Abstract
Recombinant Escherichia coli strains harboring heterologous polyhydroxyalkanoate (PHA) biosynthesis genes were shown to accumulate unusually large amounts of PHA. In the present study, integrated cellular responses of metabolically engineered E. coli to the accumulation of poly(3-hydroxybutyrate) (PHB) in the early stationary phase were analyzed at the protein level by two-dimensional gel electrophoresis. Out of 20 proteins showing altered expression levels with the accumulation of PHB, 13 proteins were identified with the aid of mass spectrometry. Three heat shock proteins, GroEL, GroES, and DnaK, were significantly up-regulated in PHB-accumulating cells. Proteins which play essential roles in protein biosynthesis were unfavorably influenced by the accumulation of PHB. Cellular demand for the large amount of acetyl coenzyme A and NADPH for the PHB biosynthesis resulted in the increased synthesis of two enzymes of the glycolytic pathway and one enzyme of the Entner-Doudoroff pathway. The expression of the yfiD gene encoding a 14.3-kDa protein, which is known to be produced at low pH, was greatly induced with the accumulation of PHB. Therefore, it could be concluded that the accumulation of PHB in E. coli acted as a stress on the cells, which reduced the cells' ability to synthesize proteins and induced the expression of various protective proteins.
Proteomics is a newly emerging research field which allows the analysis of when and under what conditions gene-encoded events (e.g., protein translation) occur (3, 11, 24). Proteome analysis by two-dimensional gel electrophoresis (2-DE) has been proposed elsewhere as a powerful tool for making genomics functional (3, 12, 21). One of the cornerstones for making proteomics a powerful tool is the development of mass spectrometry supported by the matrix-assisted safe ionization of peptide fragments and delayed extraction for the purpose of enhancing resolution power (22, 23). These extended capabilities of mass spectrometry, along with the ever-increasing amount of protein sequence data in various databases, are making protein identification and the characterization process a feasible task.
Poly(3-hydroxybutyric acid) (PHB) is an intracellular carbon and energy storage material synthesized by numerous microorganisms, usually when growth is impaired by the depletion of a specific nutrient in the presence of excess carbon source (13, 14, 33). PHB has been drawing much attention because of its complete biodegradability and the possibility of producing it from renewable resources (13, 14). In Ralstonia eutropha and Alcaligenes latus, PHB is synthesized from acetyl coenzyme A (CoA) in three sequential reaction steps catalyzed by β-ketothiolase, acetoacetyl-CoA reductase, and PHB synthase (13, 16, 30). The second reaction catalyzed by the reductase requires NADPH as a cofactor. A metabolically engineered Escherichia coli strain constitutively expressing the heterologous PHB biosynthesis genes has been suggested elsewhere to be a good candidate for PHB production due to fast growth, a large amount of PHB accumulation, and the availability of well-established high-cell-density culture techniques (8, 13, 18). Even though recombinant E. coli has been successfully employed for the high-level production of PHB (37), whether the overall cellular physiology is altered due to the expression of heterologous PHB biosynthesis genes and the accumulation of PHB granules in the cytoplasm remains unclear.
In this study, we analyzed and compared the proteomes of a metabolically engineered E. coli strain under PHB-producing and non-PHB-producing conditions. Proteome expression patterns of recombinant E. coli were resolved on 2D gels, and the variations in the relative expression levels of particular proteins were examined using a software-aided protein quantification tool.
MATERIALS AND METHODS
Bacterial strain, plasmid, and growth condition.
The E. coli strain used in this study was XL1-Blue (supE44 hsdR17 recA1 gyrA96 thi relA1 lac F′ [proAB+ laclq lacZΔM15 Tn10(Tetr)]). Plasmid pJC4, which contains the A. latus PHB biosynthesis genes, has been described previously (8). The PHB biosynthesis genes are constitutively expressed in E. coli (8). However, these enzymes cannot be detected on the 2D gel due to a low expression level (31). As a control plasmid, pJC4Δphb was constructed by deleting the PHB operon from pJC4. E. coli XL1-Blue, recombinant E. coli XL1-Blue(pJC4), and recombinant E. coli XL1-Blue(pJC4Δphb) were grown to early stationary phase in 250-ml flasks containing 100 ml of Luria-Bertani (LB) medium or LB medium plus 20 g of glucose per liter in a shaking incubator at 200 rpm at 37°C. Ampicillin was added at a concentration of 50 mg/liter when cultivating recombinant E. coli XL1-Blue(pJC4) and recombinant E. coli XL1-Blue(pJC4Δphb).
Analytical procedures.
Cell growth was monitored by measuring the absorbance at 600 nm (optical density at 600 nm; DU Series 600 spectrophotometer; Beckman, Fullerton, Calif.). The PHB concentration was determined by measuring the concentration of 3-hydroxybutyric acid methyl ester, which was prepared by methanolysis of PHB, with a gas chromatograph (Donam Co., Seoul, Korea) equipped with a fused silica capillary column (Supelco SPB-5; 30 m by 0.32 mm in inside diameter; 0.25-μm film; Bellefonte, Pa.) using benzoic acid as an internal standard (6). The cell concentration, defined as dry cell weight per liter of culture broth, was determined as previously described (37). The PHB content (wt%) was defined as the percent ratio of PHB concentration to cell concentration.
Glucose and acetic acid concentrations were measured with a high-performance liquid chromatograph (Hitachi chromatography system; Tokyo, Japan) equipped with an Aminex HPX-87H column (300 by 7.8 mm; Bio-Rad Laboratories, Hercules, Calif.) and a refractive index detector (L-3300; Hitachi chromatography system). The column was eluted isocratically with 5 mM H2SO4.
2-DE.
The 2-DE was carried out using a Protean II xi 2-D cell (Bio-Rad Laboratories) following the procedures described previously (10, 12) with slight modifications as follows. Carrier ampholyte solutions having a 4:1 ratio (vol/vol) of Bio-lyte pH 5 to 7 to Bio-lyte pH 3 to 10 (Bio-Rad) were used for the formation of a pH gradient in an isoelectric focusing (IEF) tube gel. Culture broth was centrifuged for 5 min at 3,500 × g and 4°C. The pellet was washed four times with TE solution (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) and was resuspended in double-distilled water followed by four cycles of sonication (each for 10 s at 10% of maximum output; high-intensity ultrasonic liquid processors; Sonics & Material, Inc., Newtown, Conn.). Soluble protein was obtained by the centrifugation of cell extract at 10,000 × g and 4°C for 20 min. After the protein quantification by the Bradford assay using bovine serum albumin as a standard (5), protein samples (300 μg) were dried down by vacuum centrifugation, suspended in IEF denaturation buffer [9 M urea, 0.5% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 10 mM dithiothreitol, 0.2% (wt/vol) Bio-lyte pH 3 to 10, 0.001% (wt/vol) bromophenol blue; final volume, 200 μl], and were carefully loaded into the IEF tube gel with a syringe. Then, the loaded tube gels were placed on sodium dodecyl sulfate–12% polyacrylamide gels prepared by a standard protocol (12). Coomassie brilliant blue R-250 (Bio-Rad) was used for protein staining (10). After overnight destaining in a solution composed of 40% (vol/vol) methanol and 10% (vol/vol) acetic acid, gels were scanned using a GS710 calibrated imaging densitometer (Bio-Rad). Melanie II software (Bio-Rad) was used to automate the process of finding protein spots within the image and to quantify the density of the spots on a volume basis (i.e., values were calculated from the integration of spot optical intensity over the spot area). To check the reproducibility and to estimate standard deviations, protein samples taken from duplicate cultures were analyzed in duplicate 2-D gels.
Peptide mass fingerprinting.
Samples for the matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry analysis were prepared as described previously (28) with some modifications described below. Trypsin digestion was carried out overnight at 37°C in a stationary incubator. A relatively high concentration of trypsin (25 ng/μl; sequencing grade; Boehringer Mannheim Co., Mannheim, Germany) was used to enhance trypsin autolysis. Two standard peaks of 805.417 (m/z) and 2,163.056 (m/z) were generated from this trypsin autolytic reaction. Other peptide fragment peaks, which are usually placed between these two standard peaks, were calibrated using the values of these two peaks. In-gel-digested peptide fragments were extracted from gel pieces by the addition of 100 μl of 60% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid (TFA) solution, followed by vortexing for 1 h. After the transfer of supernatant solution into a new Eppendorf tube, the acetonitrile-TFA solution was added for the final extraction of remaining peptide fragments. After these two supernatant solutions were combined, solute materials including peptide fragments were dried down by vacuum centrifugation. To eliminate impurities, such as salt molecules, gel particles, and traces of Coomassie brilliant blue, the samples were passed through the ZipTip column (Millipore Co., Bedford, Mass.) in which C18 resin is fixed at the end of the tip. α-Cyano-4-hydroxycinnamic acid, saturated in a solution composed of 50% (vol/vol) acetonitrile and 0.1% (vol/vol) TFA, was used as a matrix for MALDI-TOF. This matrix was incorporated with contaminant-free peptide fragments, placed on the sample plate, and crystallized with the peptide sample by air drying. The MALDI-TOF mass spectrometry system used was the Voyager Biospectrometry system (PerSeptive Biosystems, Inc., Framingham, Mass.). Laser intensity for the ionization of samples was optimized between 2,400 and 2,600. The accelerating voltage for the flight of ionized particles was 21,000 V, and the delayed extraction time was 150 ns.
The ProteinProspector server (http://prospector.ucsf.edu/ucsfhtml3.2/msfit.htm) was used for the identification of protein spots by querying the trypsin-digested peptide fragment data. To maintain the highest certainty of protein identification, mass tolerance was set within 50 ppm. The reference database used for the identification of target proteins was SWISS-PROT (http://www.expasy.ch/sprot).
RESULTS
Cell growth and PHB accumulation.
Flask cultures of recombinant E. coli XL1-Blue(pJC4) were grown in LB medium or LB medium containing 20 g of glucose per liter. Recombinant E. coli XL1-Blue(pJC4) could efficiently produce PHB in the latter, while it did not in the former (8). Flask cultures of E. coli XL1-Blue without the plasmid and of recombinant E. coli XL1-Blue(pJC4Δphb) were also made as controls. Table 1 shows the cell and soluble protein concentrations at the time of harvesting (early stationary phase) for proteome analysis. The PHB concentration and PHB content obtained for XL1-Blue(pJC4) in LB medium plus 20 g of glucose per liter at the time of proteome analysis were 0.89 g/liter and 68%, respectively. For both wild-type and recombinant strains, the acetic acid concentrations at the time of harvesting were 0 and 2 g/liter when grown in LB medium and LB medium plus 20 g of glucose per liter, respectively.
TABLE 1.
Comparison of cell and extracted soluble protein concentrations at the time of harvesting for proteome analysis
| Strain | Growth medium | Cell concn (g [DCW]b/liter) | Extracted soluble protein concna (mg/ml) |
|---|---|---|---|
| XL1-Blue | LB | 1.43 ± 0.070 | 0.623 ± 0.010 |
| LB plus 20 g of glucose per liter | 1.15 ± 0.124 | 0.443 ± 0.030 | |
| XL1-Blue(pJC4) | LB | 1.08 ± 0.018 | 0.483 ± 0.001 |
| LB plus 20 g of glucose per liter | 1.31 ± 0.035 | 0.353 ± 0.015 | |
| XL1-Blue(pJC4Δphb) | LB | 1.07 ± 0.025 | 0.473 ± 0.022 |
| LB plus 20 g of glucose per liter | 1.12 ± 0.051 | 0.457 ± 0.043 |
Soluble proteins were extracted by sonication, and the amounts of proteins were determined by the Bradford assay as described in Materials and Methods.
DCW, dry cell weight.
Proteome analysis.
Proteome expression profiles of XL1-Blue and XL1-Blue(pJC4) grown in two media (LB medium and LB medium plus 20 g of glucose per liter) are shown in Fig. 1. To compare the relative expression levels of cellular proteins, the similar amounts of proteins were loaded on 2D gels. Cellular proteins possessing pIs between 4.7 and 6.2 could be nicely separated in our 2D gels. The overall profiles of synthesized proteins within this range were quite reproducible and distinctive enough to be compared and matched even by naked eyes.
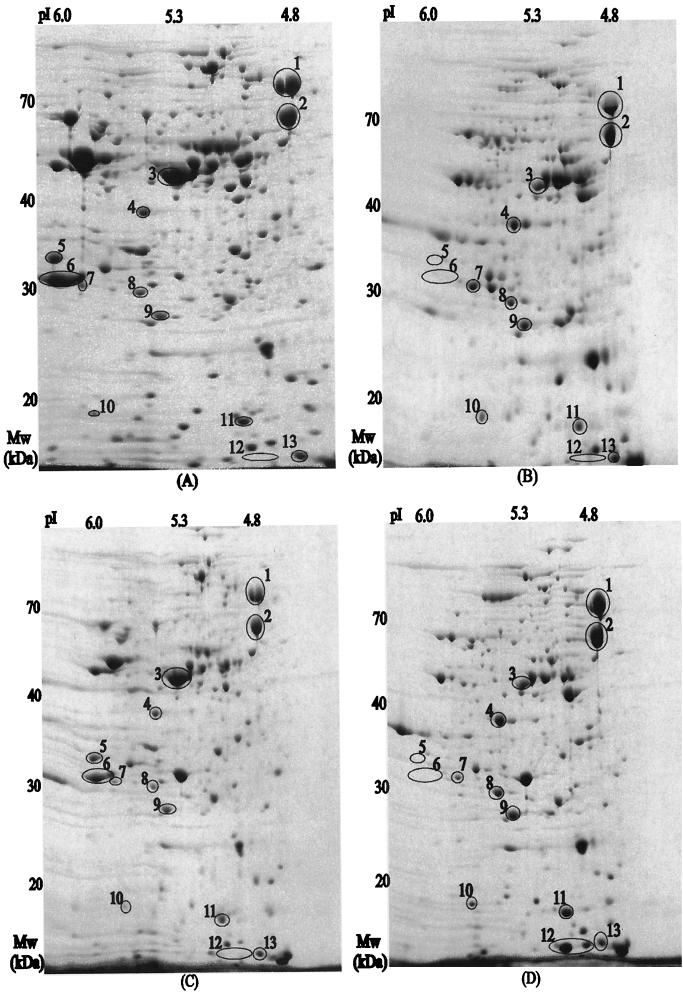
FIG. 1.
Proteome expression profiles of E. coli XL1-Blue grown in LB medium (A), E. coli XL1-Blue grown in LB medium plus 20 g of glucose per liter (B), recombinant E. coli XL1-Blue harboring pJC4 grown in LB medium (C), and recombinant E. coli XL1-Blue harboring pJC4 grown in LB medium plus 20 g of glucose per liter (D). The horizontal axes represent the isoelectric points, and the vertical axes represent the molecular masses in kilodaltons.
Completely destained gels were scanned, and 20 proteins showing altered expression levels under four different conditions were selected for a further identification process. Excised protein spots were subjected to MALDI-TOF mass spectrometry; a list of proteins identified by peptide mass fingerprinting is shown in Table 2. Out of 20 spots, 13 protein spots were identified exactly by database search. Out of five possible candidates listed by database search, the first ranked protein with the molecular weight search (MOWSE) score (22) greater than 1.0×e+3 was assigned to be the protein spot of interest. The amount of protein in each spot was determined by Melanie II software (Bio-Rad) (Fig. 2). In order to examine the physiological roles of these proteins, they were categorized (Table 2) according to their functions in the cell (26).
TABLE 2.
Categorized functions of identified proteins showing different expression levels under PHB-producing versus non-PHB-producing conditions
| Spot no. | Category | Gene | Proteina | pl/mol massb |
|---|---|---|---|---|
| Cell process | ||||
| 1 | Chaperoning | dnaK | DnaK, chaperone for Hsp70 | 4.81/69.6 |
| 2 | Chaperoning | groEL | GroEL, chaperone for Hsp60 | 4.85/57.3 |
| 11 | Chaperoning | groES | GroES, chaperone for Hsp10 | 5.15/10.4 |
| 6 | Transport-binding | rbsB | RbsB, d-ribose periplasmic binding protein | 6.85/30.9 |
| Energy metabolism | ||||
| 4 | Glycolysis | fba | Fba, fructose-bisphosphate aldolase, class II | 5.52/39.2 |
| 7 | Glycolysis | gpmA | GpmA, phosphoglycerate mutase | 5.85/28.5 |
| 8 | Glycolysis | tpiA | TpiA, triosephosphate isomerase | 5.64/27.0 |
| 9 | Entner-Doudoroff pathway | eda | Eda, 2-oxo-3-deoxy-6-phosphogluconate aldolase (2-keto-4-hydroxyglutarate aldolase) | 5.57/22.3 |
| Central intermediary metabolism | ||||
| 5 | General | kba | Kba, d-tagatose-1,6-bisphosphate aldolase | 5.98/31.1 |
| Macromolecular metabolism | ||||
| 3 | Protein translation and modification | tufA | EF-Tu, protein chain elongation factor | 5.3/43.3 |
| Other | ||||
| 10 | Adaptation | dps | Dps, DNA protection protein during starvation | 5.72/18.7 |
| 13 | Broad regulatory functions | relB | RelB, negative regulator of translation | 4.81/9.1 |
| 12 | Unknown function | yfiD | YfiD, 14.3-kDa hypothetical protein | 5.09/14.3 |
Proteins were identified by the peptide fingerprinting method. The ProteinProspector server was used for the identification of proteins, and SWISS-PROT was used as a reference database.
Molecular masses are shown in kilodaltons.
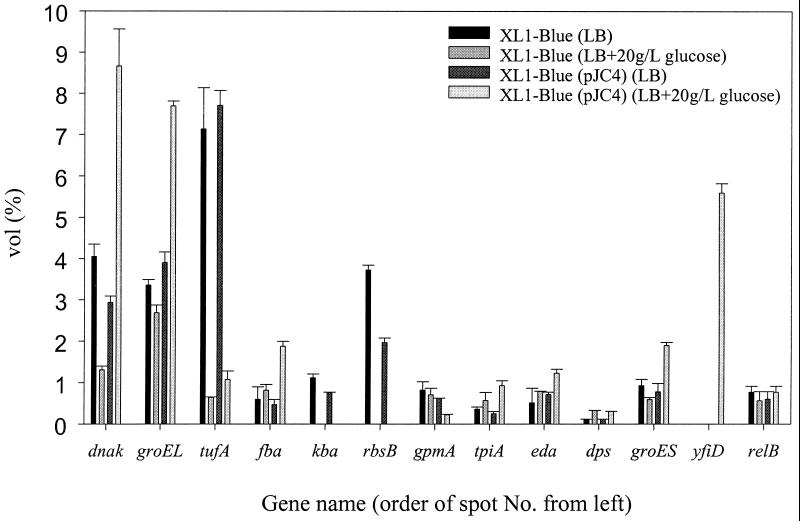
FIG. 2.
Quantification of identified protein spots showing altered expression level from Fig. 1. Spot intensities were measured and normalized as described in Materials and Methods. Error bars represent the standard deviations. Vol% is the percentage of relative values calculated from the integration of spot intensity over the spot area. For the symbol key, top to bottom corresponds to left to right for each group of bars.
When the proteomes of XL1-Blue and XL1-Blue(pJC4) grown in LB medium were compared, it could be seen that the presence of pJC4 did not significantly alter the proteome except for RbsB, the expression level of which was twice as high in plasmid-free XL1-Blue. When the proteomes of XL1-Blue grown in LB medium and those of XL1-Blue grown in LB medium plus 20 g of glucose per liter were compared, heat shock proteins including GroEL, GroES, and DnaK were down-regulated in the presence of glucose. On the other hand, Dps, a DNA protection protein, was up-regulated in the presence of glucose. More changes included the significant down-regulation of EF-Tu and the disappearance of Kba and RbsB spots in the presence of 20 g of glucose per liter. Reduced synthesis of EF-Tu suggests that the protein synthesis capacity of E. coli cells is much reduced in the presence of 20 g of glucose per liter, which is a relatively high concentration for E. coli. In addition, when the proteomes of XL1-Blue(pJC4Δphb) grown in LB medium and those of XL1-Blue grown in LB medium plus 20 g of glucose per liter were compared, it was found that the presence of this backbone plasmid without the PHB biosynthesis genes did not significantly alter the proteome except for Dps, the expression level of which was threefold higher in the presence of glucose (data not shown).
One of the most distinguishable variations upon the accumulation of PHB was the significantly increased synthesis of heat shock proteins GroEL, GroES, and DnaK. Out of three identified enzymes of the glycolytic pathway, Fba and TpiA showed up-regulated synthesis in PHB-producing cells, while the synthesis of GpmA was down-regulated. One identified enzyme of the Entner-Doudoroff pathway, Eda, showed up-regulated synthesis with the PHB accumulation. The synthesis level of the 14.3-kDa protein in the suppressor ribosomal mutant B-uracil nucleic acid glycosylase (SRMB-UNG) intergenic region encoded by yfiD was significantly up-regulated with the PHB accumulation. These changes are truly due to the accumulation of PHB because the proteomes of XL1-Blue (pJC4Δphb) did not show these alterations.
DISCUSSION
Recombinant E. coli harboring a plasmid containing the heterologous PHB biosynthesis genes was found to be able to accumulate a large amount of PHB (8, 13, 18, 37). It was expected that this metabolically engineered E. coli strain would undergo physiological changes upon the accumulation of PHB, which is not a normal metabolite of E. coli. We attempted to examine these physiological changes by analyzing the proteomes of recombinant E. coli XL1-Blue under two different culture conditions: PHB-producing and non-PHB-producing conditions. The proteomes of XL1-Blue without the plasmid and XL1-Blue harboring backbone plasmid were also analyzed for comparison.
Glucose effect.
EF-Tu is one of the most abundant cytosolic proteins and plays a crucial role in protein biosynthesis. It was reported previously that EF-Tu interacts with various cellular macromolecules such as tRNAs charged with amino acids; another type of protein chain elongation factor, EF-Ts; and ribosomes to make the translational process proceed properly (35). According to Fig. 1 and 2, it can be seen that the expression level of EF-Tu decreased significantly in cells grown in the presence of glucose. Since cellular protein synthesis is reduced, more catabolic intermediates seem to be available for PHB biosynthesis under this condition. This was supported by the finding that a large amount of PHB was accumulated only when glucose was present in the medium (17, 18). The synthesis of Dps was twofold greater in the presence of glucose. Dps has been shown previously to be expressed when cells produce acetic acid (27, 36). This is in agreement with our finding that 2 g of acetic acid per liter was produced only in the presence of glucose.
PHB accumulation effect.
In general, heat shock proteins are synthesized in order to protect cells from external stresses such as sudden increase of temperature, UV irradiation, virus infection, organic solvents, and others. Expression of heterologous genes can also trigger heat shock response (9). Based on our experimental observation of increased expression of three heat shock proteins (GroEL, GroES, and DnaK) in PHB-producing cells, PHB accumulation in recombinant E. coli can be considered as a stress on the cells inducing heat shock response. Furthermore, the takeover of the cytosolic space by PHB granules would disturb the normal intracellular architecture such as chromosome attachment and consequently would result in heat shock response. Bacteria naturally accumulating PHB synthesize phasin protein, which covers the surface of PHB granules. E. coli does not naturally produce PHB and therefore does not have the phasin gene (39). Therefore, the hydrophobic PHB granules are in direct contact with intracellular biomolecules including DNA, RNA, and proteins. Obviously, this will become a major stress on the cells for several possible reasons including denaturation of proteins on the surface of PHB granules. This unfavorable condition generated by PHB accumulation along with the reduced synthesis of EF-Tu seems to have resulted in the further reduction of protein synthesis.
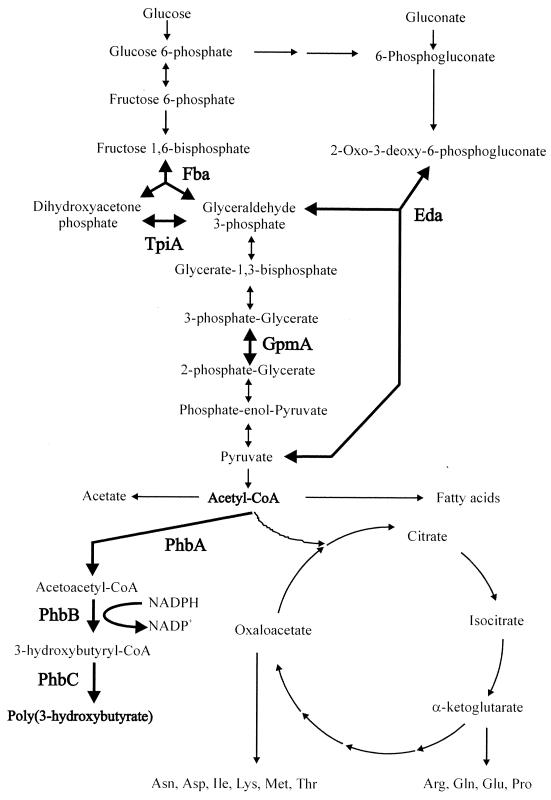
The PHB biosynthesis pathway competes for acetyl-CoA with three other metabolic pathways (Fig. 3) (16, 17). Consumption of acetyl-CoA for the synthesis of PHB results in the reduced availability of acetyl-CoA for other metabolic pathways: tricarboxylic acid cycle, acetate formation pathway, and fatty acids synthetic pathway. Considering that 10 out of 20 amino acids are synthesized from the intermediary metabolites of the tricarboxylic acid cycle (Fig. 3), the reduced synthesis of amino acids may result in impaired synthesis of proteins. This is obviously linked to the reduced synthesis of EF-Tu, resulting in overall reduction of cellular protein content under PHB-accumulating conditions (Table 1).
FIG. 3.
Central metabolic pathways of recombinant E. coli producing PHB. Several competing metabolic pathways leading to the synthesis of PHB, acetate, citrate, and fatty acids from acetyl-CoA are shown. Four identified enzymes, Fba, TpiA, GpmA, and Eda, and the reaction steps that they catalyze are shown in boldface. The reaction pathways leading to the synthesis of PHB along with the enzymes involved are also shown in boldface.
The synthesis of Fba and TpiA was up-regulated in PHB-accumulating cells compared with non-PHB-accumulating cells. As shown in Fig. 3, these two enzymes catalyze the formation of glyceraldehyde-3-phosphate, the first three-carbon metabolite of glycolysis. It has been reported previously that 96% of triose phosphate exists as dihydroxyacetone phosphate at equilibrium (34). When glyceraldehyde-3-phosphate is used by subsequent reactions, dihydroxyacetone phosphate is converted to glyceraldehyde-3-phosphate accordingly. It is therefore possible to conclude that the purpose of the increased expression of TpiA resulting in the enhanced rate of conversion of dihydroxyacetone phosphate to glyceraldehyde-3-phosphate is to provide a three-carbon metabolite, which is eventually used up for PHB synthesis. The Eda catalyzes the final reaction of the Entner-Doudoroff pathway to supplement glyceraldehyde-3-phosphate and pyruvate (Fig. 3) in a coordinated way with the above two enzymes of the glycolysis pathway to increase the amount of acetyl-CoA. It is likely that metabolic demand for the large amount of acetyl-CoA and NADPH results in this elevated expression profile in the PHB-producing cells. Conversely, the reason why XL1-Blue(pJC4) cultured in LB medium did not accumulate PHB may be the shortage of acetyl-CoA and NADPH for PHB accumulation.
One of the most intriguing results was that the yfiD gene product was detected at a high level only in the PHB-producing cells. It was first thought that there were two different proteins (two spots at spot 12). However, the results of the database search showed that these two spots represent the same protein. Most proteins in E. coli are present in a single charged form. Some proteins, however, are present in multiple charged isoforms. For example, several outer membrane proteins of E. coli were shown to be resolved into at least two charged isoforms (20). Unfortunately, we have been unable to conclusively establish the nature of this charge variation, although we have observed some deamidated peptides that may contribute to protein heterogeneity. Although the biological function of this 14.3-kDa protein in the SRMB-UNG intergenic region (127 amino acids) remains unclear, this protein has been reported to be a homologue of pyruvate formate lyase. The YfiD was reported previously to be expressed under acidic conditions (4). The pHs of the culture media at the time of proteome analysis were 4.86, 4.77, and 4.74 for XL1-Blue, XL1-Blue(pJC4Δphb), and XL1-Blue(pJC4), respectively, when they were grown in LB medium plus 20 g of glucose per liter. Normally, E. coli maintains the cytoplasmic pH about 1 to 1.5 U higher than the external pH of acidic to neutral range (9). Recombinant E. coli cells accumulating a large amount of PHB were shown previously to become fragile with altered cell envelope structures (17). Therefore, it seems that these PHB-accumulating cells may not be able to achieve cellular homeostasis, and their cytoplasm becomes acidic. This is most likely why the yfiD gene was expressed only under PHB-accumulating conditions.
Since PHB accumulation acts as a stress on the cells, a fermentation strategy should be developed so that cells do not synthesize PHB too early during the cultivation. This strategy was already successfully practiced for the high-level production of PHB (16). The findings that the expression levels of several enzymes in the glycolytic pathway were adjusted to provide more acetyl-CoA and NADPH are also of great importance to consider during metabolic pathway engineering of cells for enhanced PHB production. Metabolic flux analysis can be complementary to this approach.
It should be noted that the identification of protein spots in the 2D gel is currently a limiting step in proteomic research. For example, we failed to assign functions for 7 out of 20 spots after MALDI-TOF analysis. Even though we were able to analyze only 13 proteins that showed different expression patterns on the 2D gel, a number of physiological changes caused by PHB accumulation could be explained. As the number of identifiable protein spots increases, we will be able to understand global physiological changes under various environmental conditions, and this valuble information will be utilized toward strain improvement and the construction of various databases regarding metabolism.
ACKNOWLEDGMENTS
We thank Jong Shin Yu at the Korea Basic Science Institute for his generous help in the MALDI-TOF analysis.
This work was supported by the National Research Laboratory program of the Korean Ministry of Science and Technology and by the First Young Scientist's Award to S. Y. Lee by the President of Korea.
REFERENCES
- 1.Alexeyev A A, Bakhlanova I V, Zaitev E N, Lanzov V A. Genetic characteristics of new recA mutants of Escherichia coli K-12. J Bacteriol. 1996;178:2018–2024. doi: 10.1128/jb.178.7.2018-2024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almiron M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 3.Blackstock W P, Weir M P. Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999;17:121–127. doi: 10.1016/s0167-7799(98)01245-1. [DOI] [PubMed] [Google Scholar]
- 4.Blankenhorn D, Philips J, Slonczewski J L. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J Bacteriol. 1999;181:2209–2216. doi: 10.1128/jb.181.7.2209-2216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Braunegg G, Sonnleitner B, Lafferty R M. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol. 1978;6:29–37. [Google Scholar]
- 7.Cayrol C, Petit C, Raynaud B, Capdevielle J, Guillemot J C, Defais M. Recovery of respiration following the SOS response of Escherichia coli requires recA-mediated induction of 2-keto-4-hydroxyglutarate aldolase. Proc Natl Acad Sci USA. 1995;92:11806–11809. doi: 10.1073/pnas.92.25.11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J, Lee S Y, Han K B. Cloning of the Alcaligenes latus polyhydroxyalkanoates biosynthesis genes and use of these genes for enhanced production of poly(3-hydroxybutyrate) in Escherichia coli. Appl Environ Microbiol. 1998;64:4897–4903. doi: 10.1128/aem.64.12.4897-4903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross C A. Functions and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 10.Harrington M G, Gudeman D, Zewert T, Yun M, Hood L. Analytical and micropreparative two-dimensional electrophoresis of proteins. Methods Companion Methods Enzymol. 1991;3:98–108. [Google Scholar]
- 11.Hochstrasser D F. Proteome in perspective. Clin Chem Lab Med. 1998;36:825–836. doi: 10.1515/CCLM.1998.146. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser D F, Harrington M G, Hochstrasser A C, Miller M J, Merril C R. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem. 1988;173:424–435. doi: 10.1016/0003-2697(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 13.Lee S Y. Bacterial polyhydroxyalkanoates. Biotechnol Bioeng. 1996;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Lee S Y. Plastic bacteria? Progress and prospects for polyhydroxyalkanoate production in bacteria. Trends Biotechnol. 1996;14:431–438. [Google Scholar]
- 15.Lee S Y, Chang H N. Effect of complex nitrogen source on the synthesis and accumulation of poly(3-hydroxybutyric acid) by recombinant Escherichia coli in flask and fed-batch cultures. J Environ Polym Degrad. 1994;2:169–176. [Google Scholar]
- 16.Lee S Y, Chang H N. Production of poly(3-hydroxybutyric acid) by recombinant Escherichia coli strains: genetic and fermentation studies. Can J Microbiol. 1995;41:207–215. doi: 10.1139/m95-189. [DOI] [PubMed] [Google Scholar]
- 17.Lee S Y, Lee K M, Chang H N, Steinbüchel A. Comparison of recombinant Escherichia coli strains for synthesis and accumulation of poly(3-hydroxybutyric acid) and morphological changes. Biotechnol Bioeng. 1994;44:1337–1347. doi: 10.1002/bit.260441110. [DOI] [PubMed] [Google Scholar]
- 18.Lee S Y, Yim K S, Chang H N, Chang Y K. Construction of plasmids, estimation of plasmid stability, and use of stable plasmid for the production of poly(3-hydroxybutyric acid) by recombinant Escherichia coli. J Biotechnol. 1994;32:203–211. doi: 10.1016/0168-1656(94)90183-x. [DOI] [PubMed] [Google Scholar]
- 19.Little J W, Mount D W. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 20.Molloy M P, Herbert B R, Slade M B, Rabilloud T, Nouwens A S, Williams K L, Gooley A A. Proteomic analysis of the Escherichia coli outer membrane. Eur J Biochem. 2000;267:2871–2881. doi: 10.1046/j.1432-1327.2000.01296.x. [DOI] [PubMed] [Google Scholar]
- 21.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 22.Pappin D J, Hojrup P, Bleasby A J. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 23.Patterson S D. Mass spectrometry and proteomics. Physiol Genomics. 2000;2:59–65. doi: 10.1152/physiolgenomics.2000.2.2.59. [DOI] [PubMed] [Google Scholar]
- 24.Persidis A. Proteomics. Nat Biotechnol. 1998;16:393–394. doi: 10.1038/nbt0498-393. [DOI] [PubMed] [Google Scholar]
- 25.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riley M, Labedan B. Escherichia coli gene products: physiological functions and common ancestries. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2118–2202. [Google Scholar]
- 27.Roe A J, McLaggan D, Davidson I, O'Byrne C, Booth I R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenfeld J, Capdevielle J, Guillemot J, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 29.Schellhorn H E, Stones V L. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J Bacteriol. 1992;174:4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert P, Steinbüchel A, Schlegel H G. Cloning of Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol. 1988;170:5837–5847. doi: 10.1128/jb.170.12.5837-5847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sim S J, Snell K D, Hogan S A, Stubbe J, Rha C, Sinskey A J. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat Biotechnol. 1997;15:63–67. doi: 10.1038/nbt0197-63. [DOI] [PubMed] [Google Scholar]
- 32.Slater S C, Voige W H, Dennis D E. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthetic pathway. J Bacteriol. 1988;170:4431–4436. doi: 10.1128/jb.170.10.4431-4436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinbüchel A, Wiese S. Bacterial and other biological systems for polyester production. Trends Biotechnol. 1998;16:419–427. doi: 10.1016/s0167-7799(98)01194-9. [DOI] [PubMed] [Google Scholar]
- 34.Stryer L. Glycolysis. In: Stryer L, editor. Biochemistry. 4th ed. New York, N.Y: W. H. Freeman & Co.; 1995. pp. 483–508. [Google Scholar]
- 35.Talens A, Boon K, Kraal B, Bosch L. Translational activities of EF-Tu (G222D), which cannot be reconciled with classical scheme of the protein chain elongation cycle. Biochem Biophys Res Commun. 1996;225:961–967. doi: 10.1006/bbrc.1996.1279. [DOI] [PubMed] [Google Scholar]
- 36.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, Lee S Y. Production of poly(3-hydroxybutyrate) by fed-batch culture of filamentation-suppressed recombinant Escherichia coli. Appl Environ Microbiol. 1997;63:4756–4769. doi: 10.1128/aem.63.12.4765-4769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellner F, Jörgensen S T, Diderichsen B, Karlstrom O H. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 1985;4:1059–1066. doi: 10.1002/j.1460-2075.1985.tb03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wieczorek R, Pries A, Steinbüchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol. 1995;177:2425–2435. doi: 10.1128/jb.177.9.2425-2435.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]