ABSTRACT
Saccharibacteria Nanosynbacter lyticus strain TM7x is a member of the broadly distributed candidate phylum radiation. These bacteria have ultrasmall cell sizes, have reduced genomes, and live as epibionts on the surfaces of other bacteria. The mechanisms by which they establish and maintain this relationship are not yet fully understood. The transcriptomes of the epibiont TM7x and its host bacteria Schaalia odontolytica strain XH001 were captured across the establishment of symbiosis during both the initial interaction and stable symbiosis. The results showed a dynamic interaction with large shifts in gene expression for both species between the initial encounter and stable symbiosis, notably in transporter genes. During stable symbiosis, the host XH001 showed higher gene expression for peptidoglycan biosynthesis, mannosylation, cell cycle and stress-related genes, whereas it showed lower expression of chromosomal partitioning genes. This was consistent with the elongated cell shape seen in XH001 infected with TM7x and our discovery that infection resulted in thickened cell walls. Within TM7x, increased pili, type IV effector genes, and arginine catabolism/biosynthesis gene expression during stable symbiosis implied a key role for these functions in the interaction. Consistent with its survival and persistence in the human microbiome as an obligate epibiont with reduced de novo biosynthetic capacities, TM7x also showed higher levels of energy production and peptidoglycan biosynthesis, but lower expression of stress-related genes, during stable symbiosis. These results imply that TM7x and its host bacteria keep a delicate balance in order to sustain an episymbiotic lifestyle.
IMPORTANCE Nanosynbacter lyticus type strain TM7x is the first cultivated member of the Saccharibacteria and the candidate phyla radiation (CPR). It was discovered to be ultrasmall in cell size with a highly reduced genome that establishes an obligate epibiotic relationship with its host bacterium. The CPR is a large, monophyletic radiation of bacteria with reduced genomes that includes Saccharibacteria. The vast majority of the CPR have yet to be cultivated, and our insights into these unique organisms to date have been derived from only a few Saccharibacteria species. Being obligate parasites, it is unknown how these ultrasmall Saccharibacteria, which are missing many de novo biosynthetic pathways, are maintained at a high prevalence within the human microbiome as well as in the environment.
KEYWORDS: Nanosynbacter, RNAseq, Saccharibacteria, TM7x, actinomycetes, gingivitis, microbiome, oral microbiome, periodontitis, transcriptome
INTRODUCTION
Saccharibacteria Nanosynbacter lyticus type strain TM7x has an ultrasmall cell size (200 to 300 nm) and a highly reduced genome. It was originally isolated from the human oral cavity as an epibiont on a bacterial host (1), Schaalia odontolytica strain XH001 (formerly Actinomyces odontolyticus strain XH001) (2). N. lyticus strain TM7x later became the first cultivated species of a large bacterial group discovered from filtered groundwater, denoted as the candidate phyla radiation (CPR), a monophyletic group with reduced genomes, also now referred to as the Patescibacteria superphylum (1, 3–5). Genomes from the yet-to-be-cultivated CPR members are noted for being reduced with minimal biosynthetic capabilities (3, 4, 6). Recent work has demonstrated a high prevalence and a large genomic diversity of Saccharibacteria in the human microbiome and across mammals (5, 7, 8). The currently recognized broad presence of these bacterial epibionts generates much interest in understanding the mechanisms underlying their obligate symbiosis and, specifically, in how these ultrasmall bacteria with limited biosynthetic capacity survive and persist in their diverse niches.
Despite being obligate epibionts with reduced de novo biosynthetic capacity, the mammalian-associated Saccharibacteria have remarkable genomic diversity. One of the six major groups (G1 to G6) (5, 9), the G1, uniquely consists of both mammalian members and environmental representatives that share ~60% of their protein-coding genes (5). Furthermore, N. lyticus strain TM7x and other recently isolated G1 strains from the oral cavity have retained a highly syntenic gene arrangement with their environmental counterparts (1, 5, 7, 10). All oral Saccharibacteria genomes appear to have undergone further reductive evolution during their adaptation and evolution in mammals. As with other members of the CPR, TM7x is missing a number of major pathways, including the tricarboxylic acid cycle and the de novo synthesis of nucleotides and amino acids. However, of the recently fully-closed genomes of sequenced isolates (10, 11) from the G1 group, TM7x has the most streamlined genome of the oral Saccharibacteria. An initial analysis of the TM7x genome predicted 739 genes (1), 692 of which are protein encoding, meaning that TM7x is densely coded with a percentage coding base count of 94%. This results in short intergenic regions with no currently validated small RNAs, although only one global transcriptome study at a single time point has been reported to date (1).
Actinomyces are short rods that can typically form hyphal branched structures and grow in fastidious anaerobic conditions (12). They prefer complex media containing rich biological ingredients (e.g., brain heart infusion broth), although some strains can grow on defined media. Our previous in vitro experiments illustrated that XH001 shared most of these qualities. However, its optimal growth condition is microaerophilic (2% oxygen) rather than aerobic or anaerobic (13). When in an established coculture with TM7x, under optimal conditions, XH001 has a reduced growth rate and inhibited cell division (14). We also illustrated that TM7x tends to induce host cell stress in XH001, although at low enough levels to leave the host viable.
Through extensive in vitro studies on the dynamics of the XH001/TM7x interaction, it was found that while TM7x is unable to grow without a host bacterium, free-floating TM7x could be isolated from a coculture and could infect a new host horizontally (14). It was also observed that naive bacterial hosts, meaning those that were never part of an established coculture, underwent unusual growth behavior when infected with isolated TM7x. The initial encounter was first followed by a killing phase, in which host numbers decreased dramatically while large numbers of TM7x infected single host cells (14). This was followed by a recovery phase, in which host numbers returned to original levels while the number of TM7x on each host cell decreased. Finally, after 5 to 7 passages, the coculture reached a stable symbiosis between TM7x and the host XH001 and no longer showed drastic killing. Explorations of the TM7x-host range found that the epibiont was able to consistently grow on a single group of phylogenetically related Schaalia species, revealing this strain’s restricted host-range (15). While there were variations in the timing and extent of the killing phase, most hosts capable of establishing symbiosis with TM7x followed the same pattern as XH001: initial encounter, killing phase, recovery phase, and stable symbiosis.
To better understand the dynamics between XH001/TM7x across these different phases toward the establishment of stable symbiosis, we investigated the temporal transcriptional profiles of both species after the initiation of infection. This work characterizes the initial encounter as well as the stable symbiosis, showing the temporal expression dynamics for a member of the CPR. Extensive expression changes were found between the initial encounter and the stable symbiosis in both XH001 and TM7x, including changes in energy metabolism, transporters, and cell wall metabolism, enabling further insights into the lifestyle of this unique, mammalian-associated, ultrasmall epibiont with a highly reduced genome.
RESULTS AND DISCUSSION
Experimental design and RNA sequencing.
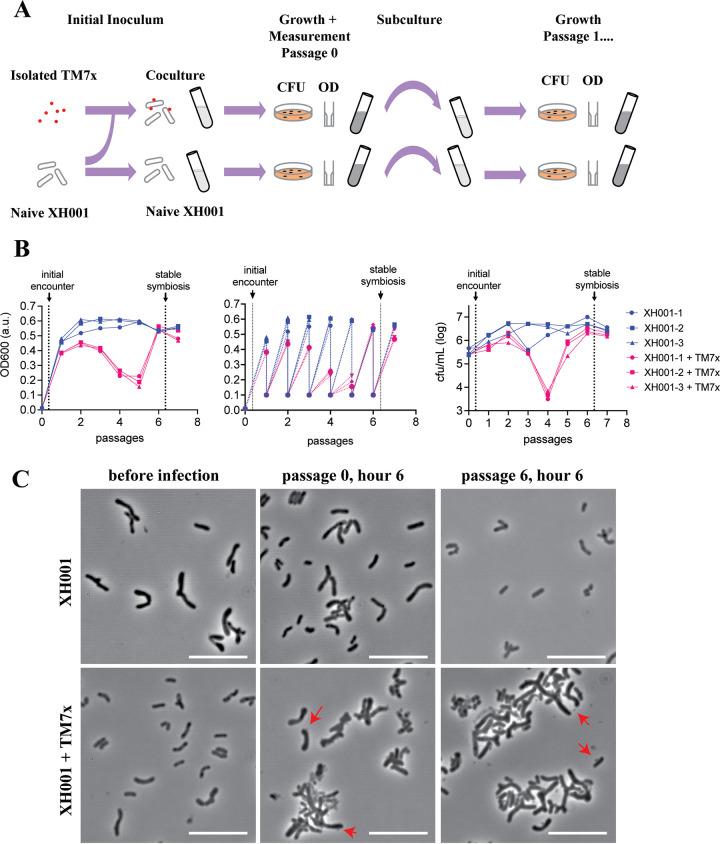
To produce samples for RNA sequencing, free-floating TM7x cells were isolated from an established XH001/TM7x coculture. A naive XH001 strain (XH001n) was exposed to isolated TM7x at a ratio of roughly 1:1 and passaged six times. For each passage, the coculture was grown for 24 h and then subcultured at a ratio of 1:10 into fresh media. As a control, XH001n was grown through the same passaging regime, using exactly the same protocol and time scale. XH001 numbers were determined at the subculture points, using both the optical cell density at 600 nm (OD600) and colony forming units (CFU) (Fig. 1A). Due to their small size, the TM7x numbers have little effect on OD600, and TM7x does not form CFU without a host bacterium (14). The TM7x-exposed XH001, though not the unexposed control, underwent a killing phase and a recovery phase, and by passage 6, the coculture had advanced to the stable symbiosis phase (Fig. 1B).
FIG 1.
Experimental design. (A) Schematic of culture setup, growth, and measurement for the coculture and naive cultures. Red dots represent TM7x, while the XH001 are shown as rod shaped tubes. Optical density at 600 nm (OD600) and colony forming units (CFU) were quantified at each step of the passage and are represented in panel B. (B) Growth measurements for passages using both OD600 (left and middle) and CFU (right) over seven passages (x axis). The middle panel shows the OD600 values for the dilutions as well as at the end of each passage. Three individual XH001n cultures are shown in blue circles, squares, and triangles, while cultures of XH001 with TM7x added are shown in red circles, squares, and triangles. Sampling points for the initial encounter (passage 0 at 6 h) and stable symbiosis (passage 6 at 6 h) phases for the transcriptomics sequencing are indicated by arrows. (C) Phase contrast images of XH001n and XH001/TM7x cultures at indicated sampling times are shown. Instances of TM7x infecting XH001 on the surface are indicated by red arrows. Scale bars are 10 μm.
Samples for both the XH001/TM7x coculture and the XH001n monoculture were collected from the following passaging time points: 6 h into passage 0, (termed the “initial encounter phase”), and 6 h into passage 6, (termed the “stable symbiosis phase”) (Fig. 1B). During the initial encounter, TM7x still attached to the surface of the host bacteria. Later, during the stable symbiosis phase, even more TM7x could be found on the host bacteria, and the host bacteria had an elongated morphology (Fig. 1C). For each sample, total RNA was extracted, rRNA was depleted, and the libraries were sequenced using the Illumina Hiseq platform (see Materials and Methods). The paired end reads were trimmed of low-quality sequences at a quality cutoff of 30. Trimmed sequences were mapped to a fused XH001 plus TM7x reference sequence. Mapped reads were split into XH001, with an average of 27,000,000 mapped reads per sample, and TM7x, at 880,000 reads. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (16) and are accessible through GEO Series accession number GSE196744.
Functional level changes during the initial encounter and stable symbiosis phases.
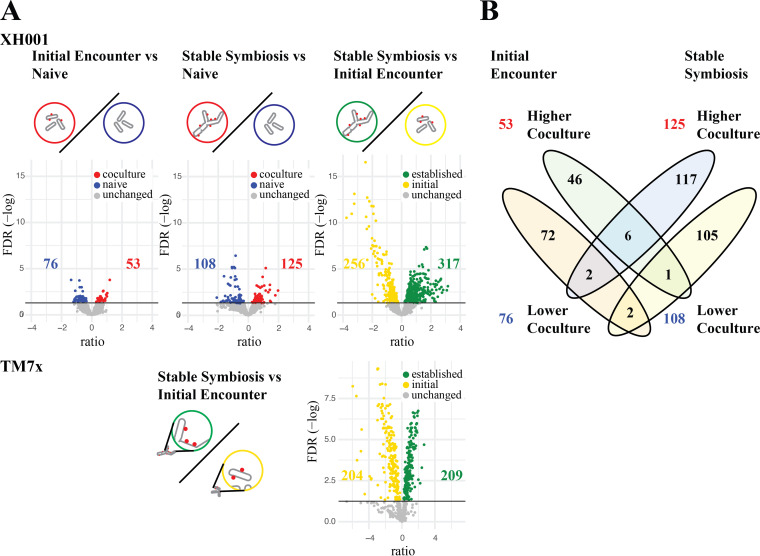
Expression ratios and false discovery rates (FDR) were calculated using voom/limma (17). Results for all genes are given in Table S1. Comparisons using a significance cutoff of FDR 0.05 are shown in Table 1 and Fig. 2.
TABLE 1.
Significant gene expression differences, comparing phases to controls and between phasesa
| Comparisons | Differential expression | Unchanged | Differential expression |
|---|---|---|---|
| XH001 coculture versus XH001n | Lower with TM7x | Unchanged | Higher with TM7x |
| Initial encounter | 76 (9/0) | 1807 (2/0) | 53 (3/0) |
| Stable symbiosis | 125 (32/0) | 1703 (52/0) | 108 (11/0) |
| XH001 initial versus stable symbiosis | Higher in initial phase | Unchanged | Higher in stable symbiosis phase |
| Stable symbiosis versus initial encounter | 256 (64/9) | 1363 (4/0) | 317 (104/15) |
| TM7x initial versus stable symbiosis | Higher in initial phase | Unchanged | Higher in stable symbiosis phase |
| Stable symbiosis versus initial encounter | 204 (128/27) | 279 (44/14) | 209 (72/2) |
The numbers of genes with nonlog ratios of ≥2/≥5 are listed in parentheses.
FIG 2.
Significant differences between conditions. (A) Volcano plots of the log2 ratio of expression levels and the −log of the FDR. Shown are all three XH001 comparisons: initial encounter coculture versus naive, stable symbiosis coculture versus naive, and stable symbiosis coculture versus initial encounter coculture. Colored dots indicate genes that made the 0.05 FDR cutoff. Blue, lower in coculture; red, higher in coculture; yellow, higher in initial encounter coculture; green, higher in stable symbiosis coculture. The number of significant differences are shown on the plots. The TM7x comparison of the stable symbiosis coculture versus the initial encounter coculture is also shown. (B) Venn diagram comparing significantly different genes for the initial encounter versus naive and the stable symbiosis versus naive comparisons.
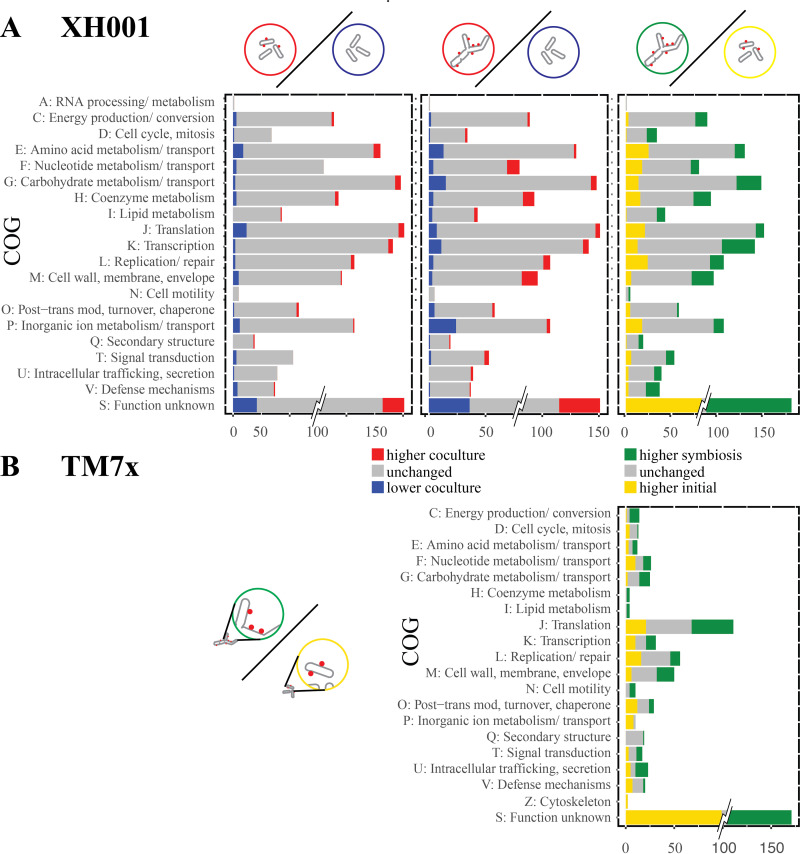
Our first analysis focused on changes in host bacterium gene expression during the initial encounter. XH001n cells 6 h after challenge with TM7x, allowing for initial binding and host response during the encounter phase, were compared to XH001n alone at the same time point (initial encounter versus naive control). As seen in Table 1 and Fig. 2, relatively few significant differences were detected in this comparison. 53 genes showed increased RNA expression in XH001/TM7x compared to the XH001n control and 76 showed decreased expression. This is understandable, given that XH001 and TM7x had only a short time to interact. Genes were assigned to categories of orthologous genes (COGs) using eggNOG-mapper genome-wide functional annotations as well as manually curated annotations. As seen in Fig. 3, the largest number of statistically significant differentially expressed genes, 30%, was in COG S: function unknown. For the other COGs, the largest groups showing increased expression in XH001/TM7x compared to XH001n were E: amino acid metabolism and transport, J: translation, G: carbohydrate metabolism and transport, and K: transcription. Decreased expression was seen in J: translation, E: amino acid metabolism and transport, P: inorganic ion metabolism and transport, and M: cell wall, membranes, and envelope biogenesis. That some of the largest numbers of increased and decreased genes were seen in amino acid metabolism and transport and in translation implied a shift rather than an overall increase or decrease in these functional categories (Fig. 3A).
FIG 3.
Clusters of orthologous groups (COG). The number of unchanged (gray) and significantly differentially expressed (colored) genes for each COG are shown for both the XH001 (top row) and TM7x (bottom row) comparisons. To prevent the large number of genes in cluster S (unknown function) from dominating the scale, only the significant differences are shown at full value. XH001 contains 621 genes annotated as S: unknown function, and TM7x contains 260.
Our second analysis focused on host expression in the stable symbiosis phase, comparing XH001/TM7x with XH001n 6 h after subculturing into passage 6 (stable symbiosis versus naive control). This comparison detected additional significant differences between XH001/TM7x and XH001n, beyond those that were seen during the initial encounter. 108 genes were found to have increased, while 125 decreased. A COG analysis showed increased expression in the XH001/TM7x coculture for M: cell wall, membranes, and envelope biogenesis, F: nucleotide metabolism and transport, and H: coenzyme metabolism. Decreases were seen for P: inorganic ion metabolism and transport, G: carbohydrate metabolism and transport, E: amino acid metabolism and transport, J: translation, and K: transcription (Fig. 3B).
There were some consistencies between the first two comparisons. Inorganic ion metabolism and transport were decreased in the cocultures at both phases. However, COG G: carbohydrate metabolism and transport shifted from more genes with increased expression in the coculture during the initial encounter to more genes with decreased expression in the stable symbiosis. The results supported the presence of a distinctive shift between the XH001 response when first encountering TM7x and upon establishing a stable symbiosis. The results also indicated that that inorganic ions, amino acids, carbohydrates, and changes to the cell wall play an important role in the host/epibiont interactions.
A third analysis compared XH001/TM7x from the stable symbiosis to XH001/TM7x during the initial encounter. Significant differences were seen between these states (317 genes increased during stable symbiosis, and 256 increased during the initial encounter), indicating a substantial shift in expression within XH001 during the change to stable symbiosis. This result was emphasized by the limited overlap seen between the first two comparisons (Fig. 2B).
Most COGs showed significant differences between the initial encounter phase and the stable symbiosis phase. C: energy production and conversion, D: cell cycle, I: lipid metabolism, M: cell wall, membrane, and envelope biogenesis, and V: defense mechanisms showed predominantly more increased genes in the stable symbiosis phase. In contrast, E: amino acid metabolism and transport, F: nucleotide metabolism and transport, J: translation, L: replication and repair, and P: inorganic ion metabolism and transport showed more increased genes during the initial encounter. These results were consistent with the idea that XH001 provides nutrients and cell wall components to TM7x. The higher levels of defense mechanisms could imply higher stress for the parasitized host.
A fourth analysis looked at gene expression within the TM7x epibiont, which had not previously been investigated. Since TM7x does not grow without a host, no monoculture TM7x control was available. TM7x RNA from the XH001/TM7x stable symbiosis was compared to the XH001/TM7x initial encounter (TM7x stable symbiosis versus initial encounter). An extensive shift was seen between the phases, with a majority of genes showing significant differences. 413 out of a total of 692 genes displayed differential expression (Table 1; Fig. 2B and 3).
COG analysis showed that many functions had sets of genes that increased in the initial encounter as well as genes that increased during stable symbiosis (Fig. 3). However, several functions were skewed heavily toward increases in stable symbiosis. These included energy production and conversion, carbohydrate metabolism, coenzyme metabolism, lipid metabolism, intracellular trafficking and secretion, and cell motility. These pathways are mostly dependent on host substrates and correlate well with host functional changes. COGs with genes that mostly increased during the initial encounter were related to the cell cycle, inorganic ion metabolism and transport, and defense mechanisms.
Cell morphology, cell cycle, and cell wall/membrane.
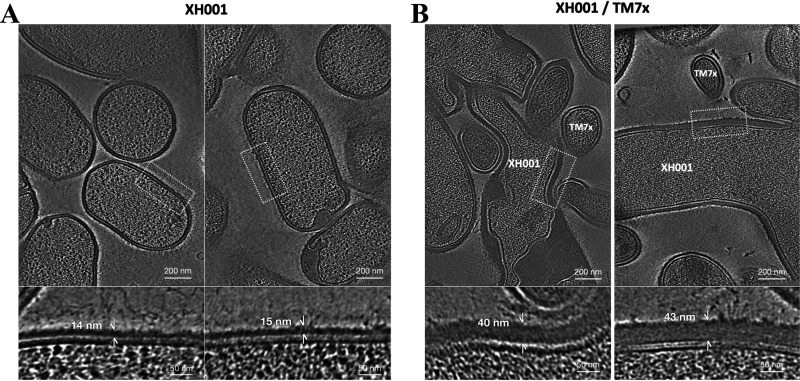
XH001 has shown drastic changes in cell division and cell shape when infected with TM7x (14, 18). During stable symbiosis, 48 to 72% of host cells were found to be associated with TM7x (14). These cells had reduced cell growth and inhibited cell division, leading to elongated and clubbed end morphology. The XH001 cells also had a sticky cell surface, forming smaller aggregates in coculture (18). It was speculated that the XH001/TM7x coculture was maintained indefinitely by TM7x parasitizing new hosts from the dividing non-TM7x-associated cells (14). Using cryoEM measurements, we found a thickening of the cell wall from ~15nm to more than 40 nm during stable symbiosis (Fig. 4, lower panels). Given these observations, XH001 would be expected to show changes in genes for DNA replication, the cell cycle, and cell wall/membrane biosynthesis during infection. These processes are also intimately related to each other in bacteria (19).
FIG 4.
Cell wall of XH001 observed by cryo-FIB milling and cryo-ET imaging. Representative section selected from the tomograms corresponding to XH001 cells in monoculture (A) and in established XH001/TM7x symbiosis coculture (B). The cell wall thickness was measured based on the density plot along the cell wall using IMOD and ImageJ. Top: Scale bars are 200 nm. Bottom: Scale bars are 50 nm.
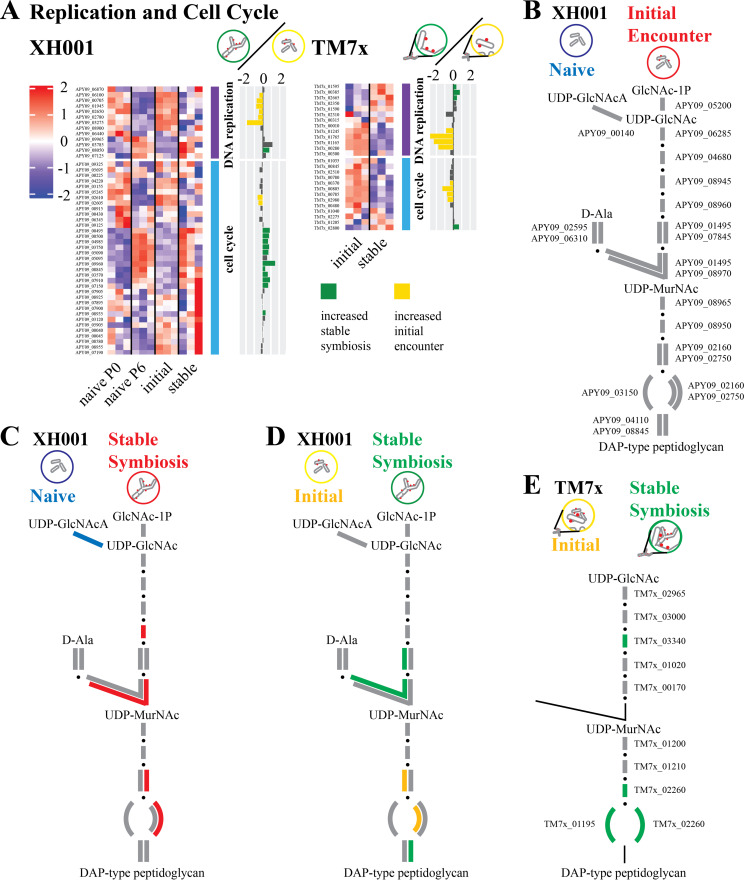
The COG analysis (Fig. 3) indicated few differences for COG L: replication and repair genes in the first two comparisons and displayed a predominance of increased genes during the initial encounter compared to stable symbiosis. Focusing on the genes for DNA replication, the first two comparisons of XH001/TM7x with XH001n showed little difference for DNA replication (Fig. 5A; Table S2). However, reduced RNA expression for DNA replication genes was seen during the stable symbiosis compared to the initial encounter. This was consistent with the observed reduction of cell growth and the inhibition of cell division seen in the TM7x infected cells during stable symbiosis (14).
FIG 5.
DNA replication, cell cycle, and peptidoglycan biosynthesis. (A) Genes for DNA replication and the cell cycle. Heatmaps of z score-adjusted sequence counts for the three biological replicates for each condition. Bar plots show the log2 ratio between the stable symbiosis coculture and the initial encounter coculture. Green indicates significantly increased during the stable symbiosis phase, and yellow indicates significantly increased in the initial encounter phase. (B–E) A schematic of the peptidoglycan biosynthesis pathway for XH001 and TM7x. Due to its small genome, TM7x has fewer predicted genes in these pathways. Steps with alternative pathways are shown as separate connections. Steps with multiple subunits are shown with multiple lines. Red, increased in XH001/TM7x versus XH001n; blue, decreased in XH001/TM7x versus XH001n; green, increased in stable symbiosis; yellow, increased in initial encounter; gray, statistically unchanged. (B) Initial encounter versus naive. The XH001 APY09 gene designations are given. (C) Stable symbiosis versus naive. (D) Stable symbiosis versus initial encounter. (E) TM7x stable symbiosis versus initial encounter. The TM7x gene designations are given. Steps that currently have no predicted gene but are expected to exist are shown in thin black lines.
The picture was more complex when looking at genes for cell division control (Fig. 5A; Table S2). When comparing the initial encounter to naive, ftsK (APY09_09125) was decreased in XH001/TM7x. However, for stable symbiosis versus naive, two genes, including ftsZ (APY09_08935), were increased in XH001/TM7x, while one decreased. As seen in the COG analysis for COG D: cell cycle and mitosis (Fig. 3), despite DNA replication genes generally being increased in the initial encounter compared to stable symbiosis, cell cycle genes showed increased levels during stable symbiosis versus the initial encounter. Out of 34 putative cell cycle genes, 11 were increased in the stable symbiosis phase, including ftsZ (APY09_08935) and a possible ftsK (APY09_09960). Interestingly, the genes that significantly increased in the initial encounter were chromosome partitioning genes (APY09_02605, APY09_02610). The combination of decreased levels of DNA replication genes and increased levels of cell cycle genes during stable symbiosis, compared to the initial encounter, may result in the elongated and hyphal morphology seen in the infected XH001 (13). The lack of complete cell division would be consistent with reduced genome replication and chromosome partitioning, while genes ordinarily associated with the cell cycle might produce the elongated cell structures (13).
Over the course of infection, TM7x numbers increased on the host, reaching large numbers during the killing phase and reducing to a moderate number by the stable symbiosis phase. DNA replication and cell cycle genes in TM7x would be predicted to show increased expression in the initial encounter phase compared to the stable symbiosis phase. TM7x showed mixed results for DNA replication. However, consistent with the growth observed during the initial infection, 4 of the 13 putative cell cycle genes were increased during the initial encounter phase, including ftsZ (TM7x_00785) and ftsK (TM7x_00370). Only ftsA (TM7x_02800) was increased in the stable symbiosis phase.
In keeping with cryoEM showing a thickened cell wall/membrane (Fig. 4), COG M: cell wall, membrane, and envelope biogenesis (Fig. 3) showed more significantly increased genes over the course of the epibiont infection. XH001 has a rhamnose cell wall (1). In terms of rhamnose biosynthesis, a putative dTDP-4-dehydrorhamnose epimerase/reductase (APY09_00110) was increased during stable symbiosis compared to XH001n (Table S2). However, the only significant difference between stable symbiosis and the initial encounter encoded the first step, glucose-1-phosphate thymidylyltransferase (APY09_00105), which had increased expression during the initial encounter.
A closer look at the peptidoglycan biosynthesis genes showed no significant differences in the initial encounter versus naive comparison (Fig. 5B). Stable symbiosis versus naive showed three increased genes (APY09_02750, APY09_08960, APY09_08970) in the presence of TM7x (Fig. 5C; Table S2), implying a possible increase in peptidoglycan production. However, stable symbiosis versus the initial encounter (Fig. 5D) showed mixed results, with two genes increased during the stable symbiosis phase (APY09_01495, APY09_08845) and one increased in the initial encounter phase (APY09_02160). In TM7x, genes for the final synthesis steps were increased during stable symbiosis versus the initial encounter (TM7x_01195, TM7x_02260, TM7x_03340). This implied possibly higher peptidoglycan synthesis and cell division for TM7x during stable symbiosis, as well.
While TM7x encodes peptidoglycan synthesis genes, its reduced genome does not encode the production of the UDP-GlcNAc building block for the pathway. Presumably, the epibiont obtains UDP-GlcNAc from the host. Despite increased peptidoglycan gene RNA in XH001 and TM7x during stable symbiosis (Fig. 5E), XH001 did not show increases in the pathway leading to UDP-GlcNAc. However, XH001 has an alternative pathway by which to consume UDP-GlcNAc by converting it to UDP-GlcNAcA. This gene (APY09_06140) was decreased in the stable symbiosis versus naive comparison (Fig. 5D). A reduction in the alternative use might increase the flow of UDP-GlcNAcA into peptidoglycan synthesis.
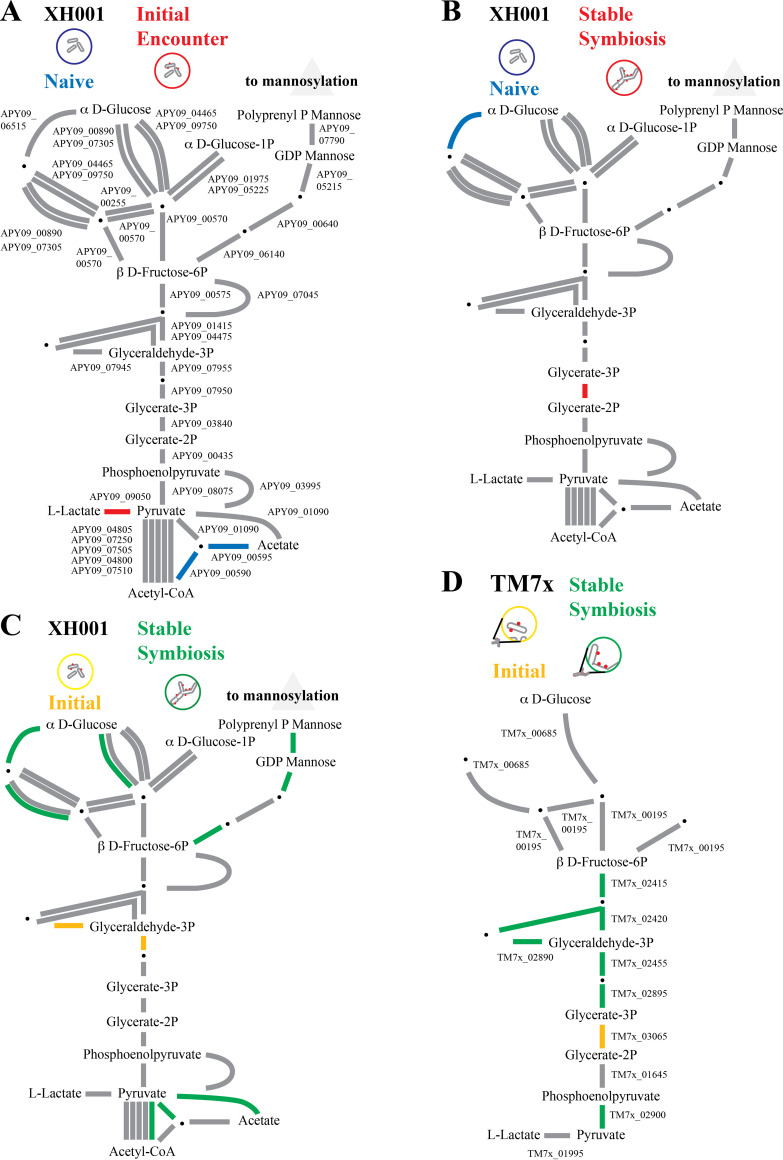
Mannosylation, a glycosylation process found in all domains of life, can use GDP-mannose and polyprenyl phosphate mannose as mannose donors (20). As seen in Fig. 6 (Table S2), instead of proceeding through glycolysis to pyruvate, an alternative use for β-d-fructose-6P in XH001 is the production of GDP-mannose and polyprenyl phosphate mannose. There were no significant differences detected in the first two comparisons, but comparing the stable symbiosis versus the initial encounter showed an increase in expression for genes in the pathway (APY09_05215, APY09_06140, APY09_07790) to these mannose sources. This implied increased mannosylation during stable symbiosis. Mannose may also be added to polysaccharides, and cell surface polysaccharides could contribute to the thickened cell walls/membranes.
FIG 6.
Glycolysis and mannosylation substrates. A schematic of the glycolysis pathway and polyprenol phosphate mannose for XH001 and TM7x. Due to its small genome, TM7x has fewer predicted genes in these pathways. Steps with alternative pathways are shown as separate connections. Steps with multiple subunits are shown with multiple lines. Red, increased in XH001/TM7x versus XH001n; blue, decreased; green, increased in stable symbiosis; yellow, increased in initial encounter; gray, statistically unchanged. (A) Initial encounter versus naive. The XH001 APY09 gene designations are given. (B) Stable symbiosis versus naive. (C) Stable symbiosis versus initial encounter. (D) TM7x stable symbiosis versus initial encounter. The TM7x gene designations are given.
Energy metabolism.
COG C: energy production and conversion indicated few differences between XH001/TM7x and XH001n in the first two comparisons, but some increase in expression was observed when comparing the stable symbiosis to the initial encounter. To better understand these results, we took a more detailed look at glycolysis. Fig. 6 shows the glycolysis pathway for XH001 and TM7x. TM7x has a more limited repertoire of glycolysis genes. Comparing the initial encounter to XH001n showed few differences (Fig. 6A; Table S2). However, two of the genes leading from pyruvate to acetate as an end product (APY09_00590, APY09_00595) had decreased expression, while l-lactate dehydrogenase (APY09_01090) leading to l-lactate was increased in XH001/TM7x. This implied a shift in end products from acetate to l-lactate. A metabolic flux model of TM7x and the XH001 host indicated that l-lactate might play an important role in the coculture metabolism (21). A shift toward l-lactate production during the initial encounter would be consistent with the model.
Stable symbiosis versus naive control (Fig. 5B) also showed few significant differences. The shift from acetate to l-lactate seen in the initial encounter was no longer observed during stable symbiosis, possibly indicating a shift in nutrient transfer during the stable symbiosis phase. Only 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (APY09_03840) showed a significant increase in XH001/TM7x.
In contrast, comparing XH001/TM7x during stable symbiosis to the initial encounter implied lower levels for pyruvate production in the stable symbiosis phase than in the initial encounter phase (Fig. 6C). Even though COG C showed more genes increased in the stable symbiosis, between β-d-fructose-6P and pyruvate, the only significant differences were triosephosphate isomerase (APY09_07945) and 6-phosphofructokinase (APY09_07955), both of which were decreased during stable symbiosis. However, despite the decreases in these genes, stable symbiosis showed increased expression of α d-glucose-6P (APY09_00890, APY09_06515). It is unclear why genes for the glycolysis pathway substrate would be increased during stable symbiosis while the glycolysis genes themselves would be increased during the initial encounter. As mentioned in the previous section, this might be explained by a routing of the higher levels of α d-glucose-6P into mannosylation, rather than glycolysis, during stable symbiosis.
The TM7x results comparing stable symbiosis with the initial encounter are interesting, given the changes seen in the host (Fig. 6D). Most of the glycolysis pathway genes were increased during stable symbiosis. This is unsurprising, as TM7x would be expected to derive its energy primarily from its host, and this interaction should be optimal during stable symbiosis. However, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, the only XH001 glycolysis gene increased in stable symbiosis, was decreased in TM7x during stable symbiosis (TM7x_03065). One possible explanation for this would be an exchange of nutrients between the host and the epibiont, with TM7x effectively relying on XH001 for this step.
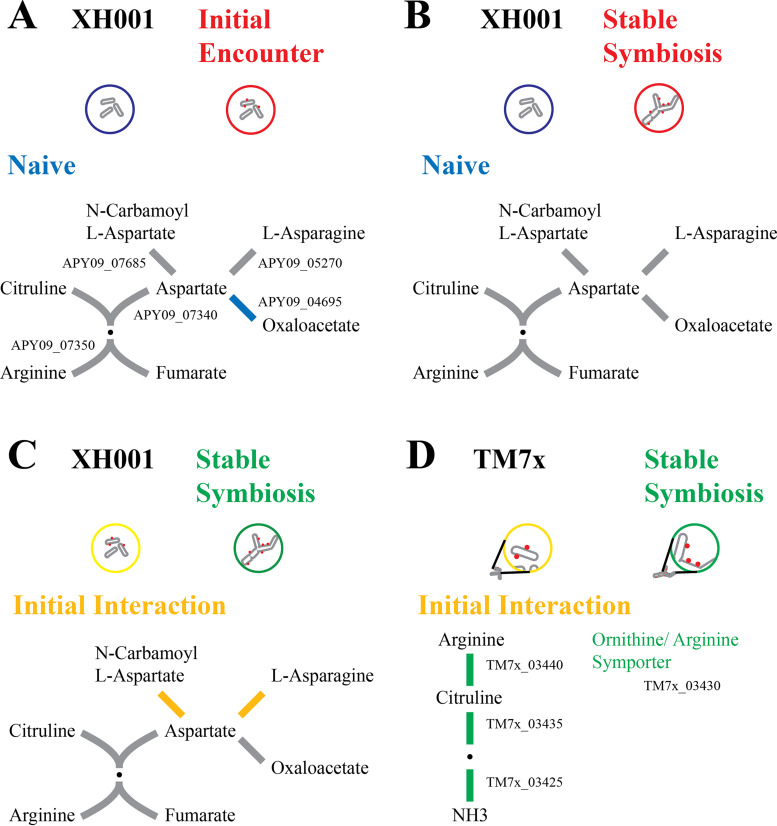
Arginine.
Members of the CPR are noted for their limited biosynthetic capabilities, including missing pathways for de novo amino acid biosynthesis (3, 4). However, TM7x encodes several genes in the arginine pathway that only occur in the mammalian-associated members of Saccharibacteria and has been verified to utilize arginine (22). Fig. 7 shows the pathway for XH001 and TM7x. Relatively few genes in the XH001 arginine pathway showed significant differences in the first three comparisons (Fig. 7A–C; Table S2), giving no strong indication of an overall change in arginine biosynthesis in the host. However, TM7x showed a significant increase in all of the genes for arginine biosynthesis/catabolism (TM7x_03425, TM7x_03435, TM7x_03440) as well as an ornithine/arginine symporter (TM7x_03430) during stable symbiosis, implying a role for arginine in the long-term epibiont interaction (Fig. 7D). This is consistent with the recent finding that TM7x can metabolize arginine and that it uses the arginine to stay viable and generate ATP in the absence of its host bacteria (22).
FIG 7.
Arginine metabolism. A schematic of the arginine pathway for XH001 and TM7x. Red, increased in XH001/TM7x versus XH001n; blue, decreased in XH001/TM7x versus XH001n; green, increased in stable symbiosis; yellow, increased in initial encounter; gray, statistically unchanged. (A) Initial encounter versus naive. The XH001 APY09 gene designations are given. (B) Stable symbiosis versus naive. (C) Stable symbiosis versus initial encounter. (D) TM7x stable symbiosis versus initial encounter. The TM7x gene designations are given.
Autoinducer.
TM7x can induce higher levels of biofilm formation in its host XH001, an effect that is dependent on the autoinducer AI-2 synthase-encoding gene luxS and lsrB, which encodes the AI-2 transporter (23). lsrB (APY09_02520) showed possible differences in the first two comparisons, initial encounter versus naive and stable symbiosis versus naive, but they did not make the significance cutoff (Table S2). However, luxS (APY09_06105) was increased in the stable symbiosis versus naive control comparison, consistent with a role in long-term associations between the species. A putative autoinducer 2 receptor-encoding gene was found in TM7x (TM7x_02705). Expression of this gene was increased in TM7x during the initial encounter rather than during stable symbiosis.
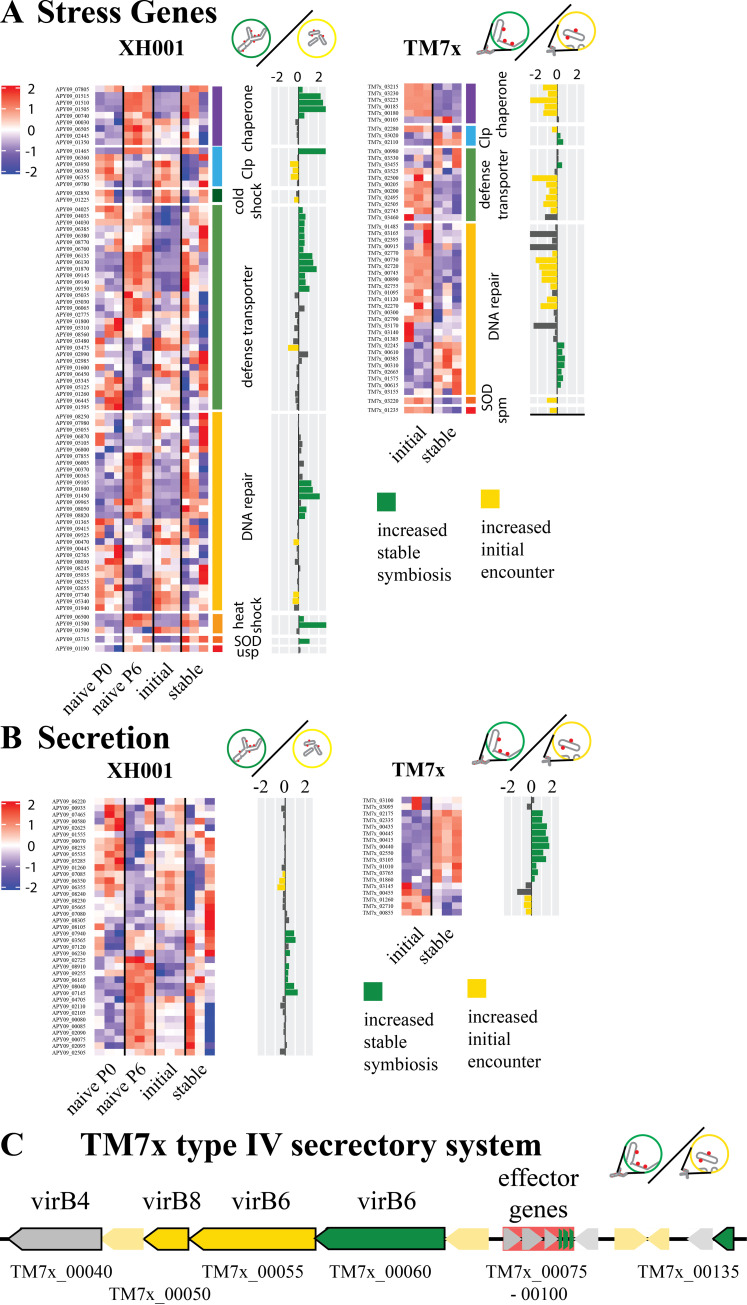
Stress.
Previous studies indicated that the presence of TM7x caused increased stress in XH001 (13). As seen in Fig. 8A, the transcriptome results did not show consistent increases in stress genes for the cocultures. For the first comparison, initial encounter versus naive control, three stress related genes were increased in the presence of TM7x: RNA-binding SsrA-binding protein (APY09_00030), DNA repair exodeoxyribonuclease III (APY09_00470), and a putative defense-involved transporter (APY09_03475) (Table S2). However, five genes, heat shock chaperone (APY09_07805) and four putative transporters involved in defense (APY09_04025, APY09_04030, APY09_04035, APY09_08770), had significantly decreased expression.
FIG 8.
Stress and secretory systems. (A and B) Heatmaps of z score-adjusted sequence counts for the three biological replicates for each condition. Bar plots show the log2 ratio between the stable symbiosis coculture and the initial encounter coculture. Green indicates significantly higher during stable symbiosis, and yellow indicates significantly higher in the initial encounter phase. (A) Genes involved in stress responses broken down into subcategories: chaperones, heat shock proteins, cold shock proteins, clp proteases, defense transporters, DNA repair, sodium oxide dismutase (SOD), universal stress protein (usp), and spermidine synthase (spm). (B) Genes for secretory systems, excluding the type IV system from TM7x. (C) A schematic of the type IV secretory system region of TM7x. Black outlines indicate predicted type IV component genes. Putative effector genes are backed in red. Green indicates significantly increased during stable symbiosis, and yellow indicates significantly increased in the initial encounter.
The second comparison, stable symbiosis versus naive control, possibly implied reduced stress in XH001 in the presence of TM7x. Repair protein recF (APY09_02655) and a defense-associated transporter (APY09_02990) showed increases in the coculture. However, two important chaperones, dnaJ (APY09_06505) and groEL (APY09_01350), as well as a putative heat shock inducible repressor (APY09_06500) were decreased with TM7x. Additionally, DNA repair genes DNA-3-mehtyladenine glycosylase (APY09_07855) and recombination and repair protein recO (APY09_06005), as well as a defense transporter (APY09_05030), were also significantly decreased.
The third comparison, stable symbiosis versus initial encounter, indicated higher stress in the stable symbiosis phase. A total of 27 stress related genes had increased expression levels in the established coculture, including clpB (APY09_01465), dnaK (APY09_01515), groES (APY09_00740), grpE (APY09_01510), heat inducible repressor (APY09_06500), heat shock protein hspR (APY09_011500), and sodium oxide dismutase (APY09_03715). In contrast, only 8 had increased expression during the initial interaction. These included cold shock protein (APY09_01225) and both putative clpP (APY09_06350, APY09_06355) and clpC (APY09_03950) (Fig. 8A).
Establishing itself as an epibiont appeared to reduce stress for TM7x. In the TM7x stable symbiosis versus initial encounter comparison, 22 stress-related genes were increased in the initial encounter. These included sodium oxide dismutase (TM7x_03220), spermidine synthase (TM7x_01235), and 8 genes involved in DNA repair. In contrast, only 10 genes were increased during stable symbiosis. The majority of those were also within the DNA repair group, though clpB and clpP (TM7x_03020, TM7x_02110) were also increased. This indicated that genes within this function are being differentially regulated and warrants further investigation.
Secretion systems and appendage genes.
TM7x has minimal biosynthetic capabilities and cannot replicate without a host (1), implying that nutrient transfer is likely to play a key role in the symbiosis between host and epibiont. Host bacterial and TM7x secretion systems, therefore, could be of significant interest. COG U: intracellular trafficking and secretion (Fig. 3) showed few differences in the first two comparisons and mostly increased expression during stable symbiosis compared to the initial encounter (Fig. 8B; Table S2). Comparing XH001 gene expression during table symbiosis versus the initial encounter showed 8 increased genes during stable symbiosis and 3 increased genes during the initial encounter out of 39 genes with predicted secretory functions. The eight (APY09_03565, APY09_06165, APY09_06230, APY09_07145, APY09_07940, APY09_08040, APY09_08910, APY09_09255) were individual genes from different pathways, including predicted allantoin permease (APY09_06230), branched-chain amino acid transporter (APY09_08040), signal peptidase II (APY09_08910), and preprotein translocase subunit SecG (APY09_07940). The three increased during the initial encounter were a signal recognition particle protein (APY09_07085) and two putative ATP-dependent Clp protease proteolytic subunits (APY09_06350, APY09_06355) which have broad effects rather than focused secretory functions.
For TM7x, a majority of the predicted secretion system genes showed significant differences between stable symbiosis and the initial encounter. TM7x showed 11 genes with increased expression during stable symbiosis, including a predicted signal peptidase I (TM7x_00435), preprotein translocase subunits secA and secY (TM7x_01010, TM7x_01860), a general secretion pathway protein PilB/GspE family ATPases (TM7x_02335), three pilus assembly proteins (TM7x_03105, TM7x_00445, TM7x_00415), and the twitching motility protein pilT (TM7x_00440). Microscopy has shown pili like structures extending from CPR members to other cells (4). The increased pilus gene expression during stable symbiosis implied that pilus-like structures may play a role in long-term epibiont association. The three genes increased during the initial encounter showed some overlap with the genes increased in stable symbiosis, including a predicted signal peptidase I (TM7x_00855), preprotein translocase subunit secG (TM7x_02710), and DNA processing protein dprA (TM7x_01260).
Pathogens and symbionts do transfer effector molecules into hosts to modify host cell behavior, typically through a specialized apparatus, such as a type IV secretory system (24). TM7x is predicted to have a type IV secretion system as well as six putative effector genes, a system that is maintained across all mammalian and environmental Saccharibacteria as well as other CPR members (5, 7). Fig. 8C shows the significant differences between the stable symbiosis phase and the initial encounter phase for the type IV secretory region. Out of five predicted type IV secretion protein genes, two showed significant increases during stable symbiosis, a predicted virB2 (TM7x_00135) and virB6 (TM7x_00060), while two were increased during the initial encounter, another predicted virB6 (TM7x_00055) and virB8 (TM7x_00050). However, half of the predicted effector genes (TM7x_00090-00100) were increased during stable symbiosis, consistent with genes for altering the host cell to maintain the coculture.
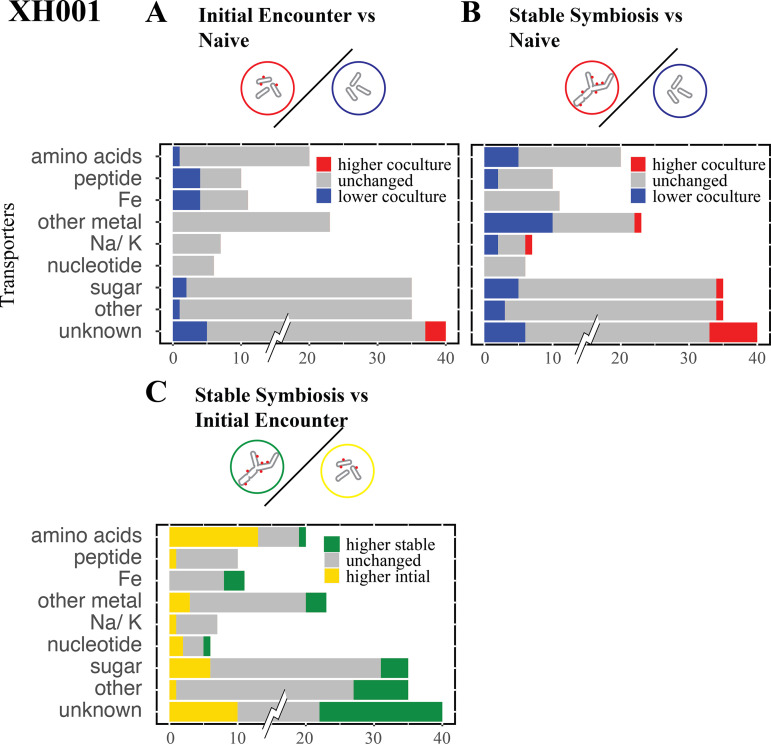
Transporters.
XH001 contains 296 genes that are putatively annotated as transporters. A combination of consensus among multiple annotation platforms and a BLAST analysis was used to group the transporters by possible substrate specificity (see Materials and Methods), though many genes could only be given a general annotation as transporters. The initial encounter showed few significant differences compared to the naive control (Fig. 9A; Table S2). The majority of altered genes were decreased in XH001/TM7x and included peptide, iron, and some sugar transporters. Genes increased in XH001/TM7x only had general transporter predictions. This might indicate that XH001 limits transport when encountering TM7x. When comparing the stable symbiosis phase to the naive control (Fig. 9B), the number of significantly different genes increased. There was a shift from decreased iron transport to decreased expression for transporters of other metals.
FIG 9.
Transporters. The number of unchanged and significantly differentially expressed genes for the transporter groups are shown for the XH001 comparisons. Groups: amino acids, peptides, iron (Fe), noniron metal transporters, sodium/potassium (Na/K), sugar, other, and genes predicted to be transporters but without an identifiable substrate (unknown). To prevent the large number of genes in the unknown category (95) from dominating the scale, only the significant differences are shown at full value. (A) Initial encounter versus naive control. (B) Stable symbiosis versus naive control. (C) Stable symbiosis versus initial encounter.
Comparing XH001/TM7x during stable symbiosis to the initial encounter reinforced the presence of a shift in expression of many transporter genes (Fig. 9B). A majority of amino acid transporters were increased in the initial encounter. Some noniron metal transporters were also increased, including possible transporters of cobalt (APY09_02405, APY09_02880) and manganese (APY09_05850). In contrast, iron transporters were increased during stable symbiosis. Sugar transporters showed mostly increased levels during the initial encounter, though several were increased in stable symbiosis, possibly indicating a shift in sugar metabolism.
TM7x has 52 putative transport genes, 23 of which showed significant increases in the initial encounter, including a predicted sodium transporter (TM7x_01545), lipid A export permease (TM7x_02745), a putative copper/silver translocating ATPase (TM7x_01845), two putative magnesium transporters (TM7x_03415, TM7x_03420), macrolide ABC transporter (TM7x_00205), multidrug ABC transporter (TM7x_02495), and a possible AI-2 receptor (TM7x_02705) discussed previously. Interestingly, there were no obvious iron transporters in the annotation. Given the importance of iron acquisition, TM7x either has a novel iron transporter that is not found in the database, or it could be acquiring iron from the host bacteria directly. We observed that 17 genes increased during stable symbiosis. Most of these were involved in secretion and were discussed in the previous section. However, these did include a putative polysaccharide permease (TM7x_03740) and an H(+) transporting ATPase (TM7x_01440). As mentioned previously, an arginine/ornithine antiporter (TM7x_03430), which could play role in host-selection (14), was also increased in the stable symbiosis phase.
Proteomics comparison.
RNA sequencing results are generally interpreted as if they reflect the resulting levels for the encoded proteins, even though it is well-known that RNA transcriptomics and proteomics do not correlate completely (25). To support the RNA data, we compared the transcriptomic results with a proteomic data set comparing XH001 with and without TM7x (Fig. S2; Table S3). Correlation between the current RNAseq data set and the generated proteomic data set investigating TM7x and XH001 indicates that many of the 1,936 total proteins that we detected were observed to change similarly between these analyses (Table S4). Genes with both RNA and proteomics support are primary candidates for experimental validation in future studies. Examples include genes for rhamnose (APY09_00110) and peptidoglycan biosynthesis (APY09_02750), stress-associated molecular chaperone dnaJ (APY09_06505), and repair protein recF (APY09_02655), as well as an α d-glucose-6P (APY09_06515) involved in sugar interconversion.
Conclusions.
The CPR/Patescibacteria superphylum may constitute a quarter of bacterial diversity, and its members are likely to live as epibionts on the surfaces of other bacteria (4), as has been conclusively demonstrated for cultivated Saccharibacteria Nanosynbacter lyticus type strain TM7x and its host bacterium Schaalia odontolytica XH001 (1), as well as within recent cultures of additional epibiont-bacterial host pairs (10, 11, 26, 27). Yet, little is understood about how these organisms establish and maintain stable symbiosis with their hosts. In prior studies, we have learned that as TM7x infects its host bacterium, there are distinct temporal phases, including an initial interaction phase, a killing phase, a recovery phase, and a stable symbiosis phase (14). To further investigate this interaction, we used RNAseq to examine both the epibiont and its host bacterium during their initial encounter and after having established a stable symbiosis. The study showed a dynamic interaction at the transcript level between the epibiont and the host during the establishment of the stable symbiosis.
Our data suggest that metabolites play an important role in the process. Of the few differences seen between the initial infected and naive host, several centered on enzymes related to metabolites. There might have been a shift from acetate to l-lactate as an end product of energy metabolism. Additionally, the host showed lower expression of several iron, peptide, and sugar transporters. Comparing the stable symbiosis to the initial encounter revealed an extensive shift in transporter expression, indicating a changing interaction across the infection. TM7x also showed a significant shift in transporters between the initial encounter and the stable symbiosis. Notably, the entire TM7x arginine catabolism/biosynthesis was increased in stable symbiosis, as was an ornithine/arginine symporter. Furthermore, TM7x energy metabolism and peptidoglycan biosynthesis, which should be dependent on substrates from the host, had higher expression after establishment on the host.
The epibiont pili and type IV secretion system appeared to be involved in the interaction. TM7x encodes a type IV secretion system. The secretory machinery genes did not show consistent expression changes. However, three of the putative effector genes had increased expression during stable symbiosis, implying a role for the effectors in the long-term association of the epibiont and the host. Microscopy has also shown pili in ultrasmall cells from fractions enriched in CPR genomes possibly interacting with hosts (6). Consistent with this, several pili genes in this study showed increased expression during stable symbiosis.
Stable symbiosis has been shown to have a dramatic effect on host cell shape, resulting in an elongated and hyphal morphology (14). We have also shown a distinct phenotype in infected hosts, with thickening of the cell wall being observed (Fig. 4). Consistent with this extensive cellular change, numerous XH001 genes involved in cell wall and membrane biosynthesis were increased during stable symbiosis. Cell cycle genes also showed higher expression, with the exception of the reduced expression of the chromosome partitioning genes.
The interaction also appeared to be stressful to the host under these conditions. A previous study showed increased stress gene expression in the presence of TM7x (13). Additionally, host cells showed physiological changes, growth arrest, and an elongated cell shape (14). Consistent with these results, most of the XH001 stress related genes showed increased expression during stable symbiosis. In contrast, the majority of the stress genes of epibiont TM7x were decreased after establishment on the host. Overall, this study has revealed the gene expression of the host bacterium and the epibiont during this dynamic interaction, representing initial insights into the mechanisms of how these unique microbes with limited de novo biosynthetic capabilities may survive and persist at a high prevalence within the human microbiome as well as in the environment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Naive XH001 (XH001n) (Schaalia odontolytica, formerly Actinomyces odontolyticus subsp. Actinosynbacter strain XH001) monoculture and XH001/TM7x coculture were both grown in Bacto brain heart infusion (BHI, BD) medium at 37°C in a microaerophilic chamber (2% O2, 5% CO2, balanced with N2) (14, 15). These were determined to be the optimal growth conditions for XH001 (13) and were used for all growth in this study. However, because it is difficult to avoid sample contamination in the microaerophilic chamber, the XH001 and XH001/TM7x cultures were diluted between passages outside the chamber, on the lab benchtop, under aerobic conditions. While XH001 prefer a microaerophilic environment, they are sufficiently oxygen-tolerant such that in reduced BHI media, brief aerobic exposure does not affect their growth. Free-floating TM7x (Nanosynbacter lyticus strain TM7x HMT-952) was isolated from the XH001/TM7x coculture as described previously (14). Briefly, the XH001/TM7x coculture were filtered through a 0.45 μm filter, and TM7x cells were collected by ultracentrifugation at 80,000 × g for 90 min. Isolated TM7x cells were used freshly on the same day for infection experiments without freezing or overnight storage.
TM7x infection, passaging, and sample collection.
Starting from a single frozen stock of XH001n, three independent cultures of XH100n were inoculated into BHI broth. To ensure consistent inoculations of cells in similar growth states and relatively healthy conditions for the experiments, the cells were passaged for 24 h in BHI media twice after retrieval from the frozen stock. 24 h has been shown to be sufficient to allow XH001 to attain its stationary-phase, typically reaching an OD of 0.6 to 0.8 (13, 14). The TM7x infection assays were set up as previously described with modification (12) and are summarized in Fig. 1. By performing passaging, we tried to mimic a continuous culture as much as possible and ensure that nutrients were not a limiting factor for XH001n growth. Briefly, 115 mL of stationary-phase XH001n cells diluted to an OD600 of 0.1 were mixed at a roughly 1:1 ratio with isolated free-floating TM7x cells in triplicate (12). After 6 h of incubation, the first time point (passage 0) was sampled from each replicate (passage 0, hour 6). For each sample, 50 mL of culture were removed and centrifuged at 5,500 rpm for 10 min at 4°C. Supernatants were discarded, and the pellets were flash-frozen in liquid nitrogen. The remaining cultures were grown to the 24 h mark, and XH001 cell density measurements by OD600 and CFU by plating were determined at the end of each passage (Fig. 1B). The next passages were then started by diluting 10 to 50 mL of the cultures to 0.1 OD600 (Fig. 1B, middle panel). The volume depended on the follow-up collection and sampling. In the sixth passage, after 6 h (passage 6, hour 6), samples of 50 mL culture were collected, pelleted, and flash-frozen. We also conducted, in triplicate, a parallel experiment in which XH001n cells alone, without the addition of TM7x, underwent the same inoculation, passaging, and sampling. Throughout our experiment, we ensured the purity of our cultures by checking them under a phase-contrast microscope to look for the typical rod and hyphal morphologies of the XH001 cells infected with TM7x (Fig. 1C). We consider these to be biological replicates and did not conduct technical replicates for each separate set up. A statistical analysis of the OD600 and CFU measurements is shown in Fig. S1.
RNA isolation and sequencing.
Cell pellets were resuspended in cold PBS buffer and mixed with glass beads (ref. number 6914100-98596, MPbiomedicals LLC). Using the BeadBlaster Microtube Homogenizer (BenchMark), the mixtures were shaken vigorously 3 times for 30 s with 30 s breaks in between at a speed of 6 m/s. From each sample the total RNA was isolated using the High Pure RNA Isolation Kit (ref. number 11828665001). Isolated RNA was cleaned and concentrated with Zymo Research RNA Clean & Concentrator Kit, following the kit protocol (cat. number R1015). DNA was removed using the Ambion TURBO DNA-free Kit (cat. number AM1907). The RNA concentration was determined by the RNA broad range Qubit kit, and the nucleic acid purity was determined by Nanodrop and Qubit 3.
The total RNA was further processed by rRNA depletion with the Ribozero rRNA Removal Kit (Illumina, South Plainfield, NJ, USA), and the libraries were sequenced by GENEWIZ, LLC (South Plainfield, NJ, USA), using the Illumina HiSeq platform. The paired end reads were trimmed of low-quality sequences at a quality cutoff of 30 using BBduk 38.37 (28). The trimmed sequences were mapped to a reference database containing XH001 and TM7x reference sequences using Geneious Prime (https://www.geneious.com) with the default settings. The mapped reads were split into XH001, with an average of 27,000,000 mapped reads per sample, and TM7x, at 880,000 reads. A subset of samples checked after sequencing for rRNA (5S, 16S, and 23S) removal success contained an average 26.5% of the raw reads of rRNA.
Bioinformatics.
Expression ratios and false discovery rates (FDR) were calculated using voom/limma (17) in Degust (29), using all biological replicates, with a significance cutoff of FDR 0.05. Annotations were assigned through consensus among multiple annotation platforms (1): GenBank (30), SEED (31), prokka (32), eggNOG (33), PATRIC (34), and UNIPROT (35). BLAST analysis (36) was also used on selected genes. Categories of orthologous genes (COGs) were obtained from eggNOG (37). Pathways were derived from the KEGG database (38, 39).
Cryo-ET.
Exponential-phase XH001n monoculture and established XH001/TM7x coculture were prepared, and cells were harvested and resuspended in PBS to a final bacterial concentration of ~109 cells/mL. 100 μL of cell solution were mixed with 100 μL PBS and used for subsequent cryo-ET sample preparation.
Cryo-ET samples were prepared using copper grids with holey carbon support film (200 mesh, R2/1, Quantifoil). The grids were glow-discharged for ~30 s before depositing 5 μL of cell solution onto them. Then, the grids were blotted with Whatman filter paper from the back side for about 8 s and rapidly frozen in liquid ethane cooled with liquid nitrogen using a home-made gravity-driven plunger apparatus.
The plunge-frozen grids were clipped into cryo-FIB AutoGrids and mounted into the specimen shuttle under liquid nitrogen. Samples were milled to thin lamellae using an Aquilos cryo-FIB system (Thermo Fisher Scientific). Samples were sputter-coated with Pt to improve the overall sample conductivity, deposited with an organometallic Pt layer (4 to 5 μm thick) using a gas injection system for sample protection, and milled using a gallium ion beam at 30 kV with a stage tilt angle of approximately 17˚. The ion beam current was reduced according to the lamella thickness during the milling process to prevent beam damage and was finally polished to ~200 nm in thickness with a 30 pA ion beam current. Afterwards, a thin Pt layer was sputter-coated onto the lamella to prevent a possible charging issue during the cryo-ET imaging, and the grids were stored in liquid nitrogen.
The cryo-FIB lamellae were transferred to a 300 kV Titan Krios electron microscope (Thermo Fisher Scientific) that was equipped with a Direct Electron Detector, a Volta Phase Plate (VPP), and an energy filter (Gatan). SerialEM (40) was used to collect single-axis tilt series around 0 μm defocus with VPP, with a cumulative dose of ~90 e-/Å covering tilt angles from −51° to 51° (3° tilt step). Images were acquired with an effective pixel size of 3.384 Å at the specimen level. All recorded images were first drift corrected by the software MotionCor2 (41) and then stacked by the software package IMOD (42). The tilt series were then aligned by IMOD with the patching tracking method. Tomograms were then reconstructed in IMOD, using the aligned stacks.
A representative section was selected from the tomograms corresponding to different samples. The cell wall thickness was measured based on the density plot along the cell wall using IMOD (42) and ImageJ.
Proteomics cell growth.
The XH001n monoculture and the XH001/TM7x coculture were inoculated and propagated in BHI (brain heart infusion, BD Bacto, Sparks, MD) broth at 37°C under microaerophilic conditions (2% O2, 5% CO2, balanced with N2) overnight. Exponential-phase cells of XH001n and XH001/TM7x were harvested by centrifugation at 7,800 rpm for 10 min at 4°C and washed 3 times using FMC chemically defined medium (43) to remove BHI carryover. The washed cells were resuspended in 50 mL of FMC medium starting at an OD600nm of 0.4. After 24 h of incubation under microaerophilic conditions, the cells were spun down at 7,800 rpm for 10 min at 20°C, and the supernatants were removed. Then, the pellets were resuspended in 50 mL of fresh FMC medium. The cells were incubated under microaerophilic conditions for another 12 h. After incubation, the cells were spun down by centrifugation, and the supernatants and the cell pellets were collected and frozen with liquid nitrogen immediately, then stored at −80°C.
Cell harvesting, isolation of proteins, and tryptic digestion.
Bacterial cells were harvested by centrifugation at 6,000 × g for 5 min at 4°C and washed twice with 10 mL ice-cold PBS buffer to remove extracellular proteins and medium compounds. The resulting cell pellets were snap-frozen in liquid nitrogen and lyophilized overnight. Total cell protein was isolated from 10 mg lyophilized cells as described by Chipashvili et al. (44). Cells were suspended in 500 μL lysis buffer (2% wt/vol SDS, 50 mM Tris pH 8.0, 10 mM DTT) and disrupted in screw cap tubes filled with 0.1 mm glass pearls (Biospec Products Inc., Bartlesville, OK, USA) filled in Screw Cap Microcentrifuge Tubes (Fisher Scientific, Waltham, MA, USA) using a Bead Mill 24 (Fisher Scientific, Waltham, MA, USA) at maximum speed for 2 × 30 sec. Lysates were cleared by centrifugation for 10 min at 20,000 × g at 4°C. Protein was precipitated by mixing samples with ice-cold acetone at a 5:1 ratio and incubating the mixture for 2 h at −20°C. The protein precipitate was pelleted by centrifugation for 5 min at 20,000 × g at 4°C and washed twice with 500 μL 80% ice-cold acetone. Air-dried protein pellets were reconstituted in 50 μL rehydration buffer (8 M urea, 50 mM Tris pH 8.0, 10 mM DTT) at room temperature, and total protein was quantified by using the Micro BCA Assay (Pierce, Dallas, TX, USA). 80 μg protein were diluted in 20 μL rehydration buffer, and 55 μL of 50 mM ammonium bicarbonate (ABC) buffer was added. Proteins were alkylated by adding 20 μL of 200 mM iodoacetamide in 50 mM ABC, followed by a 30 min incubation time at RT. The alkylation reaction was quenched with 5 μL of 1 M DTT, and the mixture was incubated for 10 min at RT. The protein was digested overnight at 37°C with trypsin (10 μL of 0.1 μg/μL trypsin in ABC buffer), and an additional 10 μL trypsin (0.1 μg/μL) were added in the morning for an additional 2 h of digestion at 37°C to cleave undigested proteins.
SDB-RPS extraction of trypsinized bacterial proteins.
Tryptic digests were acidified (0.5% wt/vol TFA) and extracted by using custom-made SDB-RPS tips (CDS Analytical, Oxford, PA, USA), following the descriptions by Rappsilber et al. (45). Peptides were eluted from SBD-RPS filters with 80% (vol/vol) ACN and 5% (vol/vol) ammonium hydroxide, evaporated to dryness in a Vacufuge plus (Eppendorf, Hamburg, Germany), and taken up in 25 μL 0.1% (vol/vol) formic acid. Particles were removed from extracts by centrifugation for 10 min at 20,000 × g at 4°C before LC-MS analysis.
nanoLC-MS/MS analysis.
The LC-MS/MS analysis was performed using an Easy-nLC 1000 system connected to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with an Easy Spray ESI source for the ionization of eluting fractions. Peptide extracts were separated via a PepMap C18 (ES903, 50 μm × 50 mm, 100 A particle size) column (Thermo Fisher Scientific) guarded by an Acclaim PepMap 100 (100 μm × 2 cm) trap column. An injection volume of 5 μL peptide extract was employed for chromatographic separation, using a 150 min gradient (2 to 32% solution B) of 0.1% (vol/vol) formic acid in water (Solution A) and 0.1% formic acid (vol/vol) in acetonitrile (Solution B) at a flow rate of 200 nL/min and a column temperature of 45°C. The ion source was set to a voltage of 2,000 V (positive mode) at 275°C. Data were measured in data dependent acquisition (DDA) mode over a mass range of m/z 350 to 1,200 at 60,000 resolution, using an automatic gain control (AGC) target setting of 3.0 × 105 and 250 ms injection time. MS/MS spectra of the top 20 most abundant MS1 ions (minimum intensity: 2.0 × 104) were fragmented by HCD, using a collision energy of 30 and a 2.5 Da isolation window. The resulting product ion spectra were acquired at a resolution of 15,000, using an AGC target setting of 5 × 104, a maximum ion accumulation time of 150 ms, and a dynamic exclusion time of 40 s.
Data analysis.
Protein identification and label free quantitation was carried out with the PEAKS Studio X Pro software suite. The LC-MS data files were imported using the following parameters: merge scans (retention time window: 5 min, precursor m/z tolerance: 10 ppm, precursor mass and charge states z = 1 to 10) with other data preprocessing functions (centroiding, deisotoping, and deconvolution) being executed automatically. Protein identities were assigned by searching against the Schaalia odontolytica database (Uniprot, version 2020-07) using the following parameters: parent mass error tolerance 10 ppm; fragment mass error tolerance 0.05 Da; semispecific trypsin enzyme search; fixed modifications: carbamidomethylation (C); variable modifications: phosphorylation (STY), pyro-glu (Q), oxidation (M), deamidation (NQ), acetylation (N-terminal); maximal two missed cleavages, maximal three variable posttranslational modifications per peptide. False discovery was controlled by using the decoy-fusion method, and peptides with a false discovery rate of ≤0.5% and proteins above the significance threshold of 20 (–10lgP) were classified as identified. For the relative quantitation of proteins, the peak areas of the top 3 most abundant unique peptides were employed. Raw protein peak areas were exported as CSV tables and processed using Microsoft Excel.
Data availability.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (16) and are accessible through GEO Series accession number GSE196744.
ACKNOWLEDGMENTS
We thank Meng Shao for preparing cryo-ET samples with cryo-FIB/SEM at the Liu laboratory at Yale; Y.C. and J.L. were supported by grants 3R01DE023810-07S1, R01AI087946 and R01AI132818 from the National Institute of Allergy and Infectious Diseases (NIAID); Cryo-ET data were collected at the Yale Cryo-EM resources and funded in part by the NIH grant 1S10OD023603-01A1. This research was supported by grants from the National Institute of Dental and Craniofacial Research of the National Institutes of Health under Awards 1K99DE027719–01 (B.B.); T90DE021984, KL2TR000421 (K.A.K.); and 1R01DE023810 (X.H., W.S. and J.S.M.).
Footnotes
Supplemental material is available online only.
Contributor Information
Jeffrey S. McLean, Email: jsmclean@uw.edu.
Laurie E. Comstock, University of Chicago
REFERENCES
- 1.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA 112:244–249. 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLean JS, Liu Q, Bor B, Bedree JK, Cen L, Watling M, To TT, Bumgarner RE, He X, Shi W. 2016. Draft genome sequence of Actinomyces odontolyticus subsp. actinosynbacter strain XH001, the basibiont of an Oral TM7 epibiont. Genome Announc 4. 10.1128/genomeA.01685-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 4.Castelle CJ, Banfield JF. 2018. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 172:1181–1197. 10.1016/j.cell.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 5.McLean JS, Bor B, Kerns KA, Liu Q, To TT, Solden L, Hendrickson EL, Wrighton K, Shi W, He X. 2020. Acquisition and adaptation of ultra-small parasitic reduced genome bacteria to mammalian hosts. Cell Rep 32:107939. 10.1016/j.celrep.2020.107939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luef B, Frischkorn KR, Wrighton KC, Holman HY, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH, Comolli LR, Banfield JF. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6:6372. 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 7.McLean JS, Bor B, To TT, Liu Q, Kerns KA, Solden L, Wrighton K, He X, Shi W. 2018. Evidence of independent acquisition and adaption of ultra-small bacteria to human hosts across the highly diverse yet reduced genomes of the phylum Saccharibacteria. bioRxiv. 10.1101/258137. [DOI]
- 8.Jaffe AL, Thomas AD, He C, Keren R, Valentin-Alvarado LE, Munk P, Bouma-Gregson K, Farag IF, Amano Y, Sachdeva R, West PT, Banfield JF. 2021. Patterns of gene content and co-occurrence constrain the evolutionary path toward animal association in candidate phyla radiation bacteria. mBio 12:e0052121. 10.1128/mBio.00521-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camanocha A, Dewhirst FE. 2014. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 phyla/candidate divisions. J Oral Microbiol 6. 10.3402/jom.v6.25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bor B, Collins AJ, Murugkar PP, Balasubramanian S, To TT, Hendrickson EL, Bedree JK, Bidlack FB, Johnston CD, Shi W, McLean JS, He X, Dewhirst FE. 2020. Insights obtained by culturing Saccharibacteria with their bacterial hosts. J Dent Res 99:685–694. 10.1177/0022034520905792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamont EI, Hendrickson EL, McLean JS, He X, Bor B. 2020. Complete genome sequence of strain BB001, a novel epibiont bacterium from the candidate phylum Saccharibacteria (TM7). Microbiol Resour Announc 9. 10.1128/MRA.00810-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan J. 2013. Bergey's Manual of Systematic Bacteriology (second edition) Volume 5 and the study of Actinomycetes systematic in China. Wei Sheng Wu Xue Bao 53:521–530. [PubMed] [Google Scholar]
- 13.Bor B, Poweleit N, Bois JS, Cen L, Bedree JK, Zhou ZH, Gunsalus RP, Lux R, McLean JS, He X, Shi W. 2016. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont Actinomyces. Microb Ecol 71:243–255. 10.1007/s00248-015-0711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bor B, McLean JS, Foster KR, Cen L, To TT, Serrato-Guillen A, Dewhirst FE, Shi W, He X. 2018. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc Natl Acad Sci USA 115:12277–12282. 10.1073/pnas.1810625115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Utter DR, He X, Cavanaugh CM, McLean JS, Bor B. 2020. The saccharibacterium TM7x elicits differential responses across its host range. ISME J 14:3054–3067. 10.1038/s41396-020-00736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker JL, Bor B, Agnello M, Shi W, He X. 2017. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol 25:362–374. 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Errington J, Daniel RA, Scheffers D-J. 2003. Cytokinesis in bacteria. Microbiol Mol Biol Rev 67:52–65. table of contents. 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howlett R, Anttonen K, Read N, Smith MCM. 2018. Disruption of the GDP-mannose synthesis pathway in Streptomyces coelicolor results in antibiotic hyper-susceptible phenotypes. Microbiology (Reading) 164:614–624. 10.1099/mic.0.000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bor B, Bedree JK, Shi W, McLean JS, He X. 2019. Saccharibacteria (TM7) in the human oral microbiome. J Dent Res 98:500–509. 10.1177/0022034519831671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian J, Utter DR, Cen L, Dong PT, Shi W, Bor B, Qin M, McLean JS, He X. 2022. Acquisition of the arginine deiminase system benefits epiparasitic Saccharibacteria and their host bacteria in a mammalian niche environment. Proc Natl Acad Sci USA 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedree JK, Bor B, Cen L, Edlund A, Lux R, McLean JS, Shi W, He X. 2018. Quorum sensing modulates the epibiotic-parasitic relationship between Actinomyces odontolyticus and its Saccharibacteria epibiont, a Nanosynbacter lyticus strain, TM7x. Front Microbiol 9:2049. 10.3389/fmicb.2018.02049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padmalayam I, Karem K, Baumstark B, Massung R. 2000. The gene encoding the 17-kDa antigen of Bartonella henselae is located within a cluster of genes homologous to the virB virulence operon. DNA Cell Biol 19:377–382. 10.1089/10445490050043344. [DOI] [PubMed] [Google Scholar]
- 25.Gygi SP, Rochon Y, Franza BR, Aebersold R. 1999. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19:1720–1730. 10.1128/MCB.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murugkar PP, Collins AJ, Chen T, Dewhirst FE. 2020. Isolation and cultivation of candidate phyla radiation Saccharibacteria (TM7) bacteria in coculture with bacterial hosts. J Oral Microbiol 12:1814666. 10.1080/20002297.2020.1814666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross KL, Campbell JH, Balachandran M, Campbell AG, Cooper CJ, Griffen A, Heaton M, Joshi S, Klingeman D, Leys E, Yang Z, Parks JM, Podar M. 2019. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol 37:1314–1321. 10.1038/s41587-019-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushnell B. 2020. BBDuk. Jt Genome Inst Available online: https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-userguide/bbduk-guide/. (accessed on 25 June 2020)
- 29.Powell DR. 2015. Degust: interactive RNA-seq analysis. Zenododoi 10. [Google Scholar]
- 30.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res 44:D67–72. 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 33.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol 34:2115–2122. 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anonymous. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. 2021. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 49:D545–D551. 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 43.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11:649–655. 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chipashvili O, Utter DR, Bedree JK, Ma Y, Schulte F, Mascarin G, Alayyoubi Y, Chouhan D, Hardt M, Bidlack F, Hasturk H, He X, McLean JS, Bor B. 2021. Episymbiotic Saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe 29:1649–1662 e7. 10.1016/j.chom.2021.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download jb.00112-22-s0001.pdf, PDF file, 1.9 MB (1.9MB, pdf)
Table S1. Download jb.00112-22-s0002.xlsx, XLSX file, 1.0 MB (1MB, xlsx)
Table S2. Download jb.00112-22-s0003.xlsx, XLSX file, 0.03 MB (27.6KB, xlsx)
Table S3. Download jb.00112-22-s0004.xlsx, XLSX file, 0.2 MB (175.1KB, xlsx)
Table S4. Download jb.00112-22-s0005.xlsx, XLSX file, 0.02 MB (17.1KB, xlsx)
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (16) and are accessible through GEO Series accession number GSE196744.