Abstract
Reliable and comprehensive monitoring data are required to trace and counteract biodiversity loss. High-throughput metabarcoding using DNA extracted from community samples (bulk) or from water or sediment (environmental DNA) has revolutionized biomonitoring, given the capability to assess biodiversity across the tree of life rapidly with feasible effort and at a modest price. DNA metabarcoding can be upscaled to process hundreds of samples in parallel. However, while automated high-throughput analysis workflows are well-established in the medical sector, manual sample processing still predominates in biomonitoring laboratory workflows limiting the upscaling and standardization for routine monitoring applications. Here we present an automated, scalable, and reproducible metabarcoding workflow to extract DNA from bulk samples, perform PCR and library preparation on a liquid handler. Key features are the independent sample replication throughout the workflow and the use of many negative controls for quality assurance and quality control. We generated two datasets: i) a validation dataset consisting of 42 individual arthropod specimens of different species, and ii) a routine monitoring dataset consisting of 60 stream macroinvertebrate bulk samples. As a marker, we used the mitochondrial COI gene. Our results show that the developed single-deck workflow is free of laboratory-derived contamination and produces highly consistent results. Minor deviations between replicates are mostly due to stochastic differences for low abundant OTUs. Thus, we successfully demonstrated that robotic liquid handling can be used reliably from DNA extraction to final library preparation on a single deck, thereby substantially increasing throughput, reducing costs, and increasing data robustness for biodiversity assessments and monitoring.
Keywords: Pipetting robot, Automatization, Standardization, High-throughput sequencing
Highlights
-
•
Concept for high-throughput DNA metabarcoding on robotic platforms is presented.
-
•
Workflow performance validated using German stream biomonitoring samples.
-
•
Results support consistent and contamination-free high-throughput data generation.
-
•
Robotic workflows can reduce costs and enhance robustness of biomonitoring data.
1. Introduction
Global biodiversity loss proceeds at a fast pace [[1], [2], [3]]. With the post-2020 Global Biodiversity Framework, the United Nations Convention on Biological Diversity (CBD) currently focuses and aligns international efforts in order to meet the proclaimed vision for 2050 of “living in harmony with nature” as outlined in the Sustainable Development Goals [4]. One strategic milestone is the 2021–2030 “UN decade on ecosystem restoration”. While major drivers of biodiversity loss have been identified, a common concern is the lack of highly resolved biodiversity data that would allow to monitor change as well as to assess the specific benefits of management actions on biodiversity across the tree of life, i.e., from prokaryotes over fungi and unicellular eukaryotes to complex multicellular organisms. Traditional monitoring methods assess biodiversity based on phenotypic features using, e.g., microscopes and identification keys. Reliable species identification especially of larger eukaryotes is possible, yet with very limited temporal and spatial resolution. While citizen science approaches [5] sometimes deliver highly resolved data, this is mainly true for few selected taxonomic groups such as birds. However, for the vast majority of biodiversity, e.g., insects, available data are scarce, and therefore trends remain uncertain [6,7]. In view of the demand for reliable and scalable biodiversity assessment solutions, new molecular tools can become a game changer [8,9]. Especially the analysis of environmental DNA (eDNA) samples collected from water, sediment, soil, plants, or air has an incredible potential for high-throughput assessment of diversity [10]. Such samples can be analyzed on high-throughput sequencers in a massively parallel fashion, thereby scaling up the number of samples to be analyzed simultaneously [11]. Sequencing technologies evolved rapidly within the last decade leading to a drastic increase in data output and a decrease of per base pair costs. For DNA-based biodiversity assessments, DNA metabarcoding [12] is currently the most widely applied approach to assess the biodiversity of marine [13,14], limnic [15,16], and terrestrial ecosystems [17,18]. While many different DNA metabarcoding laboratory protocols exist [[19], [20], [21]], all DNA metabarcoding approaches can be roughly divided into a limited number of distinct workflow steps: i) pre-amplification sample processing, ii) DNA amplification and library preparation, iii) sequencing, and iv) bioinformatic analysis. In the wet lab workflow, DNA is initially isolated from the sample (e.g., organismal tissue or eDNA on filters). Depending on the sample type or taxon, the extracted DNA is PCR-amplified with primers targeting a specific DNA fragment, such as the mitochondrial cytochrome c oxidase subunit I (COI) gene [[22], [23]], different fragments of the small and large subunits of the ribosomal RNA, like the 12S [24,25] or 16S marker or the internal transcribed spacer (ITS; [26,27]). Specific tags or indexes are then added either using ligation or PCR-based strategies [28] to allow distinguishing samples during bioinformatic processing. Post-PCR steps can vary between protocols, but mostly include a normalization step, size-selection, and DNA quantification. The final library is then sequenced on one of the various available sequencing platforms, e.g., Illumina, Ion Torrent, DNBSEQ, PacBio, or Oxford Nanopore. Despite a large variety in labware and chemicals, differences and personal preferences in laboratory setups, as well as the choice of target organisms and the researcher's questions, all those protocols share this basic workflow [20]. Here, the implementation of reliable and standardized quality control measures is crucial to produce trustworthy data in large scale routine laboratory practice. All this has been realized in medical laboratories using robotic liquid handling techniques. The success of the high-throughput PCR screens for SARS-CoV-2 with millions of samples per week documented this [29,30]. In general, robotic liquid handling is used for protein folding analyses [31], cell cultivation [32], genotyping [33,34], selected reaction monitoring mass spectrometry [35], forensic analyses [36,37] (see [38] for an overview), and in particular many clinical diagnostics [[39], [40], [41], [42]] including microbiome screening [43,44]. International standards for the validation of liquid handling accuracy exist and are being refined further (e.g., ISO/IWA15; ISO/CD23783). In view of these existing automated laboratory workflows it is remarkable that the vast majority DNA metabarcoding-biodiversity assessments studies are still based on manual sample processing. To our knowledge, no established, standardized, and fully automated protocol for biodiversity assessment analyses, supported by validation datasets, has been published yet. However, in view of the global challenges with respect to the mission to halt biodiversity loss and restore ecosystems, there is an urgent need for a robust, scalable, and standardized processing workflow in analogy to medical laboratory workflows. Therefore, we here propose such a standardized and highly scalable biodiversity data generation and validation workflow that uses an automated liquid handling station (supplementary figure 1). The laboratory protocols were developed to produce robust and reproducible data, minimize manual liquid handling operations, prevent cross-contamination during liquid handling and drastically scale up the throughput by transitioning to 96-well plate-based study designs.
To validate the robustness of the automated workflows, we produced two different datasets targeting the mitochondrial cytochrome c oxidase subunit I (COI) gene. Dataset 1 consisted of morphologically identified single specimen samples processed and analyzed individually. The dataset was designed to identify potential cross-contamination, as each sample was to be expected to only represent reads from the one respective species (validation dataset). Dataset 2 consisted of a community metabarcoding analysis of freshwater invertebrates performed as part of a regulatory routine biomonitoring program in Bavaria, Germany (biomonitoring dataset). In addition, we present summary statistics on 196 samples analyzed that support the universal applicability of the workflow for high-throughput metabarcoding. We discuss our findings and derive considerations when moving towards the next decade of high-throughput genetic assessments [45], also termed Biomonitoring 2.0 [46].
2. Methods
The first dataset for validating the robustness of sample processing consisted of 42 single specimens from 42 different arthropod species. Specimens were captured in emergence traps at the stream Breitenbach in 2013 and Kleine Schmalenau in 2014 (both Germany, N 50.66206 E 9.62389; N 51.43928, E 8.13869; permissions for sampling were obtained from the respective state authorities). All specimens were morphologically identified to either species (34), genus (2), family (5) or order (1) level. In case morphological identification was carried out to a higher taxonomic level than species (e.g., order) the respective taxon was only represented by one specimen. The second dataset consisted of 60 sorted macroinvertebrate bulk samples, each containing hundreds of specimens, collected and provided by the Bavarian Environmental Agency (LfU).
2.1. Sample preparation
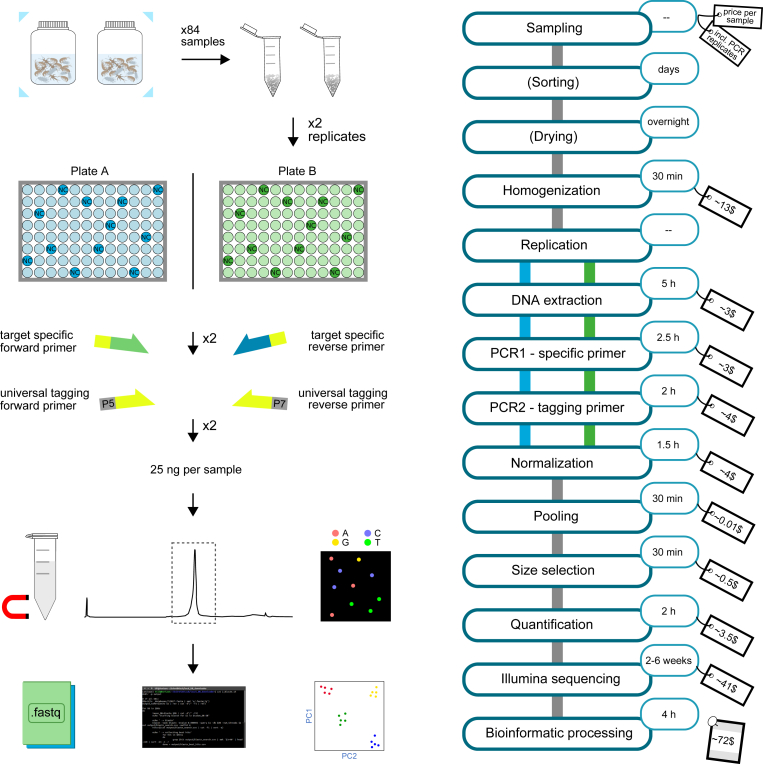
Single specimens for dataset 1 were separately dried in sterile Petri-dishes overnight. Specimens were then transferred into 2 mL twist-top tubes prefilled with 5 zirconia beads (2 mm diameter; BioSpec Products, Bartlesville, USA) and ground to tissue powder using the FastPrep-24 (MP Biomedicals, Eschwege, Germany) for 3 × 45 s at 6 m/s. Ground tissue powder was dissolved in 300 μL TNES buffer and stored at −20 °C until further processing. Bulk specimens from the regulatory biomonitoring program (dataset 2) were separated from the storage ethanol using sterile filter paper. For specimens which strongly differed in size compared to the average size of the other specimens in a sample, a part of the body (e.g., leg or abdomen) was cut off and used for further processing while the leftovers were discarded. Samples were dried overnight and transferred into sterile Turrax grinding tubes (IKA, Stauffen, Germany). Samples were ground to tissue powder for 30 min at 4000 rpm on the Ultra Turrax (IKA, Stauffen, Germany). After grinding, the tissue powder was dissolved in 10 mL TNES buffer, and subsequently 300 μL of the homogenate was transferred into 2 mL twist-top tubes and stored at −20 °C until further processing. All samples were then processed following the workflow outlined in Fig. 1. All following steps have been automatized to the maximum degree possible on a Biomek FXP Automated Workstation (Beckman Coulter, Indianapolis USA).
Fig. 1.
Blueprint of a standardized and automatized DNA metabarcoding workflow established for automated liquid handlers. Prices and DNA input depend on the actual materials used and can differ between manufacturers and custom-made materials. The time for sequencing can vary depending on third-party companies or the availability of a sequencer. The prices shown are list prices (2021) for the products used in the workflow.
2.2. DNA extraction
Replication is of critical importance for the validation of datasets. Therefore, prior to DNA extraction, each homogenized sample was split into two wells on separate plates (Fig. 1, plate A and plate B). Here, 60 μL of the dissolved tissue powder was lysed with 133 μL additional TNES buffer and 7 μL proteinase K (10 mg/mL) in a final volume of 200 μL for 3 h at 55 °C and 1000 rpm. From the point of replication until after the individual tagging of samples in the second PCR, replicates of the same sample were never simultaneously on the robotic deck to control for cross-contamination. Twelve negative controls only containing TNES buffer were placed on the plates, according to Elbrecht & Steinke [11]. DNA was extracted using a modified NucleoMag Tissue kit (Macherey Nagel, Düren, Germany, supplementary material 1). The quality of the extracted DNA was examined using a 1% agarose gel.
2.3. PCR
A fragment of the mitochondrial cytochrome c oxidase subunit 1 (COI) gene was amplified using a two-step PCR protocol, according to Zizka et al. [28]. In the first step, DNA was amplified using the Qiagen Multiplex PCR Plus Kit (Qiagen, Hilden, Germany) in 25 μL assays containing 1x Multiplex PCR Master Mix, 1x CoralLoad Dye, 100 nM of each primer (fwhF2, fwhR2n; supplementary table 5 [47]; in four length-varying versions and a universal tail attached as in Leese et al. [48]), 2.5 μL DNA, and RNase-free water. Amplification was performed using a touchdown PCR protocol with the following parameters: 95 °C for 5 min as initial denaturation, 10 cycles of 95 °C for 30 s denaturation, 68-59 °C for 90 s annealing (with 1 °C decrease per cycle), and 72 °C for 30 s elongation, followed by 20 cycles with the same conditions but 58 °C as annealing temperature, followed by a 10 min final elongation step at 68 °C. For the second step, primers matching the universal tail with an i5/i7 index and P5/P7 Illumina adapter attached were used (supplementary table 5, see also [[49], [50]]). The assay was similar to the one of the first step with the exception that only 1 μL of first step PCR product was added to the PCR reaction. The PCR protocol for the second step was 95 °C for 5 min initial denaturation, 15 cycles of 95 °C for 30 s and 72 °C for 120 s, finished with a final elongation of 68 °C for 10 min. PCR success was checked using a 1% agarose gel. For samples that showed a weak band in the gel, the second step was repeated with 20 cycles.
2.4. Normalization, pooling, and library preparation
All samples were normalized to 1.25 ng/μL with the SequalPrep Normalization Plate (Applied Biosystems, Foster City, CA, USA) using the full amount of remaining PCR product. After normalization, all samples of one dataset were pooled and concentrated with a NucleoSpin Gel and PCR Clean-up kit (Macherey Nagel, Düren, Germany). Both libraries were left side size-selected to remove remaining primer dimers with the NucleoMag kit for clean-up and size selection (Macherey Nagel, Düren, Germany) with a ratio of 0.76. Library concentrations were measured using the Fragment Analyzer (High Sensitivity NGS Fragment Analysis Kit; Advanced Analytical, Ankeny, USA). Both concentrating the library and the size selection were performed manually. The validation dataset was sequenced using the MiSeq platform with a paired-end v2 kit (read length 2 × 250 bp) at a commercial service provider (CeGaT, Tübingen, Germany). For the biomonitoring dataset, each replicate plate was sequenced on a separate run using the same sequencing platform.
2.5. Bioinformatics
Raw reads for the three libraries were received from CeGaT as demultiplexed fastq files. The quality of the raw reads was checked using FastQC [51]. Subsequently, samples were renamed using a custom Python script. Paired-end reads were merged using VSEARCH version 2.11.1 [52], allowing for 25% differences between merged pairs and a minimum overlap of 20 bp. Afterwards, primers were trimmed using cutadapt version 2.8 [53]. Reads were then filtered by length (195–215 bp threshold for target fragment) and by maximum expected error (maxee = 1), using VSEARCH. The filtered reads were dereplicated, singletons and chimeras were removed with VSEARCH. All reads were then pooled using a custom Python script and globally dereplicated. Sequences were 97% similarity clustered into operational taxonomic units (OTUs), and the consensus sequences were extracted as representative OTU sequences. The OTUs were remapped (usearch_global function, 97% similarity) to the individual sample files to create the read table. The read table was filtered by column (read abundance threshold: >0.01% of reads per sample to keep the OTU) and then by row (OTU must be present in at least one of the samples), using a custom Python script. Taxonomic assignment of OTUs was conducted using BOLDigger version 1.1.10 [54] and the Barcode of Life data system (BOLD) database [55]. The option “JAMP filter” was applied to extract the final taxonomy table. Downstream processing of both datasets and visualization were conducted using TaxonTableTools (TTT) version 1.3.0 [56]. Initially, the read table and the taxonomy table were converted to the TTT input format for both datasets (supplementary Tables 1 and 2). Prior to downstream analyses, negative controls were removed from the tables (sample-based filter tool). Read-based rarefaction curves were calculated for the biomonitoring dataset (dataset 2) with 10 repetitions and 5% increments (supplementary figure 2). Then, the number of shared OTUs between replicates was calculated and a correlation analysis between replicates (OTU correlation, Spearman correlation coefficient) was performed using the respective tools within TTT. To account for PCR stochasticity, all OTUs that were not present in both extraction replicates were discarded before samples were subsequently merged (supplementary Tables 3 and 4). Prior to the replicate merging step, a read proportions plot was calculated for the validation dataset. Additionally, the taxonomy results of the validation dataset were matched against the Global Biodiversity Information Facility (GBIF) database to detect synonyms and spelling errors (GBIF taxonomy check tool). For both datasets, basic statistics (i.e., number of OTUs and reads), taxonomic richness and taxonomic resolution were calculated. Furthermore, a Krona chart [57] was created for the biomonitoring dataset (supplementary figure 6).
2.6. Additional datasets
The here presented workflow can easily be upscaled to hundreds of samples. Thus, we included results of additional datasets (196 samples in total) produced on the Biomek FXP, using the same workflow as the second dataset. Contrary to the biomonitoring dataset, the samples included in the additional datasets were sequenced on Illumina HiSeq sequencing runs, which produce substantially more reads compared to an Illumina MiSeq platform, which was used for the biomonitoring dataset.
3. Results
3.1. Single specimen validation dataset
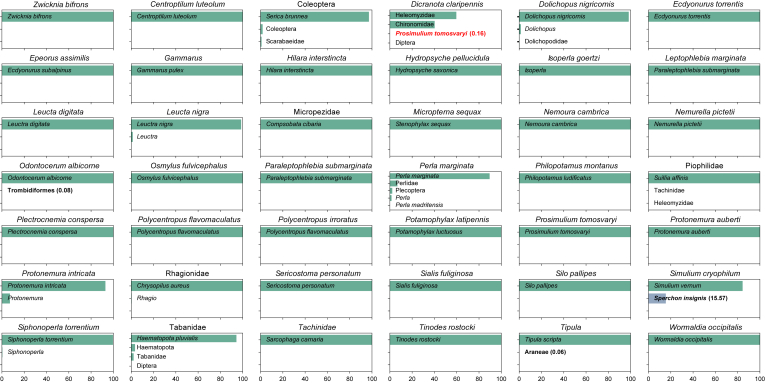
The COI validation dataset (dataset 1) consisted of 11,105,659 raw reads, of which 1664 were assigned to negative controls. After quality filtering, 10,392,660 reads remained (only 3 reads in negative controls). Reads were then pooled, dereplicated, and singletons and chimeras were removed. The 97% similarity clustering of reads resulted in 152 OTUs. After read abundance filtering (0.01% threshold), 97 OTUs remained and all OTUs but one could be assigned taxonomically. Read numbers in negative controls fell below the filtering threshold of 0.01% and were thus discarded. In total, the 96 OTUs were assigned to 6 different higher taxa, i.e., Arthropoda (89 OTUs), Ascomycota (3), Bacillariophyta (1), Basidiomycota (1), Cnidaria (1), and Nematomorpha (1). After the removal of unique OTUs (i.e., excluding OTUs that are not present in both replicates), 76 OTUs were left (99.9% of reads). Overall, 46 OTUs were assigned to species level, covering 40 different species of 2 phyla, i.e., Arthropoda and one OTU assigned to Ascomycota. Since the study focused on invertebrates, the Ascomycota hit was discarded for the downstream analyses. As every of the 42 reaction wells on the pipetting robot contained only one individual of a distinct taxon each (42 in total, based on morphological identification), possible cross-contamination between wells is readily observable in HTS sequencing data. While there were some differences in the morphological and DNA-based identification results (either morphological misidentification or lower taxonomic resolution in morphological identification), 29 cases showed only one taxonomic assignment per used individual (and well) (Fig. 2). In 9 of the remaining 13 cases, multiple taxonomic assignments per specimen and well were observed but are not conflicting (or only hint at misidentified references sequences in the database). For instance, all reads of sample 40 (morphologically identified as Leuctra nigra) were either assigned to (i) Leuctra nigra or (ii) Leuctra (genus only). Most reads of the morphologically-identified pediciid dipteran Dicranota claripennis (sample 38) clustered in two OTUs that had low similarity (∼90%) to reference sequences in BOLD belonging to specimens from several dipteran families (Heleomyzidae, Chironomidae, Limoniidae, Pediciidae etc.). Albeit finally assigned to two different families (Heleomyzidae and Chironomidae), the two OTUs most likely derive from the same species as the distance between OTUs is fairly low (2.44%). As D. claripennis is not present in the BOLD database its morphological ID can neither be confirmed nor rejected. A false morphological identification at family level can be excluded. In three more cases reads of either Trombidiformes (0.08% of the respective reads), Araneae (0.06%), or Sperchon insignis (Trombidiformes, 15.57%) were found additionally to the target taxon. While all three mentioned taxa were not part of the 42 used taxa, only in one case reads (0.16% of the respective sample) assigned to a used taxon (sample 30: Prosimulium tomosvaryi) were detected in a different sample (sample 38: Dicranota claripennis).
Fig. 2.
Relative read proportions and their assigned taxonomy of the validation dataset samples. Hits classified as co-amplification are highlighted in bold. One potential dataset internal cross-contamination is highlighted in bold and red (sample: Dicranota claripennis, hit: Prosimulium tomosvaryi). Relative read proportions are written in parenthesis.
3.2. Biomonitoring dataset
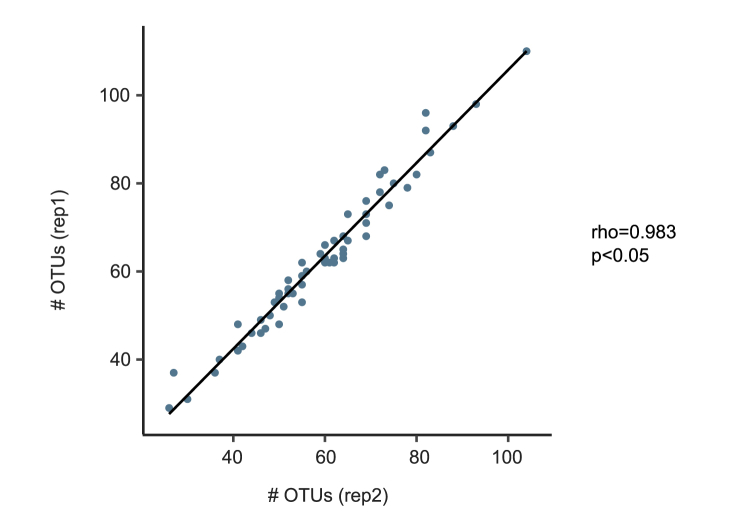
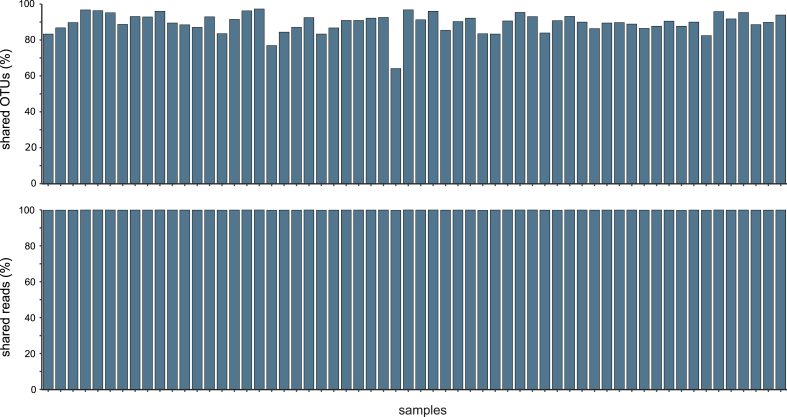
The freshwater invertebrate COI biomonitoring dataset (dataset 2) consisted of 18,478,511 raw reads, of which 29,991 were assigned to negative controls. After bioinformatic processing and before OTU clustering, 16,160,528 reads remained (200 reads in negative controls). Subsequently, reads were pooled, dereplicated, singletons and chimeras were removed, and the remaining reads clustered into 1720 OTUs. After the read abundance filtering (0.01% threshold), 972 OTUs remained, and all but one OTU were compared against the BOLD database. Here, the negative controls contained 13 reads in total, assigned to five arthropod species (Baetis rhodani, Halesus radiatus, Isoperla grammatica, Nemoura flexuosa, Platambus maculatus), but were all discarded after applying the replicate consistency filter. All samples showed a sufficient sequencing depth in the read-based rarefaction analysis (supplementary figure 2). A strong positive correlation was observed for the number of OTUs per replicate (r = 0.983, p <0.05; Fig. 3). All but two samples shared at least 80% of their OTUs between extraction replicates (average of 89.63%; Fig. 4). The non-shared OTUs were exclusively those that accounted for very low, i.e., <0.01% of reads in the respective sample (Fig. 5). The average of reads assigned to OTUs and shared between replicates was 99.93%. After the removal of unique OTUs, 888 OTUs (99.93% of reads) remained and were assigned to 6 phyla and 413 species, with 559 OTUs assigned to species level (i.e., in several cases, more than one OTU was assigned to the same species). Most reads accounted for Arthropoda (99% of all reads), while less than 1% of the reads were assigned to Annelida, Bryozoa, Mollusca, Nematoda and Rotifera. Within the arthropods, the most represented orders were Trichoptera (25% of all reads), Plecoptera (24%), Diptera (21%), and Ephemeroptera (17%).
Fig. 3.
Spearman correlation analysis of OTUs between replicates of the biomonitoring dataset samples.
Fig. 4.
Relative proportions of shared OTUs (average 89.63%) and shared reads (99.93%) between PCR replicates of the biomonitoring dataset.
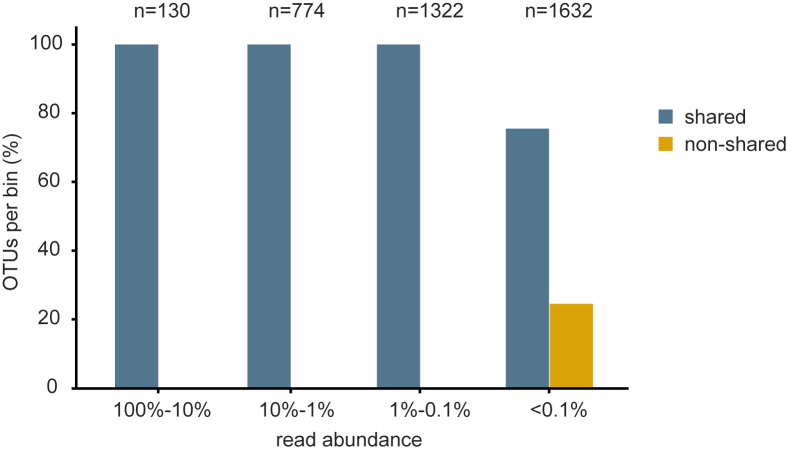
Fig. 5.
Relative proportions of shared and non-shared OTUs per bin (i.e., 100%–10%, 10%–1%, 1%–0.1%, and <0.1%) between replicates of the biomonitoring dataset. The number of total OTUs per bin is given above the bars.
3.3. Additional datasets
In the additional datasets, we observed that with increasing sequencing depth the amount of low abundant OTUs that are not shared among extraction replicates increased, leading to 58.16% of shared OTUs on average. However, the average number of shared reads between replicates remained high with 97.59%.
4. Discussion
Effective quality control mechanisms are needed in workflows of high-throughput genetic analyses to control for potential sources of contamination in laboratory protocols [58]. This is of particular importance when working with single-deck solutions that do not separate pre- and post-PCR steps, which was also the case of this study. Proper negative control sample layouts are efficient to control for quality. Here we propose 12 negative controls per plate, one per row and column of a 96-well plate following the suggestions of [11]. We could show that only a very small fraction of reads (i.e., 13 reads in total for the biomonitoring dataset and 0.00009% of filtered reads) was assigned to the negative controls after bioinformatic processing, in particular as a result of the 0.01% detection threshold. These few remaining reads were only found in one replicate and were thus excluded. Thereby, we could rule out cross-contamination during the lab processing on the robot. We highly recommend using sufficient numbers of negative controls in relation to the number of samples, as discussed above.
Furthermore, another key step for quality assurance was to split replicates into two fully independent plates that are never opened at the same time after splitting. By doing so, and by accepting only OTUs that occur in both independent replicates (possibly rejecting many low-abundant OTUs), a robust and reliable QA/QC mechanism is established. Through physical replicate splitting, it is in addition possible to control for sufficient homogenization. The biomonitoring dataset confirmed this with remarkably high numbers of shared OTUs between the replicates. The extraction replicates shared an average of 89.6% of their OTUs, supporting the reproducibility of the robotic workflow. The 10.4% of the OTUs that were not shared between replicates only accounted for 0.07% of the reads, highlighting that the used high-throughput design delivers robust results except for very rare and small species, which is congruent with results from hundreds of additional samples processed in our lab so far (supplementary figures 3, 4 and 5). However, in complex environmental samples, many small specimens may exist, and skewed rank abundance curves will always leave the rare species to be shared only to a limited degree. Here, adding positive control samples of known taxon composition and differing template concentrations will further allow to control for sufficient sequencing depth in such cases.
To specifically test for potential contamination during the automated workflow, we designed the single specimen validation dataset to detect potential cross-contamination between samples that would not be detected in conventional setups, such as the biomonitoring dataset. Although we used many negative controls in the proposed deck setup (i.e., 12 of 96 wells accounting for negative controls), both systematic errors and errors by chance could slip through the precaution measurements. The design of the validation dataset allows detecting these cross-contaminations that, for example, could occur during sample handling by the robot (i.e., by dripping into other wells, aerosols from the lab or by spillovers during plate shaking). Here, our results of the validation dataset showed that the robot protocol and proposed workflow is not prone to cross-contamination. We were able to declare 41 of 42 single specimen samples contamination-free. In three cases, we found OTUs assigned to Araneae, Trombidiformes and the water mite Sperchon insignis. All these hits are likely to originate from ectoparasites that were present in the sample, i.e., by being attached to their host during the time of sampling. By applying DNA metabarcoding on whole single specimens, it is evident that ectoparasites will be detected alongside their host. Furthermore, none of the included single specimens of the validation dataset were part of the Araneae order, which rules out dataset derived cross-contamination. Thus, only one case of potential cross-contamination was flagged. Here, the species Prosimulium tomosvaryi was detected in two independent samples in both replicates. The first sample was expected to show Prosimulium tomosvaryi, as it was the according morphological identification. The second sample can be regarded as a cross-contamination. Nevertheless, this bias originates most likely not from the robotic workflow. Since all specimens derive from one bulk sample, it is highly likely that during sample collection parts of the Prosimulium tomosvaryi specimen got in contact with the Dicranota claripennis specimen. Particularly during the conservation process in ethanol, specimens are known to get entangled with their extremities, which inevitably become fragile and often break during subsequent sample sorting. Thus, in this particular case, a claw or leg of Prosimulium tomosvaryi could have been transferred along with the Dicranota claripennis specimen, since they were both present in the original bulk sample. Overall, this single and still only potential case of cross-contamination represents only 0.16% of all reads of the sample. Hits with such low abundances would probably not even be detected in a bulk sample analysis like the biomonitoring dataset and thus raise no concern on the robustness of the workflow.
Generally, when programming new protocols for liquid handling workstations, particular attention needs to be paid to the minimum pipetting volume. In this study, a minimum volume of 2 μL for the Span-8 was used, while the 96-well head was able to handle volumes down to 1 μL. For the handling of viscous liquids (e.g., enzymes, tissue dissolved in buffers), wide bore tips should be considered since they hardly ever clog. While amplicon sequencing and metabarcoding works reliably even in 5 μL PCR reaction volumes [44,50], individual applications may use increased volumes due to sample inhibition or minimum pipetting volumes possible with automated workstations.
4.1. Standardization of biomonitoring
The standardization of genetic methods to be included in routine biomonitoring workflows is a central challenge for the upcoming decade. First steps to establish standards in the field of biodiversity have already been taken by forming a new expert committee on biodiversity by the International Organization for Standardization (ISO/TC 331). The presented standards aim to develop terms and definitions to be used globally, methodologies for impact analysis, frameworks for defining strategies, as well as action plans, monitoring and reporting tools and guidelines. Many national ISO members have already declared participation in the ISO/TC 331 committee, such as France (secretariat), Brazil, China, Germany, or Russia. Similar efforts for standardization in the field of aquatic biodiversity have been made by the European Committee for Standardization (CEN) by creating a technical body for DNA and eDNA analysis (TC 230/WG28). This formal working group aims for the standardization of novel environmental DNA and other ecogenomic analyses for aquatic ecosystems. These international standardization efforts in the field of biomonitoring highlight the need for more sophisticated, robust and standardized methodologies, as presented in this study and demand for inter-lab ring test validations [59]. Similar to medical and commercial toxicological labs, we think that the implementation of automated liquid handlers will be a crucial step to produce reliable and comparable data to meet the globally upcoming demand of environmental monitoring programs. The main advantage is that any organismal group can be targeted with metabarcoding, i.e., while we presented solutions for macroinvertebrates, the same protocols can also be applied for other target groups (bacteria, microalgae, fungi etc.; see e.g. [23,60]). Furthermore, through the ongoing optimization of laboratory workflows (both custom and commercial), the costs of DNA-based biomonitoring can even be further reduced in the future and thus increase its applicability in biomonitoring campaigns. Given the high initial cost, often >200k $, and sometimes substantial maintenance costs for several automated robotic platforms, it is obvious that such devices should be installed at central core facilities, biomonitoring centers, and commercial service providers that can perform thousands of analyses. The formal standardization of published automated liquid handler protocols will thus be the next step required to unlock the potential for standardized biodiversity assessments and biomonitoring.
4.2. A zoo of different robots - how does our concept work out?
A variety of different automated liquid handling machines exists, and it is not our aim to promote a particular model. Although for the implementation of automated DNA metabarcoding applications mostly basic liquid handling operations are required, the choice of brand and model can seemingly have extensive consequences. However, the solutions and protocols presented here for a Biomek FXP can be easily adopted to the many other currently available as well as future automated liquid handling robots. The here outlined main protocol steps are based on standard labware (i.e., 96-well plate, 2 mL screw cap tube) and for special cases, blueprint or commercial alternatives are available (i.e., 50 mL Falcon tube holder) or can easily be adapted with alternative solutions. Every automated liquid handling robot model that meets the basic requirement of i) a high pipetting accuracy down to 1 μL, ii) compatibility to the 96 or 384-well plate format, and iii) customizable workflows is suitable for such standardized and robust DNA metabarcoding analyses. Especially open platforms such as the EvoBot [61], an open-source, modular liquid handling robot, highlight the potential of future widespread applications in routine biomonitoring. Such platforms are directly compatible with the here presented workflow and simply require some fine tuning and adaptations. To ensure accurate and reliable metabarcoding data for biomonitoring, the application of a validation dataset (similar to dataset 1, or even just using synthetically designed template DNA), checking for cross-contamination via many negative controls, and consistency checks between two or more independent replicates will ensure high-quality analysis of the workflow design irrespective of the robot model used.
5. Conclusion
We here demonstrated that straightforward automated liquid handling technology can be used reliably in a simple and standardized manner from DNA extraction to final library preparation. By this, costs and throughput for biodiversity assessments and biomonitoring can be standardized and upscaled substantially. In analogy to the medical sector, access to protocols as well as formal standardization are the next steps required to unlock the full technology's potential for environmental monitoring.
Author contributions
D.B., T.M. and A.B. conceived and designed the study. D.B. and T.M. wrote the pipetting robot protocols. A.B. and F.L. provided input to the pipetting robot workflow and supervised the project. D.B. and M.W. prepared the samples and processed the libraries. T.M. and D.B. processed and analyzed the sequencing data. T.M., D.B., A.B., M.W. and F.L. wrote the paper. All authors read and approved the final manuscript.
Data accessibility
All data are accessible via ENA accession number PRJEB45792.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study is a part of the GeDNA project funded by the German Environment Agency (FKZ 3719 24 2040). All members are part of COST Action DNAqua-Net (CA15219). D..B is supported by a grant of the German Research Foundation (DFG; LE 2323/9-1). We thank Kristin Stolberg (LfU) for discussions and permission to use the metabarcoding data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2021.100122.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- 1.Pimm S.L., Jenkins C.N., Abell R., Brooks T.M., Gittleman J.L., Joppa L.N., Raven P.H., Roberts C.M., Sexton J.O. The biodiversity of species and their rates of extinction, distribution, and protection. Science. 2014;344(6187) doi: 10.1126/science.1246752. [DOI] [PubMed] [Google Scholar]
- 2.IPBES . Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services. IPBES secretariat; Bonn, Germany: 2019. [Google Scholar]
- 3.Almond R.E.A., Grooten M., Petersen T. Living Planet Report 2020—Bending the Curve of Biodiversity Loss. WWF; Gland, Switzerland: 2020. [Google Scholar]
- 4.UN, General Assembly . UN; 2015. Transforming Our World: the 2030 Agenda for Sustainable Development. [Google Scholar]
- 5.Kelly R., Fleming A., Pecl G.T., Julia von Gönner, Bonn Aletta. Citizen science and marine conservation: a global review. Phil. Trans. Biol. Sci. 2020;375(1814):20190461. doi: 10.1098/rstb.2019.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klink R., Bowler D.E., Gongalsky K.B., Swengel A.B., Gentile Alessandro, Chase Jonathan M. Meta-analysis reveals declines in terrestrial but increases in freshwater insect abundances. Science. 2020;368(6489):417–420. doi: 10.1126/science.aax9931. [DOI] [PubMed] [Google Scholar]
- 7.Jähnig S.C., Baranov V., Altermatt F., Peter C., Friedrichs-Manthey M., Geist J., He F., et al. Revisiting global trends in freshwater insect biodiversity. WIREs Water. 2021;8(2) doi: 10.1002/wat2.1506. [DOI] [Google Scholar]
- 8.Deiner K., Bik H.M., Mächler E., Seymour M., Lacoursière-Roussel A., Altermatt F., Simon C., et al. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 2017;26(21):5872–5895. doi: 10.1111/mec.14350. [DOI] [PubMed] [Google Scholar]
- 9.Hobern D. “BIOSCAN: DNA barcoding to accelerate taxonomy and biogeography for conservation and Sustainability1.”. Genome, April. 2020 doi: 10.1139/gen-2020-0009. [DOI] [PubMed] [Google Scholar]
- 10.Pawlowski J., Kelly-Quinn M., Altermatt F., Apothéloz-Perret-Gentil L., Pedro B., Boggero A., Borja A., et al. The future of biotic indices in the ecogenomic era: integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018;637–638(October):1295–1310. doi: 10.1016/j.scitotenv.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Elbrecht V., Steinke D. Scaling up DNA metabarcoding for freshwater macrozoobenthos monitoring. Freshw. Biol. 2019;64(2):380–387. doi: 10.1111/fwb.13220. [DOI] [Google Scholar]
- 12.Taberlet P., Coissac E., Pompanon F., Brochmann C., Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012;21(8):2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x. [DOI] [PubMed] [Google Scholar]
- 13.Aylagas E., Borja Á., Muxika I., Rodríguez-Ezpeleta N. Adapting metabarcoding-based benthic biomonitoring into routine marine ecological status assessment networks. Ecol. Indicat. 2018;95(December):194–202. doi: 10.1016/j.ecolind.2018.07.044. [DOI] [Google Scholar]
- 14.Zaiko A., Pochon X., Garcia-Vazquez E., Olenin S., S. A., Wood Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: management and surveillance of non-indigenous species. Frontiers in Marine Science. 2018;5:322. [Google Scholar]
- 15.Elbrecht V., Peinert B., Leese F. Sorting things out: assessing effects of unequal specimen biomass on DNA metabarcoding. Ecology and Evolution. 2017;7(17):6918–6926. doi: 10.1002/ece3.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bush A., M. W.A., Zacchaeus G., Compson D.L., Peters T.M., Porter, Shokralla S., Michael T., Wright G., Hajibabaei M., Donald J., Baird DNA metabarcoding reveals metacommunity dynamics in a threatened boreal wetland wilderness. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117(15):8539–8545. doi: 10.1073/pnas.1918741117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beng K.C., Tomlinson K.W., Shen X.H., Surget-Groba Y., Hughes A.C., Corlett R.T., Ferry Slik J.W. The utility of DNA metabarcoding for studying the response of arthropod diversity and composition to land-use change in the tropics. Sci. Rep. 2016;6(1):24965. doi: 10.1038/srep24965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter T.M., Morris D.M., Basiliko N., Hajibabaei M., Doucet D., Bowman S., Erik J., Emilson S., et al. Variations in terrestrial arthropod DNA metabarcoding methods recovers robust beta diversity but variable richness and site indicators. Sci. Rep. 2019;9(1):18218. doi: 10.1038/s41598-019-54532-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leese F., Bouchez A., Abarenkov K., Altermatt F., Borja Á., Bruce K., Ekrem T., et al. In: Bohan David A., Dumbrell Alex J., Woodward Guy, Jackson Michelle., editors. vol. 58. Next Generation Biomonitoring: Part 1. Academic Press; 2018. Why we need sustainable networks bridging countries, disciplines, cultures and generations for aquatic biomonitoring 2.0: a perspective derived from the DNAqua-Net COST action; pp. 63–99. (Advances in Ecological Research). [Google Scholar]
- 20.Piper A.M., Batovska J., Cogan N.O.I., Weiss J., Cunningham J.P., Rodoni B.C., Blacket M.J. Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience. 2019;8(8) doi: 10.1093/gigascience/giz092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M., Laurence J., Clarke, Susan C.B., Jordan G.J., Burridge C.P. A practical guide to DNA metabarcoding for entomological ecologists. Ecol. Entomol. 2020;45(3):373–385. doi: 10.1111/een.12831. [DOI] [Google Scholar]
- 22.Leray M., J. Y., Yang, Christopher, Meyer P., Suzanne C., Mills, Agudelo Natalia, Vincent Ranwez, Boehm Joel T., Machida Ryuji J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 2013;10(1):34. doi: 10.1186/1742-9994-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macher J., Vivancos A., Piggott J.J., Centeno F.C., Matthaei C.D., Leese F. Comparison of environmental DNA and bulk-sample metabarcoding using highly degenerate cytochrome c oxidase I primers. Molecular Ecology Resources. 2018;18(6):1456–1468. doi: 10.1111/1755-0998.12940. [DOI] [PubMed] [Google Scholar]
- 24.Miya M., Sato Y., Fukunaga T., Sado T., Poulsen J.Y., Sato K., Minamoto T., et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. Royal Society Open Science. 2015;2(7):150088. doi: 10.1098/rsos.150088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänfling B., Handley L.L., Daniel S., Read, Christoph H., Jianlong L., Paul N., Rosetta C., Blackman, Anna O., Winfield I.J. Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 2016;25(13):3101–3119. doi: 10.1111/mec.13660. [DOI] [PubMed] [Google Scholar]
- 26.Hollingsworth P.M. vol. 108. 2011. Refining the DNA Barcode for land plants; pp. 19451–19452. (Proceedings of the National Academy of Sciences). 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.China Plant BOL Group. Li D.-Z., Gao L.-M., Li H.-T., Wang H., Ge X.-J., Liu J.-Q., et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should Be incorporated into the core Barcode for seed plants. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108(49):19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zizka V.M.A., Vasco E., Jan-Niklas M., Florian L. Assessing the influence of sample tagging and library preparation on DNA metabarcoding. Molecular Ecology Resources. 2019;19(4):893–899. doi: 10.1111/1755-0998.13018. [DOI] [PubMed] [Google Scholar]
- 29.European Centre for Disease Prevention and Control . vol. 6. December, 2020. (Sequencing of SARS-CoV-2). TECHNICAL REPORT. [Google Scholar]
- 30.Wang Y., Kang H., Liu X., Tong Z. Combination of RT-QPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J. Med. Virol. 2020;92(6):538–539. doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An P., Winters D., Walker K.W. Automated high-throughput dense matrix protein folding screen using a liquid handling robot combined with microfluidic capillary electrophoresis. Protein Expr. Purif. 2016;120(April):138–147. doi: 10.1016/j.pep.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann R., Severitt J.C., Roddelkopf T., Junginger S., Thurow K. Biomek cell workstation: a variable system for automated cell cultivation. J. Lab. Autom. 2016;21(3):439–450. doi: 10.1177/2211068215599786. [DOI] [PubMed] [Google Scholar]
- 33.Seipp M.T., Herrmann M., Wittwer C.T. Automated DNA extraction, quantification, dilution, and PCR preparation for genotyping by high-resolution melting. J. Biomol. Tech.: J. Biochem. (Tokyo) 2010;21(4):163–166. [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkening S., Tekkedil M.M., Lin G., Fritsch E.S., Wu W., Gagneur J., Lazinski D.W., Camilli A., Steinmetz L.M. Genotyping 1000 yeast strains by next-generation sequencing. BMC Genom. 2013;14(1):90. doi: 10.1186/1471-2164-14-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu M., Zhang P., Geng-Spyropoulos M., Moaddel R., Semba R.D., Ferrucci L. A robotic protocol for high-throughput processing of samples for selected reaction monitoring assays. Proteomics. 2017;17(6):1600339. doi: 10.1002/pmic.201600339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grubb J.C., Katie M., Horsman-Hall, Karen L., Sykes V., Rebecca A., Schlisserman, Covert Vanessa M., Han Na, Rhee, Ban Jeffrey D., Greenspoon Susan A. Implementation and validation of the teleshake unit for DNA IQTM robotic extraction and development of a large volume DNA IQTM method. J. Forensic Sci. 2010;55(3):706–714. doi: 10.1111/j.1556-4029.2010.01345.x. [DOI] [PubMed] [Google Scholar]
- 37.Stangegaard M., Hansen T.M., Hansen A.J., Morling N. “Automated addition of chelex solution to tubes containing trace items.” forensic science international: genetics supplement series. Progress in Forensic Genetics. 2011;14(1):e163–e164. doi: 10.1016/j.fsigss.2011.08.082. 3. [DOI] [Google Scholar]
- 38.Tegally H., James Emmanuel San, Giandhari J., de Oliveira T. Unlocking the efficiency of genomics laboratories with robotic liquid-handling. BMC Genom. 2020;21(1):729. doi: 10.1186/s12864-020-07137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nassar A.F., Wisnewski Adam V., Raddassi Khadir. Automation of sample preparation for mass cytometry barcoding in support of clinical Research: protocol optimization. Anal. Bioanal. Chem. 2017;409(9):2363–2372. doi: 10.1007/s00216-017-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brüggemann M., Kotrová M., Knecht H., Bartram J., Boudjogrha M., Bystry V., Fazio G., et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia. 2019;33(9):2241–2253. doi: 10.1038/s41375-019-0496-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexovič M., Urban P.L., Tabani H., Sabo J. Recent advances in robotic protein sample preparation for clinical analysis and other biomedical applications. Clin. Chim. Acta. 2020;507(August):104–116. doi: 10.1016/j.cca.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Messner C.B., Vadim D., Daniel W., Laura M., Matthew W., Anja F., Textoris-Taube K., et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Systems. 2020;11(1):11–24. doi: 10.1016/j.cels.2020.05.012. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comeau A.M., Douglas G.M., Langille M.G.I. Microbiome helper: a custom and streamlined workflow for microbiome Research. mSystems. 2017;2(1) doi: 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minich J.J., Humphrey G., Rodolfo A.S., Jon Sanders B., Austin S., Allen E.E., Knight R. High-throughput miniaturized 16S RRNA amplicon library preparation reduces costs while preserving microbiome integrity. mSystems. 2018;3(6) doi: 10.1128/mSystems.00166-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordier T., Alonso-Sáez L., Apothéloz-Perret-Gentil L., Aylagas E., Bohan D.A., Bouchez A., Anthony C., et al. Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap. Mol. Ecol. 2021;3(13):293–295. doi: 10.1111/mec.15472. n/a (n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baird D.J., Hajibabaei M. Biomonitoring 2.0: a new paradigm in ecosystem Assessment made possible by next-generation DNA sequencing. Mol. Ecol. 2012;21(8):2039–2044. doi: 10.1111/j.1365-294X.2012.05519.x. [DOI] [PubMed] [Google Scholar]
- 47.Vamos E., Elbrecht V., Leese F. Short COI markers for freshwater macroinvertebrate metabarcoding. Metabarcoding and Metagenomics. 2017;1(September) doi: 10.3897/mbmg.1.14625. [DOI] [Google Scholar]
- 48.Leese F., Sander M., Buchner D., Elbrecht V., Haase P., Vera M., Zizka A. Improved freshwater macroinvertebrate detection from environmental DNA through minimized nontarget amplification. Environmental DNA. 2021;3(1):261–276. doi: 10.1002/edn3.177. [DOI] [Google Scholar]
- 49.Buchner D., Beermann A.J., Leese F., Weiss M. Cooking small and large portions of ‘biodiversity-soup’: miniaturized DNA metabarcoding PCRs perform as good as large-volume PCRs. Ecology and Evolution. 2021;11(13):9092–9099. doi: 10.1002/ece3.7753. [DOI] [Google Scholar]
- 50.Buchner D., Haase P., Leese F. Wet grinding of invertebrate bulk samples – a scalable and cost-efficient protocol for metabarcoding and metagenomics. Metabarcoding Metagenomics. 2021;5 doi: 10.3897/mbmg.5.67533. [DOI] [Google Scholar]
- 51.Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 52.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 54.Buchner D., Leese F. “BOLDigger – a Python package to identify and organise sequences with the Barcode of life data systems. Metabarcoding and Metagenomics. 2020;4(June) doi: 10.3897/mbmg.4.53535. [DOI] [Google Scholar]
- 55.Ratnasingham S., Hebert P.D.N. BOLD: the Barcode of life data system (Www.Barcodinglife.Org) Mol. Ecol. Notes. 2007;7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macher T.H., Beermann A.J., Leese F. TaxonTableTools - a comprehensive, platform-independent graphical user interface software to explore and visualise DNA metabarcoding data. Molecular Ecology Resources n/a (n/a) 2021 doi: 10.1111/1755-0998.13358. [DOI] [PubMed] [Google Scholar]
- 57.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a web browser. BMC Bioinf. 2011;12(1):385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sepulveda A.J., Hutchins Patrick R., Forstchen M., Mckeefry M.N., Swigris A.M. The elephant in the lab (and field): contamination in aquatic environmental DNA studies. Frontiers in Ecology and Evolution. 2020;8 doi: 10.3389/fevo.2020.609973. [DOI] [Google Scholar]
- 59.Blackman R.C., Elvira M., Florian A., Amanda A., Pedro B., Pieter B., Bastian E., et al. “Advancing the use of molecular methods for routine freshwater macroinvertebrate biomonitoring – the need for calibration experiments. Metabarcoding and Metagenomics. 2019;3(November) doi: 10.3897/mbmg.3.34735. [DOI] [Google Scholar]
- 60.Li F., Peng Y., Fang W., Altermatt F., Xie Y., Yang J., Zhang X. Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers. Environ. Sci. Technol. 2018;52(20):11708–11719. doi: 10.1021/acs.est.8b03869. [DOI] [PubMed] [Google Scholar]
- 61.Faiña A., Nejati B., Kasper S. EvoBot: an open-source, modular, liquid handling robot for scientific experiments. Appl. Sci. 2020;10(3):814. doi: 10.3390/app10030814. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are accessible via ENA accession number PRJEB45792.