists were not performed. Possible intermediary transmission steps include the burn unit environment, and health care personnel. Randomized trials have shown limited impact of interventions aimed at preventing spread from HCP to patients for Gram-negative bacteria (24, 25). In contrast, many outbreaks of Gram-negative bacterial infections in intensive care units have been linked to environmental sources. In a French ICU study, contaminated tap water was a predictor (OR 1.76, 95% CI 1.09 to 2.84) of acquiring P. aeruginosa during ICU stay (26). Sink traps were shown to be the source of an ICU outbreak of OXA-48-producing Serratia marcescens (27). However, another study showed frequent and prolonged colonization of hospital sinks with carbapenemase-producing Enterobacterales without any documented transmission to patients (28).
The C. freundii isolates were found to produce KPC-31, the most common ceftazidime-avibactam-resistant variant of KPC-3, with a D179Y amino acid substitution in the Ω loop of the enzyme (29, 30). Of interest, two copies of blaKPC-31 were present on two distinct plasmids. Inactivation of a single copy failed to restore susceptibility to ceftazidime-avibactam. The coexistence of more than one copy of resistance genes may improve the survival of bacteria under antibiotic therapy and increase the further spread of the resistance plasmids. We postulate that the additional ~106 kb IncFIB(pHCM2) type plasmid in the second isolate may have been acquired after transmission. Our study further demonstrated that CRISPR-Cas9-mediated plasmid curing method provide a useful tool to dissect the relative contribution of different plasmids to the antimicrobial resistance in clinical MDR strains. In addition, resistance gene or plasmid curing can be potentially used as a novel approach to resensitize resistant strains to antibiotics (31).
This study has several limitations. This was a cross-sectional study without longitudinal samples, therefore the timing of acquisition of MDR GNB could not be established. As a cross-sectional sample, we screened patients at various time points since admission. Nonetheless, our study provides evidence of high rates of colonization with MDR GNB in burn patients and evidence of transmission in this setting. We collected a relatively small sample size during the short time period. The environment may be an important intermediary for MDR GNB transmission in the BICU, however, environmental sampling was beyond the scope of the current study.
In conclusion, intestinal carriage of MDR Gram-negative bacteria was very common in this cohort of patients admitted to the burn ICU. Pseudomonal isolates were the most frequently encountered and several of these were resistant to carbapenems. We also found evidence for transmission of a ceftazidime-avibactam-resistant C. freundii strain between two patients. These findings reinforce the need for strict antimicrobial stewardship and infection prevention measures in this highly vulnerable population.
MATERIALS AND METHODS
Patients.
During the study period of August to December of 2019, patients who were admitted to the burn intensive care unit (BICU) of the North Carolina Jaycee Burn Center were included in the study. The BICU has 20 beds and approximately 650 annual admissions. Patients with cutaneous burns, with inhalational injury only, and with nonburn severe skin disorders (e.g., Stevens-Johnson syndrome) were all eligible. Clinical data were collected from the electronic health record. This study was approved by the Institutional Review Board.
Stool samples.
Stool samples were collected using the BioWipe method in all patients and additional whole stool samples when available. The BioWipe collection method was previously described (32). Briefly, a 100 × 160 mm square of soft, absorbent synthetic fiber material attached to a plastic backing layer (Fisher Scientific, USA) is used before cleaning with toilet paper after a bowel movement. The collected stool sample is placed onto the surface of an absorbent pad (3M Petroleum Sorbent Pads, Fisher Scientific, USA) containing modified Cary Blair transport media followed by elution with 20 mL mix of Phosphate Buffer Saline solution (PBS) and 0.1% Tween 80 (vol/vol). For whole stool samples, 100 mg of whole stool was suspended in 5 mL of PBS prior to processing.
Microbiology.
For both BioWipe and whole stool samples, 100 μL aliquots were plated on MacConkey agar plates supplemented with 1 mg/L of cefotaxime and mSuperCarba (Chromagar, Springfield, NJ) agar plates in duplicate, and incubated at 37°C for 24 ± 3 h. Selected colonies were streaked onto a new plate of the same selective media. Five colonies were selected from each type of selective media plate. The plates were incubated for 24 ± 3 h at 37°C. Purified colonies were transferred into Tryptic Soy Broth (TSB) nonselective media and incubated for 24 ± 3 h at 37°C. Matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry was used for species identification. Antimicrobial susceptibility testing was performed using agar dilution in triplicate. Breakpoints from the Clinical and Laboratory Standards Institute (CLSI) were used. Targeted PCR to detect the carbapenemases blaKPC, blaNDM, blaOXA-48-like, blaIMP, blaVIM was performed as previously described (33). Multilocus sequencing typing (MLST) of P. aeruginosa was preformed and ST type assigned according to the allelic profiles in the MLST database (https://pubmlst.org/organisms/pseudomonas-aeruginosa) (34). Quality control was performed for all microbiology procedures.
Whole-genome sequencing and analysis.
Whole-genome sequencing of two KPC-31-producing Citrobacter freundii isolates was carried out using the Illumina Novoseq 6000 sequencing platform (Illumina Inc., San Diego, CA), with 2 × 150 bp paired-end reads. These isolates were selected on the basis of phenotypic resistance to carbapenems and ceftazidime-avibactam. The raw data were filtered using Trimmomatic v0.39, followed by assembly using Spades v3.14 (35, 36). In addition, one isolate was subject long-read sequencing using the Oxford Nanopore Technologies (ONT) MinION sequencing. Hybrid assembly was conducted using Unicycler v0.4.9. with the default settings (37). Antimicrobial resistance genes were determined using AMRFinderPlus v3.10.20 and ResFinder v4.0, while plasmid replicons were analyzed using PlasmidFinder v2.1 (38, 39). Core single nucleotide polymorphism (SNP) distance was determined using methods described previously (40). In brief, trimmed, paired-end sequences from each genome were mapped to hybrid assemblies of the first C. freundii isolate, using snippy (https://github.com/tseemann/snippy), and SNPs in the repeated regions were excluded. The complete genomes sequences of the C. freundii 59174 were deposited in GenBank bioproject accession no. PRJNA549322.
Plasmid curing.
CRISPR-Cas mediated plasmid curing was conducted as previously described (31). Two guide RNAs (gRNA) were designed to target the replicon genes of the two blaKPC-31 harboring plasmids, p59174-43.6 (n20 sequences, GTACTGGATCAATCCCCACG) and p59174-80.5 (n20 sequences, AGTCATTATCCATATCCAGG), respectively.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI143910 (to D.v.D.). In addition, research reported in this publication was supported in part by the National Institutes of Health under Award Number R01AI090155 (B.K.), and in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
R.A.B. received Grant/Research Support from Achaogen, Allecra, Entasis, Merck, Roche, Shionogi, and Wockhardt. D.v.D. is a consultant for Actavis, Tetraphase, Sanofi-Pasteur, MedImmune, Astellas, Merck, Allergan, T2Biosystems, Roche, Achaogen, Neumedicine, Shionogi, Pfizer, Entasis, QPex, Wellspring, Karius, Union, Melinta, and Utility. The other authors have no conflicts of interest to disclose.
REFERENCES
- 1.Lachiewicz AM, Hauck CG, Weber DJ, Cairns BA, van Duin D. 2017. Bacterial infections after burn injuries: impact of multidrug resistance. Clin Infect Dis 65:2130–2136. 10.1093/cid/cix682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Duin D, Strassle PD, DiBiase LM, Lachiewicz AM, Rutala WA, Eitas T, Maile R, Kanamori H, Weber DJ, Cairns BA, Napravnik S, Jones SW. 2016. Timeline of health care-associated infections and pathogens after burn injuries. Am J Infect Control 44:1511–1516. 10.1016/j.ajic.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Souverein D, Euser SM, Herpers BL, Kluytmans J, Rossen JWA, Den Boer JW. 2019. Association between rectal colonization with highly resistant Gram-negative rods (HR-GNRs) and subsequent infection with HR-GNRs in clinical patients: a one year historical cohort study. PLoS One 14:e0211016. 10.1371/journal.pone.0211016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Group WHOPPLW. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 5.Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. 2021. Infectious Diseases Society of America Guidance on the treatment of extended-spectrum β-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin Infect Dis 72:1109–1116. 10.1093/cid/ciab295. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States. US Department of Health and Human Services. www.cdc.gov/DrugResistance/Biggest-Threats.html. [Google Scholar]
- 7.van Duin D, Arias CA, Komarow L, Chen L, Hanson BM, Weston G, Cober E, Garner OB, Jacob JT, Satlin MJ, Fries BC, Garcia-Diaz J, Doi Y, Dhar S, Kaye KS, Earley M, Hujer AM, Hujer KM, Domitrovic TN, Shropshire WC, Dinh A, Manca C, Luterbach CL, Wang M, Paterson DL, Banerjee R, Patel R, Evans S, Hill C, Arias R, Chambers HF, Fowler VG, Kreiswirth BN, Bonomo RA, Multi-Drug Resistant Organism Network I. 2020. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 20:731–741. 10.1016/S1473-3099(19)30755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong D, van Duin D. 2017. Novel beta-lactamase inhibitors: unlocking their potential in therapy. Drugs 77:615–628. 10.1007/s40265-017-0725-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Duin D, Bonomo RA. 2016. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation beta-lactam/beta-lactamase inhibitor combinations. Clin Infect Dis 63:234–241. 10.1093/cid/ciw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Duin D, Lok JJ, Earley M, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Doi Y, Kaye KS, Fowler VG, Paterson DL, Bonomo RA, Evans S, Antibacterial Resistance Leadership G. 2018. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 66:163–171. 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wunderink RG, Giamarellos-Bourboulis EJ, Rahav G, Mathers AJ, Bassetti M, Vazquez J, Cornely OA, Solomkin J, Bhowmick T, Bishara J, Daikos GL, Felton T, Furst MJL, Kwak EJ, Menichetti F, Oren I, Alexander EL, Griffith D, Lomovskaya O, Loutit J, Zhang S, Dudley MN, Kaye KS. 2018. Effect and safety of meropenem-vaborbactam versus best-available therapy in patients with carbapenem-resistant Enterobacteriaceae infections: the TANGO II Randomized Clinical Trial. Infect Dis Ther 7:439–455. 10.1007/s40121-018-0214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motsch J, Murta de Oliveira C, Stus V, Koksal I, Lyulko O, Boucher HW, Kaye KS, File TM, Brown ML, Khan I, Du J, Joeng HK, Tipping RW, Aggrey A, Young K, Kartsonis NA, Butterton JR, Paschke A. 2020. RESTORE-IMI 1: a multicenter, randomized, double-blind trial comparing efficacy and safety of imipenem/relebactam vs colistin plus imipenem in patients with imipenem-nonsusceptible bacterial infections. Clin Infect Dis 70:1799–1808. 10.1093/cid/ciz530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. 2016. Clinical outcomes, drug toxicity and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis 63:1615–1618. 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathers AJ, Stoesser N, Sheppard AE, Pankhurst L, Giess A, Yeh AJ, Didelot X, Turner SD, Sebra R, Kasarskis A, Peto T, Crook D, Sifri CD. 2015. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother 59:1656–1663. 10.1128/AAC.04292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smolle C, Cambiaso-Daniel J, Forbes AA, Wurzer P, Hundeshagen G, Branski LK, Huss F, Kamolz LP. 2017. Recent trends in burn epidemiology worldwide: a systematic review. Burns 43:249–257. 10.1016/j.burns.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neely CJ, Maile R, Wang MJ, Vadlamudi S, Meyer AA, Cairns BA. 2011. Th17 (IFNgamma- IL17+) CD4+ T cells generated after burn injury may be a novel cellular mechanism for postburn immunosuppression. J Trauma 70:681–690. 10.1097/TA.0b013e31820d18a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thaden JT, Park LP, Maskarinec SA, Ruffin F, Fowler VG, Jr, van Duin D. 2017. Results from a 13-year prospective cohort study show increased mortality associated with bloodstream infections caused by Pseudomonas aeruginosa compared to other bacteria. Antimicrob Agents Chemother 61:e02671-16. 10.1128/AAC.02671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norbury W, Herndon DN, Tanksley J, Jeschke MG, Finnerty CC. 2016. Infection in burns. Surg Infect (Larchmt) 17:250–255. 10.1089/sur.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armour AD, Shankowsky HA, Swanson T, Lee J, Tredget EE. 2007. The impact of nosocomially-acquired resistant Pseudomonas aeruginosa infection in a burn unit. J Trauma 63:164–171. 10.1097/01.ta.0000240175.18189.af. [DOI] [PubMed] [Google Scholar]
- 21.Devrim İ, Kara A, Düzgöl M, Karkıner A, Bayram N, Temir G, Şencan A, Sorguç Y, Gülfidan G, Hoşgör M. 2017. Burn-associated bloodstream infections in pediatric burn patients: time distribution of etiologic agents. Burns 43:144–148. 10.1016/j.burns.2016.07.030. [DOI] [PubMed] [Google Scholar]
- 22.Wanis M, Walker SAN, Daneman N, Elligsen M, Palmay L, Simor A, Cartotto R. 2016. Impact of hospital length of stay on the distribution of Gram negative bacteria and likelihood of isolating a resistant organism in a Canadian burn center. Burns 42:104–111. 10.1016/j.burns.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Decraene V, Ghebrehewet S, Dardamissis E, Huyton R, Mortimer K, Wilkinson D, Shokrollahi K, Singleton S, Patel B, Turton J, Hoffman P, Puleston R. 2018. An outbreak of multi-drug resistant Pseudomonas aeruginosa in a burns service in the north of England: challenges of infection prevention and control in a complex setting. J Hosp Infect 100:e239–e245. 10.1016/j.jhin.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Harris AD, Morgan DJ, Pineles L, Magder L, O'Hara LM, Johnson JK. 2021. Acquisition of antibiotic-resistant Gram-negative bacteria in the benefits of universal glove and gown (BUGG) cluster randomized trial. Clin Infect Dis 72:431–437. 10.1093/cid/ciaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derde LPG, Cooper BS, Goossens H, Malhotra-Kumar S, Willems RJL, Gniadkowski M, Hryniewicz W, Empel J, Dautzenberg MJD, Annane D, Aragao I, Chalfine A, Dumpis U, Esteves F, Giamarellou H, Muzlovic I, Nardi G, Petrikkos GL, Tomic V, Marti AT, Stammet P, Brun-Buisson C, Bonten MJM, Team MWS. 2014. Interventions to reduce colonisation and transmission of antimicrobial-resistant bacteria in intensive care units: an interrupted time series study and cluster randomised trial. Lancet Infect Dis 14:31–39. 10.1016/S1473-3099(13)70295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venier AG, Leroyer C, Slekovec C, Talon D, Bertrand X, Parer S, Alfandari S, Guerin JM, Megarbane B, Lawrence C, Clair B, Lepape A, Perraud M, Cassier P, Trivier D, Boyer A, Dubois V, Asselineau J, Rogues AM, Thiebaut R, group Ds. 2014. Risk factors for Pseudomonas aeruginosa acquisition in intensive care units: a prospective multicentre study. J Hosp Infect 88:103–108. 10.1016/j.jhin.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Regev-Yochay G, Smollan G, Tal I, Pinas Zade N, Haviv Y, Nudelman V, Gal-Mor O, Jaber H, Zimlichman E, Keller N, Rahav G. 2018. Sink traps as the source of transmission of OXA-48-producing Serratia marcescens in an intensive care unit. Infect Control Hosp Epidemiol 39:1307–1315. 10.1017/ice.2018.235. [DOI] [PubMed] [Google Scholar]
- 28.Lemarie C, Legeay C, Mahieu R, Moal F, Ramont C, Kouatchet A, Eveillard M. 2021. Long-term contamination of sink drains by carbapenemase-producing Enterobacterales in three intensive care units: characteristics and transmission to patients. J Hosp Infect 112:16–20. 10.1016/j.jhin.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Gaibani P, Re MC, Campoli C, Viale PL, Ambretti S. 2020. Bloodstream infection caused by KPC-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: epidemiology and genomic characterization. Clin Microbiol Infect 26:516 e511–e514. [DOI] [PubMed] [Google Scholar]
- 30.Livermore DM, Warner M, Jamrozy D, Mushtaq S, Nichols WW, Mustafa N, Woodford N. 2015. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 59:5324–5330. 10.1128/AAC.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao M, He Y, Zhang H, Liao XP, Liu YH, Sun J, Du H, Kreiswirth BN, Chen L. 2020. CRISPR-Cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 64:e00843-20. 10.1128/AAC.00843-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sozzi E, Bartelt L, Xiao J, Kanumuambidi T, Naziripour A, Ruegsegger L, Brown D, Williams F, Zhu Y, Zhu XB, Prakash T, Wood B, Srivastava JC, Stallard MA, Marshall SH, Rudin SD, Sobsey MD, Bonomo RA, van Duin D. 2022. The BioWipe: a non-invasive method to detect intestinal carriage of multi-drug resistant GRAM-negative bacteria. J Chemother 34:203–205. 10.1080/1120009X.2021.2008643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Duin D, Perez F, Rudin SD, Cober E, Hanrahan J, Ziegler J, Webber R, Fox J, Mason P, Richter SS, Cline M, Hall GS, Kaye KS, Jacobs MR, Kalayjian RC, Salata RA, Segre JA, Conlan S, Evans S, Fowler VG, Jr, Bonomo RA. 2014. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 58:4035–4041. 10.1128/AAC.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. 2021. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykasenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, Salcedo S, Cober E, Li L, Kanj SS, Gao H, Munita JM, Ordoñez K, Weston G, Satlin MJ, Valderrama-Beltrán SL, Marimuthu K, Stryjewski ME, Komarow L, Luterbach C, Marshall SH, Rudin SD, Manca C, Paterson DL, Reyes J, Villegas MV, Evans S, Hill C, Arias R, Baum K, Fries BC, Doi Y, Patel R, Kreiswirth BN, Bonomo RA, Chambers HF, Fowler VG, Arias CA, van Duin D, Multi-Drug Resistant Organism Network I. 2022. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis 22:401–412. 10.1016/S1473-3099(21)00399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


