ABSTRACT
All known group A streptococci [GAS] are susceptible to β-lactam antibiotics. We recently identified an invasive GAS (iGAS) variant (emm43.4/PBP2x-T553K) with unusually high minimum inhibitory concentrations (MICs) for ampicillin and amoxicillin, although clinically susceptible to β-lactams. We aimed to quantitate PBP2x variants, small changes in β-lactam MICs, and lineages within contemporary population-based iGAS. PBP2x substitutions were comprehensively identified among 13,727 iGAS recovered during 2015-2021, in the USA. Isolates were subjected to antimicrobial susceptibility testing employing low range agar diffusion and PBP2x variants were subjected to phylogenetic analyses. Fifty-five variants were defined based upon substitutions within an assigned PBP2x transpeptidase domain. Twenty-nine of these variants, representing 338/13,727 (2.5%) isolates and 16 emm types, exhibited slightly elevated β-lactam MICs, none of which were above clinical breakpoints. The emm43.4/PBP2x-T553K variant, comprised of two isolates, displayed the most significant phenotype (ampicillin MIC 0.25 μg/ml) and harbored missense mutations within 3 non-PBP genes with known involvement in antibiotic efflux, membrane insertion of PBP2x, and peptidoglycan remodeling. The proportion of all PBP2x variants with elevated MICs remained stable throughout 2015-2021 (<3.0%). The predominant lineage (emm4/PBP2x-M593T/ermT) was resistant to macrolides/lincosamides and comprised 129/340 (37.9%) of isolates with elevated β-lactam MICs. Continuing β-lactam selective pressure is likely to have selected PBP2x variants that had escaped scrutiny due to MICs that remain below clinical cutoffs. Higher MICs exhibited by emm43.4/PBP2x-T553K are probably rare due to the requirement of additional mutations. Although elevated β-lactam MICs remain uncommon, emm43.4/PBP2x-T553K and emm4/PBP2x-M593T/ermT lineages indicate that antibiotic stewardship and strain monitoring is necessary.
KEYWORDS: penicillin binding protein 2×, transpeptidase domain, clinical susceptibility, group A streptococcal lineages, PBP2x catalytic site, amino substituted β-lactams
INTRODUCTION
Group A streptococci (GAS) have never been reported with clinically defined resistance to β-lactam antibiotics, possibly due to core genomic limitations or limited mechanisms for horizontal gene transfer (1). In pneumococci, complex mutational pathways contribute to β-lactam resistance (2–4), with mutations affecting the penicillin binding protein 2× (PBP2x) transpeptidase domain driving low-level nonsusceptibility to β-lactam antibiotics and higher-level resistance requiring additional mutations within pbp1a and pbp2b (5–9). Within group B streptococci (GBS), substitutions within PBP2x are also associated with low-level nonsusceptibility to β-lactams (10–15). The PBP2x transpeptidase domain is highly conserved in bacteria, with three conserved motifs critical in the polarity and conformation of the catalytic site that covalently binds to β-lactam antibiotics (16, 17). Substitutions within or near these motifs in GAS are rare (18), contrasting with pneumococcal clinical isolates where such substitutions are common (19, 20).
For pneumococci and GBS, our genome sequence-based detection of PBP variants effectively detects strains with nonsusceptibility or reduced susceptibility (13–15, 20–23). However, iGAS with MICs approaching clinical nonsusceptibility have not been detected during more than 25 years of Active Bacterial Core surveillance (ABCs, unpublished). Nonetheless, genomic-based surveillance led us to discover the first GAS strain confirmed to exhibit reduced β-lactam susceptibility (24). This strain (emm43.4/PBP2x-T553K), represented by two nearly isogenic clinical isolates, expressed MICs for ampicillin and amoxicillin of at least 4–8-fold above wild-type levels, although still minimally below clinical resistance levels. While there have been additional retrospective discoveries of GAS PBP2x variants exhibiting reduced susceptibility to β-lactams (25–27), no GAS strains have been described with MICs for the key amino-substituted β-lactams approaching those of strain emm43.4/PBP2x-T553K (24). Here, we quantitively assess PBP2x sequences and associated phenotypes from ongoing population-based invasive GAS (iGAS) surveillance, as well as routine outbreak strain characterization, within the United States.
RESULTS
PBP2x substitutions and decreased β-lactam susceptibility.
Among 13,727 isolates, 55 Pbp2x transpeptidase types were observed with 1 to 4 substitutions relative to the predominant wild-type PBP2x1 (Table 1). Of the 13,727 isolates, 2,660 were initially subjected to conventional broth microdilution testing and were representative of 46 of the 55 PBP2x types. None of these isolates exhibited MICs indicative of clinical nonsusceptibility to β-lactams (28). Thirty of the 55 variant PBP2x types, representing 340 (2.5%) of the isolates and accounting for 17 emm types, exhibited reduced susceptibility to β-lactam(s). Except for one type, PBP2x43 (T553K), the 30 PBP2x types were associated with broth dilution MICs at least 4-fold below MICs indicative of clinical nonsusceptibility. Phenotypic differences between the different PBP2x types were small and affected different patterns of β-lactams; however, they were reproducible using Etests capable of measuring very low MICs (Table 1). Most of the associations of individual substitutions with reduced susceptibility were concordant with recent independent data from GAS isolated in earlier years (26, 29).
TABLE 1.
Summary of β-lactam antibiotic susceptibility testing of PBP2x amino acid substitution strains (n = 115) and PBP2x wild-type strains (n = 18)a
| PEN (0.002–32μg/mL) |
AMP (0.016–256μg/mL) |
TAX (0.002–32μg/mL) |
FOX (0.016–256μg/mL) |
MER (0.002–32μg/mL) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBP2x | Total observed | PBP2x substitution | Isolates tested | Predominant MIC | MIC range | Predominant MIC | MIC range | Predominant MIC | MIC range | Predominant MIC | MIC range | Predominant MIC | MIC range |
| 1 | 10712 | Wild type | 18 | 0.008 | 0.004–0.008 | 0.016 | <0.016–0.016 | 0.012 | 0.006–0.012 | 0.38 | 0.38–0.5 | 0.003 | 0.002–0.006 |
| 43 | 2 | T553K b | 2 | 0.012 | 0.25 | 0.023 | 0.5–0.75 | 0.003 | |||||
| 10 | 198 | M593T b | 5 | 0.012 | 0.023 | 0.016–0.023 | 0.012 | 0.012–0.016 | 1 | 0.5−1.0 | 0.006 | 0.006–0.008 | |
| 46 | 4 | S562T,M593T,P676S | 4 | 0.008–0.012 | 0.016–0.023 | 0.012 | 0.008–0.016 | 0.5 | 0.5–1 | 0.006 | 0.003–0.006 | ||
| 7 | 11 | P601L b | 5 | 0.012 | 0.012–0.016 | 0.023 | 0.023 | 0.016–0.023 | 0.5 | 0.38–0.5 | 0.008 | 0.008–0.012 | |
| 16 | 17 | S562T,P601L | 6 | 0.016 | 0.008–0.016 | 0.023 | 0.016–0.032 | 0.032 | 0.012–0.032 | 0.5 | 0.012 | 0.004–0.012 | |
| 28 | 1 | M593V,P601L | 1 | 0.032 | 0.047 | 0.023 | 0.5 | 0.023 | |||||
| 53 | 1 | S562T,M593T,P601L,P676S | 1 | 0.032 | 0.047 | 0.032 | 0.75 | 0.012 | |||||
| 20 | 11 | P601H b | 3 | 0.012 | 0.008–0.012 | 0.023 | 0.016–0.023 | 0.008 | 0.008–0.012 | 0.25 | 0.19–0.25 | 0.003–0.006 | |
| 21 | 1 | S562T,P601H | 1 | 0.012 | 0.023 | 0.006 | 0.25 | 0.004 | |||||
| 25 | 2 | M593L b | 2 | 0.012 | 0.016–0.023 | 0.012 | 0.75 | 0.004–0.006 | |||||
| 50 | 3 | S562T,M342I | 3 | 0.008 | 0.008–0.012 | 0.016 | 0.012 | 0.012–0.016 | 0.5–1.5 | 0.003 | 0.003–0.004 | ||
| 32 | 2 | M342I b | 1 | 0.012 | 0.016 | 0.008 | 0.75 | 0.003 | |||||
| 44 | 1 | T341A,S562T | 1 | 0.008 | 0.016 | 0.008 | 0.75 | 0.004 | |||||
| 55 | 1 | T341S,S562T | 1 | 0.016 | 0.023 | 0.012 | 1.5 | 0.006 | |||||
| 47 | 23 | K410E | 12 | 0.006 | 0.006–0.008 | 0.016 | 0.008–0.012 | 0.75 | 0.5–1 | 0.002–0.003 | |||
| 14 | 26 | I502V,A397V | 2 | 0.006–0.008 | 0.016 | 0.012 | 0.75 | 0.006 | |||||
| 18 | 22 | A397V | 2 | 0.006 | 0.016 | 0.012 | 0.75 | 0.006 | |||||
| 52 | 1 | G403E | 1 | 0.012 | 0.016 | 0.016 | 0.38 | 0.004 | |||||
| 34 | 1 | F274L b | 1 | 0.012 | 0.023 | 0.012 | 1.5 | 0.004 | |||||
| 35 | 1 | F274Lb,S562T | 1 | 0.012 | 0.023 | 0.012 | 1 | 0.004 | |||||
| 22 | 1 | G288S b | 1 | 0.012 | 0.023 | 0.016 | 1.5 | 0.006 | |||||
| 23 | 1 | I463L | 1 | 0.008 | 0.016 | 0.012 | 0.75 | 0.004 | |||||
| 27 | 1 | I458V | 1 | 0.008 | 0.016 | 0.012 | 0.75 | 0.004 | |||||
| 45 | 2 | P676T | 2 | 0.008–0.012 | 0.016 | 0.012 | 0.5–1 | 0.003–0.004 | |||||
| 30 | 1 | N357H,I502V,P676S | 1 | 0.012 | 0.023 | 0.012 | 0.75 | 0.004 | |||||
| 36 | 1 | I502N,G600D | 1 | 0.012 | 0.023 | 0.016 | 0.5 | 0.004 | |||||
| 39 | 1 | S562T,G643R | 1 | 0.012 | 0.023 | 0.016 | 0.5 | 0.006 | |||||
| 6 | 1 | S562T,L602F | 1 | 0.008 | 0.016 | 0.012 | 0.75 | 0.003 | |||||
| 54 | 1 | G647D | 1 | 0.012 | 0.023 | 0.016 | 0.5 | 0.003 | |||||
| 56 | 1 | D125Y,Y572H | 1 | 0.023 | 0.032 | 0.012 | 1.5 | 0.003 | |||||
| 2 | 306 | D353A,P676S | 2 | 0.006–0.008 | 0.016 | 0.012 | 0.38 | 0.006 | |||||
| 3 | 1309 | S562T | 5 | 0.008 | 0.006–0.008 | 0.016 | 0.012 | 0.008–0.012 | 0.5 | 0.38–0.5 | 0.004 | 0.002–0.004 | |
| 8 | 740 | I502V,P676S | 2 | 0.008 | 0.016 | 0.016 | 0.5 | 0.004 | |||||
| 24 | 106 | V626I | 6 | 0.008 | 0.006–0.012 | 0.016 | 0.016 | 0.006–0.016 | 0.5 | 0.38–0.75 | 0.003 | 0.002–0.004 | |
| 4 | 114 | G600Db | 2 | 0.006–0.008 | 0.016 | 0.008–0.012 | 0.25–0.38 | 0.004–0.006 | |||||
| 9 | 34 | S562T,P676S | 2 | 0.006 | 0.016 | 0.008–0.012 | 0.38–0.5 | 0.003 | |||||
| 15 | 19 | T649I | 2 | 0.008 | 0.016 | 0.012–0.016 | 0.5 | 0.006 | |||||
| 12 | 16 | M593Vb | 2 | 0.006–0.008 | 0.016 | 0.008 | 0.38 | 0.006 | |||||
| 42 | 7 | P676S | 7 | 0.008 | 0.006–0.012 | 0.016 | 0.012 | 0.006–0.012 | 0.38–0.75 | 0.003 | 0.002–0.004 | ||
| 13 | 4 | D353A | 2 | 0.006–0.008 | 0.016 | 0.008–0.012 | 0.38–0.5 | 0.004–0.016 | |||||
| 11 | 2 | T369K,S562T | 2 | 0.008 | 0.016 | 0.012 | 0.5 | 0.006 | |||||
| 31 | 3 | I502V | 2 | 0.006–0.008 | 0.016 | 0.008–0.012 | 0.5 | 0.003–0.004 | |||||
| 48 | 2 | M593I,G643R | 1 | 0.008 | 0.016 | 0.012 | 0.5 | 0.003 | |||||
| 26 | 1 | T307I | 1 | 0.008 | 0.016 | 0.012 | 0.75 | 0.003 | |||||
| 29 | 1 | T665I | 1 | 0.004 | 0.016 | 0.006 | 0.25 | 0.002 | |||||
| 33 | 1 | T346A | 1 | 0.004 | 0.016 | 0.006 | 0.19 | 0.002 | |||||
| 37 | 1 | I540L | 1 | 0.006 | <0.016 | 0.008 | 0.5 | 0.003 | |||||
| 38 | 1 | V642A | 1 | 0.008 | 0.016 | 0.008 | 0.38 | 0.004 | |||||
| 40 | 1 | S562T,E614K | 1 | 0.006 | 0.016 | 0.008 | 0.38 | 0.003 | |||||
| 41 | 1 | R632H | 1 | 0.006 | 0.016 | 0.006 | 0.38 | 0.002 | |||||
| 49 | 2 | S363I | 1 | 0.008 | 0.016 | 0.006 | 0.5 | 0.003 | |||||
| 51 | 1 | Q559H | 1 | 0.008 | 0.016 | 0.012 | 0.5 | 0.004 | |||||
| 57 | 1 | A453S | 1 | 0.004 | 0.016 | 0.006 | 0.38 | 0.002 | |||||
| 58 | 1 | S562T,Q659H | 1 | 0.006 | 0.016 | 0.012 | 0.38 | 0.003 | |||||
| 60 | 1 | G543D | 1 | 0.004 | 0.016 | 0.008 | 0.38 | 0.003 | |||||
MIC, MIC; PEN, penicillin; AMP, ampicillin; TAX, cefotaxime; FOX, cefoxitin; MER, meropenem. All antibiotic MICs are given in μg/mL. Penicillin, cefotaxime, and meropenem assays were done using low 0.002–32 μg/mL range, and ampicillin and cefoxitin assays were done using standard 0.016–256 μg/mL range. Boldface indicates PBP2x substitutions associated with elevated MIC values in comparison to wild-type MICs.
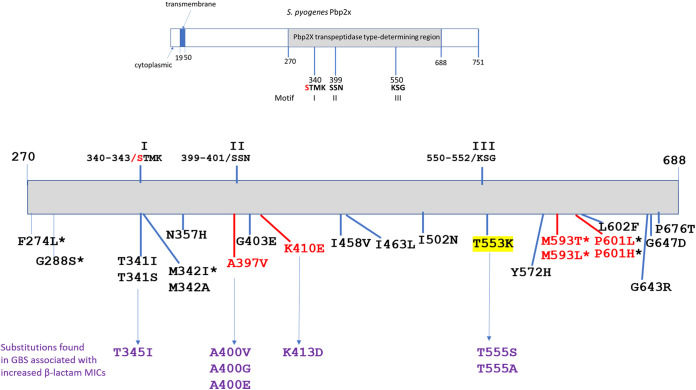
Within these 30 PBP2x types, 23 specific substitutions at one of 19 different PBP2x1 residues were observed (Table 1). Of these 19 PBP2x1 positions, 11 are conserved with the aligned GBS PBP2x protein (not shown). We recovered GBS reduced β-lactam susceptibility variants with substitutions in 4 of these 11 conserved PBP2x residues (Fig. 1) during the period of 2015 to present (13-15, unpublished). These residues are each within or closely neighboring one of the 3 conserved catalytic motifs (16–18) and correspond to the GAS PBP2x positions T341, A397, G403, and T553 (Fig. 1).
FIG 1.
Depiction of the Pbp2x full-length protein and the transpeptidase encompassing region indicating, 3 conserved catalytic site motifs, and substitutions within 19 residues (23 substitutions) associated with decreased susceptibility in this study to β-lactam antibiotics. The catalytic serine in motif I that covalently binds to β-lactam antibiotics is indicated in red. The substitution associated with the most significant MIC, T553K, is highlighted in yellow. Substitutions at 4 positions indicated in red font accounted for 322/340 (94.7%) of the isolates with reduced susceptibility to β-lactams and occurred at positions M593 (205 isolates), P601 (42 isolates), A397 (48 isolates), and K410 (23 isolates). Corresponding substitutions in the GBS PBP2x associated with reduced susceptibility or nonsusceptibility to β-lactams are indicated below. Substitutions reported in other studies (25–27, 29) to be associated with reduced β-lactam susceptibility in naturally occurring isolates or through allelic exchange experiments are indicated with an asterisk.
Five PBP2x types associated with decreased β-lactam susceptibility were present in multiple genetic backgrounds (PBP2x types 10, 7, 20, 32, and 45), with PBP2x-10 found among 4 different emm types (Table 2). Reduced susceptibility sublineages (emm type/PBP2x type combinations) accounting for multiple isolates were contained within closely related phylogenetic clusters, indicating that each likely arose from the same progenitor strain. This is shown for sublineages, including 15 or more isolates within PBP2x-10 (Fig. 2 and 4), PBP2x-14 (Fig.S1), PBP2x-18 (Fig. 3, Fig. S2), and PBP2x-47 (Fig. 3, Fig. S3). Twenty-three of the 34 sublineages were represented by single isolates (Table 2).
TABLE 2.
Geographical and yearly distributions of sublineages associated with decreased β-lactam susceptibilitya
| PBP2x type |
Emm type/CC (no. isolates) |
2015 |
2016 |
2017 |
2018 |
2019 |
Partial 2020–2021 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | Sites | Isolates | Sites | Isolates | Sites | Isolates | Sites | Isolates | Sites | Isolates | Sites | ||
| 10 | 4/39 (142) | 16 | CO, GA, MD, MN, NY |
25 | CA, CO, GA, MN, NY |
35 | CA, CO, GA, MN, NM, OR, TN |
25 | CO, GA, MD, MN, OR, TN |
29 | CA, CO, GA, MD, MN, NM OR, TN |
12 | CA, CO, MN, NM, OR, TN |
| 75/49 (37) | 3 | CO, MD TN |
3 | GA, MN | 6 | GA, MN | 15 | CO, GA, MN, NM, TN |
7 | CA, MN | 3 | CA, MN | |
| 1/28 (17) | 7 | CO | 5 | CA, CO MN |
2 | CA, NM | 3 | CA, CO | 0 | 0 | |||
| 12/36 (2) | 0 | 0 | 0 | 1 | CO | 1 | TN | ||||||
| 14 | 101/182 (26) | 5 | CO, MN | 7 | CA, CO, MN, OR |
4 | CA, MN | 5 | MN | 4 | MN | 1 | OR |
| 18 | 1/28 (22) | 0 | 6 | NY | 9 | CT, NY | 4 | NY | 3 | NY | 0 | ||
| 16 | 89/101 (17) | 2 | GA, MN | 2 | GA | 4 | GA, NM | 4 | NM | 3 | OR, TN | 2 | OR |
| 47 | 1/28 (23) | 0 | 0 | 0 | 4 | CT, MN, OR |
11 | MN, NY, OR |
8 | CT, MN, NY, OR |
|||
| 7 | 75/861 (3)/49(3) | 3 | GA, TN | 3 | GA | 0 | 0 | 0 | 0 | ||||
| 1/28 (2) | 0 | 1 | NM | 0 | 1 | GA | 0 | 0 | |||||
| 11/403 (2) | 0 | 0 | 0 | 2 | CO | 0 | 0 | ||||||
| 4/39 (1) | 1 | TN | 0 | 0 | 0 | 0 | 0 | ||||||
| 20 | 75/49 (10) | 0 | 5 | CO, NM | 4 | CO | 0 | 0 | 1 | CO | |||
| 12/36 (1) | 0 | 0 | 1 | CA | 0 | 0 | 0 | ||||||
| 46 | 73/331 (4) | 0 | 0 | 0 | 3 | CA, GA, OR |
1 | NY | 0 | ||||
| 50 | 89/101 (3) | 0 | 0 | 0 | 3 | TN, PA | 0 | 0 | |||||
| 25 | 2/55 (2) | 0 | 1 | MN | 0 | 1 | MN | 0 | 0 | ||||
| 32 | 1/28 (1) | 0 | 0 | 0 | 0 | 1 | GA | 0 | |||||
| 75/49 (1) | 0 | 1 | NM | 0 | 0 | 0 | 0 | ||||||
| 43 | 43/3 (2) | 0 | 0 | 0 | 2 | WA | 0 | 0 | |||||
| 45 | 19/769 (1) | 0 | 0 | 0 | 0 | 1 | MD | 0 | |||||
| 65/1055 (1) | 0 | 0 | 1 | CO | 0 | 0 | 0 | ||||||
| Remaining 17 types | 1,2,12,73,77,81, 82,87,89,169 |
2 | CA, NM | 6 | CA, CO, GA, MD | 2 | MN, NM | 5 | CA, MD, MN, NM, TN | 2 | NM | 0 | |
| Total | 39/1440 (2.7%) | 65/1786 (3.6%) | 68/2274 (3.0%) | 78/2412 (3.2%) | 62/2371 (2.6%) | 28/2368 (1.2%) | |||||||
ABCs, Active Bacterial Core surveillance; CA, California (ABCs); CT, Connecticut (ABCs); CO, Colorado (ABCs); GA, Georgia (ABCs); MD, Maryland (ABCs); MN, Minnesota (ABCs); NM, New Mexico (ABCs); NY, New York (ABCs); OR, Oregon (ABCs); TN, Tennessee (ABCs); PA, Pennsylvania (non ABCs); WA, Washington (non ABCs).
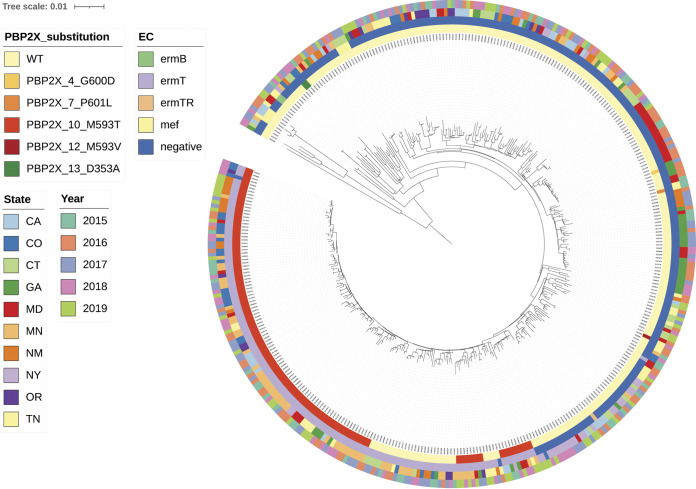
FIG 2.
Core phylogeny of emm4 isolates (all 390 recovered in 2015 to 2019 are characterized). All isolates are within clonal complex CC39. Legends for this and all other phylogenies depicted employ same scheme as follows: Innermost circle is color coded for PBP2x substitution, the second circle from inside is color coded for resistance determinants for erythromycin and clindamycin (both constitutive and inducible), the third circle from inside is depicting state, and the fourth circle from inside is depicting year of isolation. Figure depicts well established emm4/PBP2x10 sublineage recovered in multiple states and years and is the principal component of a single major branch within the overall invasive emm4 phylogeny. The emm4/ermT-positive/PBP2x1 isolates are situated on the same main branch, potentially indicating that the pbp2x10 missense mutation originated within an emm4/ermT background.
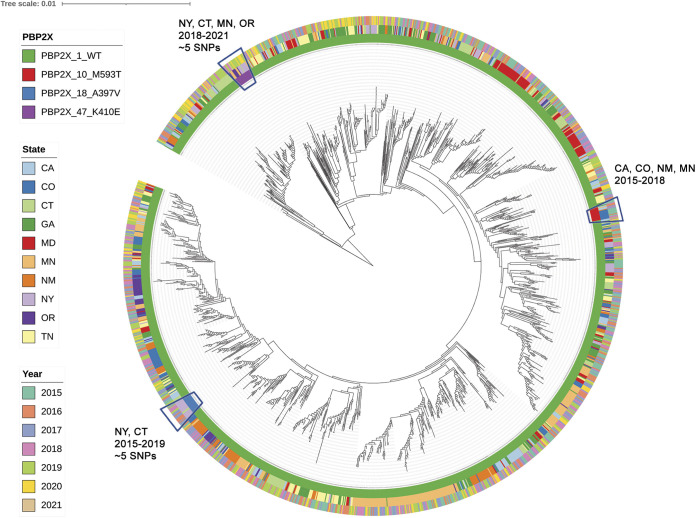
FIG 3.
Core phylogeny of emm1 sequence type ST28 lineage, average distance 45 SNPs (all 1619 characterized). PBP2x types of this lineage associated with reduced beta-lactam susceptibility and represented by 15 or more isolates were characterized. Figure depicts three distinct tight phylogenetic clusters of PBP2x10, PBP2x18, and PBP2x47 collected within multiple years and states.
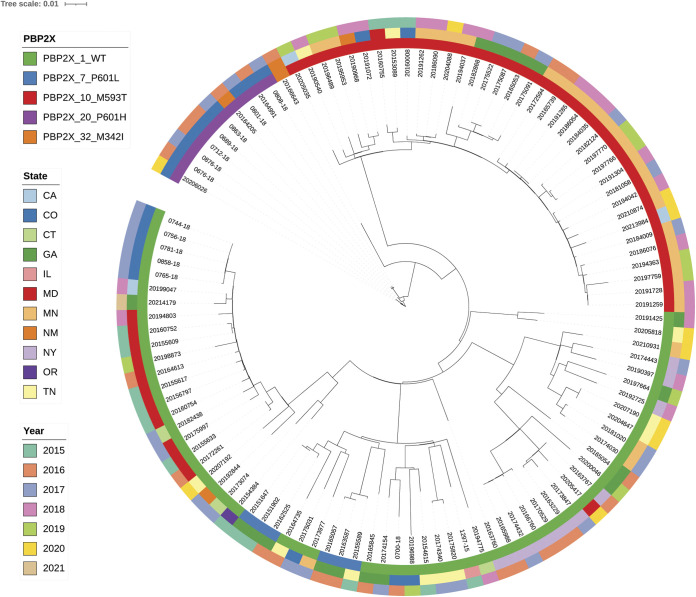
FIG 4.
Core phylogeny of emm75 sequence type ST49 lineage associated with four reduced-susceptibility PBP2x variant types (all 111 characterized). Figure depicts tight phylogenetic clusters of emm75/PBP2x10 collected during multiple years, with highest incidence in Minnesota and more recently emerged emm75/PBP2x20 within Colorado and New Mexico. Legends for this and all other phylogenies depicted employ same scheme as follows: innermost circle is color coded for PBP2x substitution, the second circle from inside is depicting state, and the third circle from inside is depicting year of isolation.
Eleven different sublineages, accounting for 9 different PBP2x types associated with reduced β-lactam susceptibility were found in multiple states and years. The emm4/PBP2x10 sublineage accounted for 116 isolates recovered throughout 2015 to 2021 within 9 of the 10 ABCs sites (Table 2, Fig. 2). Nonetheless, no sublineage revealed an obvious trend of increase during 2015 to 2021 (Table 2).
Pbp2x43 (T553K).
The most remarkable phenotype was the elevated MICs (0.25 μg/mL for ampicillin and amoxicillin obtained by both broth dilution and Etest) observed for two previously described emm43.4/PBP2x-T553K isolates from an outbreak investigation (24). The ampicillin MIC was at the breakpoint between susceptibility and resistance, and more than 10-fold higher than the basal MIC of ≤0.016 μg/mL of strains carrying PBP2x1 (Table 1) and 5- to10-fold higher than any of the other PBP2x variants (Table 1). The PBP2x-T553K isolates reproducibly displayed elevated MICs for penicillin, cefotaxime, and cefoxitin; however, these MICs were not higher than for other PBP2x variants (Table 1).
The T553K substitution was the only substitution closely neighboring the conserved PBP2x catalytic motif III (Fig. 1). In invasive GBS, corresponding substitutions T555S and T555A have been associated with reduced β-lactam susceptibility (13–15), unpublished, Fig. 1.
Fortuitously, from the same community screening, we were provided with three emm43.4/PBP2x1 isolates that were highly related to the two nearly isogenic emm43.4/PBP2x43 isolates (24). There were only 14 variable genomic positions evident among the five isolates (pairwise range of 2 to10 SNPs), six of which were genomically unlinked missense mutations exclusively shared by the two PBP2x43 isolates (Table 3). Of the six substitutions specific to the PBP2x43 variants, two included the PBP2x43 and ParCS79F substitutions (SNPs 3 and 8), and three mapped within core genes with known involvement in peptidoglycan structure and/or defense against β-lactam antibiotics (SNPs 5,13, and 11). SNP5 (YidC-G23S), corresponded to an amino acid substitution within the terminal residue of the highly predicted (despite the substitution) lipoprotein signal sequence lipoprotein cleavage site of the YidC membrane insertase/chaperone (spy0290 in NCBI accession CP000262) (30, 31). SNP13 mapped to spy1819 (accession CP000262), encoding a predicted membrane protein component of an antimicrobial efflux ABC transporter (32). The third SNP, SNP11, was a missense mutation within spy0519 (accession CP000262) encoding a GH25 class peptidoglycan hydrolase (33). In a random screening of 100 isolates among 20 emm types, these 3 non-PBP genes were invariably present, consistent with inclusion in the core GAS genome. Screened representatives within all 29 of the other PBP2x reduced-susceptibility variants carried alleles of these three genes that were identical to basally β-lactam susceptible strains of the same lineage. Subsequent to this work, we received 167 subtype emm43.4 variants, the majority of which were highly related to the two emm43.4/PBP2x-T553K isolates (average distance 10 SNPs). All of these emm43.4 isolates carried wild-type alleles of pbp2x, parC, yidC, spy1918, and spy0519.
TABLE 3.
Missense SNPs specifically associated with emm43.4/PBP2x43a
| SNP | Sequence in: emm43.4,PBP2 × 43/ emm43.4,PBP2x1 |
Amino acid substitution (protein target) | Protein function (theoretical or known association of missense derivative with antimicrobial resistance) | Relevant references |
|---|---|---|---|---|
| 3 |
AATCAGGAAAAGCACAAAT/ AATCAGGAACAGCACAAAT |
T553K (Pbp2x conserved motif) |
Peptidoglycan synthesis/cell division (Decreased Pbp2x affinity for β-lactam antibiotics) | 24 |
| 8 |
ATGGGGATTTCTCTATTTA/ ATGGGGATTCCTCTATTTA |
S79F (ParC) | DNA gyrase (fluoroquinolone-resistance) | 21 |
| 5 |
ACCTTGACAAGCTGTGTGG/ ACCTTGACAGGCTGTGTGG |
G23S (YidC signal sequence cleavage site) | Membrane transport (membrane localization of Pbp2 × 43 protein) | 30, 35 |
| 13 |
GTTAGTCAACGGTAACATC
GTTAGTCAAAGGTAACATC |
K255N (spy1819 membrane fusion protein component of resistance-nodulation-division family ABC transporter) | Antimicrobial efflux (persistence in presence of penicillin, antimicrobial efflux) | 32 |
| 11 |
GCTCCTAATGCAACCTTAG/ GCTCCTAATACAACCTTAG |
T236A (GH25 bacterial lysozyme) | 1,4-beta-N-acetylmuramidase (peptidoglycan maintenance) |
33 |
| 9 |
GGAGGACAGTCTGTGACGG
GGAGGACAGCCTGTGACGG |
P77S (Acetate CoA-transferase beta subunit) | Fatty acid biosynthesis (unknown) | Relevance unknown |
The bold/underline highlights missense SNPs in amino acid sequence.
Substitutions of positions M593 and P601.
Together, or in combination, substitutions of PBP2x positions M593 (M593T and M593L) and P601 (P601L and P601H) accounted for 9 of the 30 different PBP2x types associated with decreased β-lactam susceptibility and 246 of the 340 (72.3%) isolates (Table 1). The variety of strain backgrounds described below associated with substitutions within these 2 positions indicate that the strains described in this study are not directly related to corresponding variants described in recently published reports (25–27, 34).
PBP2x-M593T was observed in 198 isolates of 4 different lineages, including 129 (65%) emm4/ermT isolates (Fig. 2) with high-level macrolide and clindamycin-resistance. Overall, type emm4 iGAS comprised isolates with a pairwise average distance of 57 SNPs. The 129 emm4/ermT/PBP2x10 isolates had an average pairwise distance of 19.6 SNPs and comprised 129/390 (33%) of emm4 iGAS recovered during 2015 to 2019. The emm4/ermT/PBP2x-M593T variant constituted the primary component of a single major branch within the emm4 phylogeny. Since 29 emm4/ermT-positive/PBP2x1 isolates are on the same main branch, this potentially indicates that the PBP2x-M593T originated within the ermT-positive background. Strain 20154138/ermT/PBP2x1, recovered during 2015, was one of these 29 isolates and differed from its closest match (20155033/ermT/PBP2x10) by only 6 SNPs. Extensive MIC and PCR-based testing of ABCs isolates recovered during 1999 to 2008 revealed no ermT-positive emm4 strains ([35], unpublished), suggesting the first appearance of emm4/ermT strains within ABCs occurred no earlier than 2009. Macrolide-susceptible, PBP2x1 isolates differed by as few as 18 SNPs from ermT-positive isolates of both types PBP2x1 and PBP2x10, further suggesting relatively recent emergence of emm4/ermT/PBP2x10 (Fig. 2).
PBP2x-M593T strains with reduced β-lactam susceptibility were also evident within tight clusters of emm1/PBP2x10 (Fig. 3) and emm75/Pbp2x10 (Fig. 4) isolates recovered during multiple years, with highest incidence in Colorado and Minnesota, respectively (Table 2).
The two emm12/PBP2x-M593T isolates from this study were recovered during 2018 and 2020. A small cluster of closely related (avg SNP distance = 10) type emm73/PBP2x46 (M593T, S562T, P676S) isolates had the same β-lactam MICs as PBP2x10 isolates (Table 1). The M593T substitution was also recently reported among isolates with reduced β-lactam susceptibility in other countries, including Iceland (26) and Japan (27). These included a large cluster of mefA-positive emm12 variants recovered in Iceland during 2000–2005 (26); however, M593T substitutions were not reported among emm1, emm4, emm12, and emm73, and there were only two macrolide-susceptible emm12/M593T variants recovered in this study during 2018 and 2020–2021 (Table S1, Table 2). The S562T and P676S substitutions were each found within various PBP2x types, including several not associated with decreased β-lactam susceptibility, that collectively accounted for 2431 isolates (Table 1). Two unrelated isolates (Table 2) carrying the P676T substitution (PBP2x45) revealed slightly higher MICs for penicillin and cefoxitin. An isolate with the M593L substitution (PBP2x25), also had decreased β-lactam susceptibility, also consistent with a recent study (29).
P601 substitutions (P601L and P601H) were evident among 9 different lineages (42 isolates and 6 PBP types), including PBP2x7, PBP2x16, PBP2x53, PBP2x20, PBP2x21, and PBP2x28 (Table 2, Fig. 2, 4, and Fig. S4, S5, S6). The P601L substitution was the only substitution, either alone or in combination with M593T, that was associated with increased MICs for 4 or all 5 of the antimicrobials assayed (Table 1). The P601L substitution was present in 7 different lineages during 2015 to 2021, including 17 emm89/ST101 isolates (Fig. S4 and S5). A recent survey of emm types 1, 28, and 89 reported this substitution within multiple U.S. isolates and a single Finland isolate during 2008 to 2013 (25, 26).
The combined M593T and P601L substitutions evident in the single PBP2x53 variant were reproducibly associated with MICs for penicillin and ampicillin that were 2-fold higher than for the singly occurring substitutions (Table 1). The PBP2x53 variant is likely to have arisen from a point mutation of pbp2x46 within an emm73 precursor strain (Table 2, Fig. S6). Although substitution M593V (PBP2x12), was not independently associated with decreased β-lactam susceptibility, occurrence of M593V together with P601L (PBP2x28) was associated with increased penicillin and ampicillin MICs compared to P601L alone.
PBP2x16 (S562T,P601L) was associated with increased MICs to four of the five β-lactams tested and was found in encapsulated emm89/ST101 (21) strains recovered from five states during 2015 to 2019 (Fig. S5). Seven of these isolates were recovered from NM in 2017 to 2018 and shared near identity (differed by 0 to 3 SNPs) (Fig. S4). The remaining eight isolates were more distantly related. In contrast to other analyses (25, 29), we did not detect reduced β-lactam susceptibility in multiple isolates carrying the G600D substitution. A single isolate carrying PBP2x36 (G600D in combination with I502N) had slightly elevated MICs to penicillin and ampicillin (Table 1).
Substitutions within motif I.
PBP2x T341 and M342 occur within motif I (STMK) that includes the catalytic serine. The three substitutions at these two positions (T341A,T341S, M342I) were found within four different PBP2x types associated with reduced β-lactam susceptibility, although they only accounted for seven isolates. The reduced susceptibility associated with PBP2x-M342I has been corroborated in other studies (25, 29). In GBS we have encountered a substitution corresponding to PBP2x-T341A (T345I) that is associated with reduced β-lactam susceptibility (unpublished). The pneumococcal PBP2x-T341A equivalent (T338A) was highlighted in resistant mutants and PBP2x structural analyses (16, 17).
Substitutions neighboring motif II.
We detected three different substitutions (K401E/PBP2x47, A397V/PBP2x14 and PBP2x18, and G403E/PBP2x52) among 72 isolates that exhibited β-lactam MIC increases. In GBS we have also noted decreased β-lactam susceptibility in variants containing substitutions in the positions corresponding to A397 and G403 (Fig. 1) (13–15). GAS isolates with the PBP2x-A397V substitution (26 emm101/PBP2x14 isolates and 22 emm1/PBP2x18 isolates), only exhibited 2- to 3-fold increased MICs for cefoxitin. The A397V substitution was also associated with slightly increased MICs to cefotiam (second generation cephalosporin) and ceftibuten (third generation cephalosporin) in a GAS isolate recovered in Japan (27). Phylogenetic analysis revealed that the 22 emm1 isolates sharing PBP2x-A397V were highly similar (Fig. S2), with an average 5 SNPs pairwise difference. Except for two isolates from CT, all were recovered in NY during 2016 to 2019, consistent with recent transmission (Fig. S2). In contrast, emm101/ST182 GAS sharing type PBP2x14 (I502V,A397V) were recovered in five states during 2015 to 2019 and appeared to be both temporally related and unrelated strains (Fig. S1). PBP2x-K410E, conferring reduced susceptibility for cefoxitin, was detected primarily among nearly genetically identical (average 5 SNPs) emm1/ST28 strains found in four states (NY, CT, MN, OR) during 2018 to 2019 (Fig. 3, Fig. S3).
Remaining substitutions associated with reduced β-lactam susceptibility.
Substitutions within 11 additional positions mapping within the transpeptidase region (F274L, G288S, N357H, I458V, I463L, I502N, Y572H, L602F, G643R, G646D, and P676D), each corresponded to a single PPB2x type, except F274L (also in combination with S562T), and were associated with reduced β-lactam susceptibility. Other than PBP2x-P676T, found in single isolates of two lineages, these were restricted to a single isolate.
Substitutions mapping outside the transpeptidase domain.
We also screened all substitutions outside our query region (residues 270 to 688) for potential reduced susceptibility to β-lactam antibiotics. Of 20 variants (1,867 isolates) with substitution(s) within the N-terminal region (residues 1 to 269) and of 10 variants (1,215 isolates) with substitutions mapping in the C-terminal region (residues 689–751), a subset of 461 (15%) isolates that accounted for each PBP transpeptidase type were subjected to conventional MIC and Etest assays. No substitutions within these regions were associated with increased β-lactam MICs. The entire compilation of distribution of the combined PBP2x transpeptidase types/flanking substitutions from the data set is provided in Table S1.
DISCUSSION
The advent of genome sequence-based strain surveillance has provided a detection mechanism for subtle or rare antimicrobial resistance features that are difficult to discern with traditional phenotypic testing. During 2015 to 2021, we detected only 340/13,727 iGAS isolates (2.5%) that had slightly decreased susceptibility, and only two isolates with MICs at clinically defined cutoffs. Presently we see no trend of increases in decreased susceptibilities to β-lactams among iGAS; however, this was a short surveillance period. Although clinically defined β-lactam antibiotic resistance in S. pyogenes had not been reported, we theorized that should it ever occur within our surveillance catchment areas that we would detect it through our WGS bioinformatics pipeline (21). Detection of PBP2x-T553K variants led to discovery of an unprecedented GAS MIC (0.25 μg/mL) for amino substituted β-lactam antibiotics at the breakpoint of clinical nonsusceptibility (24).
The rarity of GAS higher level MICs for β-lactams might be indicative of high fitness costs associated with altering PBP2x substrate binding properties. We postulate that at least three of the shared substitutions within the two PBP2X-T553K variants were positively selected to alleviate fitness costs and/or directly contribute to the resistance phenotype. Each of these three SNPs conferred a missense mutation within a distinct core gene with documented involvement with antimicrobial resistance or peptidoglycan modification. The high relatedness of the PBP2x-T553K variants to subtype emm43.4 isolates subsequently recovered through ABCs attests to the high clustering nature of iGAS, where most cases represent highly related cluster isolates (36). The fact that none of the 167 subtype emm43.4 isolates shared mutant alleles with emm43.4/PPB2x-T553K that were associated with resistance to β-lactams or fluoroquinolones is consistent with powerful selective effects of the extensive antimicrobial treatment histories associated with both of the emm43.4/T553K iGAS cases (24). YidC, found in all life forms, plays essential roles in inserting and assembling proteins into bacterial cell membranes (37). In Escherichia coli the YidC protein is required for the functional assembly of active PBP2x (FtsI) into the cell membrane (30). The substitution within the GAS PBP2x-T553K mutants occurs at the leader peptide processing junction of the unprocessed YidC and could affect the amount of the functional processed form of YidC produced to assemble more effectively a functional PBP2x-T553K derivative within the cell membrane. SNP13 mapped within a GAS gene encoding a membrane component of an ABC-type transporter involved in drug efflux that was highly upregulated in penicillin-persister strains (32). Finally, SNP11 occurred within the ubiquitous GH25 class of bacterial lysozymes that serve essential roles in cell wall maintenance (33). Although the contributions of these three unlinked, non-PBP mutations to the MICs or fitness of the PBP2x43 isolates are not proven, their known involvement in β-lactam resistance and peptidoglycan structure is unlikely to be coincidental and may have roles that alleviate fitness costs or contribute to the resistance phenotypes observed within the PBP2x-T553K mutants. The markedly elevated MICs of type PBP2x43 isolates for amino-substituted antibiotics ampicillin and amoxicillin compared to other β-lactams is consistent with previous observations in pneumococci where the GAS PBP2x-T553 counterpart (T550) residue was found to be important in substrate specificity (38, 39).
Other than for PBP2x43, the increases of β-lactam MICs in iGAS associated with pbp2x missense mutations have been small and well below clinical nonsusceptibility breakpoints (25–27). The few mutant PBP2x types that have shown widespread emergence do not show trending increases in ABCs. Still, the emm4/ermT/PBP2x10 (M593T) variant is concerning. This variant combines increased MICs for β-lactam antibiotics with high-level macrolide/lincosamide resistance and is circulating within 9 of the 10 ABCs sites. Although not yet detected within the emm4/ermT background, the combination of M593T and P601L seen within type PBP2x53 is associated with somewhat higher MICs and that have apparently arisen in a stepwise manner within an emm73 lineage. Further, the P601L substitution has been shown to increase GAS strain fitness in a mouse model of necrotizing fasciitis in the presence of subtherapeutic levels of penicillin (40). The potential of PBP2x variants to confer increased fitness in disease and during asymptomatic carriage is ample reason to continue monitoring for subtle increases in β-lactam MICs and potential emergence of resistant strains. Although not currently detected within ABCs, the emm43/PBP2x-T553K variant that exhibits amino-substituted β-lactam MICs at the breakpoints that define nonsusceptibility is cause for concern. This strain showed no growth defects in in vitro growth and potentially has a fitness advantage in carriage and disease (24).
The PBP2x substitutions that were not associated with decreases in antimicrobial susceptibility also require monitoring, as these variants could still have survival advantages in the presence of β-lactams. For example, a missense mutation mapping within the transglycosylase domain of the bifunctional transglycosylase/transpeptidase PBP1b was described in pneumococci associated with increased propensities to cause meningitis and was found to persist longer in the presence of β-lactams (41). PBP2x is an essential class B PBP that lacks a transglycosylase region, and in pneumococcal clinical isolates the substitutions within the broad transpeptidase domain that affect conformation or polarity of its active site cavity most commonly affect MICs to β-lactams (16, 17). Nonetheless, substitutions not directly affecting the binding of β-lactams to the active site of PBP2x can decrease susceptibility to β-lactams (42).
In summary, we see no significant emergence of decreased β-lactam susceptibility in iGAS during multistate facilitated population-based strain surveillance. Ours and other recent data (25–27) indicate that these PBP2x variants are not new but do indicate that until now methods to detect increased GAS β-lactam MICs have lacked adequate sensitivity. These data strongly indicate the need for strict stewardship of these antibiotics and for continued monitoring of GAS strains for the potential emergence of strains that cross clinical breakpoints for nonsusceptibility. β-lactam antibiotics remain the frontline antimicrobials of choice for GAS disease. There is little doubt that the PBP2x variants described here and elsewhere have arisen and disseminated throughout the United States as a result of continued β-lactam selective pressure (29).
MATERIALS AND METHODS
Isolates.
All iGAS isolates were characterized as previously described (21, 34, 36). The surveillance areas included 10 sites during 2015 to 2021, with populations ranging from 33.7 to 34.6 million residents (https://www.cdc.gov/abcs/reports-findings/surv-reports.html). Isolates were recovered from five full years of population-based iGAS surveillance (2015 to 2019) and 2 years (2020 to 2021) that is currently incompletely compiled. An invasive case is defined as illness with isolation of GAS from either a normally sterile site or a wound culture accompanied by necrotizing fasciitis or streptococcal toxic shock syndrome, in a resident of the surveillance site. GAS received from state laboratory requests for assistance in outbreak investigations (24) were also characterized.
Sequencing and bioinformatics.
Genomic sequences were generated with Illumina instruments (21, 34) and deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProjects accession numbers PRJNA395240 and PRJNA559889. Our bioinformatics pipeline was employed ([34], https://github.com/BenJamesMetcalf/GAS_Scripts_Reference).
Full-length (751 residues) PBP2x sequences were extracted from all isolates, with PBP2x transpeptidase types assigned based upon amino acids 270 – 688 (Table S2). Each type was assigned a number for comparison with full-length reference PBP2x-1 (GenBank accession AE004092). In all isolates, substitutions flanking the type-determining region were combined with transpeptidase type and recorded.
Single-nucleotide polymorphisms (SNPs) were determined for core genomes employing kSNP3.0 with kmer size of 19 (43). Pairwise comparisons were generated employing Mega7 (44). Phylogenetic trees were generated using iTOL (45) from core genomic matrices with the maximum likelihood method based upon the general time-reversible model.
Susceptibility testing.
MICs were determined for ampicillin, penicillin G, cefotaxime, cefoxitin, and meropenem by broth microdilution and by the agar diffusion gradient method (Etest, bioMérieux) for selected isolates. MICs were recorded independently by three investigators. Broth dilution panels employed much higher minimum concentrations for ampicillin (0.03 μg/mL), penicillin G (0.015 μg/mL), cefotaxime (0.015 μg/mL), cefoxitin (2 μg/mL), and meropenem (0.06 μg/mL) compared to the Etest ranges shown in Table 1.
The CDC determined that this surveillance project was not human subjects research; therefore, CDC Institutional Review Board approval was not required.
ACKNOWLEDGMENTS
We are indebted to all the hospitals and laboratories participating in the Active Bacterial Core Surveillance component of the Emerging Infections Programs network, a collaboration of the CDC, state health departments, and universities. We are grateful to the Minnesota Department of Public Health laboratory for susceptibility testing of all the isolates recovered in Minnesota. We acknowledge the following members of the Active Bacterial Core surveillance team and others for their contributions at the study sites: Paul Cieslak (OR), Ghinwa Dumyati (NY), Monica Farley (GA), Ruth Lynfield (MN), Alison Muse (NY), Sue Petit (CT), Bill Schaffner (TN), Nancy Spina (NY), James Watt (CA), Rachel Herlihy (CO).
This study used the S. pyogenes MLST website (http://pubmlst.org/spyogenes/) at the University of Oxford (K. A. Jolley and M. C. J. Maiden, BMC Bioinformatics 11:595, 2010, https://doi.org/10.1186/1471-2105-11-595). The development of this site has been funded by the Wellcome Trust.
Major funding for this work was provided through support from the CDC Advanced Molecular Detection (AMD) initiative and the CDC Emerging Infection Program.
We have no conflicts of interest to report.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Horn DL, Zabriskie JB, Austrian R, Cleary PP, Ferretti JJ, Fischetti VA, Gotschlich E, Kaplan EL, McCarty M, Opal SM, Roberts RB, Tomasz A, Wachtfogel Y. 1998. Why have group A streptococci remained susceptible to penicillin? Report on a symposium. Clin Infect Dis 26:1341–1345. 10.1086/516375. [DOI] [PubMed] [Google Scholar]
- 2.Hakenbeck R, Tarpay M, Tomasz A. 1980. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 17:364–371. 10.1128/AAC.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hakenbeck R, Tornette S, Adkinson NF. 1987. Interaction of non-lytic beta-lactams with penicillin-binding proteins in Streptococcus pneumoniae. J Gen Microbiol 133:755–760. 10.1099/00221287-133-3-755. [DOI] [PubMed] [Google Scholar]
- 4.Hakenbeck R, Grebe T, Zähner D, Stock JB. 1999. beta-lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol Microbiol 33:673–678. 10.1046/j.1365-2958.1999.01521.x. [DOI] [PubMed] [Google Scholar]
- 5.Barcus VA, Ghanekar K, Yeo M, Coffey TJ, Dowson CG. 1995. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol Lett 126:299–303. 10.1111/j.1574-6968.1995.tb07433.x. [DOI] [PubMed] [Google Scholar]
- 6.Coffey TJ, Daniels M, McDougal LK, Dowson CG, Tenover FC, Spratt BG. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother 39:1306–1313. 10.1128/AAC.39.6.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krauss J, van der Linden M, Grebe T, Hakenbeck R. 1996. Penicillin-binding proteins 2x and 2b as primary PBP targets in Streptococcus pneumoniae. Microb Drug Resist 2:183–186. 10.1089/mdr.1996.2.183. [DOI] [PubMed] [Google Scholar]
- 8.Grebe T, Hakenbeck R. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob Agents Chemother 40:829–834. 10.1128/AAC.40.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albarracín Orio AG, Piñas GE, Cortes PR, Cian MB, Echenique J. 2011. Compensatory evolution of PBP mutations restores the fitness cost imposed by β-lactam resistance in Streptococcus pneumoniae. PLoS Pathog 7:e1002000. 10.1371/journal.ppat.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahesh S, Hensler ME, Van Sorge NM, Gertz RE, Schrag S, Nizet V, Beall BW. 2008. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob Agents Chemother 52:2915–2918. 10.1128/AAC.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura K, Suzuki S, Wachino J-i, Kurokawa H, Yamane K, Shibata N, Nagano N, Kato H, Shibayama K, Arakawa Y. 2008. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob Agents Chemother 52:2890–2897. 10.1128/AAC.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano N, Nagano Y, Toyama M, Kimura K, Shibayama K, Arakawa Y. 2014. Penicillin-susceptible group B streptococcal clinical isolates with reduced cephalosporin susceptibility. J Clin Microbiol 52:3406–3410. 10.1128/JCM.01291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metcalf BJ, Chochua S, Gertz RE, Hawkins PA, Ricaldi J, Li Z, Walker H, Tran T, Rivers J, Mathis S, Jackson D, Glennen A, Lynfield R, McGee L, Beall B. Jr., Active Bacterial Core surveillance team . 2017. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the United States. Clin Microbiol Infect 23:574.e7–14. 10.1016/j.cmi.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 14.McGee L, Chochua S, Li Z, Mathis S, Rivers J, Metcalf B, Ryan A, Alden N, Farley MM, Harrison LH, Snippes Vagnone P, Lynfield R, Smelser C, Muse A, Thomas AR, Schrag S, Beall BW. 2021. Multistate, population-based distributions of candidate vaccine targets, clonal complexes, and resistance features of invasive group B Streptococci within the United States, 2015–2017. Clin Infect Dis 72:1004–1013. 10.1093/cid/ciaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi M, McGee L, Chochua S, et al. 2020. Low but increasing prevalence of reduced beta-lactam susceptibility among invasive group B streptococcal isolates, US population-based surveillance, 1998–2018. Open Forum Infect Dis 8:ofaa634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon E, Mouz N, Duee E, Dideberg O. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme Form: implication in drug resistance. J Mol Biol 299:477–485. 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 17.Pernot L, Chesnel L, Le Gouellec A, Croizé J, Vernet T, Dideberg O, Dessen A. 2004. PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to β-lactam antibiotics. J Biol Chem 279:16463–16470. 10.1074/jbc.M313492200. [DOI] [PubMed] [Google Scholar]
- 18.Hayes A, Lacey JA, Morris JM, Davies MR, Tong SYC. 2020. Restricted sequence variation in Streptococcus pyogenes penicillin binding proteins. mSphere 5:e00090-20. 10.1128/mSphere.00090-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laible G, Spratt BG, Hakenbeck R. 1991. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol 5:1993–2002. 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, Walker H, Hawkins PA, Tran T, Whitney CG, McGee L, Beall BW. 2016. Penicillin-binding protein transpeptidase signatures for tracking and predicting β-lactam resistance levels in Streptococcus pneumoniae. mBio 7:e00756-16. 10.1128/mBio.00756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chochua S, Metcalf BJ, Li Z, Rivers J, Mathis S, Jackson D, Gertz RE, Srinivasan V, Lynfield R, Van Beneden C, McGee L, Beall B. 2017. Population and whole genome sequence based characterization of invasive group A Streptococci recovered in the United States during 2015. mBio 8:e01422-17. 10.1128/mBio.01422-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metcalf BJ, Gertz RE, Gladstone RA, Walker H, Sherwood LK, Jackson D, Li Z, Law C, Hawkins PA, Chochua S, Sheth M, Rayamajhi N, Bentley SD, Kim L, Whitney CG, McGee L, Beall B. 2016. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect 22:60.e9–60.e29. 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, Walker H, Hawkins PA, Tran T, McGee L, Beall BW, on behalf of the Active Bacterial Core surveillance team . 2017. Validation of β-lactam minimum inhibitory concentration predictions for pneumococcal isolates with newly encountered penicillin binding protein (PBP) sequences. BMC Genomics 18:621. 10.1186/s12864-017-4017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vannice KS, Ricaldi J, Nanduri S, Fang FC, Lynch JB, Bryson-Cahn C, Wright T, Duchin J, Kay M, Chochua S, Van Beneden CA, Beall B. 2020. Streptococcus pyogenes pbp2x mutation confers reduced susceptibility to β-lactam antibiotics. Clin Infect Dis 71:201–204. 10.1093/cid/ciz1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser JM, Beres SB, Zhu L, Olsen RJ, Vuopio J, Hyyryläinen H-L, Gröndahl-Yli-Hannuksela K, Kristinsson KG, Darenberg J, Henriques-Normark B, Hoffmann S, Caugant DA, Smith AJ, Lindsay DSJ, Boragine DM, Palzkill T. 2020. Reduced in vitro susceptibility of Streptococcus pyogenes to beta-lactam antibiotics associated with mutations in the pbp2x gene is geographically widespread. J Clin Microbiol 58:e01993-19. 10.1128/JCM.01993-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southon SB, Beres SB, Kachroo P, Saavedra MO, Erlendsdóttir H, Haraldsson G, Yerramilli P, Pruitt L, Zhu L, Musser JM, Kristinsson KG. 2020. Population genomic molecular epidemiological study of macrolide-resistant Streptococcus pyogenes in Iceland, 1995 to 2016: identification of a large clonal population with a pbp2x mutation conferring reduced in vitro β-lactam susceptibility. J Clin Microbiol 58:e00638-20. 10.1128/JCM.00638-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda T, Suzuki R, Jin W, Wachino J-I, Arakawa Y, Kimura K. 2021. Isolation of group A Streptococci with reduced In Vitro β-lactam susceptibility harboring amino acid substitutions in penicillin-binding proteins in Japan. Antimicrob Agents Chemother 65:e0148221. 10.1128/AAC.01482-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2018. M100 performance standards for antimicrobial susceptibility testing, 29th ed CLSI, Wayne, PA. [Google Scholar]
- 29.Beres SB, Zhu L, Pruitt L, Olsen RJ, Faili A, Kayal S, Musser JM. 2022. Integrative reverse genetic analysis identifies polymorphisms contributing to decreased antimicrobial agent susceptibility in Streptococcus pyogenes. mBio 13:e0361821. 10.1128/mbio.03618-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Sousa Borges A, de Keyzer J, Driessen AJM, Scheffers D-J. 2015. The Escherichia coli membrane protein insertase YidC assists in the biogenesis of penicillin binding proteins. J Bacteriol 197:1444–1450. 10.1128/JB.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teufel F, Armenteros JJA, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, Winther O, Brunak S, Heijne GV, Nielsen H. 2021. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nature Biotechnology 40:1023–1025. 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martini CL, Coronado AZ, Melo MCN, Gobbi CN, Lopez ÚS, de Mattos MC, Amorim TT, Botelho AMN, Vasconcelos ATR, Almeida LGP, Planet PJ, Zingali RB, Figueiredo AMS, Ferreira-Carvalho BT. 2021. Cellular growth arrest and efflux pumps are associated with antibiotic persisters in Streptococcus pyogenes induced in biofilm-like Environments. Front Microbiol 12:716628. 10.3389/fmicb.2021.716628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32:259–286. 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Rivers J, Mathis S, et al. 2020. Genomic surveillance of Streptococcus pyogenes strains causing invasive disease, United States, 2016–2017. Front Microbiol 11:1547. 10.3389/fmicb.2020.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodbury RL, Klammer KA, Xiong Y, Bailiff T, Glennen A, Bartkus JM, Lynfield R, Van Beneden C, Beall BW, Active Bacterial Core Surveillance Team . 2008. Plasmid-borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrob Agents Chemother 52:1140–1143. 10.1128/AAC.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metcalf B, Nanduri S, Chochua S, Li Y, Fleming-Dutra K, McGee L, Beall B. 2022. Cluster transmission drives invasive group A Streptococcus disease within the US and is focused on communities experiencing disadvantage. J Infect Dis Online ahead of print. 10.1093/infdis/jiac162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samuelson JC, Chen M, Jiang F, Möller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637–641. 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 38.Sifaoui F, Kitzis MD, Gutmann L. 1996. In vitro selection of one-step mutants of Streptococcus pneumoniae resistant to different oral beta-lactam antibiotics is associated with alterations of PBP2x. Antimicrob Agents Chemother 40:152–156. 10.1128/AAC.40.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mouz N, Di Guilmi AM, Gordon E, Hakenbeck R, Dideberg O, Vernet T. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae: role in the specificity for beta-lactam antibiotics. J Biol Chem 274:19175–19180. 10.1074/jbc.274.27.19175. [DOI] [PubMed] [Google Scholar]
- 40.Olsen RJ, Zhu L, Musser JM. 2020. A single amino acid replacement in penicillin binding protein 2x in Streptococcus pyogenes significantly increases fitness on subtherapeutic benzylpenicillin treatment in a mouse model of necrotizing myositis. Am J Pathol 190:1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Metcalf BJ, Chochua S, Li Z, Walker H, Tran T, Hawkins PA, Gierke R, Pilishvili T, McGee L, Beall BW. 2019. Genome-wide association analyses of invasive pneumococcal isolates identify a missense bacterial mutation associated with meningitis. Nat Commun 10:178. 10.1038/s41467-018-07997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maurer P, Todorova K, Sauerbier J, Hakenbeck R. 2012. Mutations in Streptococcus pneumoniae penicillin-binding protein 2x: importance of the C-terminal penicillin-binding protein and serine/threonine kinase-associated domains for beta-lactam binding. Microb Drug Resist 18:314–321. 10.1089/mdr.2012.0022. [DOI] [PubMed] [Google Scholar]
- 43.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877e8–282878. 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870e4–181874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I, Bork P. 2021. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download aac.00802-22-s0001.xlsx, XLSX file, 5.3 MB (5.3MB, xlsx)
Table S2. Download aac.00802-22-s0002.xlsx, XLSX file, 0.01 MB (13.2KB, xlsx)
Supplemental legends. Download aac.00802-22-s0003.pdf, PDF file, 0.9 MB (904.4KB, pdf)