Abstract
Pseudomonas azelaica HBP1 degrades the toxic substance 2-hydroxybiphenyl (2-HBP) by means of three enzymes that are encoded by structural genes hbpC, hbpA, and hbpD. These three genes form a small noncontiguous cluster. Their expression is activated by the product of regulatory gene hbpR, which is located directly upstream of the hbpCAD genes. The HbpR protein is a transcription activator and belongs to the so-called XylR/DmpR subclass within the NtrC family of transcriptional activators. Transcriptional fusions between the different hbp intergenic regions and the luxAB genes of Vibrio harveyi in P. azelaica and in Escherichia coli revealed the existence of two HbpR-regulated promoters; one is located in front of hbpC, and the other one is located in front of hbpD. Northern analysis confirmed that the hbpC and hbpA genes are cotranscribed, whereas the hbpD gene is transcribed separately. No transcripts comprising the entire hbpCAD cluster were detected, indicating that transcription from PhbpC is terminated after the hbpA gene. E. coli mutant strains lacking the structural genes for the RNA polymerase ς54 subunit or for the integration host factor failed to express bioluminescence from PhbpC- and PhbpD-luxAB fusions when a functional hbpR gene was provided in trans. This pointed to the active role of ς54 and integration host factor in transcriptional activation from these promoters. Primer extension analysis revealed that both PhbpC and PhbpD contain the typical motifs at position −24 (GG) and −12 (GC) found in ς54-dependent promoters. Analysis of changes in the synthesis of the hbp mRNAs, in activities of the 2-HBP pathway enzymes, and in concentrations of 2-HBP intermediates during the first 4 h after induction of continuously grown P. azelaica cells with 2-HBP demonstrated that the specific transcriptional organization of the hbp genes ensured smooth pathway expression.
The hbp genes allow Pseudomonas azelaica strain HBP1 to metabolize the toxic compounds 2-hydroxybiphenyl (2-HBP) and 2,2′-dihydroxybiphenyl (2,2′-DHBP) (21, 22, 44). The hbp system consists of three structural genes, hbpC, hbpA, and hbpD, which encode the enzymes for the first steps of 2-HBP degradation (Fig. 1), and of the regulatory gene hbpR (20, 44). Expression of the 2-HBP pathway is tightly regulated, and the respective enzyme activities can only be measured when cells are induced with 2-HBP or 2,2′-DHBP (20, 22). By using knockout studies and complementation assays we identified the HbpR protein as the key regulator for 2-HBP pathway expression (20).
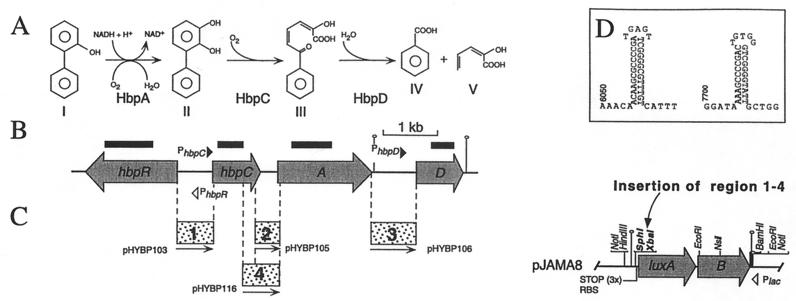
FIG. 1.
(A) Initial metabolism of 2-HBP and 2,2′-DHBP in P. azelaica HBP1. The enzymes responsible for catalyzing the different reactions are indicated below each conversion step. HbpA catalyzes the NADH-dependent ortho hydroxylation of 2-HBP (I) to 2,3-dihydroxybiphenyl (II) (21, 22, 48). HbpC catalyzes the cleaving of 2,3-dihydroxybiphenyl at a meta position, which results in 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid (III) (22, 44). This compound is then hydrolyzed by HbpD to benzoic acid and 2-hydroxy-2,4-pentadienoic acid (IV and V) (22). (B) Genetic organization of the hbp genes. Shaded arrows, orientations and sizes of the genes; solid line, noncoding DNA; black bars, DNA fragments used as probes in the Northern analysis. Hairpin-like structures indicate the relative locations of predicted terminators. (C) (Left) Fragments used for transcriptional fusions with luxAB in pJAMA8 (resulting plasmid names are indicated). (Right) Restriction map of plasmid pJAMA8. The unique SphI and XbaI sites used for cloning are in boldface. The locations of other relevant restriction sites and the orientation of the lac promoter are indicated. RBS, ribosome binding site; STOP (3×), stop codons in all three reading frames. Solid line, vector-derived DNA. (D) ρ-independent transcription terminator structures predicted from the DNA sequence downstream of hbpA (position 6050 of the sequence with GenBank accession no. U73900) and hbpD (position 7700).
On the basis of sequence comparisons, the HbpR protein belongs to the NtrC family of prokaryotic transcriptional activators (20). Members of this family specifically bind to (nearly) palindromic DNA sequences located around 100 to 200 bp upstream of their target promoters (the so-called bacterial enhancer-like elements or upstream activating sequences [UASs]) (reviewed in references 25 and 29). Transcriptional activation by NtrC-type regulators occurs through specific biochemical or physiological stimuli which may change the protein's conformation and which may provoke multimerizations, eventually triggering an ATPase activity (29, 34, 38). The ATPase activity is needed for catalyzing the formation of the open transcriptional complex by ς54-containing RNA polymerase (RNAP) (25) at promoters with a −24 (GG)/−12 (GC) motif (26). Histone-like proteins such as integration host factor (IHF) and protein HU may assist in the process of transcriptional activation. IHF binds DNA specifically while introducing strong hinge-like bends of 140° or greater, whereas HU binds DNA aspecifically and increases the flexibility of the bound DNA (reviewed in reference 31). IHF and HU are capable of establishing a particular geometry at the promoter DNA which may enable a bound NtrC-type activator at the UASs to contact promoter-bound RNAP-ς54 (12, 18, 37). For XylR and its Pu promoter, IHF was even shown to promote a better recruitment of ς54-RNAP to the −24/−12 promoter by providing additional contacts between the α subunit of the holoenzyme and an otherwise-distant cis element (UP-like element) (8, 10, 41).
One subclass within the NtrC family, the XylR/DmpR subclass, is formed by regulatory proteins which are activated by direct interaction with aromatic effector compounds without the need for a sensor kinase component (reviewed in reference 45). These NtrC-type monocomponent regulators exhibit a modular design. The N-terminal A domain recognizes the effector, the central C domain is essential for the different steps needed in transcriptional activation (ATP binding and hydrolysis, oligomerization, and contacting RNAP-ς54), and the C-terminal D domain binds to the DNA at the UASs by means of a helix-turn-helix motif (reviewed in reference 29). In the current model for activation, the A domain acts as a specific interdomain inhibitor which occludes the otherwise constitutive ATPase activity of the central C domain (13, 32, 33). The binding of an effector molecule leads to a conformational change in the A domain which is transmitted through a short flexible interdomain linker hinge region, the Q linker (52), in such a way that the inhibition of ATPase activity of the C domain is released (45).
Based on sequence homology and the capability to interact directly with 2-HBP and other aromatic effectors, HbpR could be assigned to the XylR/DmpR subclass (20). Within this group, HbpR takes a distinct position since it is activated by bicyclic structures, such as 2-HBP and 2,2′-DHBP and the structural analogs 2-aminobiphenyl and 2-hydroxybiphenylmethane (20). Monoaromatic compounds are not effectors for HbpR-mediated transcriptional activation (20).
Here we report on the transcriptional organization of the hbpCAD genes, which is rather unusual for Pseudomonas catabolic genes. By using promoter fusion studies, primer extension experiments, and Northern analysis, we discovered two separately regulated operons within the hbpCAD gene cluster. The expression of both operons is mediated by HbpR and requires RNAP-ς54 and IHF for full activation. From observations of the first stages of induction of the 2-HBP pathway in chemostat-grown P. azelaica cells, we discuss how the present transcriptional organization effects expression of the hbp genes for achieving 2-HBP degradation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. P. azelaica HBP1 is able to use 2-HBP and 2,2′-DHBP as a sole source of carbon and energy (21). P. azelaica strains HBP104 (20), HBP107, HBP108, and HBP118 originate from strain HBP1 and contain transcriptional fusions between the different intergenic regions of the hbp structural genes and the luxAB genes (Fig. 1C) integrated on the chromosome in monocopy.
TABLE 1.
Bacterial strains used in this work
| Strain | Relevant genotype or characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| CC118λpir | Host strain to propagate plasmids with an R6K origin of replication | 17 |
| DH5α | Host strain in routine cloning experiments | Gibco BRL, Life Technologies |
| N99 | galK | 28 |
| A5196 | Cmr Kmr; N99 hupA16::Kan hupB11::Cat; derivative of strain N99 lacking the α and β subunits of HU | 28 |
| A5475 | N99 ΔhimA82 hip157; derivative of strain N99 lacking the α and β subunits of IHF | 28 |
| ET8000 | rbs lacZ::IS1 gyrA hutCK(Con) | 27 |
| ET8045 | Tcr; ET8000 rpoN208::Tn10; derivative of strain ET8000 lacking the ς54 subunit | 27 |
| P. azelaica | ||
| HBP1 | 2-HBP+; wild type | 21 |
| HBP104 | Kmr; 2-HBP+; contains mini-Tn5 (region 1 [PhbpC]-luxAB res-npt-res) of plasmid pHYBP104 | 20 |
| HBP107 | Kmr; 2-HBP+; contains mini-Tn5 (region 2-luxAB res-npt-res) of plasmid pHYBP107 | This work |
| HBP108 | Kmr; 2-HBP+; contains mini-Tn5 (region 3 [PhbpD]-luxAB res-npt-res) of plasmid pHYBP108 | This work |
| HBP118 | Kmr; 2-HBP+; contains mini-Tn5 (region 4-luxAB res-npt-res) of plasmid pHYBP118 | This work |
| HBP104121 | Kmr Smr Tcr; 2-HBP−; derivative of strain HBP104 with disrupted hbpR | 20 |
| HBP108121 | Kmr SmrTcr; 2-HBP−; derivative of strain HBP108 with disrupted hbpR | This work |
Escherichia coli strains were grown at 37°C on Luria-Bertani medium (42). P. azelaica strains were routinely grown at 30°C on Pseudomonas mineral medium (MM) (16) containing 10 mM succinate or 2.9 mM 2-HBP. When required the media were supplemented with antibiotics as described before (20).
Chemostat cultivation of P. azelaica HBP1 and RNA isolation.
P. azelaica HBP1 was continuously cultivated in a 2.5-liter reactor (MBR, Wetzikon, Switzerland). Culture conditions were, in short, a dilution rate of 0.085 h−1 under carbon-limited conditions, a temperature of 30°C, an operating volume of the reactor of 1.2 liters, pH 6.8, and a stirring velocity of 450 rpm. To all media, silicon antifoam was added at a final concentration of 100 ppm. Growth medium for noninducing conditions was based on MM containing 20 mM glucose, on which the cells were grown for about eight volume changes before being induced. Optical density at 600 nm of the culture at steady state was 2.6. Induction was achieved by adding 0.24 ml from a 2.5 M 2-HBP solution in dimethyl sulfoxide (DMSO) and simultaneously shifting the feed medium to MM with 20 mM glucose and 0.5 mM 2-HBP.
Total RNA from P. azelaica HBP1 was isolated from the chemostat 1 min before shifting to 2-HBP-containing medium and 3, 7, 13, 23, and 30 min after the shift. Three-milliliter samples were taken directly from the chemostat, and cells were immediately pelleted by centrifugation (15,000 × g, 30 s) at 4°C. Total RNA was then isolated as described previously (6) and treated with DNase I (RNase free; Boehringer GmbH, Mannheim, Germany). RNA concentrations were determined spectrophotometrically by measuring the optical density at 260 nm in a Uvikon 800 spectrophotometer (Kontron Instruments AG, Zürich, Switzerland).
Recombinant DNA techniques, DNA sequencing, and Southern analysis.
Plasmid DNA isolations, ligations, transformations, PCR, and other DNA manipulations were carried out according to well-established procedures (4, 42) or as described previously (20). Double-stranded template sequencing was performed with primers that were labeled with fluorescent dye IRD-800 or IRD-700 at the 5′ end, as described elsewhere (40).
Chromosomal insertions of mini-Tn5 derivatives or homologous recombined DNA into the P. azelaica chromosome were verified by Southern analysis. DNA fragments were radioactively labeled by using a randomly primed DNA labeling kit (Roche Schweiz AG, Rotkreuz, Switzerland) in the presence of [α-32P]dATP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
Northern analysis.
For Northern analysis, either 1 (for probing with hbpC, hbpA, or hbpD gene fragments) or 8 μg (for probing with hbpR DNA) of RNA was denatured for 1 h at 50°C in the presence of 10 mM sodium phosphate buffer (pH 7.0)–50% DMSO–0.89 M glyoxylate in a total volume of 44.6 μl by standard procedures (4). After 0.1 volume of RNA loading buffer (50% sucrose, 15 mg of bromophenol blue)/ml was added, the glyoxylated RNA mixture was subsequently fractionated in a 1% agarose gel prepared in 10 mM sodium phosphate buffer (pH 7.0), with continuous buffer circulation. RNAs were transferred to Hybond-N membranes by blotting overnight in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
The following hbp gene fragments were used as probes (Fig. 1B): for hbpR, a 0.89-kb SspI-NsiI fragment from plasmid pHYBP111 (20); for hbpC, a 0.47-kb DNA fragment amplified in the PCR (for the location of the fragment, see Fig. 1B); for hbpA, a 0.74-kb HindIII-EcoRI fragment from plasmid pHBP160 (44); for hbpD, a 0.40-kb DNA fragment amplified by PCR.
Primer extension analysis.
Primer extension reactions were carried out in 0.5-ml reaction tubes which were placed into a Crocodile II thermocycler (Appligene, Illkirch, France). First 2 pmol of primer (for hbpC, 5′-CGC AGG CCA AGA CTG ACA CCG G-3′, 122 bp downstream of the hbpC start codon; for hbpD, 5′-CCA CCA TGC AGC ATG ATC ACG G-3′, 122 bp downstream of the hbpD start codon) was mixed with 6 μg of total RNA in a total volume of 5 μl and covered with 1 drop of mineral oil (Sigma). Both primers were labeled at the 5′ end with fluorescent dye IRD-800 (MWG Biotech, Ebersberg, Germany). After an annealing step for 5 min at 68°C, the mixture was cooled to 42°C, and 3 μl of reverse transcriptase mixture was added. Final concentrations during the primer extension reaction were 50 mM Tris-HCl (pH 8.3), 50 mM NaCl, 8 mM MgCl2, 1 mM dithiothreitol, 0.6 mM (each) deoxynucleotide (Amersham), 5% (vol/vol) DMSO, and 18 U of avian myeloblastosis virus reverse transcriptase (Amersham). After incubation for 1 h at 42°C the samples were denatured for 3 min at 95.5°C and loaded on a sequence gel next to samples from a sequence reaction performed on plasmid pHBP130 (44) with the same primers.
Construction of luxAB-based promoter-probe plasmids.
A 704-bp DNA fragment containing the intergenic region between the hbpR and hbpC genes (region 1 in Fig. 1C) was obtained by PCR with P. azelaica HBP1 total DNA as described before (20). A 455-bp DNA fragment containing the intergenic region between the hbpC and hbpA genes (region 2 in Fig. 1C) and an 840-bp DNA fragment with the intergenic region between the hbpA and hbpD genes (region 3 in Fig. 1C) were obtained by PCR with P. azelaica HBP1 total DNA using primer pairs 5′-GCA TGC CAC TTG GGA GGT CAA GCG C-3′, 68 bp upstream of the hbpC stop codon, and 5′-TCT AGA CAT AGC GCC AGC CGG ACC-3′, 59 bp downstream of the hbpA start codon and 5′-GCA TGC GTA ACC GGT TGG TGA ACC-3′, 3 bp upstream of the hbpA stop codon, and 5′-TCT AGA TCC ATT CAA TGA GCC TGC C-3′, 3 bp downstream of the hbpD start codon, respectively. The cloning of the PCR-generated DNA fragments into pT7Blue(R) T vector (Novagen) resulted in plasmids pHYBP101 (region 2) and pHYBP102 (region 3). The inserts of plasmids pHYBP101 and pHYBP102 were sequenced and confirmed to be identical with the original hbp sequence. The inserts were then recovered as SphI-XbaI fragments and ligated into luxAB-based promoter-probe vector pJAMA8 (20), as outlined in Fig. 1C. After transformation this resulted in plasmids pHYBP105 and pHYBP106, respectively. The hbpC-hbpA intergenic region of plasmid pHYBP105 was extended in plasmid pHYBP116 with a 0.24-kb upstream DNA region by exchanging the 0.16-kb SphI-SalI fragment of pHYBP105 for the 0.4-kb NarI-SalI fragment from plasmid pHBP130 (SphI and NarI were made blunt by treatment with T4 DNA polymerase) (Fig. 1C). By using the unique NotI sites at the flanks, all luxAB fusions were recovered and exchanged with the 3.2-kb NotI fragment present in Tn5 delivery vector PCK218 (24). This resulted in plasmids pHYBP104 (PhbpC-luxAB), pHYBP107 (region 2-luxAB), pHYBP108 (PhbpD-luxAB), and pHYBP118 (region 4-luxAB).
Testing hbp-lux promoter-probe constructs in E. coli.
All the different hbp-lux promoter-probe plasmids were cotransformed in E. coli with a plasmid expressing either the hbpR gene or an hbpR gene with an internal frameshift mutation (hbpRΔ). Plasmid pHYBP124 was obtained by cloning a 2.8-kb SalI-NruI DNA fragment from pHYBP122 (20), comprising the hbpR gene plus its own promoter, into pACYC184 (11) (digested with SalI and NruI). Plasmid pHYBP131 is similar to pHYBP124, except for having the hbpR gene inserted into the chloramphenicol resistance gene of pACYC184. Plasmid pHYBP125 (containing hbpRΔ) was created by first cloning a 2.8-kb NotI-XbaI fragment from pHYBP110 (20) into pUC28 (7) to give plasmid pHYBP123. Subsequently, the insert in pHYBP123 was retrieved as a 2.8-kb SalI-NruI fragment and cloned into pACYC184 (digested with SalI and NruI) to produce plasmid pHYBP125.
Single chromosomal insertion of hbp-lux promoter-probe constructs in P. azelaica.
By using mini-Tn5 delivery, the hbp-lux promoter-probe fusions of plasmids pHYBP104, pHYBP107, pHYBP108, and pHYBP118 were inserted into the chromosome of P. azelaica, as described previously (20). Selection for P. azelaica exconjugants was done on MM plates with 50 μg of kanamycin/ml and 2.9 mM 2-HBP. Proper insertion of the constructs was verified by Southern hybridization of the P. azelaica exconjugants (data not shown).
In P. azelaica HBP108 (containing the hbpD′::lux fusion) the hbpR gene was disrupted by single recombination as described earlier (20). Obtained P. azelaica HBP108 recombinants were checked for proper disruption of the hbpR gene by PCR and Southern hybridizations (the resulting strain is referred to as HBP108121; Table 1).
Enzyme assays.
Induction experiments with luxAB-harboring E. coli and P. azelaica strains were performed by in MM at 30°C as described before (20). Expression of luciferase was analyzed by measuring bioluminescence on whole cells at a final n-decanal concentration of 2 mM in a MicroLumat LB 96 P luminometer (Berthold AG, Regensdorf, Switzerland) as described previously (47).
Activities of the HbpA, HbpC, and HbpD enzymes were measured in cell extracts prepared from 3-ml samples taken from chemostat-grown cultures of P. azelaica HBP1. Preparation of cell extract and enzyme assays for 2-hydroxybiphenyl-3-monooxygenase (HbpA), 2,3-dihydroxybiphenyl dioxygenase (HbpC), and 2-hydroxy-6-oxo-6-phenyl-2,4-dienoic acid hydrolase (HbpD) were carried out as described previously (20).
HPLC analysis.
The disappearance of 2-HBP and the formation of 2-HBP intermediates during induction of chemostat-grown cells of P. azelaica HBP1 with 0.5 mM 2-HBP were determined by high-pressure liquid chromatography (HPLC) analysis with a Gynkotek (Germering, Germany) HPLC system. The system consisted of a Gina 50 automated-injection module, a M480 G gradient pump, an on-line degasser, and a UVD 340 S photodiode array detector. The column used was a Nucleosil 100-5 C18 reversed-phase column. The mobile phase was prepared by mixing 65% of solution A (containing 10 mM H3PO4 at pH 3.0 in water) and 35% of solution B (which is 90% [vol/vol] methanol and 10% solution A). The flow rate was 0.6 ml/min. Samples were taken from the chemostat culture, cells were spun down by centrifugation, and the supernatant was acidified and stored at −20°C. Immediately before HPLC injection the supernatants were filtered through 0.2-μm-pore-size filters to remove cell debris.
RESULTS
Expression of the hbpCAD genes is mediated from two separate promoters.
Since the hbpC, hbpA, and hbpD genes displayed rather abnormally large intergenic regions (Fig. 1B), we investigated whether all three genes would be expressed from one promoter or from more. To identify possible promoters for hbpCAD expression, transcriptional fusions between different hbp intergenic regions (Fig. 1C, regions 1 to 4) and the luxAB genes of Vibrio harveyi were constructed. These fusions were then placed randomly into the chromosome of P. azelaica HBP1 by mini-Tn5 transposition. Southern analysis confirmed that all strains had acquired the transcriptional fusions by a proper transposition and not by homologous recombination of the complete Tn5-bearing plasmids at the hbp locus (data not shown).
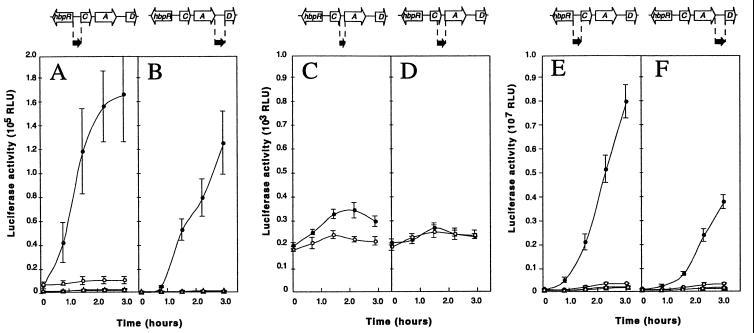
After induction with 2-HBP for 3 h, P. azelaica strain HBP104 (carrying the hbpC′::luxAB fusion) showed a 17-fold increase in bioluminescence compared to that for uninduced conditions (Fig. 2A). This confirmed our previous results that a promoter activated by HbpR was located upstream of hbpC (20). In contrast, strain HBP107 (hbpA′::luxAB) showed only a slight increase in bioluminescence activity 3 h after the addition of 2-HBP; this corresponds to a maximum induction factor of 1.6 (Fig. 2C). Furthermore, absolute luminescence activities of strain HBP107 were 45-fold lower than those of strain HBP104. From this we concluded that no, or at most a very weak, promoter was present in the region between hbpC and hbpA. To make sure that no additional promoters, activated in the presence of 2-HBP, were located further upstream, the hbpC-hbpA intergenic region was extended to include 0.3 kb of the 3′ part of the hbpC gene (Fig. 1C, region 4). A P. azelaica strain with this hbpC-hbpA′::luxAB fusion (HBP118) displayed basically the same bioluminescence levels as strain HBP107, and no induction with 2-HBP was detected (Fig. 2D). The next region we examined, i.e., the region upstream of hbpD, again showed promoter activity (Fig. 2B). Luciferase activity after 3 h of induction with 2-HBP was 75% of that observed with the hbpC′::luxAB fusion (Fig. 2A).
FIG. 2.
Promoter activity from hbp-derived intergenic regions 1 to 4 (Fig. 1) in P. azelaica (A to D) and E. coli (E and F), with a functional hbpR (circles) or with a disrupted hbpR (triangles). Experiments were carried out with 0.2 mM 2-HBP (solid symbols) or with DMSO only (open symbols). (A) P. azelaica HBP104 (hbpC′::luxAB) and HBP104121 (HBP104 with disrupted hbpR). (B) P. azelaica HBP108 (hbpD′::luxAB) and HBP108121 (HBP108 with disrupted hbpR). (C) P. azelaica HBP107 (hbpA′::luxAB). (D) P. azelaica HBP118 (hbpC-A′::luxAB). (E) E. coli DH5α with pHYBP103 (hbpC′::luxAB) plus pHYBP124 (intact hbpR) or with pHYBP103 plus pHYBP125 (disrupted hbpR). (F) E. coli DH5α with pHYBP106 (hbpD′::luxAB) plus pHYBP124 or with pHYBP106 plus pHYBP125. Solid arrows, positions and orientations of the intergenic regions with respect to the luxAB fusion. Note that the scale of luciferase activity in panels A and B is different from that in panels C to F. Error bars, standard deviations in two independent experiments each carried out in triplicate. RLU, relative light units.
In derivatives of P. azelaica strains HBP104 (hbpC′::luxAB) and HBP108 (hbpD′::luxAB) in which the hbpR gene was disrupted by homologous recombination, no induction of luciferase activity in the presence of 2-HBP was measurable (Fig. 2A and B). This indicated that expression of not only hbpC, as previously reported (20), but also hbpD was controlled by HbpR.
Interestingly, the induction of luciferase activity from the hbpD promoter showed a distinct lag, whereas that from the hbpC promoter started almost immediately after 2-HBP was added to the cells (Fig. 2A and B). For both promoter constructs we observed a maximal luciferase activity, which for PhbpC occurred at 2-HBP concentrations around 0.5 mM but which for PhbpD occurred at 2 mM 2-HBP. However, at higher 2-HBP concentrations, responses from both promoters decreased sharply. This might be caused either by a direct down regulation of the promoter response or by general toxic effects of 2-HBP or its metabolite 2,3-dihydroxybiphenyl on cellular metabolism or on the luciferase system itself (30, 50).
To exclude the possibility that the role of HbpR in activating transcription from two promoters upstream of hbpC and hbpD was indirect, we repeated the induction studies with E. coli containing a plasmid with hbpC′::luxAB or with hbpD′::luxAB plus a compatible plasmid with hbpR (pHYBP124). Luciferase activity was induced with 2-HBP from both hbpC′::luxAB and from hbpD′::luxAB in E. coli (Fig. 2E and F). This confirmed the direct role of HbpR in transcriptional activation from PhbpC and PhbpD. In contrast, a plasmid with hbpA′::luxAB did not show 2-HBP-dependent light emission in E. coli (not shown). When a plasmid with a frameshift in the open reading frame of hbpR was provided (pHYBP125) instead of a functional hbpR, none of the hbp::luxAB transcriptional fusions led to 2-HBP-dependent light emission in E. coli. In cells without functional HbpR, addition of 2-HBP even reduced background bioluminescence levels by 20 (from PhbpC) and 31% (from PhbpD).
Two separate transcripts are formed from the hbpCAD genes.
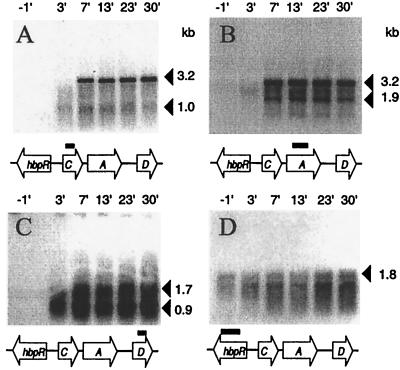
The lengths of transcripts that appeared upon the induction of the hbpCAD genes were estimated by Northern analysis with RNA isolated from a chemostat culture of wild-type strain HBP1 just before and after a shift to medium with 0.5 mM 2-HBP. For each of the hbp structural genes, transcripts were detectable only after induction with 2-HBP (Fig. 3). No apparent differences in the time of appearance of the hbp mRNAs, except that of hbpR, were detectable on Northern hybridization. Within 7 min after the induction start, hbpC-specific transcripts appeared, with estimated lengths of 1.0 and 3.2 kb (Fig. 3A). The longer (3.2-kb) transcript was also visible on blots hybridized with the hbpA probe. In addition, blots hybridized with a hbpA gene fragment showed a smaller, 1.9-kb transcript (Fig. 3B). Two clear hbpD-specific transcripts with estimated sizes of 0.9 and 1.7 kb were detected (Fig. 3C). RNA isolated only 3 min after induction with 2-HBP showed transcripts which were still incomplete. For example, the hbpCA transcript had a size of around 2.5 kb after 3 min (Fig. 3A and B). This demonstrated that the 1.0-kb transcript seen in Fig. 3A after 3 min encompasses hbpC, whereas the 1.9-kb transcript containing hbpA only cannot be seen yet, since it has to be formed from the complete 3.2-kb hbpCA transcript. In contrast, hybridization with the hbpR probe resulted in a 1.8-kb transcript, which was visible both before and after the shift, although the band intensities increased about twofold after induction with 2-HBP (Fig. 3D).
FIG. 3.
Northern analysis of the different hbp transcripts. RNA was isolated from a carbon-limited glucose-grown chemostat culture of P. azelaica HBP1, 1 min before and 3, 7, 13, 23, and 30 min after a shift to an immediate pulse of 0.5 mM 2-HBP and subsequent medium change to glucose plus 0.5 mM 2-HBP. From each sample, approximately 1 (A to C) or 8 μg (D) of RNA was blotted in each lane and hybridized with radioactively labeled probes against hbpC (A), hbpA (B), hbpD (C), or hbpR (D). Black bars, locations and sizes of the gene-specific probes. Transcript sizes are marked on the right. The presence of two bands in panel D is an artifact caused by the abundance of 16S rRNA migrating slightly below the hbpR transcript.
PhbpC and PhbpD are ς54-dependent promoters.
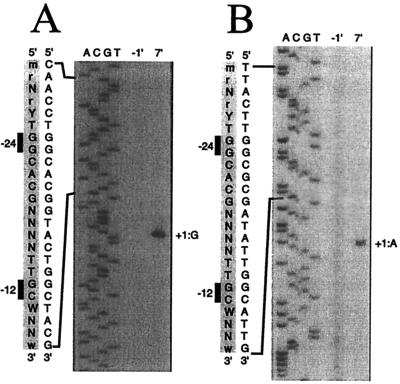
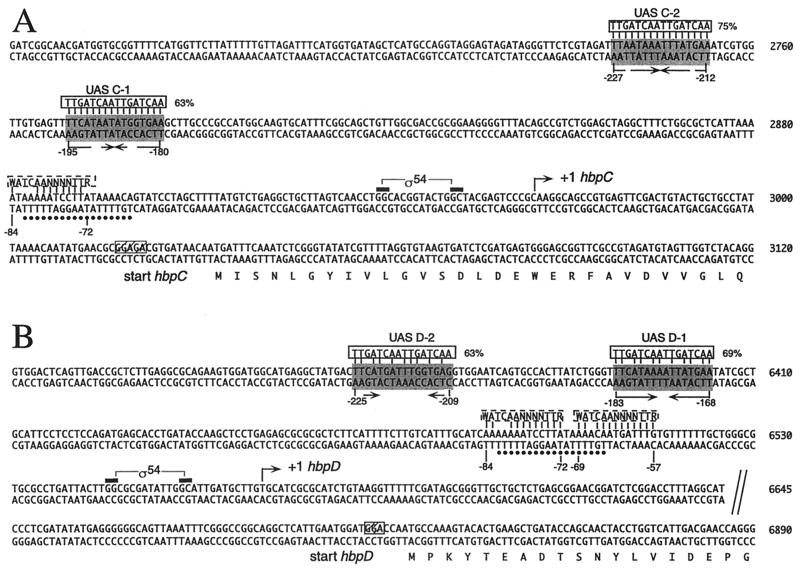
The in vivo transcriptional start sites of the hbpC and hbpD genes in P. azelaica HBP1 were determined by primer extension analysis with RNA isolated from a chemostat culture of strain HBP1 7 min after induction with 0.5 mM 2-HBP. A clear extension product could be seen for the hbpC gene after induction with 2-HBP; this product was not visible under uninduced conditions (Fig. 4A). This transcript corresponded to a transcriptional start site 68 bp upstream of the ATG start codon of the hbpC gene (Fig. 5A). For the hbpD gene, a specific cDNA was detected after 2-HBP induction; the cDNA corresponded to a transcriptional start site 133 bp upstream of the ATG start codon of the hbpD gene (Fig. 4B and 5B). This confirmed that the regions upstream of hbpC and hbpD contained a promoter activated in the presence of 2-HBP. Upstream of the transcriptional start sites of the hbpC and hbpD genes, −24 (GG)/−12 (GC) motifs, which are typical for ς54-dependent promoters, were found (Fig. 4 and 5).
FIG. 4.
Mapping of the in vivo transcriptional start sites of the hbpC (A) and the hbpD (B) genes by primer extension analysis of RNA isolated from a glucose-grown chemostat culture of P. azelaica HBP1 1 min before and 7 min after induction with 0.5 mM 2-HBP. The primer extension products were run next to products of sequence reactions performed with the same primer. +1, transcriptional start site. An expanded view of the (complementary) nucleotide sequence (5′-to-3′ direction) surrounding the ς54-dependent promoter is shown at the left of the panels. Also shown is an alignment with a consensus sequence for −24/−12 promoters (R = A or G; Y = C or T; M = A or C; W = A or T; N = any nucleotide) (5).
FIG. 5.
Sequences of the DNA regions upstream of the hbpC (A) and the hbpD (B) genes. The base pair numbering corresponds to that of the GenBank entry under accession no. U73900. Bent arrows, transcriptional start sites (+1) for the hbpC and hbpD genes and the direction of transcription; black bars, −24 and −12 elements in ς54-dependent promoters PhbpC and PhbpD; shaded boxes, putative UASs for HbpR binding (arrows underneath, palindromic structures within these DNA regions). Alignments with the consensus sequence for XylR/DmpR-type UASs (5′-TTGATCAATTGATCAA-3′; solid boxes) as proposed by Pérez-Martín and de Lorenzo (36) are shown at the top of putative UASs with numbers indicating the percentages of identity between the two DNA motifs. Dashed boxes, alignments of DNA regions similar to the consensus IHF binding site (5′-WATCAANNNNTTR-3′, where W = A or T, R = A or G, and N = any nucleotide) (15); dots, 18-bp-long DNA stretch overlapping the putative IHF sites and in common to both the hbpC and the hbpD promoters. The numbers below putative UASs and IHF sites indicate the relative positions with respect to the transcriptional start site. The N-terminal parts of HbpC and HbpD are indicated below the corresponding sequences. Hatched boxes, putative ribosome binding sites.
To establish if RNAP-ς54 was indeed involved in the transcriptional activation from PhbpC and PhbpD, induction experiments were carried out with E. coli strains devoid of the rpoN gene (i.e., E. coli ET8045). No induction was detected in E. coli ET8045 harboring plasmids pHYBP124 (with hbpR) and pHYBP103 (with hbpC′::luxAB) or plasmids pHYBP124 and pHYBP106 (with hbpD′::luxAB), whereas luciferase activity increased as expected after exposure of E. coli ET8000 to 2-HBP (Table 2). This indicated that RNAP-ς54 is the holoenzyme which is responsible for transcription from the PhbpC and PhbpD promoters.
TABLE 2.
Activation from PhbpC and PhbpD in dependency of ς54
| Strain | Plasmidsd | Luciferase activity (relative light units) ± SDa with:
|
Induction factorc | |
|---|---|---|---|---|
| No inducer | 2-HBPb | |||
| ET8000 | pHYBP103, pHYBP124 | 7.48E+04 ± 1.27E+04 | 1.49E+06 ± 2.21E+05 | 20 |
| pHYBP106, pHYBP124 | 1.89E+05 ± 9.49E+03 | 1.34E+06 ± 1.26E+05 | 7.1 | |
| ET8045e | pHYBP103, pHYBP124 | 8.50E+04 ± 5.00E+03 | 8.11E+04 ± 7.94E+03 | 0.95 |
| pHYBP106, pHYBP124 | 2.39E+05 ± 1.57E+04 | 2.49E+05 ± 1.02E+04 | 1.1 | |
All values were obtained in two independent experiments, each performed in triplicate.
Assay concentration for 2-HBP was 0.2 mM.
Induction factor is the quotient of the bioluminescence measured with and without the inducer.
pHYBP103 contains hbpC::luxAB; pHYBP106 contains hbpD::luxAB; pHYBP124 contains hbpR.
RpoN mutant.
IHF is required for transcription from PhbpC and PhbpD.
To study if additional factors were needed for transcriptional activation from PhbpC and PhbpD, we carried out similar induction experiments with E. coli strains lacking the structural genes for HU or for IHF (Table 3). In the absence of IHF, the observed induction factors obtained for the expression from PhbpC (1.2) and PhbpD (0.96) were much lower than those in the presence of IHF (ratios of 20 and 2.9, respectively). This indicated that transcription from PhbpC and PhbpD upon induction with 2-HBP was restricted in the absence of IHF. Without HU, expression from PhbpC in the presence of 2-HBP was fourfold lower than with HU but the obtained induction factors were virtually the same (21 without HU and 20 with HU). Hence, transcription from PhbpC seemed not to be affected by the absence of HU. 2-HBP-induced expression from PhbpD in an HU-negative background, however, decreased ninefold compared to that in a wild-type background. The observed induction factor was reduced about twofold (from 2.9 to 1.5) in an HU-negative background.
TABLE 3.
Activation from PhbpC and PhbpD in dependency of IHF and HU
| Straind | Plasmidse | Luciferase activity (relative light units) ± SDa with:
|
Induction factorc | |
|---|---|---|---|---|
| No inducer | 2-HBPb | |||
| N99 | pHYBP103, pHYBP131 | 1.08E+05 ± 7.70E+03 | 2.13E+06 ± 5.06E+05 | 20 |
| pHYBP106, pHYBP131 | 1.26E+05 ± 1.53E+04 | 3.72E+05 ± 4.95E+04 | 2.9 | |
| A5475 | pHYBP103, pHYBP131 | 6.22E+04 ± 9.73E+03 | 7.21E+04 ± 1.49E+04 | 1.2 |
| pHYBP106, pHYBP131 | 5.18E+04 ± 1.26E+04 | 4.96E+04 ± 1.71E+04 | 0.96 | |
| A5196 | pHYBP103, pHYBP131 | 2.61E+04 ± 1.34E+03 | 5.56E+05 ± 4.88E+04 | 21 |
| pHYBP106, pHYBP131 | 2.74E+04 ± 3.80E+03 | 4.06E+04 ± 4.54E+03 | 1.5 | |
All values were obtained in two independent experiments, each performed in triplicate.
Assay concentration for 2-HBP was 0.2 mM.
Induction factor is the ratio of the bioluminescence measured with and without the inducer.
E. coli strains A5196 and A5475 are derivatives of E. coli N99 without HU protein and IHF, respectively.
Plasmid pHYBP103 contains hbpC::luxAB, pHYBP106 contains hbpD::luxAB, and pHYBP131 contains hbpR.
Induction dynamics of the 2-HBP pathway.
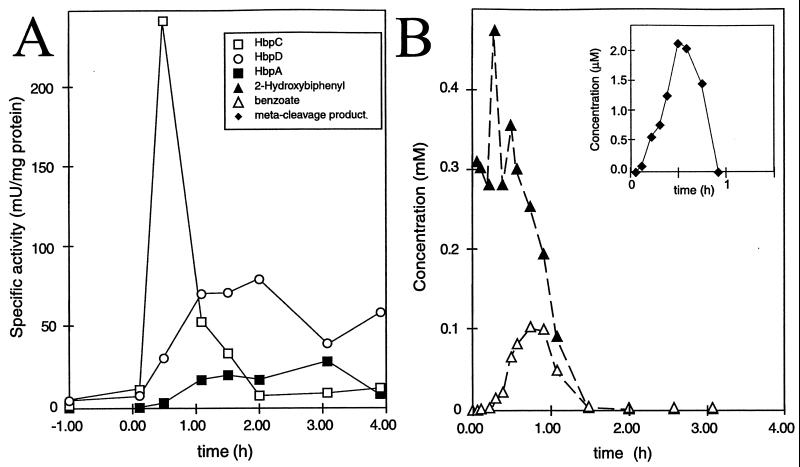
In order to determine the effectiveness of the overall induction of the 2-HBP pathway, we induced chemostat-grown cells of P. azelaica HBP1 with 2-HBP and analyzed changes in mRNA abundance, in specific enzyme activities of the 2-HBP pathway, and in metabolites during the first 4 h after induction. Steady-state cultures grown under carbon limitation with glucose displayed only basal levels of activity of the three 2-HBP-specific enzymes. After addition of 2-HBP to the chemostat to achieve an immediate concentration of 0.5 mM (and, therefore, a temporary release of carbon limitation), we observed a very fast formation of specific mRNAs for hbpCA and hbpD (Fig. 3). After approximately 1.5 h, the mRNA levels decreased to much lower levels (not shown). Specific activities of HbpC, HbpA, and HbpD started to increase from about 5 min after induction (Fig. 6). HbpC activity peaked after around 30 min at a maximum of 250 mU/mg of protein and then rapidly decreased. This decrease could be partially caused by reactions which irreversibly inhibited the enzyme (23). Activities increased much more steadily for HbpA and HbpD and did not decrease during the first 4 h after induction (Fig. 6A).
FIG. 6.
Temporary changes in enzyme activities of the HBP pathway (A) and in concentrations of 2-HBP and its intermediates (B) upon induction of a glucose-grown steady-state continuous culture of P. azelaica with 2-HBP. Time zero is the time of addition of 2-HBP to the culture. (By inset) Concentrations of 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid, the meta cleavage product. Only very low concentrations of this compound could be measured (note the difference in scale). The meta cleavage product, however, is unstable in solution at pH 7 (22).
After a short lag of about 30 min, 2-HBP started to disappear from the chemostat (Fig. 6B). The product of the first reaction, 2,3-dihydroxybiphenyl, which is formed by HbpA, could not be detected in the chemostat, probably because of very rapid conversion by an excess of HbpC enzyme. Transient accumulation of the meta cleavage product (inset of Fig. 6B) occurred, indicating that for a short time period hydrolysis of the meta cleavage product was rate limiting. Also, a temporary accumulation of benzoate, which is one of the products of the reaction catalyzed by HbpD, was measured. This indicated that the enzymes for the conversion of benzoate were absent during growth without 2-HBP. After 2 h, neither substrate nor intermediates could be detected in the supernatant of the continuous culture, even though 2-HBP was continuously supplied with the fresh medium.
DISCUSSION
Previously, we had determined that the HbpR protein, which is encoded by a 1,710-bp large open reading frame closely linked and oriented oppositely to the hbpCAD genes, is the main transcriptional activator of the 2-HBP pathway (20). As we now discovered, HbpR actually activates transcription from two separate promoters in this gene cluster. One of these (PhbpC) is located upstream of the hbpC gene, whereas the other one appears to be located upstream of the hbpD gene (PhbpD). Our evidence that HbpR is activating expression from both promoters in the hbp cluster is the following. Disruption of the hbpR gene in P. azelaica strains containing either a hbpC′::luxAB or a hbpD′::luxABfusion integrated in monocopy on the chromosome led to complete abolishment of luciferase expression in the presence of 2-HBP. Furthermore, we could show that the presence of an intact hbpR was required in E. coli to activate transcription from both hbpC and the hbpD promoter. In contrast to what was found for the regions upstream of the hbpC and hbpD genes, no promoter regulated by HbpR could be identified upstream of the hbpA gene.
The synthesis of different transcripts for the hbpCAD genes, originating from PhbpC and PhbpD upon induction with 2-HBP, was confirmed by Northern analysis. Transcripts encompassed either hbpC and hbpA (3.2 kb), as expected when starting from PhbpC, or hbpD alone (1.7 kb), when transcribed from PhbpD. Northern analysis also suggested that an effective cleavage or processing of the 3.2-kb hbpCA mRNA took place between hbpC and hbpA, resulting in the formation of an hbpC-specific (1.0-kb) and an hbpA-specific (1.9-kb) transcript. The occurrence of an hbpA-specific transcript of 1.9 kb and the absence of a specific promoter directly in front of hbpA are evidence against the possibility that a weak terminator would be present directly downstream of hbpC. The smaller 0.9-kb hbpD-specific transcript, which was observable in Northern hybridizations, might have been generated from early termination at a 27-bp-long ρ-independent terminator structure 21 bp downstream of the hbpD gene (Fig. 1D). A transcript comprising all three hbpCAD genes was not found, although some material of larger size weakly hybridized with either the hbpC, hbpA, or hbpD probe (Fig. 3). This indicates that a transcription terminator must be present downstream of the hbpA gene. The DNA sequence in this region showed a 30-bp-long inverted repeat 45 bp downstream of the hbpA gene, which might function as a ρ-independent terminator (Fig. 1D).
Activation of both the hbpC and the hbpD promoters was dependent on alternative sigma factor ς54. This became evident from mapping the transcriptional start sites of the hbpCA and hbpD transcripts and by studying expression of both promoters in E. coli mutants lacking the ς54 subunit. This is in line with other pathways regulated by XylR/DmpR subclass members, such as AphR (2), DmpR (46), MopR (43), TbuT (9), TouR (3), and XylR (reviewed in reference 39). In addition to the ς54 factor, IHF was found to be important for optimal transcriptional activation from PhbpC and PhbpD in E. coli. Examination of the DNA sequence upstream of PhbpC and PhbpD revealed one and two regions, respectively, with significant homology to the consensus IHF-binding site (15) (Fig. 5). These putative IHF-binding sites are centered on an 18-bp-long DNA stretch common to both promoters. IHF was also needed for maximum expression from the XylR-responsive Pu promoter (1, 35, 37) and for optimum in vivo expression from the DmpR-regulatable Po promoter (49). Other consensus sequence features between the hbpC and the hbpD promoters point to possible binding sites for HbpR (Fig. 5). The base pairs of these two 16-bp regions are between 63 and 75% identical to those of the consensus binding site for XylR/DmpR (36).
It was surprising to find that luciferase expression from the PhbpD promoter occurred significantly later than that from the PhbpC promoter when tested both in P. azelaica and in E. coli (Fig. 2). However, since mRNA for hbpD was not being formed any later than hbpC mRNA in P. azelaica HBP1 upon induction (Fig. 3), we must assume that the slower induction of luciferase activity from the PhbpD promoter is a translational effect. Luciferase expression from PhbpD was also lower than that from PhbpC (after the same induction time and with the same 2-HBP concentration). Although it is known that the luciferase gene may alter the promoter configuration (14), a comparison of the amounts of specific mRNA formed suggested that less hbpD mRNA than hbpCA mRNA was synthesized upon induction with 2-HBP (not shown). This may point to a suboptimal local geometry of the PhbpD promoter and explain why we found a weak but evident coassisting role of HU in the activation from PhbpD (Table 3). Aberrant spacing of the two HbpR binding sites at the PhbpD promoter might be responsible for this. From extensive work on the XylR regulator protein and its activation of the Pu promoter in Pseudomonas putida, it is known that the spacing and orientation of the two XylR-binding sites are very important for expression of Pu. Optimal expression is obtained only when both sites are oriented on the same side of the DNA helix (1, 36). The centers of the two XylR binding sites in the Pu and Ps promoters and of the DmpR binding sites in the Po promoter are separated by three complete DNA helical turns (36). For the PhbpC promoter three helical turns (assuming one DNA helical turn corresponds to 10.5 nucleotides) separated the centers of the putative HbpR-binding sites (Fig. 5B). However, the spacing for the putative HbpR-binding sites in the PhbpD promoter was four helical turns. In addition, the HbpR-binding sites at the PhbpD promoter seem rotated by 51° (when assuming no bends in the DNA) with respect to the −12/−24 region.
The hbpCA and hbpD genes of P. azelaica HBP1 are organized unusually for a catabolic gene cluster regulated by an XylR/DmpR-type activator protein (51). Only three structural genes make up the cluster, and these genes have rather large intergenic distances (315 bp between hbpC and hbpA and 831 bp between hbpA and hbpD). This may point to a relatively recent insertion of a DNA fragment containing the hbpA gene into an ancestor cluster with the hbpC and hbpD genes. Newly arranged gene clusters are interesting to study, since they may show peculiarities in the organization of transcription and particular adaptive features ensuring proper expression of the complete pathway. For the 2-HBP pathway, one of the most important tasks is to prevent the substances 2-HBP and its intermediates from becoming toxic to the cell. Both 2,3-dihydroxybiphenyl and the meta cleavage product of 2,3-dihydroxybiphenyl are product inhibitors for enzymatic activities of HbpA (48) and HbpC (23), respectively. Furthermore, 2,3-dihydroxybiphenyl may auto-oxidize to quinones which interfere with the electron transport chain (19, 50). Several features of the hbp transcriptional organization seem to be responsible for accomplishing proper induction of the 2-HBP pathway and preventing buildup of toxic intermediates. For example, the activity of HbpA (2-HBP monooxygenase) is kept low compared to that of HbpC (the extradiol dioxygenase) (20) and HbpC activity appears much faster than HbpA or HbpD activity (Fig. 6). Therefore, no accumulation of 2,3-dihydroxybiphenyl is seen in a continuous culture pulsed with 2-HBP. By having hbpC transcribed first and possibly by processing the hbpCA transcript, cells ensure that HbpC is synthesized faster than HbpA.
If at any point during evolution a fragment containing hbpA became inserted into a gene cluster with hbpC and hbpD, this would have disrupted proper transcription of hbpD, since a transcription terminator seems to be present downstream of hbpA. This might be the reason for the presence of a second separate promoter in front of hbpD, which ensures that transcription of hbpD starts simultaneously with that of hbpC. However, since translation of the hbpD mRNA seems less effective, the meta cleavage product of 2,3-dihydroxybiphenyl can accumulate rapidly. This appears to be the one feature which is not “smoothly” controlled by the cells, since the meta cleavage product can irreversibly inhibit activity of the HbpC extradiol dioxygenase (23). Therefore, fast and intensive transcription of hbpC serves two purposes, i.e., lowering the concentration of toxic 2,3-dihydroxybiphenyl and replenishing the inactivated enzyme.
ACKNOWLEDGMENTS
We thank V. de Lorenzo (Centro Nacional de Biotecnologia, CSIC, Madrid, Spain) for kindly supplying us plasmid pCK218 and strains N99, A5196, and A5475. Further we thank R. Dixon (Nitrogen Fixation Laboratory, John Innes Centre, Norwich, United Kingdom) for providing us with strains ET8000 and ET8045. The help of Christoph Werlen with the chemostat cultivations is gratefully acknowledged.
The work of M.C.M.J. was supported by grant 5001-044754 from the Swiss Priority Program Environment.
REFERENCES
- 1.Abril M-A, Buck M, Ramos J L. Activation of the Pseudomonas TOL plasmid upper pathway operon. J Biol Chem. 1991;266:15832–15838. [PubMed] [Google Scholar]
- 2.Arai H, Akahira S, Ohishi T, Maeda M, Kudo T. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology. 1998;144:2895–2903. doi: 10.1099/00221287-144-10-2895. [DOI] [PubMed] [Google Scholar]
- 3.Arenghi F L, Pinti M, Galli E, Barbieri P. Identification of the Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase regulatory gene (touR) and of its cognate promoter. Appl Environ Microbiol. 1999;65:4057–4063. doi: 10.1128/aem.65.9.4057-4063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 5.Barrios H, Valderrama B, Morett E. Compilation and analysis of ς54-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann B, Snozzi M, Zehnder A J B, van der Meer J R. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J Bacteriol. 1996;178:4367–4374. doi: 10.1128/jb.178.15.4367-4374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benes V, Hostomsky Z, Arnold L, Paces V. M13 and pUC vectors with new unique restriction sites for cloning. Gene. 1993;130:151–152. doi: 10.1016/0378-1119(93)90360-f. [DOI] [PubMed] [Google Scholar]
- 8.Bertoni G, Fujita N, Ishihama A, de Lorenzo V. Active recruitment of ς54-RNA polymerase to the Pu promoter of Pseudomonas putida: role of IHF and αCTD. EMBO J. 1998;17:5120–5128. doi: 10.1093/emboj/17.17.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrne A M, Olsen R H. Cascade regulation of the toluene-3-monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J Bacteriol. 1996;178:6327–6337. doi: 10.1128/jb.178.21.6327-6337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmona M, de Lorenzo V, Bertoni G. Recruitment of RNA polymerase is a rate-limiting step for the activation of the ς54 promoter Pu of Pseudomonas putida. J Biol Chem. 1999;274:33790–33794. doi: 10.1074/jbc.274.47.33790. [DOI] [PubMed] [Google Scholar]
- 11.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lorenzo V, Herrero M, Metzke M, Timmis K N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the ς54-dependent Pu promoter of TOL plasmid. EMBO J. 1991;10:1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández S, de Lorenzo V, Pérez-Martín J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg Å J, Pavitt G D, Higgins C F. Use of transcriptional fusions to monitor gene expression: a cautionary tale. J Bacteriol. 1994;176:2128–2132. doi: 10.1128/jb.176.7.2128-2132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman D I. Integration host factor: a protein for all reasons. Cell. 1988;55:545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- 16.Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. [Google Scholar]
- 17.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoover T R, Santero E, Porter S, Kustu S. The integration host factor stimulates interaction of RNA polymerase with NifA, the transcriptional activator for nitrogen fixation operons. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 19.Irons R D, Sawahata T. Phenols, catechols, and quinones. In: Anders M W, editor. Bioactivation of foreign compounds. Orlando, Fla: Academic Press; 1985. pp. 259–281. [Google Scholar]
- 20.Jaspers M C M, Suske W A, Schmid A, Goslings D A M, Kohler H-P E, van der Meer J R. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J Bacteriol. 2000;182:405–417. doi: 10.1128/jb.182.2.405-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler H-P E, Kohler-Staub D, Focht D D. Degradation of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1. Appl Environ Microbiol. 1988;54:2683–2688. doi: 10.1128/aem.54.11.2683-2688.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohler H-P E, Schmid A, van der Maarel M. Metabolism of 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1: production and consumption of 2,2′,3-trihydroxybiphenyl. J Bacteriol. 1993;175:1621–1628. doi: 10.1128/jb.175.6.1621-1628.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler H-P E, van der Maarel M J E C, Kohler-Staub D. Selection of Pseudomonas sp. strain HBP1 Prp for metabolism of 2-propylphenol and elucidation of the degradative pathway. Appl Environ Microbiol. 1993;59:860–866. doi: 10.1128/aem.59.3.860-866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 26.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacNeil T, MacNeil D, Tyler B. Fine-structure deletion map and complementation analysis of the glnA-glnL-glnG region in Escherichia coli. J Bacteriol. 1982;150:1302–1313. doi: 10.1128/jb.150.3.1302-1313.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendelson I, Gottesman M, Oppenheim A B. HU and integration host factor function as auxiliary proteins in cleavage of phage lambda cohesive ends by terminase. J Bacteriol. 1991;173:1670–1676. doi: 10.1128/jb.173.5.1670-1676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa Y, Moore G A. Cytotoxic effects of postharvest fungicides, ortho-phenylphenol, thiabendazole and imazalil, on isolated rat hepatocytes. Life Sci. 1995;57:1433–1440. doi: 10.1016/0024-3205(95)02106-s. [DOI] [PubMed] [Google Scholar]
- 31.Nash H A. The HU and IHF proteins: accessory factors for complex protein-DNA assemblies. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 149–179. [Google Scholar]
- 32.Ng L C, O'Neill E, Shingler V. Genetic evidence for interdomain regulation of the phenol-responsive ς54-dependent activator DmpR. J Biol Chem. 1996;271:17281–17286. doi: 10.1074/jbc.271.29.17281. [DOI] [PubMed] [Google Scholar]
- 33.Pérez-Martín J, de Lorenzo V. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc Natl Acad Sci USA. 1995;92:9392–9396. doi: 10.1073/pnas.92.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Martín J, de Lorenzo V. ATP binding to the ς54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell. 1996;86:331–339. doi: 10.1016/s0092-8674(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Martín J, de Lorenzo V. In vitro activities of an N-terminal truncated form of XylR, a ς54-dependent transcriptional activator of Pseudomonas putida. J Mol Biol. 1996;258:575–587. doi: 10.1006/jmbi.1996.0270. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Martín J, de Lorenzo V. Physical and functional analysis of the prokaryotic enhancer of the ς54-promoters of the TOL plasmid of Pseudomonas putida. J Mol Biol. 1996;258:562–574. doi: 10.1006/jmbi.1996.0269. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Martín J, Timmis K N, de Lorenzo V. Co-regulation by bent DNA. J Biol Chem. 1994;269:22657–22662. [PubMed] [Google Scholar]
- 38.Porter S C, North A K, Wedel A B, Kustu S. Oligomerization of NtrC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 39.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 40.Ravatn R, Studer S, Zehnder A J B, van der Meer J R. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J Bacteriol. 1998;180:5505–5514. doi: 10.1128/jb.180.21.5505-5514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Schirmer F, Ehrt S, Hillen W. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol. 1997;179:1329–1336. doi: 10.1128/jb.179.4.1329-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid A. Ph.D. thesis. Stuttgart, Germany: Universität Stuttgart; 1997. [Google Scholar]
- 45.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 46.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sticher P, Jaspers M C M, Stemmler K, Harms H, Zehnder A J B, van der Meer J R. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63:4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suske W A, Held M, Schmid A, Fleischmann T, Wubbolts M G, Kohler H-P. Purification and characterization of 2-hydroxybiphenyl 3-monooxygenase, a novel NADH-dependent, FAD-containing aromatic hydroxylase from Pseudomonas azelaica HBP1. J Biol Chem. 1997;272:24257–24265. doi: 10.1074/jbc.272.39.24257. [DOI] [PubMed] [Google Scholar]
- 49.Sze C C, Moore T, Shingler V. Growth phase-dependent transcription of the ς54-dependent Po promoter controlling the Pseudomonas-derived (methyl)phenol dmp operon of pVI150. J Bacteriol. 1996;178:3727–3735. doi: 10.1128/jb.178.13.3727-3735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tayama S, Nakagawa Y. Effect of scavengers of active oxygen species on cell damage caused in CHO-K1 cells by phenylhydroquinone, an o-phenylphenol metabolite. Mutat Res. 1994;324:121–131. doi: 10.1016/0165-7992(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 51.van der Meer J R. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek. 1997;71:159–178. doi: 10.1023/a:1000166400935. [DOI] [PubMed] [Google Scholar]
- 52.Wootton J C, Drummond M H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]