Abstract
The quantification of developmental potential is critical for determining developmental stages and identifying essential molecular signatures in single-cell studies. Here, we present FitDevo, a novel method for inferring developmental potential using scRNA-seq data. The main idea of FitDevo is first to generate sample-specific gene weight (SSGW) and then infer developmental potential by calculating the correlation between SSGW and gene expression. SSGW is generated using a generalized linear model that combines sample-specific information and gene weight learned from a training dataset covering scRNA-seq data of 17 previously published datasets. We have rigorously validated FitDevo’s effectiveness using a testing dataset with scRNA-seq data from 28 existing datasets and have also demonstrated its superiority over current methods. Furthermore, FitDevo’s broad application scope has been illustrated using three practical scenarios: deconvolution analysis of epidermis, spatial transcriptomic data analysis of hearts and intestines, and developmental potential analysis of breast cancer. The source code and related data are available at https://github.com/jumphone/fitdevo.
Keywords: developmental potential, scRNA-seq, deconvolution analysis, spatial transcriptomic data analysis, breast cancer
Introduction
Single-cell RNA sequencing (scRNA-seq) technology is now widely used for deciphering cell molecular activities [1, 2]. Meanwhile, computational approaches are becoming more and more indispensable for taking advantage of scRNA-seq data [3, 4]. In a single-cell study of developmental biology, data analysis typically starts by using well-established computational frameworks, such as Seurat [5–8] and Scanpy [9], to preprocess data and conduct dimension reduction [e.g. principal component analysis (PCA), uniform manifold approximation and projection (UMAP) and Monocle, etc.] [10–13], which is then followed by determining the developmental potential (DP) using prior information, such as known marker genes [14, 15]. Identified marker genes, however, are often not available when novel developmental systems are investigated, thus encouraging researchers to explore other accessible features that are correlated with the developmental process, including RNA velocity (e.g. Velocyto and Dynamo) [16, 17], principal component (PC) polarity (VECTOR) [18], network property (e.g. SCENT and CCAT) [19, 20] and transcriptional diversity (CytoTRACE) [21].
For RNA velocity and VECTOR, users must reanalyze intronic reads and calculate over 100 PCs, respectively [16, 18]. Those procedures are time-consuming, and sometimes may not be possible because of the limitation of sequencing platforms or the lack of captured cells. Therefore, more convenient and independent methods are urgently needed to infer the DP with scRNA-seq data. By now, several methods are constructed without using intronic reads or too many PCs. In 2017, Teschendorff et al. proposed a method named SCENT which uses the entropy of cell’s transcriptome in the context of a given network to estimate DP [19]. In 2020, Gulati et al. reported that a cell’s transcriptional diversity (the number of detected genes) is positively correlated with the cell’s DP and therefore built a method named CytoTRACE [21]. In addition, the authors also showed that using sample-specific information (sample-specific signatures correlated with transcriptional diversity) can significantly improve the performance of inferring DP. In the same year, Teschendorff et al. published an ultra-fast method named CCAT [20]. In the study of CCAT, the authors find that the correlation (Pearson Correlation Coefficient, PCC) between gene expression and gene’s network degree is an approximate estimate, but not equivalent, of cell entropy. Therefore, they use the network degree as gene weight (GW), and use the correlation between GW and gene expression to represent the cell’s DP. CCAT is shown to have better performance than previously published methods, including SCENT [19], CytoTRACE [21], scEnergy (in the package named scEpath) [22], StemID [23], cmEntropy [24] and SLICE [25]. In summary, calculating the correlation between GW and gene expression is an effective way to infer DP. However, CCAT does not use sample-specific information, and the network degree may not be the best choice for determining GW.
To evaluate the performance of different methods in inferring DP, people need to know the correct developmental order of cells. In the study of CytoTRACE, the authors introduce two types of differentiation labels: timepoint label and phenotype (cell type) label [21]. The timepoint label is objective and has been widely used to indicate the correct developmental order, whereas phenotype label is often subjective due to the artificial labeling of phenotype (cell type). In the study of CCAT, to conduct a discrimination test, the authors only keep cells with starting and ending labels [20]. The benchmark dataset of CCAT is reliable in testing the performance of different methods, but may not be suitable for being designated as training dataset because it does not have cells in the transition phase. What’s more, existing methods (e.g. SCENT, CCAT, CytoTRACE, etc.) are shown to have good performance in their benchmark datasets [19–21]. However, it is still unclear whether those methods meet the requirement of practical scenarios, such as deconvolution analysis (in silico flow cytometry) [26, 27], spatial transcriptomic data analysis [28, 29] and cancer cell’s DP analysis [30, 31].
Here, we present FitDevo, a novel method for inferring DP using scRNA-seq data. The main idea of FitDevo is first to generate sample-specific gene weight (SSGW) and then infer DP by calculating the PCC between SSGW and gene expression. SSGW is generated using a generalized linear model (GLM), which combines sample-specific information and GW. In this study, GW is learned from 17 samples having a timepoint label (training dataset, collected by the study of CytoTRACE). In this study, the term ‘sample’ indicates the constructed sample that contains all scRNA-seq data of a previously published dataset. We test the performance of FitDevo using 28 samples (testing dataset, collected by the study of CCAT) and show that FitDevo outperforms previous methods. Furthermore, we prepare three practical scenarios that users may encounter when conducting single-cell developmental studies, including deconvolution analysis of epidermis, spatial transcriptomic data analysis of heart and intestine and DP analysis of breast cancer. After applying FitDevo, CCAT and CytoTRACE to those three practical scenarios, FitDevo achieves the best performance. FitDevo is implemented in R. Source code and benchmark datasets of this study are available at https://github.com/jumphone/fitdevo.
Results
The generation of GW
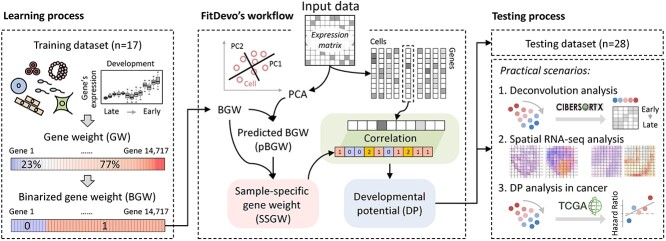
The overall study design of the computational framework and applications is summarized schematically in Figure 1. The first step of this study is the generation of GW. According to the study of CCAT, defining a cell’s DP as the correlation between GW and gene expression can achieve promising performance in their benchmark dataset [20]. The authors of CCAT define GW as a gene network degree. However, existing networks are not designed for inferring a cell’s DP, which may restrict the performance of CCAT. Here, to accurately infer cell’s DP, we propose a supervised workflow for generating GW (Figure 2A). Firstly, we download scRNA-seq data collected by the authors of CytoTRACE [21] and build a training dataset that only contains samples with a timepoint label (Supplementary Table 1 and Material and Methods). In total, our training dataset includes 17 samples (in this study, the term ‘sample’ indicates the constructed sample that contains all scRNA-seq data of a previously published dataset) and covers a wide range of developmental scenarios (Supplementary Table 1). Secondly, for each gene in each sample (genes expressed in < 9 samples are removed), we calculate a PCC between the gene’s expression and the reverse order of timepoint label. In this step, we build a PCC matrix having 14 717 rows (genes) and 17 columns (samples). Thirdly, to reduce the variance of different genes’ PCCs, we calculate standardized PCCs for each gene by dividing the root-mean-square (RMS) (Figure 2A). Finally, we define our GW as the averaged standardized PCC of all 17 samples. In total, we get GW of 14 717 genes. The GW value of a gene is supposed to represent its contribution to DP. For instance, NANOG is a well-known marker for embryonic stem cells [32] and has a high GW value (0.516). Another case in point is ENO2, also known as the neuron-specific enolase, which is a marker for mature neuron cells [33] and has a low GW value (−0.309). In the following study, the term ‘GW’ indicates the GW generated by our supervised workflow except otherwise stated.
Figure 1.
The overall study design of the computational framework and applications. For details about this study, refer to the Results and the Material and Methods sections.
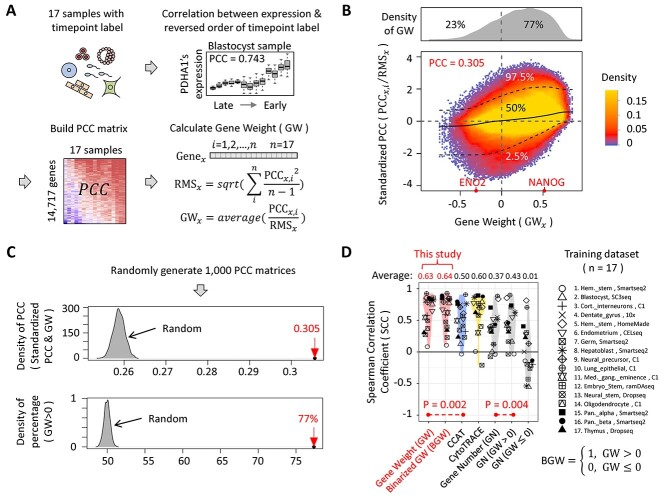
Figure 2.
The generation of GW and the discovery of BGW. (A) The workflow of generating GW. ‘PCC’ stands for Pearson Correlation Coefficient. ‘RMS’ stands for Root-Mean-Square that is calculated by using the ‘scale’ function in R. In this study, the term ‘sample’ indicates the constructed sample that contains all scRNA-seq data of a previously published dataset. (B) The scatter plot describing the relationship between GW values and standardized PCCs. For each gene, we build two vectors. The first vector (y axis) has 17 elements that are the standardized PCCs of 17 training samples, whereas the second vector (x axis) has 17 elements sharing the same value (the GW value of the given gene). The upper dot-line, middle line and lower dot-line are the 97.5%, 50% and 2.5% quintiles of standardized PCCs, respectively. The color on the scatter plot indicates the density of points, whereas the upper gray panel shows the density plot of GW values. (C) Two characteristics of GW. We calculate 1000 random GW sets by randomly generating 1000 PCC matrices. The gray areas represent the distribution of statistics of the random GW sets, whereas the red arrows highlight the statistics of the real GW set. (D) The comparison results using training dataset. ‘GN’ means that we define DP as the number of detected genes. ‘GN (GW > 0)’ and ‘GN (GW ≤ 0)’ mean that we define DP as the number of detected genes with positive GW and negative GW (there is no GW that is equal to 0), respectively.
Two characteristics of GW are shown in Figure 2B. First, GW is positively correlated (PCC = 0.305) with standardized PCC. Because GW is defined as the average of standardized PCCs, there is no wonder that they are positively correlated. However, the correlation value is significantly higher than that of randomly generated PCC matrices (ranging from 0.254 to 0.262, 1000 times) (Figure 2C), indicating that, for each gene, different samples tend to have similar correlation values between gene’s expression and the reverse order of timepoint label. Second, most values of GW are larger than 0. We notice that the percentage of positive values in GW (77%) is significantly higher than that of randomly generated PCC matrices (ranging from 48% to 51%, 1000 times) (Figure 2C), which is consistent with the study of CytoTRACE to some extent [21]. The authors of the CytoTRACE study found that the number of detected genes is positively correlated with the cell’s DP, implying that most genes’ expression values tend to be positively correlated with the reverse order of the timepoint label.
Following the study of CCAT [20], we define a cell’s DP as the correlation (PCC) between GW and gene expression. In addition, as performed by the study of CytoTRACE [21], we calculate the rank correlation (Spearman Correlation Coefficient, SCC) between inferred DP and the reverse order of timepoint label to evaluate the performance of different methods. After applying GW to our training dataset, GW is shown to have significantly higher SCCs than CCAT (average SCC of GW: 0.634, CCAT: 0.501, P-value = 0.002) (Figure 2D), demonstrating the effectiveness of GW in the training dataset. In addition, when building the training dataset, we have removed a zebrafish sample due to the limited number of homologous genes between zebrafish and mammals (Material and Methods). Nevertheless, we can still apply GW to that zebrafish sample, and get a positive SCC (0.228), indicating the versatility of GW. Considering all 17 samples in our training dataset are directly derived from the study of CytoTRACE, we would like to check the necessity and sufficiency of each sample. To check the necessity of each sample, we remove samples one-by-one and generate 17 sets of GW. The average SCCs of those 17 sets of GW range from 0.580 to 0.642 (Supplementary Figure 1), suggesting that removing either of the 17 samples will not strongly affect the performance. To check the sufficiency of each sample, we use individual samples one-by-one to generate 17 sets of GW. The average SCCs of those 17 sets of GW range from −0.065 to 0.497 (Supplementary Figure 2), suggesting that using one single sample cannot achieve acceptable performance. Finally, we randomly remove 1–15 samples to test the robustness of GW. We find that using no <13 samples can achieve a pretty high average SCC (around 0.63), demonstrating that training samples are almost saturated and the performance of GW won’t be dramatically affected by removing a small number (<5) of training samples (Supplementary Figure 3). And notably, even the quality of the training datasets (e.g. sequencing depth, gene coverage, etc.) has been observed to be different in various sequencing platforms, we find that the performance of GW is not significantly affected.
The discovery of binarized gene weight (BGW)
As shown in Figure 2D, GW only has a slightly higher average SCC (0.634) than CytoTRACE (0.601). In CytoTRACE, two types of information are used to infer a cell’s DP, including gene number (GN) and sample-specific information. Considering that the sample-specific information can significantly improve its performance [20, 21], we directly compare using GW with using GN alone. In the training dataset, GN shows a much lower average SCC (0.377) than GW (0.634). What’s more, when separately using the number of genes with positive and negative GW, we get relatively higher (0.436) and nearly zero (0.012) average SCCs, respectively (Figure 2D). Although using the number of genes with positive GW can achieve a higher average SCC (0.436) than GN (0.377), it is still not as good as GW (0.634), implying that using genes with positive GW alone is insufficient to infer cell’s DP accurately. To simultaneously use the information of genes with positive and negative GW, we replace GW with the binarized value of GW (defined as BGW) (threshold is 0) and apply BGW to our training dataset. Interestingly, BGW is shown to have an even higher average SCC (0.647) than GW (0.634) (Figure 2D), suggesting that using the binarized value of GW, rather than continuous value, is enough for inferring DP. In addition, we have tested a list of thresholds (e.g. −0.3, 0, 0.3, etc.) to generate BGW, and setting the threshold at 0 achieves the best performance (Supplementary Figure 4). Besides, in our training dataset, most samples (14 out of 17) are mouse samples, driving us to test the performance of BGW in human samples. Therefore, we download scRNA-seq data from Human Cell Landscape (HCL) and organize the data into a matrix that contains expression profiles of 1336 cell types with fetal and adult labels (Material and Methods) [34]. As shown in Supplementary Figures 5 and 6, fetal cell types have significantly higher inferred DP than adult cell types (P-value<2.2×10−16), validating the effectiveness of BGW in human samples. In addition, we have also found that BGW is positively correlated with gene network degree and gene conservation rate, which is shown in Supplementary Figure 5.
The calculation of SSGW and the validation of FitDevo
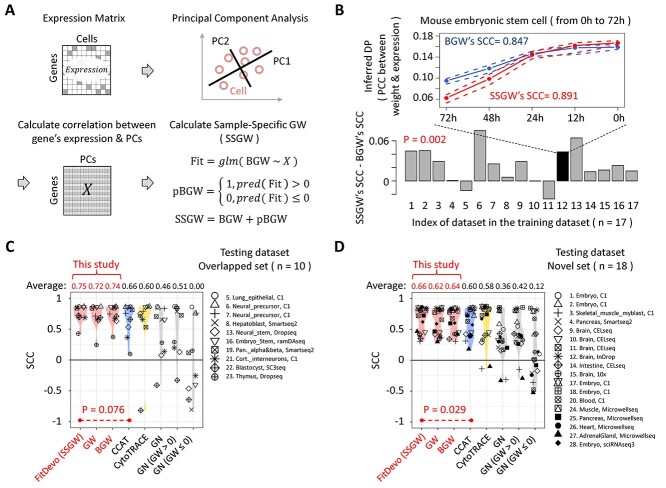
According to the study of CytoTRACE, sample-specific information can significantly improve the performance of inferring DP [21]. We therefore build a workflow to calculate SSGW (Figure 3A) and evaluate its performance. Firstly, we use a well-established computational framework named Seurat [5–8] to preprocess data and conduct PCA. Next, we calculate the correlation between gene expression and the top 50 (default number in Seurat) PCs to build a gene-PC correlation matrix. Then, we use a general linear model (GLM) to predict BGW based on the gene-PC correlation matrix (Figure 3A). Finally, we define SSGW as the sum of BGW and predicted BGW (pBGW) (we use an example to illustrate the difference between BGW and pBGW, see Supplementary Figure 7). To demonstrate the usefulness of SSGW, we apply SSGW to our training dataset. As shown in Figure 3B, SSGW has a higher average SCC than BGW in 14 out of 17 samples, and the SCCs of SSGW (average SCC: 0.669) are significantly higher than that of BGW (0.647) (P-value = 0.002). For example, in a mouse sample of embryonic stem cells [35], the DP inferred by SSGW is more positively correlated with the reverse order of timepoint label than that of BGW (SSGW:0.891, BGW:0.846). The above results indicate that SSGW can further improve the performance of BGW. It is worth noting that SSGW is designed for data optimization: SSGW is defined as ‘BGW + pBGW’ rather than ‘pBGW’ alone, because the performance of using pBGW alone can be influenced by the insufficient number of used PCs (Supplementary Figures 8 and 18). After applying top 3, 5, 10 and 50 PCs to generate pBGW and SSGW, we find that the average SCCs of pBGW range from 0.615 to 0.683, whereas the average SCCs of SSGW range from 0.658 to 0.669, suggesting that the performance of SSGW is more independent of the number of used PCs than pBGW.
Figure 3.
The calculation of SSGW and the validation of FitDevo. (A) The workflow of calculating SSGW. ‘X’ stands for the gene-PC correlation matrix. ‘glm’ stands for the generalized linear model. ‘pred’ stands for the ‘predict’. (B) The difference between SSGW’s SCCs and BGW’s SCCs. On the line chart, the upper dot-line, middle line and lower dot-line are the 75%, 50% and 25% quantile of the inferred DP (PCC between GW and gene expression), respectively. (C) and (D) show the comparison results using overlapped (overlapped with training dataset) and novel testing samples, respectively. We name the method of using SSGW as FitDevo. We only compare FitDevo with CCAT and CytoTRACE as they have better performance than other methods [20, 21]. Considering the testing dataset only has cells with starting and ending labels, following the study of CCAT, we also use the AUC to measure the performance (Supplementary Figure 11).
In the following study, we name the method of using SSGW as FitDevo. The performance of FitDevo is validated by using a testing dataset that contains 28 samples collected by the study of CCAT [20] (Supplementary Table 1 and Material and Methods). Notably, this testing dataset only contains cells with starting and ending labels, which is different from our training dataset. Of those 28 samples, 10 samples are already covered by the training dataset, whereas 18 samples are not. Here, we separately present the results of 10 overlapped samples and 18 novel samples. As shown in Figure 3C and D, FitDevo shows the highest average SCC among all used methods in 10 overlapped samples (0.753 against 0.007–0.741) and 18 novel samples (0.660 against 0.126–0.641) (after parameter tuning of CCAT and CytoTRACE, FitDevo still shows the best performance, see Supplementary Figure 9). In addition, we notice that FitDevo has significantly higher SCCs than BGW in novel samples (P-value = 0.019) (Supplementary Figure 10), indicating that the use of sample-specific information can further improve the performance in novel developmental scenarios. Considering the testing dataset only has cells with starting and ending labels, following the study of CCAT [20], we also use the area under the curve (AUC) to measure the performance. In Supplementary Figure 11, FitDevo consistently shows the highest averaged AUC among all used methods in 10 overlapped samples (0.942 against 0.501–0.934) and 18 novel samples (0.923 against 0.570–0.911). The study of CCAT has already shown that CCAT outperforms other published methods [20], and we also find that FitDevo outperforms those published methods (Supplementary Figure 8). Furthermore, we make a computational scalability evaluation and show that FitDevo can achieve high performance with a relatively high computational speed (Supplementary Figure 12).
FitDevo facilitates tissue-specific deconvolution analysis
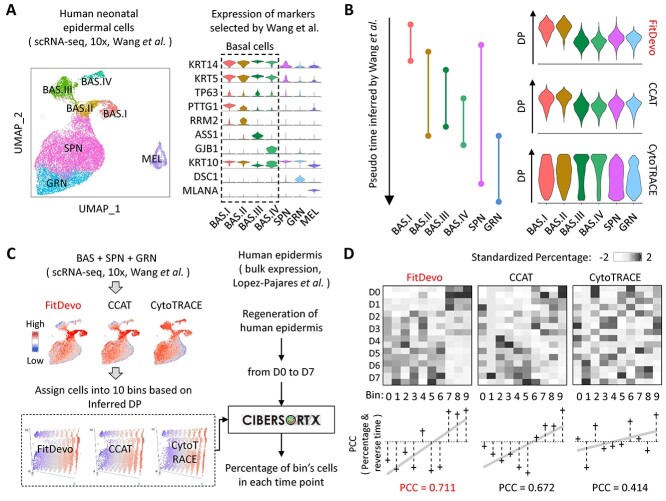
Since cells can be classified into different developmental stages based on the inferred DP, the accurate inference of the cell’s DP is essential for deconvolution analysis in developmental studies. Here, we collect both scRNA-seq and bulk expression data from human epidermal studies. The study of scRNA-seq data contains samples of human epidermal tissues, but does not have a timepoint label [36]. The study of bulk data contains samples derived from a time-course epidermal regeneration experiment (from day0 to day7) [37]. Because the single-cell study does not provide a cell type label [36], we reanalyze the scRNA-seq data and annotate cells using markers provided by the original research (e.g. KRT14, KRT5, TP63, etc.) (Figure 4A). As shown in Figure 4B, the inferred DP of FitDevo and CCAT shares a similar pattern with the pseudo time order inferred by the original study (BAS.I and BAS.II have higher DP than others), whereas the inferred DP of CytoTRACE shows a different pattern (BAS.III and BAS.IV have higher DP than others). It suggests that FitDevo and CCAT may be more suitable for analyzing epidermal differentiation than CytoTRACE. However, since the pseudo time order of the original study is predicted using the computational method, we still need to use the timepoint label of bulk data to further evaluate the performance.
Figure 4.
The deconvolution analysis of epidermis. (A) The UMAP and violin plot of human epidermal cells. On the UMAP, ‘BAS’, ‘SPN’, ‘GRN’ and ‘MEL’ stand for basal cells, spinous cells, granular cells and melanocytes, respectively. All marker genes and cell types are selected and annotated based on the original research [36]. (B) The pseudo time inferred by the original research and the DP inferred by FitDevo, CCAT and CytoTRACE. (C) The workflow of conducting deconvolution analysis using CIBERSORTx. First, we infer cells’ DP using the scRNA-seq data and assign cells into 10 bins based on the inferred DP. Then, we use CIBERSORTx to estimate the percentage of each bin in each bulk sample. In addition, we notice that the outcome of CIBERSORTx may depend on a lot of adjustable parameters, and therefore apply a correlation-based method to conduct deconvolution analysis (Supplementary Figure 14). (D) The results of deconvolution analysis. We use ‘scale’ function in R to get the standardized percentage for each column. In this plot, the ‘reverse time’ stands for the reverse order of timepoint label. The result of correlation-based method is shown in Supplementary Figure 14. ‘D0 to D7’ stands for ‘Day0 to Day7’.
To use bulk data’s timepoint label, we first assign all epidermal cells of scRNA-seq data into 10 bins based on the inferred DP (bin0 to bin9, bin9 has the highest DP). Next, we use CIBERSORTx [27] to estimate each bin’s percentage in each bulk sample (Figure 4C). Because bins with large serial numbers (e.g. bin9) have higher DP than bins with small serial numbers (e.g. bin0), there should be a strong positive correlation (bin’s PCC) between the estimated percentage and the reverse order of timepoint label when the bin’s serial number is large (e.g. bin9). Then, we calculate the correlation (global PCC) between the bin’s serial number and the bin’s PCC, and use the global PCC to evaluate the final performance of different methods. As shown in Figure 4D, FitDevo gets a higher global PCC (0.711) than CCAT (0.672) and CytoTRACE (0.414), suggesting the outperformance of FitDevo in deconvolution analysis of the epidermis (using SCC shows similar result, see Supplementary Figure 13). In addition, we notice that the outcome of CIBERSORTx may depend on a lot of adjustable parameters, and therefore apply a correlation-based method to conduct deconvolution analysis. FitDevo consistently shows the best performance (Supplementary Figure 14).
Inferring DP with spatial transcriptomic data by FitDevo
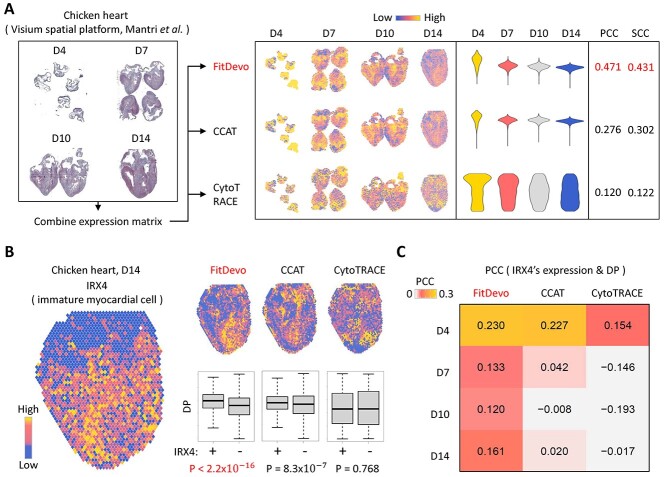
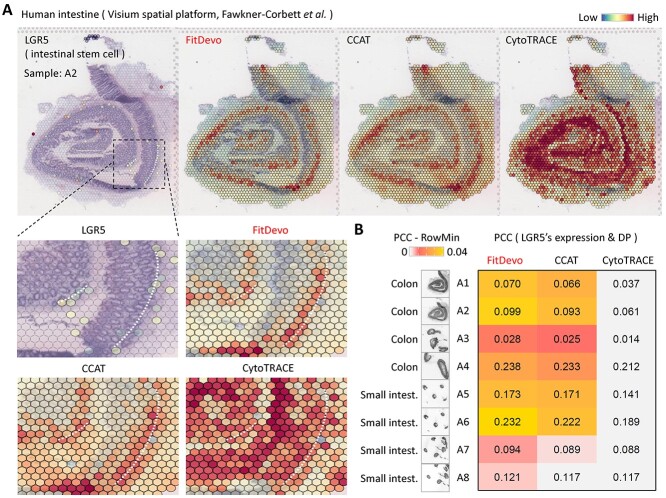
In spatial RNA-seq data, several adjacent cells are defined as a spot and sequenced together, which brings difficulties in inferring DP. To test the performance of inferring DP in spatial data, we download spatial RNA-seq data from studies of chicken hearts [28] and human intestines [29]. In the study of chicken heart, the authors have generated spatial data of chicken heart at four time points (D4, D7, D10 and D14) (Figure 5A). They identified a signature, IRX4, for labeling immature myocardial cells that should have higher DP than others [28]. First, we apply FitDevo, CCAT and CytoTRACE to the spatial RNA-seq data of chicken hearts. FitDevo gets the highest correlation (SCC and PCC) between inferred DP and the reverse order of timepoint label (SCC of FitDevo: 0.431, CCAT: 0.302, CytoTRACE: 0.122) (Figure 5A). Then, we test the difference of inferred DP between IRX4 positive and negative spots and find that FitDevo shows the most significant difference in all four time points (positive spots have higher DP than negative spots) (for other markers and time points, see Supplementary Figure 15). For example, in the sample of D14, the P-value of FitDevo is <2.2 × 10−16, which is much smaller than that of CCAT (8.3 × 10−7) and CytoTRACE (0.768) (Figure 5B). Furthermore, we calculate the correlation between IRX4’s expression and inferred DP. As shown in Figure 5C, in all four time points, FitDevo achieves the highest correlation value. In the study of human intestines, the authors have collected eight samples from different locations of the intestine [29] (Figure 6A and Supplementary Figure 16). As shown in Figure 6B, in all eight samples, FitDevo gets the highest correlation between the inferred DP and a well-known marker of intestinal stem cells (LGR5 [38, 39]). Taken together, the above results indicate the high effectiveness of FitDevo in inferring DP using spatial RNA-seq data.
Figure 5.
The spatial transcriptomic data analysis of hearts. (A) The data analysis workflow with the spatial transcriptomic data of chicken hearts. (B) The expression value of IRX4, a marker for immature myocardial cells [28] and the DP, inferred by FitDevo, CCAT and CytoTRACE, on the spatial image. Results of other markers and developmental stages are provided in Supplementary Figure 15. (C) The correlation between the expression of IRX4 and the DP inferred by FitDevo, CCAT and CytoTRACE. ‘D4’, ‘D7’, ‘D10’ and ‘D14’ stand for ‘Day4’, ‘Day7’, ‘Day10’ and ‘Day14’, respectively.
Figure 6.
The spatial transcriptomic data analysis of intestines. (A) The expression value of LGR5, a marker for intestinal stem cells [38, 39], and the DP, inferred by FitDevo, CCAT and CytoTRACE, on the spatial image of the human intestine (A2). The results of other samples are shown in Supplementary Figure 16. We use the white dot-line to indicate the potential layer of intestinal stem cells. (B) The correlation between the expression of LGR5 and the DP inferred by FitDevo, CCAT and CytoTRACE. ‘RowMin’ stands for the minimum value of each row.
The DP analysis of cancerous tissue
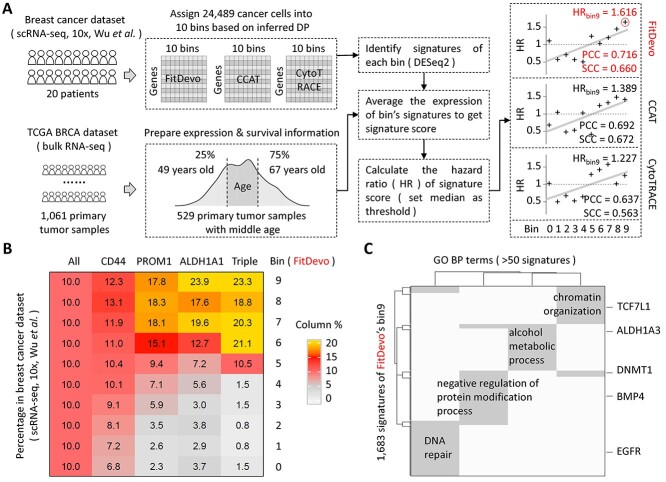
Cancer tissues are revealed as heterogeneous populations with different developmental origins and distinct functions in tumor progression. Here, we would like to check whether FitDevo can be applied to quantify the DP of cells in breast cancer tissues. Firstly, we download scRNA-seq data of 20 breast cancer patients from a recent atlas study conducted by Wu et al. [40]. Then, we apply FitDevo to all cancer cells (labeled by the original study) to infer the cell’s DP (Figure 7A). Finally, we assign cancer cells into 10 bins based on the inferred DP (bin0 to bin9, bin9 has the highest DP). As shown in Figure 7B, bins with large serial numbers (bin7, bin8 and bin9) have significantly more cells expressing well-known cancer stemness markers (CD44, PROM1 and ALDH1A1) [41–44] than those bins with small serial numbers (bin0, bin1 and bin2), suggesting that FitDevo can quantify cancer cell’s DP in breast cancer. Results of more stemness markers are shown in Supplementary Figure 17.
Figure 7.
The developmental potential analysis of breast cancer. (A) The workflow of DP analysis with the scRNA-seq and TCGA bulk data of breast cancer. Data from patients with middle age (>49 and < 67 years old) are selected to analyze. We use DESeq2 to identify the signatures of each bin. We use DESeq2 [52] to identify signatures of each bin in the scRNA-seq data of breast cancer. Because the computational efficiency of DESeq2 is low when dealing with tens of thousands of single cells, we generate pseudo-bulk samples to improve the efficiency. Firstly, we merge all 24 489 cancer cells into 2450 pseudo-bulk samples by aggregating the read counts of neighbor cells. For each bin, we get 245 pseudo-bulk samples (around 10 cells per pseudo-bulk sample). Then, we use DESeq2 to get signatures of each bin by comparing the expression values between the given bin and other bins. The cutoffs of foldchange, adjusted P-value and baseMean are 2, 0.05 and 0.5, respectively. Considering the generation of pseudo-bulk sample requires a random seed when defining neighbor cells, we therefore use three different random seeds to generate three sets of signatures and define the bin’s signatures as the intersection of those three sets. (B) The percentage of the marker gene (CD44, PROM1 and ALDH1A1) positive cells in each bin of FitDevo. ‘ALL’ indicates the percentage of all cells in each bin. The sum of each column is 100. ‘Triple’ stands for triple-positive (CD44, PROM1 and ALDH1A1) cells. Results of other markers are provided in Supplementary Figure 17. (C) The gene set (GO BO) enrichment results with the signatures of FitDevo’s bin9. ‘GO’ and ‘BP’ stand for gene ontology and biological process, respectively. We only show the terms covering > 50 signatures of FitDevo’s bin9.
Because the study of scRNA-seq data does not have patient survival information, we use the bulk data in the cancer genome atlas (TCGA) to testify the clinical relevance of the DP inferred by FitDevo (Figure 7A). Firstly, we download well-organized bulk expression data and survival information of 1061 TCGA breast cancer patients from the UCSC Xena database (http://xena.ucsc.edu/). We notice that the range of patient’s age is quite wide (ranging from 26 to 90 years old). Considering the mechanisms of tumor progression in very young and old patients may be different, we only choose patients with middle age (>49 and < 67 years old). Next, we use scRNA-seq data to identify each bin’s signatures, and use the patient’s bulk expression data to calculate each bin’s signature score. For each bin, the signature score is defined as the average expression of the bin’s signatures. Finally, we calculate each bin’s hazard ratio (HR) using each bin’s signature score (set median as the threshold). As shown in Figure 7A, the bin’s serial number is positively correlated with the bin’s HR (PCC: 0.716), and bin9 achieves the highest HR (1.616), demonstrating that bins with large serial numbers tend to have higher clinical relevance than bins with small serial number. It should be noticed that bulk tumor samples may contain normal breast stem cell, or other stem cells, thus bin9 may not strictly represent the breast cancer stem cell. Detailed cell-type-specific information and experimental characterization are needed to tease them out. Furthermore, after applying CCAT and CytoTRACE to this practical scenario, we only get smaller correlation values (CCAT: 0.692, CytoTRACE: 0.637) and lower bin9’s HRs (CCAT: 1.389, CytoTRACE: 1.227), suggesting the superiority of FitDevo in identifying clinically important signatures. In addition, we have also used SCC to measure the correlation between the bin’s serial number and the bin’s HR, and find that FitDevo (0.660) and CCAT (0.672) consistently have higher SCCs than that of CytoTRACE (0.563). Although CCAT shows a slightly higher SCC than that of FitDevo, the average of SCC and PCC of FitDevo (0.688) is still higher than that of CCAT (0.680).
What’s more, we have conducted gene set enrichment analysis using signatures of FitDevo’s bin9, and get four biological processes, including ‘DNA repair’, ‘negative regulation of protein modification process’, ‘alcohol metabolic process’ and ‘chromatin organization’ (Figure 7C). Many signatures in those four biological processes have been associated with breast cancer’s ‘stemness’ by previous studies, such as EGFR [45], BMP4 [46], DNMT1 [47], ALDH1A3 [48] and TCF7L1 [49]. For instance, Choi et al. reported that BMP4 can enhance the epithelial–mesenchymal transition and the cancer stem cell properties of breast cancer cells [46]. Another case in point is DNMT1 that plays an important role in mammary and cancer stem cell maintenance and tumorigenesis [47]. The above results imply the possibility of using FitDevo to identify potential therapeutic targets of breast cancer, which still needs further investigation.
Discussion
As we know, the biggest concern of the supervised method is about its effectiveness in dealing with novel situations. Therefore, we have conducted a series of analyses to illustrate the versatility of our method. Firstly, we have found that, for each gene, different samples tend to have similar correlation values between gene expression and the reverse order of timepoint label, suggesting that our GW may be a general template for a wide range of developmental scenarios. Secondly, we have shown that BGW can identify fetal cell types from adult cell types in HCL, suggesting the effectiveness of BGW in dealing with human samples that are not covered by our training dataset. Thirdly, we have used different benchmark datasets to train and test FitDevo separately and have shown that FitDevo has the best performance among current methods, indicating FitDevo’s superiority in dealing with novel samples. Fourthly, we have illustrated that FitDevo is able to infer the spot’s DP using spatial RNA-seq data, which further demonstrates its broad application scope. Finally, we have applied FitDevo to quantify tumor cell DP in breast cancer and have shown the high clinical relevance of signatures identified by using FitDevo. As such, we have rigorously validated the effectiveness of our method in dealing with novel situations.
The main innovative points of FitDevo are in its algorithm design: the generation of GW, the discovery of BGW and the calculation of SSGW. The underlying rationales of its high performance can be summarized into two points: (i) Unlike previous methods, FitDevo is a supervised method. The effective learning process is the key point for accurately inferring the DP. The high performance of GW and BGW in our testing dataset indicates the effectiveness of our learning process. In addition, we use the novel tissues (not covered by training dataset) in HCL to show that our BGW is able to identify fetal cell types from adult cell types, which further illustrates the effectiveness of our learning process. (ii) The use of sample-specific information further improves the performance of FitDevo. In addition to use the testing dataset to show the contribution of sample-specific information, we have also explored the difference between BGW and pBGW. By investigating a heart sample in HCL database, we find that pBGW is more effective in identifying fetal cells from adult cells, suggesting that pBGW is more suitable for accurately inferring the DP. However, the computational speed of FitDevo is not the best among all tested methods, and we will try to improve its efficiency in future studies. In addition, current FitDevo is only focusing on the one-dimensional problem, and people can combine FitDevo with other trajectory tools to investigate the multiple branches of developmental trajectories, which still requires further exploration and validation.
Besides algorithm design, the construction of benchmark datasets is also crucial for bioinformatics studies. In this study, our training and testing datasets are derived from previous studies [20, 21], which eliminates the possibility of ‘cherry-picking’. We have also provided three practical scenarios to demonstrate the practical value of DP inferred by the in silico method. In this study, we aim to provide novel ideas about the computational algorithm design and have used a series of benchmarks and practical scenarios to validate the effectiveness of our algorithm design. By using FitDevo, we are trying to get novel biological discoveries (e.g. novel signatures in bin9 of breast cancer), but the novel biological discoveries are required to be validated by using abundant data and solid experiments, which is a major focus of our future plan. In addition, the sample size of our training dataset is relatively small (17 samples). To solve this problem, we have decided to collect more scRNA-seq data with timepoint label, which is a part of our ongoing single-cell database study. Once we get a large number of training samples, more powerful machine learning methods (e.g. deep learning) can be used to generate BGW with higher accuracy. Nevertheless, FitDevo has already shown a better performance than previous methods in both standard benchmarks and practical scenarios using current training dataset. As such, FitDevo should be of great use for accelerating both computational and experimental studies relating to the inference of DP.
Material and methods
Data preparation
Training and testing datasets
We build our training and testing datasets based on the studies of CytoTRACE [21] and CCAT [20], respectively (Supplementary Table 1). When building our training dataset, we directly get the well-organized expression matrices and differentiation labels from the authors of CytoTRACE. Considering timepoint label is objective and has been widely used to indicate the correct developmental order, our training dataset only contains samples with a timepoint label. In the paper of CytoTRACE, the authors totally describe 17 samples with a timepoint label. In those 17 samples, there are 16 mammal samples (mouse, human and macaque) and 1 zebrafish sample. Considering zebrafish and mammals only share a limited number of homologous genes, the zebrafish sample is not included in our training dataset. Meanwhile, we notice that a mouse hematopoietic sample (regarded as a sample with phenotype label in the study of CytoTRACE) has both phenotype and timepoint labels [50], and we therefore add this sample into our training dataset. In total, our training dataset contains 17 samples. When building our testing dataset, we first obtain a supplementary table, containing the identifier of each testing sample, from the author of CCAT. Then, according to that table, we search and download the expression matrices and differentiation labels one-by-one. Finally, following the study of CCAT, we only keep cells with starting and ending labels. In total, our testing dataset contains 28 samples. In those 28 samples, 10 samples are covered by our training dataset, whereas the others are novel samples. To enhance the reproducibility of our study, the expression matrices and differentiation labels of our training and testing datasets are well-organized and presented at https://github.com/jumphone/fitdevo.
Datasets in practical scenarios
Seurat [5–8] is used to process the expression matrix of scRNA-seq data. We use the option named ‘LogNormalize’ to normalize gene expression value, use the ‘vst’ method in the function named ‘FindVariableFeatures’ to find most variable genes, use the function named ‘ScaleData’ to standardize expression value, use the function named ‘RunPCA’ to conduct PCA analysis and use the function named ‘RunUMAP’ to generate UMAP. The scRNA-seq data of human epidermis are generated from five samples (41), and we use BEER [51] to remove batch effect and generate the final UMAP. Because the single-cell epidermal study does not provide cell type label, we cluster cells by using the function named ‘FindClusters’ and annotate each cluster using markers provided by the original study (e.g. KRT14, KRT5, TP63, etc.) (41). The bulk data of human epidermis are microarray data [37], and we use the function called ‘rma’ in ‘affy’ package to process the raw data. When analyzing the spatial RNA-seq data (Visium spatial platform) of chicken hearts and human intestines, spatial-related functions in Seurat [8] are used to simultaneously process the expression data and image data. The function named ‘SpatialFeaturePlot’ is used to visualize features on the spatial image. The processed bulk RNA-seq data (FPKM) of TCGA breast cancer patients are downloaded from https://xenabrowser.net/datapages/ (cohort: GDC TCGA Breast Cancer). There are 1217 samples, and 1061 of them have survival information. For scRNA-seq data of HCL, cells sharing same cell types are merged together to get the expression matrix of 1336 cell types.
Competing methods
CCAT is downloaded from https://github.com/aet21/SCENT. We use the function named ‘CompCCAT’ to run CCAT. Because CCAT does not include a normalization step, we use ‘LogNormalize’ function in Seurat to normalize data. The network named ‘net17Jan16’, provided by the authors of CCAT, is used to calculate gene network degree. The source code of CytoTRACE is downloaded from https://cytotrace.stanford.edu/. We use the function named ‘CytoTRACE’ to run CytoTRACE with default parameters. Because CytoTRACE has a normalization step, we directly use read count matrix as the input of CytoTRACE. We follow the documents of SCENT (https://github.com/aet21/SCENT), StemID (https://github.com/dgrun/StemID), cmEntropy (https://github.com/skannan4/cm-entropy-score), and SLICE (https://research.cchmc.org/pbge/slice.html) to install and apply them to our datasets.
The usage of FitDevo
A detailed instruction of FitDevo is provided at https://github.com/jumphone/fitdevo. We use the 18 novel samples in our testing dataset to test the influence of PC numbers (ranging from 3 to 70) (Supplementary Figure 18). In addition, we make a summarization table to summarize the advantage and disadvantage of FitDevo and other methods (Supplementary Figure 19).
Statistical analysis
In this study, statistical analysis is done by using functions and packages implemented in R. PCC and SCC are used to quantify the correlation level and are calculated by using ‘cor’. Student’s t-test is used to evaluate the difference and is calculated by using ‘t.test’. ‘DESeq2’ package in R is used to identify signatures [52]. AUC is calculated by using ‘pROC’ package. Survival analysis is conducted by using ‘survival’ and ‘survminer’ packages. Gene set enrichment is done by using ‘clusterProfiler’ package [53]. Heatmaps are generated by using ‘ComplexHeatmap’ package [54].
Key points
We proposed a method, named FitDevo, for accurately inferring single-cell DP using SSGW.
FitDevo was validated using a testing dataset with scRNA-seq data from 28 previously published datasets, and FitDevo was shown to outperform previous methods.
To enhance the reproducibility of our study, the expression matrices and differentiation labels of our training and testing datasets were well-organized and presented at https://github.com/jumphone/fitdevo.
The practical value of FitDevo was illustrated using three practical scenarios, including deconvolution analysis of epidermis, spatial transcriptomic data analysis of hearts and intestines and DP analysis of breast cancer.
Supplementary Material
Acknowledgement
We thank Linying Li, Dr. Yong Wang, Dr. Lei Chen and Dr. Zixiu Li for discussion. We thank Jian Yu for designing and providing cartoon logos.
Author Biographies
Feng Zhang is an assistant research scientist at the Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China. fzhang@shsmu.edu.cn
Chen Yang is a PhD student at the Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China. yang_0215@sjtu.edu.cn
Yihao Wang is a PhD student at the Department of Ophthalmology, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai 200025, China; Institute of Translational Medicine, National Facility for Translational Medicine, Shanghai Jiao Tong University, Shanghai 201109, China. yh-wang@sjtu.edu.cn
Huiyuan Jiao is a master student at the Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China. j1212128@126.com
Zhiming Wang is a master student at the Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China. james.wong@sjtu.edu.cn
Jianfeng Shen is a professor at the Department of Ophthalmology, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai 200025, China; Institute of Translational Medicine, National Facility for Translational Medicine, Shanghai Jiao Tong University, Shanghai 201109, China. jfshen@shsmu.edu.cn
Lingjie Li is a professor at the Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China. lingjie@shsmu.edu.cn
Contributor Information
Feng Zhang, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Chen Yang, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Yihao Wang, Department of Ophthalmology, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai 200025, China; Institute of Translational Medicine, National Facility for Translational Medicine, Shanghai Jiao Tong University, Shanghai 201109, China.
Huiyuan Jiao, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Zhiming Wang, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Jianfeng Shen, Department of Ophthalmology, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai 200025, China; Institute of Translational Medicine, National Facility for Translational Medicine, Shanghai Jiao Tong University, Shanghai 201109, China.
Lingjie Li, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Key Laboratory of Reproductive Medicine, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Author Contributions
L.L. supervised the study. F. Z. conceived the study, designed experiments and carried out the analysis. C.Y., Y.W., H.J. and Z.W. participated in the validation of the method. F.Z., C.Y., Y.W., H.J., Z.W., J.S. and L.L. drafted and revised the manuscript. All authors read and approved the final manuscript.
Data Availability
The source code and related data are available at https://github.com/jumphone/fitdevo.
Funding
National Natural Science Foundation of China (32100516 to F.Z.; 32070867 to L.L.; 81972667 to J.S.), National Key R&D Program of China (2021YFA1100400, to L.L., 2021YFC2701103 to J.S.), Shanghai Sailing Program (21YF1422600 to F.Z.), Natural Science Foundation of Shanghai (21ZR1435900 to L.L), Program for Oriental Scholars of Shanghai Universities (to L. L.), Startup Fund for Young Faculty at SJTU (SFYF at SJTU)(21X010501077 to F.Z.), Shanghai Collaborative Innovation Center of Cellular Homeostasis Regulation and Human Disease (to L. L.).
Competing Interests
The authors declare that they have no competing interests.
References
- 1. Griffiths JA, Scialdone A, Marioni JC. Using single-cell genomics to understand developmental processes and cell fate decisions. Mol Syst Biol 2018;14(4):e8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Suva ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell 2019;75(1):7–12. [DOI] [PubMed] [Google Scholar]
- 3. Zappia L, Theis FJ. Over 1000 tools reveal trends in the single-cell RNA-seq analysis landscape. Genome Biol 2021;22(1):301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet 2019;20(5):257–72. [DOI] [PubMed] [Google Scholar]
- 5. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184(13):3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177(7):1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36(5):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Satija R, Farrell JA, Gennert D, et al. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018;37:38–44. [DOI] [PubMed] [Google Scholar]
- 11. McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction arXiv. 2018.
- 12. Qiu X, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017;14(10):979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32(4):381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marques S, Zeisel A, Codeluppi S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 2016;352(6291):1326–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haber AL, Biton M, Rogel N, et al. A single-cell survey of the small intestinal epithelium. Nature 2017;551(7680):333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature 2018;560(7719):494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qiu X, Zhang Y, Martin-Rufino JD, et al. Mapping transcriptomic vector fields of single cells. Cell 2022;185(4):690–711.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang F, Li X, Tian W. Unsupervised inference of developmental directions for single cells using VECTOR. Cell Rep 2020;32(8):108069. [DOI] [PubMed] [Google Scholar]
- 19. Teschendorff AE, Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell's transcriptome. Nat Commun 2017;8:15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teschendorff AE, Maity AK, Hu X, et al. Ultra-fast scalable estimation of single-cell differentiation potency from scRNA-Seq data. Bioinformatics 2021;37(11):1528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gulati GS, Sikandar SS, Wesche DJ, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020;367(6476):405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin S, MacLean AL, Peng T, et al. scEpath: energy landscape-based inference of transition probabilities and cellular trajectories from single-cell transcriptomic data. Bioinformatics 2018;34(12):2077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grun D, Muraro MJ, Boisset J-C, et al. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 2016;19(2):266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kannan S, Farid M, Lin BL, et al. Transcriptomic entropy benchmarks stem cell-derived cardiomyocyte maturation against endogenous tissue at single cell level. PLoS Comput Biol 2021;17(9):e1009305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo M, Bao EL, Wagner M, et al. SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res 2017;45(7):e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12(5):453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37(7):773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mantri M, Scuderi GJ, Abedini-Nassab R, et al. Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat Commun 2021;12(1):1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fawkner-Corbett D, Antanaviciute A, Parikh K, et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 2021;184(3):810–826.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tirosh I, Venteicher AS, Hebert C, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016;539(7628):309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galen P, Hovestadt V, Wadsworth II MH, et al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 2019;176(6):1265–1281.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res 2007;17(1):42–9. [DOI] [PubMed] [Google Scholar]
- 33. Isgro MA, Bottoni P, Scatena R. Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv Exp Med Biol 2015;867:125–43. [DOI] [PubMed] [Google Scholar]
- 34. Han X, Zhou Z, Fei L, et al. Construction of a human cell landscape at single-cell level. Nature 2020;581(7808):303–9. [DOI] [PubMed] [Google Scholar]
- 35. Hayashi T, Ozaki H, Sasagawa Y, et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun 2018;9(1):619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S, Drummond ML, Guerrero-Juarez CF, et al. Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nat Commun 2020;11(1):4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lopez-Pajares V, Qu K, Zhang J, et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell 2015;32(6):693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007;449(7165):1003–7. [DOI] [PubMed] [Google Scholar]
- 39. Fernandez Vallone V, Leprovots M, Ribatallada-Soriano D, et al. LGR5 controls extracellular matrix production by stem cells in the developing intestine. EMBO Rep 2020;21(7):e49224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu SZ, al-Eryani G, Roden DL, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet 2021;53(9):1334–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang X, Powell K, Li L. Breast cancer stem cells: biomarkers, identification and isolation methods, regulating mechanisms, cellular origin, and beyond. Cancers (Basel) 2020;12(12):3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gyan E, Owiredu WKBA, Fondjo LA, et al. A review of the racial heterogeneity of breast cancer stem cells. Gene 2021;796-797:145805. [DOI] [PubMed] [Google Scholar]
- 43. Ramos EK, Hoffmann AD, Gerson SL, et al. New opportunities and challenges to defeat cancer stem cells. Trends Cancer 2017;3(11):780–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiong S, Feng Y, Cheng L. Cellular reprogramming as a therapeutic target in cancer. Trends Cell Biol 2019;29(8):623–34. [DOI] [PubMed] [Google Scholar]
- 45. Steelman LS, Fitzgerald T, Lertpiriyapong K, et al. Critical roles of EGFR family members in breast cancer and breast cancer stem cells: targets for therapy. Curr Pharm Des 2016;22(16):2358–88. [DOI] [PubMed] [Google Scholar]
- 46. Choi S, Yu J, Park A, et al. BMP-4 enhances epithelial mesenchymal transition and cancer stem cell properties of breast cancer cells via notch signaling. Sci Rep 2019;9(1):11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pathania R, Ramachandran S, Elangovan S, et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun 2015;6:6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thomas ML, de Antueno R, Coyle KM, et al. Citral reduces breast tumor growth by inhibiting the cancer stem cell marker ALDH1A3. Mol Oncol 2016;10(9):1485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shy BR, Wu CI, Khramtsova GF, et al. Regulation of Tcf7l1 DNA binding and protein stability as principal mechanisms of Wnt/beta-catenin signaling. Cell Rep 2013;4(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kowalczyk MS, Tirosh I, Heckl D, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 2015;25(12):1860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang F, Wu Y, Tian W. A novel approach to remove the batch effect of single-cell data. Cell Discov 2019;5:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32(18):2847–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code and related data are available at https://github.com/jumphone/fitdevo.