ABSTRACT
Tuberculosis (TB) is the leading cause of death from any bacterial infection, causing 1.5 million deaths worldwide each year. Due to the emergence of drug-resistant strains of Mycobacterium tuberculosis (Mtb) there have been significant efforts aimed at developing novel drugs to treat TB. One promising drug target in Mtb is the arabinogalactan biosynthetic enzyme DprE1, and there have been over a dozen unique chemical scaffolds identified which inhibit the activity of this protein. Among the most promising lead compounds are the benzothiazinones BTZ043 and PBTZ169, both of which are currently in or have completed phase IIa clinical trials. Due to the potential clinical utility of these drugs, we sought to identify potential synergistic interactions and new mechanisms of resistance using a genome-scale CRISPRi chemical-genetic screen with PBTZ169. We found that knockdown of rv0678, the negative regulator of the mmpS5/L5 drug efflux pump, confers resistance to PBTZ169. Mutations in rv0678 are the most common form of resistance to bedaquiline and there is already abundant evidence of these mutations emerging in bedaquiline-treated patients. We confirmed that rv0678 mutations from clinical isolates confer low level cross-resistance to BTZ043 and PBTZ169. While it is yet unclear whether rv0678 mutations would render benzothiazinones ineffective in treating TB, these results highlight the importance of monitoring for clinically prevalent rv0678 mutations during ongoing BTZ043 and PBTZ169 clinical trials.
KEYWORDS: Mycobacterium tuberculosis, drug resistance mechanisms, efflux pumps, molecular genetics
INTRODUCTION
Tuberculosis (TB) is among the most difficult bacterial infections to treat, requiring multiple months of combination therapy to produce a durable cure (1, 2). Mycobacterium tuberculosis (Mtb), the causative agent of TB, is intrinsically resistant to many different classes of antimicrobial compounds (3–6). Further, Mtb can evolve acquired drug resistance mutations to every antitubercular drug in clinical use, adding another layer of difficulty to TB treatment (7–9). Current estimates suggest that roughly 5% of global TB cases are caused by multidrug-resistant (MDR) Mtb strains, accounting for roughly 500,000 newly diagnosed MDR infections each year (2, 10). In certain geographic regions, drug resistance rates can be as high as 40%, highlighting the need for new antitubercular drugs and drug combinations (2).
Drug discovery efforts over the past 2 decades have identified several Mtb proteins that are commonly found to be the target of new antitubercular compounds (11). Among these “promiscuous targets” are the trehalose dimycolate transporter MmpL3, the cytochrome bc1 oxidase subunit QcrB, and the decaprenylphosphoryl-β-d-ribofuranose-2′-epimerase subunit DprE1, which is involved in the synthesis of arabinogalactan (11–14). Many distinct chemical scaffolds have been found to inhibit each of these proteins and several of these compounds have advanced to clinical trials (15–19). Among these are the benzothiazinone (BTZ) DprE1 inhibitors, including PBTZ169 (20) and BTZ043 (21, 22) which are among the most potent antitubercular compounds discovered thus far, with MICs in the low nanomolar range (22). In addition to having potent antitubercular activity, the lead BTZ, PBTZ169, displays synergistic activity with other antitubercular drugs such as bedaquiline, presumably by increasing Mtb cell permeability and bedaquiline uptake (23, 24). PBTZ169 (Macozinone) underwent a phase IIa early bactericidal activity (EBA) clinical trial in Russia where it showed good safety, tolerability, and EBA (16, 25). BTZ043 is part of an ongoing phase IIa study in South Africa (18, 26).
Given the clinical promise of this class of drugs, we sought to better understand the bacterial genetic determinants of sensitivity and resistance to PBTZ169. By identifying genes whose inhibition sensitizes Mtb to PBTZ169, we hoped to identify new synergistic drug targets for BTZs. Further, by identifying genes whose inhibition results in PBTZ169 resistance, we hoped to identify novel sources of genetic resistance to BTZs, distinct from the mutations in the drug target which have been well characterized (21, 22, 27).
RESULTS
CRISPR interference-based chemical-genetic profiling identifies determinants of PBTZ169 sensitivity and resistance.
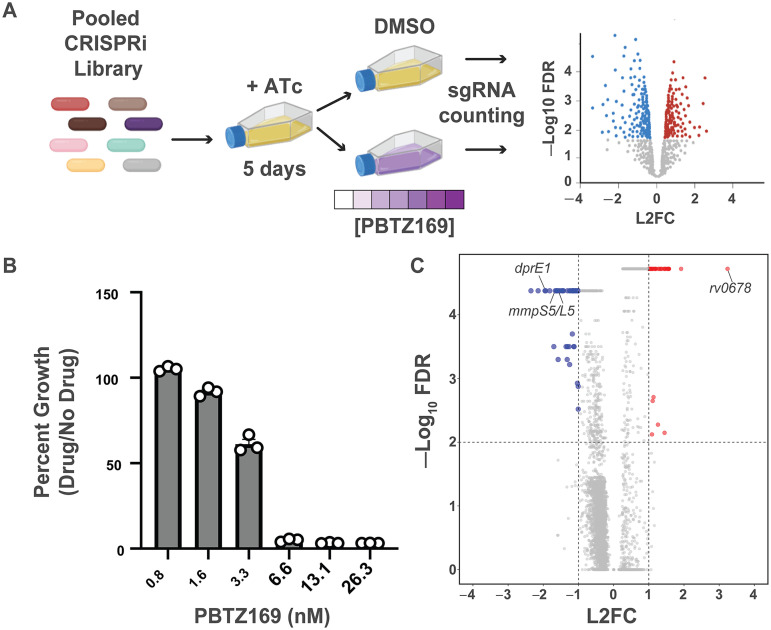
To identify the genetic determinants of PBTZ169 potency, we performed a genome-scale, chemical-genetic screen using the Mtb CRISPR interference (CRISPRi) platform described previously (28–31). This library contains 96,700 unique single guide RNAs (sgRNAs) targeting over 98% of the genes in the H37Rv Mtb genome. This approach allows for the assessment of chemical-genetic interactions for both in vitro essential and nonessential genes (30). The H37Rv CRISPRi library was treated with anhydrotetracycline (ATc) for 5 days to deplete target proteins, after which the library was split into triplicate cultures treated with either DMSO (vehicle control) or various concentrations of PBTZ169 (Fig. 1A). The 0.8 and 1.6 nM PBTZ169 concentrations (Fig. 1B) were used for downstream analysis, because these concentrations of PBTZ169 exerted selective pressure without bottlenecking (i.e., sharply reducing complexity) the CRISPRi library. The fitness of each strain was assessed by deep sequencing of the sgRNA encoding region and subsequent gene level analysis was performed using MAGeCK (32).
FIG 1.
CRISPRi chemical-genetic profiling of PBTZ169. (A) Chemical-genetic profiling workflow. The CRISPRi library contains 96,700 sgRNAs targeting 4,052/4,125 genes in the H37Rv genome. The library was predepleted for 5 days in the presence of anhydrotetracycline (ATc) prior to treatment with DMSO or various concentrations of PBTZ169 spanning the MIC. After 14 days of library outgrowth, genomic DNA was isolated from all cultures with detectable outgrowth. The sgRNA-encoding region was subjected to deep sequencing for control cultures as well as the 0.8 nM and 1.6 nM PBTZ169 cultures. Hit genes were called by MAGeCK (32). (B) Relative growth of PBTZ169-treated cultures. After 14 days of outgrowth, growth was calculated relative to the DMSO control. Individual replicates are shown. (C) Volcano plot summarizing the log2-fold change (L2FC) value and –log10 of the false discovery rate (FDR) for each gene in the PBTZ169 screen (1.6 nM). Key hit genes are highlighted.
This screen identified a total of 79 chemical-genetic hits (|L2FC > 1|, FDR < 0.01; Fig. 1C; Data Set S1). There were a total of 46 genes for which knockdown resulted in increased PBTZ169 sensitivity and 33 genes for which knockdown resulted in increased PBTZ169 resistance (Fig. 1C). Among the most sensitizing hit genes was dprE1, consistent with previous studies showing that the genetic knockdown of a drug target generally results in sensitivity to that particular drug (30, 33, 34). Also among the sensitizing hits were a number of thioredoxin genes (trxC, trxB2, sirA). Alteration of cellular redox homeostasis could alter FAD/FADH2 levels and make DprE1 more susceptible to PBTZ169-mediated inhibition (21). Further, the screen identified a number of cell wall biosynthetic enzymes (pbpA, idsA2, mmaA4) as sensitizing hits. Sensitivity of these knockdown strains to PBTZ169 may reflect the combined effects of inhibiting multiple cell envelope synthetic pathways. Alternatively, these mutants may have a compromised cell envelope, allowing PBTZ169 to reach DprE1 more easily in the mycobacterial periplasm (35, 36). However, the chemical-genetic signature of PBTZ169 is distinct from drugs like rifampicin and vancomycin, for which the envelope appears to be a relevant permeability barrier, suggesting that the activity of PBTZ169 is not limited by drug uptake (30, 37). Finally, we identified the mmpS5/L5 efflux pump genes as among the most sensitizing hits in the screen. MmpLS5/L5 is a multidrug efflux pump that extrudes a number of antitubercular drugs such as bedaquiline and clofazimine (38). These results suggest that MmpS5/L5 may also be active against PBTZ169.
This chemical-genetic screen was also used to identify BTZ resistance mechanisms independent of drug binding site mutations in DprE1 (22, 27, 39). The strongest resistance hit gene was rv0678, which showed a greater than 8-fold enrichment under PBTZ169 treatment conditions compared with the vehicle control (Fig. 1C; Supplemental file 1). Rv0678 encodes a transcription factor which negatively regulates the expression of the mmpS5/L5 efflux pump (38). Loss-of-function mutations in rv0678 are a common mechanism of resistance to the new antitubercular drug bedaquiline as well as the antileprotic/antitubercular drug clofazimine (38, 40, 41). There are several reports of rv0678 mutations arising at a high frequency in Mtb clinical isolates (41–45). Further, some rv0678 mutations predate the introduction of bedaquiline to TB treatment regimens and may have arisen in response to other selective pressures, such as clofazimine treatment of leprosy infections (43). Given the clinical prevalence of rv0678 mutations as well as the strong rv0678 signature in the PBTZ169 screen, we sought to determine whether this may be a relevant mechanism of resistance to BTZs.
Clinically prevalent mutations in rv0678 confer low-level resistance to BTZs.
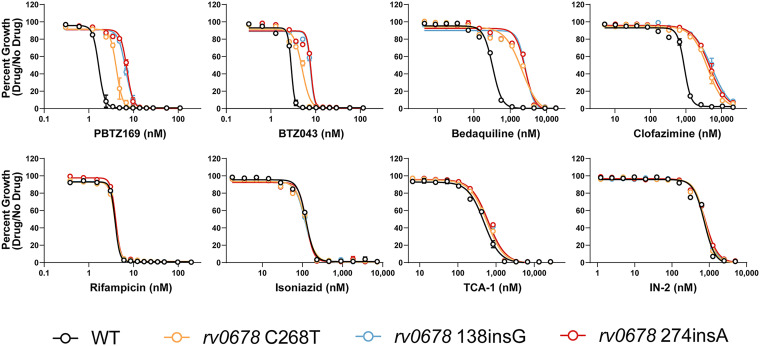
In order to assess whether mutations in rv0678 may confer cross-resistance to BTZs, we first isolated a select group of three rv0678 mutants associated with clinical bedaquiline resistance (41–44) using single-stranded DNA recombineering in H37Rv Mtb. Mutations in rv0678 were confirmed by Sanger sequencing and strains were subjected to whole-genome sequencing to ensure that there were no other mutations that are likely to influence drug susceptibility. We tested the susceptibility of all three rv0678 mutants and the wild-type strain to 16 different antitubercular drugs (Fig. 2; Fig. S1; Table 1). Consistent with the CRISPRi screening results, the rv0678 frameshift mutants showed an ~4-fold increase in IC50 to PBTZ169 and an ~3-fold increase in IC50 to BTZ043. The C268T missense mutant showed slightly smaller IC50 shifts of 2.4-fold and 1.7-fold to PBTZ169 and BTZ043, respectively. These IC50 shifts were slightly smaller than what was seen for bedaquiline but comparable to what was seen for clofazimine. The rv0678 mutants showed nearly identical susceptibility to the other drugs tested, including two chemically distinct DprE1 inhibitors, TCA-1 and IN-2 (46, 47). The lack of cross-resistance to TCA-1 and IN-2 suggest that MmpL5/S5 efflux is not relevant for all DprE1 inhibitors. Interestingly, rv0678 mutants also showed resistance to fusidic acid, an antibiotic not used in TB treatment but used widely to treat skin and soft tissue infections caused by Staphylococcus aureus (48). These results suggest that orally administered fusidic acid may be a potential selective pressure for Mtb rv0678 mutations in areas where bedaquiline and clofazimine have not been introduced (43).
FIG 2.
Antibiotic susceptibility of rv0678 mutants. Dose-response curves (mean ± s.e.m., n = 3 replicates) for the indicated Mtb strains. Results are representative of two independent experiments.
TABLE 1.
Susceptibility (IC50) of rv0678 mutants to 16 antibioticsa
| Antibiotic | Target process | Wild-type | rv0678 138insG | rv0678 274insA | rv0678 C268T | IC50 ratiob |
|---|---|---|---|---|---|---|
| PBTZ169 | Arabinogalactan | 1.7 | 6.6 | 7.1 | 4.1 | 4.2 |
| BTZ043 | Arabinogalactan | 2.9 | 7.7 | 7.8 | 5 | 2.7 |
| Bedaquiline | ATP synthase | 330 | 2700 | 2600 | 2100 | 7.9 |
| Clofazimine | Respiration | 900 | 5400 | 4700 | 4000 | 5.2 |
| Rifampicin | Transcription | 3.9 | 3.9 | 4 | 3.9 | 1.0 |
| Isoniazid | Mycolic acid | 120 | 120 | 120 | 120 | 1.0 |
| TCA-1 | Arabinogalactan | 480 | 610 | 630 | 570 | 1.3 |
| IN-2 | Arabinogalactan | 730 | 770 | 780 | 760 | 1.1 |
| Ethambutol | Arabinogalactan | 2400 | 2800 | 3200 | 3100 | 1.3 |
| Levofloxacin | DNA replication | 700 | 700 | 700 | 690 | 1.0 |
| Pretomanid | Mycolic acid | 360 | 400 | 430 | 440 | 1.2 |
| Linezolid | Translation | 1300 | 1400 | 1300 | 1200 | 1.0 |
| Streptomycin | Translation | 420 | 410 | 430 | 440 | 1.0 |
| Amikacin | Translation | 930 | 900 | 950 | 980 | 1.0 |
| Capreomycin | Translation | 1200 | 1200 | 1200 | 1200 | 1.0 |
| Fusidic Acid | Translation | 4200 | 26000 | 26000 | 17000 | 6.2 |
IC50 values reflect drug concentrations in nanomolar (nM).
The IC50 ratio was calculated by dividing the IC50 for the rv0678 274insA mutant by the IC50 for the wild-type strain.
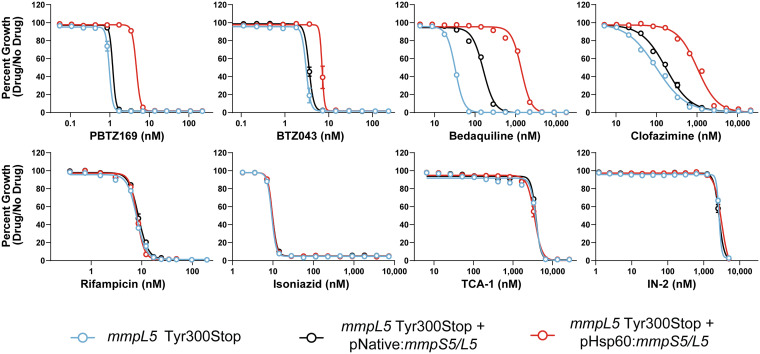
Given the phenotypes we observed with rv0678 mutants, we next sought to confirm that overexpression of the MmpS5/L5 efflux pump is responsible for the BTZ resistance in rv0678 mutants. To do this, we made use of a lineage 1 clinical isolate of Mtb harboring a nonsense mutation (Tyr300Stop) in mmpL5 (49). This strain was complemented with a functional copy of the mmpS5/L5 operon expressed from the native promoter to restore wild-type expression or by the hsp60 promoter to overexpress the efflux pump (30). Consistent with the previous results, overexpression of mmpS5/L5 resulted in increased IC50 for PBTZ169, BTZ043, bedaquiline, clofazimine, and fusidic acid, but not the other drugs tested (Fig. 3; Fig. S2; Table 2). Interestingly, while the parental mmpL5 Tyr300Stop mutant was hypersusceptible to bedaquiline, this strain was only modestly more sensitive to the BTZs, clofazimine, and fusidic acid, consistent with previous studies where gain-of-function and loss-of-function mutations may not necessarily produce opposing phenotypes of equal magnitude (50, 51).
FIG 3.
Antibiotic susceptibility of mmpL5 loss-of-function and gain-of-function strains. Dose-response curves (mean ± s.e.m., n = 3 replicates) for the indicated Mtb strains. Results are representative of two independent experiments. The Tyr300Stop mutant (blue) harbors an empty version of the mmpS5/L5 complementation vector (30).
TABLE 2.
Susceptibility of mmpL5 loss-of-function and gain-of-function strains to 16 antibioticsa
| Antibiotic | Target process | mmpL5 Tyr300Stop |
mmpL5 Tyr300Stop + pNative:mmpS5/L5 |
mmpL5 Tyr300Stop + pHsp60:mmpS5/L5 | IC50 ratiob |
|---|---|---|---|---|---|
| PBTZ169 | Arabinogalactan | 1.0 | 1.2 | 4.7 | 3.9 |
| BTZ043 | Arabinogalactan | 3.1 | 3.5 | 7 | 2 |
| Bedaquiline | ATP synthase | 33 | 160 | 1400 | 8.8 |
| Clofazimine | Respiration | 100 | 170 | 1030 | 6.1 |
| Rifampicin | Transcription | 7.2 | 7.8 | 7.3 | 0.9 |
| Isoniazid | Mycolic acid | 9.3 | 9.7 | 9.7 | 1.0 |
| TCA-1 | Arabinogalactan | 3800 | 3800 | 3500 | 0.9 |
| IN-2 | Arabinogalactan | 2700 | 2700 | 3100 | 1.1 |
| Ethambutol | Arabinogalactan | 1900 | 1900 | 2300 | 1.2 |
| Levofloxacin | DNA replication | 900 | 960 | 970 | 1.0 |
| Pretomanid | Mycolic acid | 750 | 810 | 830 | 1.0 |
| Linezolid | Translation | 1100 | 1100 | 1200 | 1.1 |
| Fusidic Acid | Translation | 1800 | 3400 | 15000 | 4.4 |
IC50 values reflect drug concentrations in nanomolar (nM).
The IC50 ratio was calculated by dividing the IC50 for the pHsp60-driven mmpS5/L5 strain by the IC50 for the pNative-driven mmpS5/L5 strain.
DISCUSSION
BTZ DprE1 inhibitors are promising antitubercular agents that are currently in early-stage clinical trials (16, 17, 52). A better understanding of the genetic factors that influence the potency of these drugs could help to inform the design of synergistic drug combinations and to diagnose the emergence of BTZ resistance. Here, we applied a genome-scale CRISPRi screen to identify the determinants of sensitivity and resistance to PBTZ169. We identified dozens of hit genes, a number of which are essential, which may be targeted to potentiate the activity of BTZs. Target-based drug discovery efforts toward these proteins may identify synergistic partners for use with BTZs. Future chemical-genetic efforts could apply a similar screening pipeline to study other non-BTZ DprE1 inhibitors (52, 53) to identify common and unique components of their chemical-genetic signatures. This may provide a global overview of the genes and pathways which restrict the activity of these compounds and allow for the design of optimized DprE1 inhibitors that fully exploit this chemically vulnerable drug target.
In addition to finding strategies to potentiate the activity of PBTZ169, we hoped to use this screen to identify resistance mechanisms for BTZs that are mediated by loss-of-function mutations. Interestingly, the strongest enriched hit gene found in our screen was rv0678, which was chosen for follow-up investigation due to the potential clinical relevance of this finding. We observed that common rv0678 mutations from Mtb clinical isolates confer low-level resistance to PBTZ169 and BTZ043. Resistance is also observed in Mtb strains directly overexpressing the mmpS5/L5 operon, suggesting that efflux of BTZs is the relevant mechanism of resistance. Consistent with our findings, another recent study isolated PBTZ169-resistant mutants on progressively higher drug concentrations and identified a number of clones harboring mutations in rv0678 (54). Although the authors do not test the individual impact of each rv0678 mutation, the results presented here suggest that these are likely to be bona fide, low-level PBTZ169 resistance mutations.
Importantly, rv0678 mutations did not promote resistance to the chemically distinct (46, 47) DprE1 inhibitors TCA-1 and IN-2. This observation argues against a role for perturbed Mtb physiology as a result of elevated MmpL5/S5 activity in promoting resistance to BTZs, rather suggesting efflux is the relevant resistance mechanism. Previous work has convincingly argued that DprE1 localizes to the periplasm, suggesting that periplasmic localization contributes to the chemical vulnerability of DprE1: inhibitors need not cross the Mtb plasma membrane to the cytosol in order to engage their target (35, 36). If this is true, how might the MmpL5/S5 efflux pump, which is localized to the plasma membrane, promote resistance to BTZs? There are several potential explanations to these findings. First, it is possible that BTZs can covalently inhibit DprE1 in the cytosol prior to Tat-mediated transport of the folded DprE1 to the periplasm. While experimental evidence for Tat-mediated DprE1 export is lacking, DprE1 is predicted to encode a Tat signal peptide (55, 56). In this scenario, MmpL5/S5 efflux of BTZs from the cytosol will hinder cytoplasmic DprE1 inhibition. Second, it is possible that MmpL5/S5 could efflux BTZs out of the periplasm to prevent DprE1 engagement, through a mechanism similar to that of the AcrB efflux pump in Escherichia coli (57). This would presumably require MmpL5/S5 interaction with mycomembrane spanning proteins to efflux BTZs outside the Mtb envelope (58–60). Along these lines, studies in Gram-negative bacteria have shown that efflux pumps can form large protein complexes that span both bacterial membranes and promote resistance to beta-lactam antibiotics, whose PBP targets also localize to the periplasm (57, 58, 61–65).
Although the exact mechanism of MmpS5/L5-mediated BTZ resistance is not clear, these results suggest that rv0678 mutations could limit the clinical efficacy of BTZs, especially in areas where bedaquiline has already been used. While the BTZ series are remarkably potent, murine pharmacokinetic data suggest that PBTZ169 accumulates to concentrations close to its MIC, particularly in caseous lesions (52). The mouse data suggest that small increases in MIC (66) may be sufficient to render PBTZ169 less effective. Future studies are necessary to test whether rv0678 mutations could contribute to BTZ treatment failure in humans.
MATERIALS AND METHODS
Mycobacterial cultures.
Mtb was grown at 37°C in Difco Middlebrook 7H9 broth or on 7H10 agar supplemented with 0.2% glycerol (7H9) or 0.5% glycerol (7H10), 0.05% Tween 80, 1X oleic acid-albumin-dextrose-catalase (OADC), and the appropriate antibiotics (kanamycin 10 to 20 μg/mL or zeocin 20 μg/mL). ATc was used at 100ng/mL in the CRISPRi screen and at 500 ng/mL for ssDNA recombineering (67). Mtb cultures were grown standing in tissue culture flasks (unless otherwise indicated) with 5% CO2. Mtb strains are derivatives of the H37Rv background, except those shown in Fig. 3 and Fig. S2. Those strains are derivatives of a Lineage 1.1 strain (ITM-2018-00084) from the Belgian Coordinated Culture Collection (BCCM) (49). This strain harbors the Tyr300Stop mutation in mmpL5. The mmpS5/L5 complementation and overexpression strains are described by Li et al. (30).
Pooled CRISPRi chemical-genetic screening.
Chemical-genetic screens were initiated by thawing 2 × 1 ml aliquots (1.0 OD600 units/mL) of the Mtb CRISPRi library (RLC12; Addgene 163954) and inoculating each aliquot into 19 mL 7H9 supplemented with kanamycin (10 μg/mL) in a vented tissue culture flask (T-75; Falcon 353136) (30). The starting OD600 of each culture was approximately 0.05. Cultures were expanded to OD600 = 1.0, pooled, and passed through a 10-μm cell strainer (pluriSelect 43–50010-03) to obtain a single-cell suspension. The single-cell suspension was then back-diluted to an OD600 of 0.05 in 4 × 25 mL cultures and treated with ATc (100 ng/mL final concentration) to initiate target predepletion. After 5 days of predepletion triplicate cultures were then inoculated at OD600 of 0.05 in 10 mL 7H9 supplemented with ATc (100 ng/mL), kanamycin (10 μg/mL), and the indicated PBTZ169 concentration or a DMSO vehicle control. In all cultures the final DMSO concentration was normalized to 0.6%. Pooled CRISPRi chemical-genetic screens were performed in vented tissue culture flasks (T-25; Falcon 353109) under standing (nonshaking) conditions. Cultures were outgrown for 14 days at 37°C, 5% CO2. ATc was replenished at 100ngml-1 at day 7. After 14 days outgrowth, OD600 values were measured for all cultures to empirically determine the MIC for each drug. The lowest three concentrations of PBTZ169-treatment were subjected to sequencing. However, the 3.3 nM condition showed evidence of library bottlenecking following sgRNA sequencing, likely because this condition was near the MIC. Downstream analysis was performed on the 0.8 nM and 1.6 nM conditions.
Genomic DNA extraction and library preparation for Illumina sequencing.
Genomic DNA was isolated from bacterial pellets using the CTAB-lysozyme method as previously described (28). After drug treatment, 10 to 20 OD600 units of the cultures were pelleted by centrifugation (10 min at 4,000 × g) and were resuspended in 1 mL PBS +0.05% Tween 80. Cell suspensions were centrifuged again for 5 min at 4,000 × g, the supernatant was removed, and pellets were frozen until processing. Upon thawing, pellets were resuspended in 800 μL TE buffer (10 mM Tris pH 8.0, 1 mM EDTA) + 15mg/mL lysozyme (Alfa Aesar J60701-06) and incubated at 37°C for 16 h. Next, 70 μL 10% SDS (Promega V6551) and 5 μL proteinase K (20 mg/mL, Thermo Fisher 25530049) were added and samples were incubated at 65°C for 30 min. Subsequently, 100 μL 5M NaCl and 80 μL 10% CTAB (Sigma-Aldrich H5882) were added, and samples were incubated for an additional 30 min at 65°C. Finally, 750 μL ice-cold chloroform was added and samples were mixed. After centrifugation at 16,100 × g and extraction of the aqueous phase, samples were removed from the biosafety level 3 facility. Samples were then treated with 25 μg RNase A (Bio Basic RB0474) for 30 min at 37°C, followed by extraction with phenol:chloroform:isoamyl alcohol (pH 8.0, 25:24:1, Thermo Fisher BP1752I-400), then chloroform. Genomic DNA was precipitated from the final aqueous layer (600 μL) with the addition of 10 μL 3M sodium acetate and 360 μL isopropanol. DNA pellets were spun at 21,300 × g for 30 min at 4°C and washed 2× with 750 μL 80% ethanol. Pellets were dried and resuspended with elution buffer (Qiagen 19086) before spectrophotometric quantification. The concentration of isolated genomic DNA was quantified using the DeNovix dsDNA high sensitivity assay (KIT-DSDNA-HIGH-2; DS-11 series spectrophotometer/fluorometer).
Next generation sequencing was performed as described by Bosch et al. (28). The sgRNA-encoding region was amplified from 500 ng genomic DNA with 17 cycles of PCR using NEBNext Ultra II Q5 master mix (NEB M0544L) as described in. Each PCR contained a pool of forward primers (0.5 μM final concentration) and a unique indexed reverse primer (0.5 μM). Forward primers contain a P5 flow cell attachment sequence, a standard Read1 Illumina sequencing primer binding site and custom stagger sequences to guarantee base diversity during Illumina sequencing. Reverse primers contain a P7 flow cell attachment sequence, a standard Read2 Illumina sequencing primer binding site and unique barcodes to allow for sample pooling during deep sequencing. Following PCR amplification, each ~230 bp amplicon was purified using AMPure XP beads (Beckman–Coulter A63882) using one-sided selection (1.2×). Bead-purified amplicons were further purified on a Pippin HT 2% agarose gel cassette (target range 180 to 250 bp; Sage Science HTC2010) to remove carry-over primer and genomic DNA. Eluted amplicons were quantified with a Qubit 2.0 fluorometer (Invitrogen), and amplicon size and purity were quality controlled by visualization on an Agilent 2100 bioanalyzer (high sensitivity chip; Agilent Technologies 5067–4626). Next, individual PCR amplicons were multiplexed into 10 nM pools and sequenced on an Illumina sequencer according to the manufacturer’s instructions. To increase sequencing diversity, a PhiX spike-in of 2.5% to 5% was added to the pools (PhiX sequencing control v3; Illumina FC-110-3001). Samples were run on the Illumina NovaSeq 6000 platform (single-read 1 × 85 cycles and 6 × i7 index cycles).
Isolation of rv0678 mutants by single-stranded DNA recombineering.
The 138insG frameshift allele was observed in a bedaquiline-treated patient and emerged as the dominant clone over the course of treatment (41). Another frameshift mutation, 274insA, was identified in multidrug-resistant TB patients with no prior evidence of bedaquiline or clofazimine treatment (43). The C268T missense mutation, which results in the Arg90Cys amino acid change, has been detected in MDR-TB patients from South Africa and Korea (42, 44). Single-stranded DNA recombineering was performed as described by Murphy et al. (67). Briefly, H37Rv Mtb was transformed with the episomal RecT recombinase-expressing plasmid pKM402 (Addgene plasmid # 107770) and was selected on 7H10 agar containing kanamycin (20 μg/mL). A single colony was picked and grown up in 7H9 + kanamycin (20 μg/mL). A single frozen stock of the RecT strain was thawed and expanded to 50 mL in 7H9 + kanamycin (20 μg/mL) in Erlenmeyer flasks under shaking conditions (37°C). After reaching an OD600 of 0.8, ATc was added at a final concentration of 500 ng/mL to induce RecT expression. After 8 additional hours of shaking incubation, sterile glycine was added to a final concentration of 200 mM to improve DNA uptake. After an additional 16 h of shaking incubation, cells were pelleted (4,000 × g) and washed a total of three times in sterile 10% glycerol. On the final wash, cells were resuspended in 5 mL of 10% glycerol. For each transformation, 400 μL of electrocompetent cells was mixed with 1 μg each ssDNA oligo of interest (see Table S1). Cells were electroporated using the Gene Pulser X cell electroporation system (Bio-Rad 1652660) set at 2,500 V, 1,000 Ω, and 25 μF and were recovered for 24 h in 15 mL of fresh 7H9 under 37°C, shaking conditions. A total of 200 μL of each recovered culture was plated on bedaquiline-containing 7H10 plates (125 ng/mL). The control strain shown in Fig. 2 and Fig. S1 was electroporated with no DNA and plated on antibiotic-free 7H10 plates. Plates were incubated for 28 days, after which colonies were picked and grown in 7H9 + bedaquiline at 500 ng/mL, due to the higher MIC of bedaquiline in liquid culture. For each strain of interest, a total of four colonies were picked and genomic DNA was extracted as described above. rv0678 was amplified and sanger sequenced. Successful recombinants were then submitted to whole-genome sequencing via Migs (Microbial Genome Sequencing Center, Pittsburgh PA). Whole-genome sequences were derived from FASTQ files and any mutations were identified using Snippy (68). Whole-genome sequencing results confirmed the presence of the desired rv0678 mutations and absence of any other mutations known to influence drug susceptibility. The negative control “wild-type” strain contained no mutations in rv0678 or other genes known to influence drug susceptibility.
Antibacterial activity measurements.
All compounds (see Table S2) were dissolved in DMSO (VWR V0231) and dispensed using an HP D300e digital dispenser in a 384-well plate format using a 2-fold dilution series. For certain drugs, concentrations near the MIC reflect a 1.41-fold dilution series to provide higher resolution. DMSO did not exceed 1% of the final culture volume and was maintained at the same concentration across all samples. Cultures were growth synchronized to late logarithmic phase (~ OD600 = 0.8) and then back-diluted to a starting OD600 of 0.01. A 50-μL cell suspension was plated in triplicate in wells containing the test compound. Plates were incubated standing at 37°C with 5% CO2. OD580 was evaluated using a Tecan Spark plate reader at 10 to 11 days postplating and percent growth was calculated relative to the DMSO vehicle control for each strain. IC50 measurements were calculated using a nonlinear fit in GraphPad Prism. For all MIC curves, data represent the mean ± s.e.m. for triplicates. Data are representative of two independent experiments.
ACKNOWLEDGMENTS
We thank members of the Rock laboratory, Véronique Dartois, Katarína Mikušová, Valerie Mizrahi, Max O’Donnell, Michelle Larsen, and James Millard for comments on the manuscript and/or helpful discussions as well as Xing Wang, Jenny Xiang, and Adrian Tan of the Weill Cornell Genomics Core for NGS. This work was supported by the Potts Memorial Foundation (M.A.D.), the Department of Defense (PR192421, J.M.R.), the Robertson Therapeutic Development Fund (J.M.R.), an NIH/NIAID New Innovator Award (1DP2AI144850-01, J.M.R.), and a joint NIH tuberculosis research units network (TBRU-N) grant (U19AI162584, J.M.R.). The illustration in Fig. 1A was generated using BioRender software.
Conceptualization: N.C.P. and J.M.R.; investigation: N.C.P.; data analysis: N.C.P., Z.A.A., and M.A.D.; writing—original draft: N.C.P. and J.M.R.; writing—review and editing: N.C.P., Z.A.A., M.A.D., and J.M.R.; funding acquisition: J.M.R.; supervision: J.M.R.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dorman SE, Nahid P, Kurbatova EV, Phillips PPJ, Bryant K, Dooley KE, Engle M, Goldberg SV, Phan HTT, Hakim J, Johnson JL, Lourens M, Martinson NA, Muzanyi G, Narunsky K, Nerette S, Nguyen NV, Pham TH, Pierre S, Purfield AE, Samaneka W, Savic RM, Sanne I, Scott NA, Shenje J, Sizemore E, Vernon A, Waja Z, Weiner M, Swindells S, Chaisson RE, AIDS Clinical Trials Group, Tuberculosis Trials Consortium . 2021. Four-month rifapentine regimens with or without moxifloxacin for tuberculosis. N Engl J Med 384:1705–1718. 10.1056/NEJMoa2033400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2021. Global tuberculosis report. WHO, Geneva, Switzerland. [Google Scholar]
- 3.Xu W, DeJesus MA, Rücker N, Engelhart CA, Wright MG, Healy C, Lin K, Wang R, Park SW, Ioerger TR, Schnappinger D, Ehrt S. 2017. Chemical genetic interaction profiling reveals determinants of intrinsic antibiotic resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:1–15. 10.1128/AAC.01334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batt SM, Minnikin DE, Besra GS. 2020. The thick waxy coat of mycobacteria, a protective layer against antibiotics and the host’s immune system. Biochem J 477:1983–2006. 10.1042/BCJ20200194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dulberger CL, Rubin EJ, Boutte CC. 2020. The mycobacterial cell envelope - a moving target. Nat Rev Microbiol 18:47–59. 10.1038/s41579-019-0273-7. [DOI] [PubMed] [Google Scholar]
- 6.Madsen CT, Jakobsen L, Buriánková K, Doucet-Populaire F, Pernodet J-L, Douthwaite S. 2005. Methyltransferase Erm(37) slips on rRNA to confer atypical resistance in Mycobacterium tuberculosis. J Biol Chem 280:38942–38947. 10.1074/jbc.M505727200. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230. 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 8.CRyPTIC Consortium. 2018. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med 379:1403–1415. 10.1056/NEJMoa1800474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt M, Bradley P, Lapierre SG, Heys S, Thomsit M, Hall MB, Malone KM, Wintringer P, Walker TM, Cirillo DM, Comas I, Farhat MR, Fowler P, Gardy J, Ismail N, Kohl TA, Mathys V, Merker M, Niemann S, Omar SV, Sintchenko V, Smith G, van Soolingen D, Supply P, Tahseen S, Wilcox M, Arandjelovic I, Peto TEA, Crook DW, Iqbal Z. 2019. Antibiotic resistance prediction for Mycobacterium tuberculosis from genome sequence data with Mykrobe. Wellcome Open Res 4:191. 10.12688/wellcomeopenres.15603.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO. 2016. WHO treatment guidelines for drug- resistant tuberculosis 2016. WHO, Geneva, Switzerland. [Google Scholar]
- 11.Lee BS, Pethe K. 2018. Therapeutic potential of promiscuous targets in Mycobacterium tuberculosis. Curr Opin Pharmacol 42:22–26. 10.1016/j.coph.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Grzegorzewicz AE, Pham H, Gundi VAKB, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SEM, Korduláková J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8:334–341. 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lechartier B, Rybniker J, Zumla A, Cole ST. 2014. Tuberculosis drug discovery in the post-post-genomic era. EMBO Mol Med 6:158–168. 10.1002/emmm.201201772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot J-P, Ewann F, Kim EH, Lee SY, Kang S, Seo MJ, Park EJ, Škovierová H, Pham H, Riccardi G, Nam JY, Marsollier L, Kempf M, Joly-Guillou M-L, Oh T, Shin WK, No Z, Nehrbass U, Brosch R, Cole ST, Brodin P. 2009. High content screening identifies decaprenyl-phosphoribose 2′ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog 5:e1000645. 10.1371/journal.ppat.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeree MJ, Heinrich N, Aarnoutse R, Diacon AH, Dawson R, Rehal S, Kibiki GS, Churchyard G, Sanne I, Ntinginya NE, Minja LT, Hunt RD, Charalambous S, Hanekom M, Semvua HH, Mpagama SG, Manyama C, Mtafya B, Reither K, Wallis RS, Venter A, Narunsky K, Mekota A, Henne S, Colbers A, van Balen GP, Gillespie SH, Phillips PPJ, Hoelscher M. 2017. High-dose rifampicin, moxifloxacin, and SQ109 for treating tuberculosis: a multi-arm, multi-stage randomised controlled trial. Lancet Infect Dis 17:39–49. 10.1016/S1473-3099(16)30274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PBTZ169. 2022. Work Gr new TB drugs. https://www.newtbdrugs.org/pipeline/compound/macozinone-mcz-pbtz-169.
- 17.OPC-167832. 2022. Work Gr new TB drugs. https://www.newtbdrugs.org/pipeline/compound/opc-167832.
- 18.BTZ043. 2022. Work Gr new TB drugs. https://www.newtbdrugs.org/pipeline/compound/btz-043.
- 19.Fernandes GFS, Thompson AM, Castagnolo D, Denny WA, Dos Santos JL. 2022. Tuberculosis drug discovery: challenges and new horizons. J Med Chem 65:7489–7531. 10.1021/acs.jmedchem.2c00227. [DOI] [PubMed] [Google Scholar]
- 20.Makarov V, Lechartier B, Zhang M, Neres J, van der Sar AM, Raadsen SA, Hartkoorn RC, Ryabova OB, Vocat A, Decosterd LA, Widmer N, Buclin T, Bitter W, Andries K, Pojer F, Dyson PJ, Cole ST. 2014. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol Med 6:372–383. 10.1002/emmm.201303575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neres J, Pojer F, Molteni E, Chiarelli LR, Dhar N, Boy-Röttger S, Buroni S, Fullam E, Degiacomi G, Lucarelli AP, Read RJ, Zanoni G, Edmondson DE, De Rossi E, Pasca MR, McKinney JD, Dyson PJ, Riccardi G, Mattevi A, Cole ST, Binda C. 2012. Structural basis for benzothiazinone-mediated killing of Mycobacterium tuberculosis. Sci Transl Med 4. 10.1126/scitranslmed.3004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarov V, Manina G, Mikusova K, Möllmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, Milano A, De Rossi E, Belanova M, Bobovska A, Dianiskova P, Kordulakova J, Sala C, Fullam E, Schneider P, McKinney JD, Brodin P, Christophe T, Waddell S, Butcher P, Albrethsen J, Rosenkrands I, Brosch R, Nandi V, Bharath S, Gaonkar S, Shandil RK, Balasubramanian V, Balganesh T, Tyagi S, Grosset J, Riccardi G, Cole ST. 2009. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324:801–804. 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechartier B, Hartkoorn RC, Cole ST. 2012. In vitro combination studies of benzothiazinone lead compound BTZ043 against mycobacterium tuberculosis. Antimicrob Agents Chemother 56:5790–5793. 10.1128/AAC.01476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupien A, Vocat A, Foo CS-Y, Blattes E, Gillon J-Y, Makarov V, Cole ST. 2018. Optimized background regimen for treatment of active tuberculosis with the next-generation benzothiazinone macozinone (PBTZ169). Antimicrob Agents Chemother 62. 10.1128/AAC.00840-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinicaltrials.gov/ct2/show/NCT03334734. Phase 2a Study of PBTZ169.
- 26.Clinicaltrials.gov/ct2/show/NCT04044001. BTZ-043 - Multiple ascending dose (MAD) to evaluate safety, tolerability and early bactericidal activity (EBA).
- 27.de Jesus Lopes Ribeiro AL, Degiacomi G, Ewann F, Buroni S, Incandela ML, Chiarelli LR, Mori G, Kim J, Contreras-Dominguez M, Park YS, Han SJ, Brodin P, Valentini G, Rizzi M, Riccardi G, Pasca MR. 2011. Analogous mechanisms of resistance to benzothiazinones and dinitrobenzamides in Mycobacterium smegmatis. PLoS One 6:e26675–7. 10.1371/journal.pone.0026675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosch B, DeJesus MA, Poulton NC, Zhang W, Engelhart CA, Zaveri A, Lavalette S, Ruecker N, Trujillo C, Wallach JB, Li S, Ehrt S, Chait BT, Schnappinger D, Rock JM. 2021. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell 184:4579–4592. 10.1016/j.cell.2021.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong AI, Rock JM. 2021. CRISPR interference (CRISPRi) for targeted gene silencing in Mycobacteria. Methods Mol Biol 2314:343–364. 10.1007/978-1-0716-1460-0_16. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Poulton NC, Chang JS, Azadian ZA, DeJesus MA, Ruecker N, Zimmerman MD, Eckartt KA, Bosch B, Engelhart CA, Sullivan DF, Gengenbacher M, Dartois VA, Schnappinger D, Rock JM. 2022. CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis. Nat Microbiol 7:766–779. 10.1038/s41564-022-01130-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNeil MB, Keighley LM, Cook JR, Cheung C-Y, Cook GM. 2021. CRISPR interference identifies vulnerable cellular pathways with bactericidal phenotypes in Mycobacterium tuberculosis. Mol Microbiol 116:1033–1043. 10.1111/mmi.14790. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554. 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson EO, LaVerriere E, Office E, Stanley M, Meyer E, Kawate T, Gomez JE, Audette RE, Bandyopadhyay N, Betancourt N, Delano K, Da Silva I, Davis J, Gallo C, Gardner M, Golas AJ, Guinn KM, Kennedy S, Korn R, McConnell JA, Moss CE, Murphy KC, Nietupski RM, Papavinasasundaram KG, Pinkham JT, Pino PA, Proulx MK, Ruecker N, Song N, Thompson M, Trujillo C, Wakabayashi S, Wallach JB, Watson C, Ioerger TR, Lander ES, Hubbard BK, Serrano-Wu MH, Ehrt S, Fitzgerald M, Rubin EJ, Sassetti CM, Schnappinger D, Hung DT. 2019. Large-scale chemical-genetics yields new M. tuberculosis inhibitor classes. Nature 571:72–78. 10.1038/s41586-019-1315-z. [DOI] [PubMed] [Google Scholar]
- 34.Koh E, Oluoch PO, Ruecker N, Proulx MK, Soni V, Murphy KC, Papavinasasundaram K, Reames CJ, Trujillo C, Zaveri A, Zimmerman MD, Aslebagh R, Baker RE, Shaffer SA, Guinn KM, Fitzgerald M, Dartois V, Ehrt S, Hung DT, Ioerger TR, Rubin EJ, Rhee KY, Schnappinger D, Sassetti CM. 2022. Chemical-genetic interaction mapping links carbon metabolism and cell wall structure to tuberculosis drug efficacy. Proc Natl Acad Sci USA 119:e2201632119. 10.1073/pnas.2201632119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer R, Neres J, Piton J, Dhar N, van der Sar A, Mukherjee R, Laroche T, Dyson PJ, McKinney JD, Bitter W, Makarov V, Cole ST. 2018. Fluorescent benzothiazinone analogues efficiently and selectively label Dpre1 in Mycobacteria and Actinobacteria. ACS Chem Biol 13:3184–3192. 10.1021/acschembio.8b00790. [DOI] [PubMed] [Google Scholar]
- 36.Brecik M, Centárová I, Mukherjee R, Kolly GS, Huszár S, Bobovská A, Kilacsková E, Mokošová V, Svetlíková Z, Šarkan M, Neres J, Korduláková J, Cole ST, Mikušová K. 2015. DprE1 is a vulnerable tuberculosis drug target due to its cell wall localization. ACS Chem Biol 10:1631–1636. 10.1021/acschembio.5b00237. [DOI] [PubMed] [Google Scholar]
- 37.McNeil MB, Chettiar S, Awasthi D, Parish T. 2019. Cell wall inhibitors increase the accumulation of rifampicin in Mycobacterium tuberculosis. Access Microbiol 1:e000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartkoorn RC, Uplekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of mmpl5 in mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trefzer C, Rengifo-Gonzalez M, Hinner MJ, Schneider P, Makarov V, Cole ST, Johnsson K. 2010. Benzothiazinones: prodrugs that covalently modify the decaprenylphosphoryl- β-D-ribose 2′-epimerase DprE1 of mycobacterium tuberculosis. J Am Chem Soc 132:13663–13665. 10.1021/ja106357w. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S, Chen J, Cui P, Shi W, Zhang W, Zhang Y. 2015. Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 70:2507–2510. 10.1093/jac/dkv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Vos M, Ley SD, Wiggins KB, Derendinger B, Dippenaar A, Grobbelaar M, Reuter A, Dolby T, Burns S, Schito M, Engelthaler DM, Metcalfe J, Theron G, van Rie A, Posey J, Warren R, Cox H. 2019. Bedaquiline microhetero resistance after cessation of tuberculosis treatment. N Engl J Med 380:2178–2180. 10.1056/NEJMc1815121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimmo C, Millard J, van Dorp L, Brien K, Moodley S, Wolf A, Grant AD, Padayatchi N, Pym AS, Balloux F, O’Donnell M. 2020. Population-level emergence of bedaquiline and clofazimine resistance-associated variants among patients with drug-resistant tuberculosis in southern Africa: a phenotypic and phylogenetic analysis. Lancet Microbe 1:e165–e174. 10.1016/S2666-5247(20)30031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villellas C, Coeck N, Meehan CJ, Lounis N, de Jong B, Rigouts L, Andries K. 2017. Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J Antimicrob Chemother 72:684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang JS, Kim KJ, Choi H, Lee SH. 2018. Delamanid, bedaquiline, and linezolid minimum inhibitory concentration distributions and resistance-related gene mutations in multidrug-resistant and extensively drug-resistant tuberculosis in Korea. Ann Lab Med 38:563–568. 10.3343/alm.2018.38.6.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ismail NA, Omar SV, Moultrie H, Bhyat Z, Conradie F, Enwerem M, Ferreira H, Hughes J, Joseph L, Kock Y, Letsaolo V, Maartens G, Meintjes G, Ngcamu D, Okozi N, Padanilam X, Reuter A, Romero R, Schaaf S, te Riele J, Variava E, van der Meulen M, Ismail F, Ndjeka N. 2022. Assessment of epidemiological and genetic characteristics and clinical outcomes of resistance to bedaquiline in patients treated for rifampicin-resistant tuberculosis: a cross-sectional and longitudinal study. Lancet Infect Dis 22:496–506. 10.1016/S1473-3099(21)00470-9. [DOI] [PubMed] [Google Scholar]
- 46.Shirude PS, Shandil RK, Manjunatha MR, Sadler C, Panda M, Panduga V, Reddy J, Saralaya R, Nanduri R, Ambady A, Ravishankar S, Sambandamurthy VK, Humnabadkar V, Jena LK, Suresh RS, Srivastava A, Prabhakar KR, Whiteaker J, McLaughlin RE, Sharma S, Cooper CB, Mdluli K, Butler S, Iyer PS, Narayanan S, Chatterji M. 2014. Lead optimization of 1,4-azaindoles as antimycobacterial agents. J Med Chem 57:5728–5737. 10.1021/jm500571f. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Sambandan D, Halder R, Wang J, Batt SM, Weinrick B, Ahmad I, Yang P, Zhang Y, Kim J, Hassani M, Huszar S, Trefzer C, Ma Z, Kaneko T, Mdluli KE, Franzblau S, Chatterjee AK, Johnson K, Mikusova K, Besra GS, Fütterer K, Jacobs WR, Schultz PG. 2013. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc Natl Acad Sci USA 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kraus CN, Burnstead BW. 2011. The safety record of fusidic acid in non-US markets: a focus on skin infections. Clin Infect Dis 52(Suppl 7):S527–S537. 10.1093/cid/cir168. [DOI] [PubMed] [Google Scholar]
- 49.Borrell S, Trauner A, Brites D, Rigouts L, Loiseau C, Coscolla M, Niemann S, De Jong B, Yeboah-Manu D, Kato-Maeda M, Feldmann J, Reinhard M, Beisel C, Gagneux S. 2019. Reference set of Mycobacterium tuberculosis clinical strains: a tool for research and product development. PLoS One 14:e0214088–12. 10.1371/journal.pone.0214088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santiago M, Lee W, Fayad AA, Coe KA, Rajagopal M, Do T, Hennessen F, Srisuknimit V, Müller R, Meredith TC, Walker S. 2018. Genome-wide mutant profiling predicts the mechanism of a Lipid II binding antibiotic article. Nat Chem Biol 14:601–608. 10.1038/s41589-018-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Claveau D, Vaillancourt JP, Roemer T, Meredith TC. 2011. High-frequency transposition for determining antibacterial mode of action. Nat Chem Biol 7:720–729. 10.1038/nchembio.643. [DOI] [PubMed] [Google Scholar]
- 52.Robertson GT, Ramey ME, Massoudi LM, Carter CL, Zimmerman M, Kaya F, Graham BG, Gruppo V, Hastings C, Woolhiser LK, Scott DWL, Asay BC, Eshun-Wilson F, Maidj E, Podell BK, Vásquez JJ, Lyons MA, Dartois V, Lenaerts AJ. 2021. Comparative analysis of pharmacodynamics in the C3HeB/FeJ mouse tuberculosis model for DprE1 inhibitors TBA-7371, PBTZ169, and OPC-167832. Antimicrob Agents Chemother 65:e0058321. 10.1128/AAC.00583-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chhabra S, Kumar S, Parkesh R. 2021. Chemical space exploration of DprE1 inhibitors using chemoinformatics and artificial intelligence. ACS Omega 6:14430–14441. 10.1021/acsomega.1c01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Li Y, Wang B, Lu Y. 2022. Identification of mutations associated with macozinone-resistant in Mycobacterium tuberculosis. Curr Microbiol 79:205. 10.1007/s00284-022-02881-x. [DOI] [PubMed] [Google Scholar]
- 55.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. 2010. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26:2811–2817. 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 56.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167–169. 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179. 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 58.Venter H, Mowla R, Ohene-Agyei T, Ma S. 2015. RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front Microbiol 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto K, Nakata N, Mukai T, Kawagishi I, Ato M. 2021. Coexpression of MmpS5 and MmpL5 contributes to both efflux transporter MmpL5 trimerization and drug resistance in Mycobacterium tuberculosis. mSphere 6:1–11. 10.1128/mSphere.00518-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danilchanka O, Mailaender C, Niederweis M. 2008. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob Agents Chemother 52:2503–2511. 10.1128/AAC.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. 10.1128/AAC.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pages JM, Lavigne JP, Leflon-Guibout V, Marcon E, Bert F, Noussair L, Nicolas-Chanoine MH. 2009. Efflux pump, the masked side of β-lactam resistance in Klebsiella pneumoniae clinical isolates. PLoS One 4:e4817. 10.1371/journal.pone.0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyer E, Dessolin J, Lustig M, Decossas M, Phan G, Cece Q, Durand G, Dubois V, Sansen J, Taveau J-C, Broutin I, Daury L, Lambert O. 2022. Molecular determinants for OMF selectivity in tripartite RND multidrug efflux systems. Antibiot (Basel, Switzerland) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh M, Sykes EME, Li Y, Kumar A. 2020. MexXY RND pump of Pseudomonas aeruginosa PA7 effluxes bi-anionic β-lactams carbenicillin and sulbenicillin when it partners with the outer membrane factor OprA but not with OprM. Microbiology (Reading) 166:1095–1106. 10.1099/mic.0.000971. [DOI] [PubMed] [Google Scholar]
- 65.Srikumar R, Li XZ, Poole K. 1997. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol 179:7875–7881. 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colangeli R, Jedrey H, Kim S, Connell R, Ma S, Venkata UDC, Chakravorty S, Gupta A, Sizemore EE, Diem L, Sherman DR, Okwera A, Dietze R, Boom WH, Johnson JL, Kenzie WRM, Alland D. 2018. Bacterial factors that predict relapse after tuberculosis therapy. N Engl J Med 379:823–833. 10.1056/NEJMoa1715849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murphy KC, Papavinasasundaram K, Sassetti CM. 2015. Mycobacterial recombineering. Methods Mol Biol 1285:177–199. 10.1007/978-1-4939-2450-9_10. [DOI] [PubMed] [Google Scholar]
- 68.Seemann T. 2020. Snippy - Rapid haploid variant calling and core genome alignment.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download aac.00904-22-s0001.xlsx, XLSX file, 0.7 MB (698KB, xlsx)
Fig. S1 and S2 and Tables S1 and S2. Download aac.00904-22-s0002.pdf, PDF file, 0.3 MB (347.5KB, pdf)