Abstract
Microplastics are a new type of environmental pollutant, and pose a serious threat to soil ecosystems. It is important to study microplastics effects on soil microorganisms to better understand their effects on terrestrial ecosystems. Therefore, we collected soil and microplastic samples from corn, pepper, peanut and cucumber fields in Shunyi District, Beijing, China, and used Illumina MiSeq high-throughput sequencing technology to analyze bacterial and fungal community composition and diversity. We focused on microplastic surface and its surrounding “rhizosphere-like” soil in the 0–10 cm (humus) and 10–20 cm (eluvial) deep horizons. Microbial richness and diversity on microplastic surface were significantly lower than those in surrounding “rhizosphere-like” soil, and microbial richness and diversity were reduced to a greater extent in the humus horizon than in the eluvial horizon. Microplastics likely enriched the microbes involved in their biodegradation. The relative abundance levels of Cyanobacteria and Basidiomycota on microplastic surfaces were significantly higher than those in surrounding “rhizosphere-like” soil, while the relative abundance levels of Acidobacteria, Chloreflexi, and Mortierellomycota were higher in “rhizosphere-like” soil. Furthermore, the relative abundance levels of pathways related to human diseases, animal pathogen, and fungal parasites were significantly higher on microplastic surfaces than in “rhizosphere-like” soil. These results show that the microbial diversity, richness, community structure and function between microplastic surfaces and surrounding “rhizosphere-like” soil are significantly different, leading to a “rhizosphere-like neighbor avoidance effect” between microplastic surfaces and the surrounding soil.
Keywords: Microplastics, Microbial diversity, Community structure, Rhizosphere, Metabolic function
Highlights
-
•
Microbial diversities were lower on microplastics than in adjacent soil.
-
•
Effect of microplastics on microbial diversity varied across different soil layers.
-
•
Microbial metabolic functions were different on microplastics and adjacent soil.
-
•
Potential plastic-degrading microbes were enriched on microplastics.
1. Introduction
Microplastics (<5 mm) are a new type of pollutant that have been raising concerns worldwide in recent years [1]. Human activities (such as using plastics for agricultural mulching, the application of soil amendments, sewage irrigation, littering.) and environmental transmission (such as flooding and atmospheric deposition) [[2], [3], [4]] have made the soil the largest storage reservoir for microplastics; their levels in soil may be 4–23 times that in the ocean [4]. China is the largest producer and consumer of plastics in the world. Its consumption of plastic film increased from 1.259 million tons in 1999 to 2.465 million tons in 2018, accounting for more than 90% of the global plastic film consumption [5]. The recycling rate of plastic film in China is less than 60%, resulting in a higher level of soil microplastic pollution in China than in other countries [6]. Microplastics have been detected in soils in places such as Zhejiang, Heilongjiang, Xinjiang, Guangxi, Shaanxi, and Hubei Province in China [[7], [8], [9], [10], [11]]. The contents of microplastics in woodland, vegetable land, and vacant land in the central suburbs of China vary from 300 to 67,500 items kg−1 [12]. The abundance of microplastics in soil with mulched crops is more than twice that of non-mulched cropped soil [7], and the amount of plastic film residues gradually increases over time, reaching a level of 502 kg hm−2 in Xinjiang, China [8].
After entering the soil, microplastics can increase soil porosity and water holding capacity while reducing soil bulk density and permeability [[13], [14], [15]], resulting in the loss of soil structural integrity and stability [16,17]. Microplastics can also change soil pH, electrical conductivity, and organic matter and nutrient contents [[18], [19], [20], [21]], thus affecting soil microbial diversity and community composition [13,14,22]. Furthermore, microplastic-induced alterations in soil physicochemical properties can affect plant physiological status, root traits, and nutrient availability, all of which indirectly affect soil microbial activities [13,14,22]. Studies have shown that microplastics can reduce the abundance of soil microorganisms, as well as affect the diversity and richness of the microbial community [1,23,24], which may also undergo an increase in its turnover rate [25]. Miao et al. [26] found that the microbial community indices for alpha diversity (richness, evenness, and diversity) are lower on microplastics than on natural substrates. Polyethylene microplastics select for Actinobacteria over Proteobacteria as the dominant phylum [27,28], and 5% (w/w) polyvinyl chloride and 1% (w/w) polyethylene microplastics significantly reduce the relative abundance levels of Sphingomonadaceae and Xanthobacteraceae, while increasing relative abundance levels of Burkholderiaceaee [22]. Soil microorganisms not only cycle soil materials, they also impact the structure and function of soil [29]. The abundance, distribution, and activity of microbial communities in soil reflect the level of soil fertility. Therefore, studying the effects of microplastics on soil microorganisms will provide important insights into the impact of microplastics on soil ecosystems.
The rhizosphere is one of the most important microbial hotspots determining the processes, dynamics and cycling of nutrients and water in terrestrial ecosystems [30]. Similar to plant-soil interactions in rhizosphere, the main processes affected by microplastic input may occur at the soil-microplastic interface (here defined as “rhizosphere-like”). Microplastics can provide new substrates for microorganisms, and microorganisms can live on microplastic surfaces for a long time and form biofilms, leading to the formation of microbial hotspots on microplastic surfaces [23]. Thus, microplastics provide a unique microenvironment of “rhizosphere-like” soil whose microbial diversity, community composition and function may differ greatly from those on the microplastics. The microbial change between microplastic surfaces and the surrounding “rhizosphere” soil is significant. This result is a “rhizosphere-like neighbor avoidance effect” between the microplastic surface and soil surrounding microplastics. However, this hasn't been reported yet. We aimed to investigate “rhizosphere-like neighbor avoidance effect” and its mechanisms to better understand the impact of microplastics on material cycling in the soil ecosystem.
In this study, microplastics and soil samples were collected from four typical farmlands (corn, pepper, peanut, and cucumber fields) in Shunyi District, Beijing, China. High-throughput sequencing technology was used to analyze the diversity and composition of microbial communities on microplastic surfaces and in surrounding “rhizosphere-like” soil. The main objectives of this study were to determine (1) the effects of microplastics on soil microbial community diversity and composition in humus and eluvial horizons, (2) the effects of microplastics on soil microbial functions in humus and eluvial horizons.
2. Materials and methods
2.1. Study site
Samples were collected in December 2019 from Shunyi District, Beijing, China. This region has a warm temperate semi-humid monsoon climate, with a mean annual temperature of 11.5 °C, mean annual relative humidity of 50%, and mean annual precipitation of 622 mm. We selected four typical farmlands, namely, corn, pepper, peanut, and cucumber fields, to collect soil and microplastic samples. The soil types of the four farmlands were fluvo-aquic. The location and soil properties of the study sites are shown in Table 1. These fields have practiced plastic mulching for >5 years. These sampled areas have no known direct sources of microplastics except for plastic mulching. Therefore, we speculate that the main source of microplastics in these fields is plastic mulching, and microplastic pollution has lasted for over 5 years.
Table 1.
Location of sampling sites and soil properties.
| Fields | Location | Soil properties |
|||||
|---|---|---|---|---|---|---|---|
| Soil organic matter /(g kg−1) |
Available phosphorus /(mg kg−1) |
Total nitrogen /(g kg−1) |
pH | Cation exchange capacity /(cmol kg−1) |
|||
| Corn | 40°11′27″N 116°35′36″E |
Humus horizon | 11.5 | 10.0 | 1.0 | 7.89 | 13.4 |
| Eluvial horizon | 10.2 | 20.0 | 1.1 | 7.76 | 11.5 | ||
| Pepper | 40°11′22″N 116°35′23″E |
Humus horizon | 12.0 | 10.3 | 1.1 | 7.96 | 10.4 |
| Eluvial horizon | 12.0 | 11.0 | 1.1 | 7.86 | 13.2 | ||
| Peanut | 40°11′24″N 116°34′33″E |
Humus horizon | 11.7 | 15.3 | 1.1 | 7.06 | 12.5 |
| Eluvial horizon | 11.3 | 12.9 | 1.0 | 7.82 | 13.5 | ||
| Cucumber | 40°11′1″N 116°35′37″E |
Humus horizon | 13.4 | 9.2 | 1.1 | 7.86 | 12.8 |
| Eluvial horizon | 12.6 | 14.8 | 1.1 | 7.84 | 13.1 | ||
2.2. Soil samples collection
Three sampling plots were established randomly in each of the fields, and five samples (10 × 10 cm) were collected randomly using an auger from each of the sampling plots. In this study, the sampling depth was determined to be 20 cm based on the tillage depth and the distribution of microplastics [2]. Each sampling plot consisted of five soil cores with a diameter of 5 cm and depths of 0–10 cm (humus horizon) and 10–20 cm (eluvial horizon). We thus collected a total of 30 soil samples from each field. After removing all plant roots and stones, soil samples of the same depth were pooled together to generate a soil sample representing the field. The samples were sealed in a sterilized sampling bag, then placed on ice for transport to the laboratory. Then, samples for microbial community analysis were placed on ice, and microplastics that were visible to the naked eye or microscope were directly picked up using sterilized forceps. Then soil attached to microplastics (0–2 mm from the surface) was collected by brushing. This soil was designated the “rhizosphere-like” soil. Microplastic samples were placed inside a sterile 5-mL centrifuge tube, and all microplastic samples and “rhizosphere-like” soil samples were stored at −80 °C until DNA extractions were performed within one week. The remaining soil samples were stored at 4 °C for analysis of soil physical and chemical properties.
2.3. Analyses of microbial community structure and diversity
Microbial communities were analyzed by Illumina high-throughput sequencing. Total genomic DNA was extracted from microplastic and soil samples using the FastDNA SPIN kits (MP Biomedicals, CA, USA) according to the manufacturer's instructions. The extracted DNA was evaluated by 1% agarose gel electrophoresis, and the quality and concentration of the extracts were determined with a spectrophotometer (NanoDrop, ND2000, Thermo Scientific, Wilmington, DE, USA). The 16S rRNA gene in bacteria was amplified using forward and reverse primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), respectively. The ITS1 gene in fungi was amplified using forward and reverse primers ITS5F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS1R (5′-GCTGCGTTCTTCATCGATGC-3′), respectively. The PCRs were performed using the following protocol: denaturation at 95 °C for 3 min, followed by 27 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, then a final extension at 72 °C for 10 min. The PCR products were purified using 2% agarose gel and the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA). Purified amplicons were sequenced in a paired end format using the Illumina MiSeq platform by Majorbio BioPharm Technology Co. Ltd (Shanghai, China).
2.4. Bioinformatic analysis
Raw sequences yielded from Illumina sequencing were processed using Quantitative Insights Into Microbial Ecology (QIIME, version 1.9.1) [31]. Briefly, paired reads were assembled and demultiplexed, and any sequences with a quality score < 20 or with truncated reads shorter than 50 bp were removed from the data set. Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using Uparse 7.0, and chimeric sequences were identified and removed using Uchime [32]. The obtained OTUs were aligned to the SILVA and UNITE reference databases to determine their taxonomic classification level (Threshold: 0.8 ∼ 1) (kingdom, phylum, class, order, family, genus, species); the non-bacterial and non-fungal reads were further removed [33]. OTU abundance information was normalized via a standard of the sample with the least sequences number. We used the output normalized data for subsequent analyses of alpha diversity and community structure of microorganisms. The functional contents of the microbiota were predicted by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) for 16S rRNA in bacteria, and FunGuild for ITS1 in fungi.
2.5. Statistical analysis
All statistical analyses were carried out in the program R software (Ver. 3.3.1). The alpha diversity, including the OTUs, Simpson, Shannon, Sobs, Chao1 and Ace indices of microbial communities were calculated with QIIME. The alpha diversity was significant differences were calculated using one-way analysis of variance with Tukey's honest significant difference test (p < 0.05). Linear discriminant analysis (LDA) coupled with effect size measurements (LEfSe) analysis was conducted to search for different species of microbial communities between soil and microplastic samples [[21], [34]]. In this study, LDA scores >4 was considered to be different species. Venn diagram was created using R software (Ver. 3.3.1), the other figures were created using Origin 2016 (OriginLab, USA).
3. Results and discussion
3.1. Effects of microplastics on microbial diversity
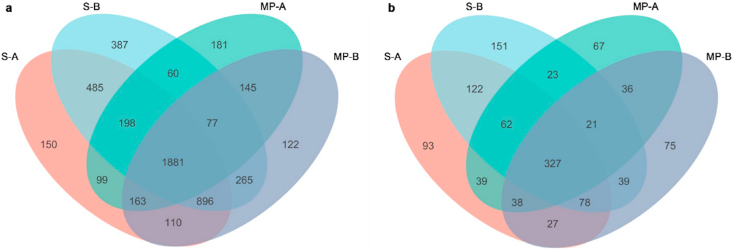
Bacterial communities of soil and microplastic samples from four typical fields consisted of 33 phyla, 93 classes, 229 orders, 396 families, 754 genera, and 1574 species; and fungal communities consisted of 11 phyla, 76 classes, 175 orders, 325 families, 504 genera, and 1198 species. The similarities and overlaps between operational taxonomic units (OTUs) in soil and microplastic samples in different soil layers were compared with a Venn diagram (Fig. 1). The numbers of bacterial OTUs in soil and microplastic samples in the humus horizon were 3982 and 2804, respectively, and those in the eluvial horizon were 4249 and 3659, respectively. The numbers of fungal OTUs in soil and microplastic samples in the humus horizon were 786 and 613, respectively, and those in the eluvial horizon were 823 and 641, respectively. Therefore, there were fewer bacterial and fungal OTUs on the microplastic surface in both humus and eluvial horizons than in surrounding “rhizosphere-like” soil; and there were more soil and microplastic OTUs in the eluvial horizon than in the humus horizon. Overall, microplastics were associated with lower numbers of bacterial and fungal OTUs compared to those in soil, which indicated that microplastics altered the diversity of soil microbial communities. In addition, the OTU ratios of bacteria and fungi in the “rhizosphere-like” soil (mean=5.12) were higher than that on microplastic surfaces (mean=4.57).
Fig. 1.
Venn diagram of bacterial (a) and fungal (b) OTU distributions in soil surrounding microplastics and on microplastic surfaces at different soil layers. S-A and MP-A represent, respectively, soil and microplastic samples in the humus horizon; S-B and MP-B represent, respectively, soil and microplastic samples in eluvial horizon.
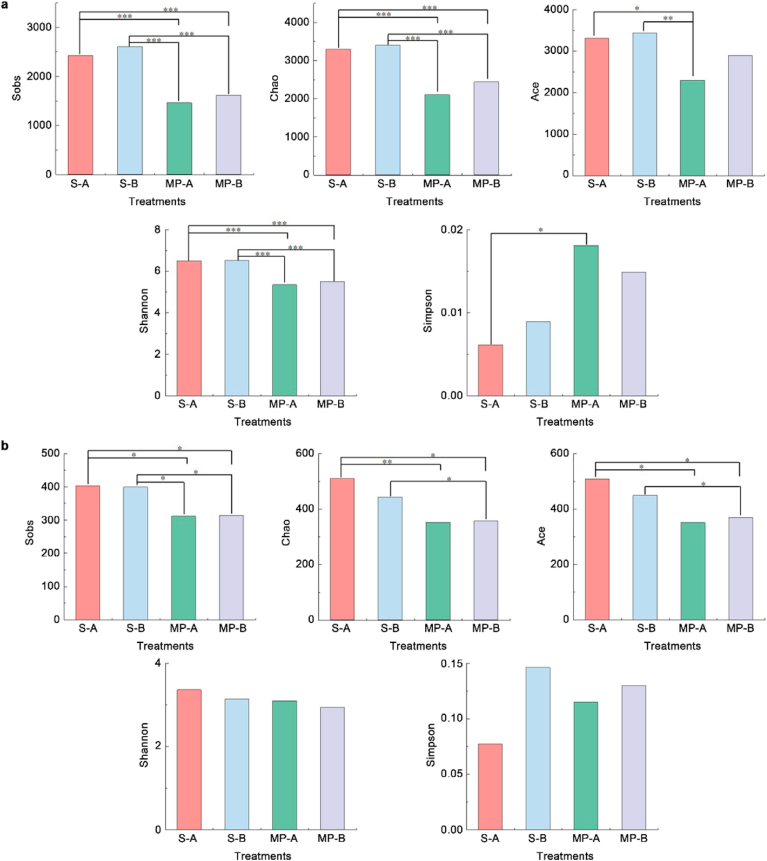
Sobs, Chao1, and Ace indices were used to evaluate microbial richness, while the Shannon and Simpson indices were used to evaluate microbial diversity. The larger Sobs, Chao1, and Ace indices are, the higher the microbial richness will be. The larger the Shannon index is, the higher the microbial diversity will be. The larger the Simpson index is, the lower the community richness will be [27,35]. The levels of coverage of bacterial and fungal diversities based on high-throughput sequencing reached over 0.96, indicating sufficient depth of sample sequencing. Fig. 2a shows that the bacterial Sobs, Chao1, Ace, and Shannon indices on the microplastic surface in the humus horizon were reduced by 39.7%, 36.4%, 30.5%, and 17.9%, respectively, compared to those in “rhizosphere-like” soil; and these indices were similarly reduced by 38.0%, 28.4%, 16.0%, and 15.6%, respectively, in the eluvial horizon. Simpson indexes of the bacterial communities on microplastic surfaces in humus and eluvial horizons were higher by 195.9% and 66.7%, respectively, compared to those of the “rhizosphere-like” soil. These results indicated that microbial diversity and richness on microplastic surfaces were lower than those in “rhizosphere-like” soil. The overall diversity and richness of bacteria in different soil horizons decreased in the following order: soil in the eluvial horizon > soil in the humus horizon > microplastics in the eluvial horizon > microplastics in the humus horizon.
Fig. 2.
Histogram of difference between the alpha diversity indices of groups in soil surrounding microplastics and on microplastic surfaces at different soil layers. (a)bacteria, (b)fungi. S-A and MP-A represent soil and microplastic samples, respectively, in the humus horizon; S-B and MP-B represent soil and microplastic samples, respectively, in the eluvial horizon. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
Fig. 2b shows that the fungal Sobs, Chao1, and Ace indices on microplastic surfaces in the humus horizon were lower by 22.7%, 30.9%, and 31.1%, respectively, compared to those in the “rhizosphere-like” soil; and these indices were lower by 21.3%, 19.2%, and 17.8%, respectively, in the eluvial horizon. These results indicated that fungal diversity and richness were lower on microplastic surfaces than in “rhizosphere-like” soil. The Shannon and Simpson indices for fungi in microplastics and soil samples did not differ significantly. Overall, the indices for fungal richness in different soil horizons decreased in the following order: soil in the eluvial horizon > soil in the humus horizon > microplastics in the eluvial horizon > microplastics in the humus horizon. These results show that fungal richness was lower on microplastic surfaces than in “rhizosphere-like” soil, and there was no significant difference in fungal diversity between microplastic surfaces and the surrounding soil.
Microbial diversity and richness were lower on microplastic surfaces than in surrounding “rhizosphere-like” soil, indicating that microplastics reduce the levels of microbial diversity and richness in soil. These results are consistent with those of Miao et al. [26], who observed that the richness, evenness, and diversity of microbial communities on microplastics are significantly lower than those on natural substrates. This may be because microplastics can compete with soil microorganisms for niches in which microorganisms can grow and move [36], thus resulting in a decrease in microbial activity, which in turn reduced microbial diversity and richness [21]. Moreover, microbial diversity and richness were reduced to greater degrees in the humus horizon, which may be caused by environmental factors (such as temperature, humidity, and ultraviolet rays) that affect the mechanism of microplastics.
3.2. Effects of microplastics on microbial community composition
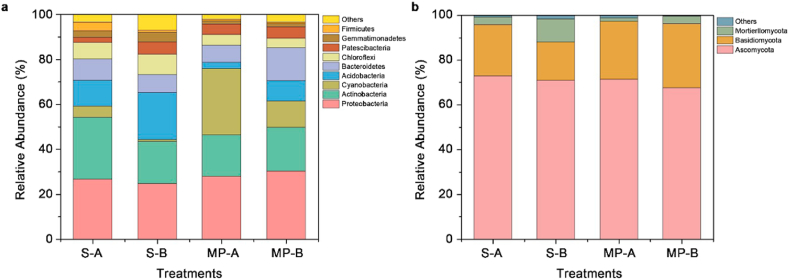
Fig. 3a shows the bacterial community composition of each sample at the phylum level. The dominant bacterial phyla were Proteobacteria (24.7%–30.3% in relative abundance, mean=27.4%), Actinobacteria (18.5%–27.4%, mean=21.0%), Cyanobacteria (2.66%–21.1%, mean=11.8%), Acidobacteria (2.66%–21.1%, mean=11.1%), Bacteroidetes (7.68%–14.6%, mean=9.87%), Chloreflexi (4.24%–9.01%, mean=6.31%), Patescibacteria (2.41%–5.40%, mean=4.43%), Gemmatimonadetes (1.29%–4.39%, mean=2.50%), and Firmicutes (0.75%–3.87%, mean=1.57%). These seven phyla accounted for more than 92% of the total bacterial community. The relative abundance levels of Cyanobacteria on microplastic surfaces in humus and eluvial horizons were higher by 497.0% and 1553.2%, respectively, compared to those in “rhizosphere-like” soil; and similarly, the abundance levels of Acidobacteria, Chloroflexi, and Gemmatimonadetes were lower by 77.1% and 57.4%, 35.8% and 53.0%, and by 55.2% and 67.6%, respectively.
Fig. 3.
Phylum level community compositions of soil bacteria (a) and fungi (b). S-A and MP-A represent soil and microplastic samples, respectively, in the humus horizon; S-B and MP-B represent soil and microplastic samples, respectively, in eluvial horizon.
The dominant fungal phyla were Ascomycota (67.59%–72.78%), Basidiomycota (17.04%–28.75%), and Mortierellomycota (1.42%–10.32%), and the relative abundance levels of these three phyla accounted for more than 98% of the total fungal community. The relative abundance levels of Basidiomycota on microplastic surfaces in humus and eluvial horizons were higher by 12.9% and 68.7%, respectively, compared to those in surrounding “rhizosphere-like” soil; while the those of Mortierellomycota were lower by 58.3% and 67.5%, respectively. Therefore, microplastics significantly decreased and increased the relative abundance levels in the soil of Mortierellomycota and Basidiomycota, respectively. This finding can be attributed to the fact that Basidiomycota is an important decomposer in soil and implicated in the decomposition of complex organic matters. Therefore, the relative abundance level of Basidiomycota on microplastic surfaces was higher than that in the surrounding soil. In addition, Mortierellomycota prefer to exist in a soil environment with high nutrient content, and microplastics may reduce soil nutrient level [21], which in turn leads to a decrease in the relative abundance of Mortierellomycota.
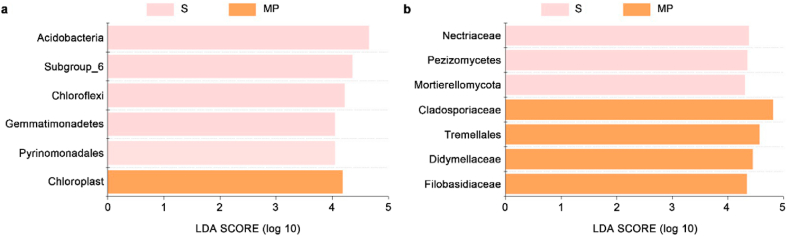
We used the LEfSe tool to perform statistical analysis from the phylum to the genus level. The results of LEfSe (Fig. 4a) show that bacterial communities in “rhizosphere-like” soil and microplastics differ significantly by six groups (LDA scores >4). Acidobacteria (phylum), Subgroup_6 (class), Chloreflexi (phylum), Gemmatimonadetes (phylum), Pyrinomonadales (order) were significantly abundant in surrounding “rhizosphere-like” soil, while Chloroplast (order) was significantly abundant on microplastic surface. Similarly, fungal communities in “rhizosphere-like” soil and on microplastic surface differed significantly by seven groups (Fig. 4b). Nectriaceae (family), Pezizomycetes (class), and Mortierellomycota (phylum) were significantly abundant in surrounding “rhizosphere-like” soil, while Cladosporiaceae (family), Tremellales (order), Didymellaceae (family), and Filobasidiaceae (family) were significantly abundant on microplastic surfaces. Four groups of fungi and one group of bacteria were detected to be significantly enriched on microplastic surfaces, indicating that microplastics have a greater impact on fungal community composition than bacteria. This may be because fungi are better adapted at degrading soil organic polymers with high chemical resistance (such as lignin or cellulose) [37].
Fig. 4.
Linear discriminant analysis Effect Size (LEfSe) results of bacterial (a) and fungal (b) taxa (LDA score >4) in soil surrounding microplastics and on microplastic surfaces. S and MP represent soil and microplastic samples, respectively.
The microbial community composition on microplastic surfaces differed from those in surrounding “rhizosphere-like” soil. These results are consistent with those of Zhang et al. [38], who observed that unique microbial communities that differed significantly from those in the surrounding soil formed on microplastic surfaces. In this study, the relative abundance levels of Cyanobacteria and Basidiomycota were significantly higher on microplastic surfaces than in the surrounding soil, and the relative abundance levels of Acidobacteria, Chloreflexi, Gemmatimonadetes, and Mortierellomycota showed an opposite trend. This may be explained by the hydrophobic surface of microplastics that provided a new substrate for heterotrophic microbial activities, thereby enriching for microbial groups involved in their biodegradation, thus affecting the overall microbial community composition [26]. These results may indicate changing the ecological functions and biogeochemical processes in the soil ecosystem.
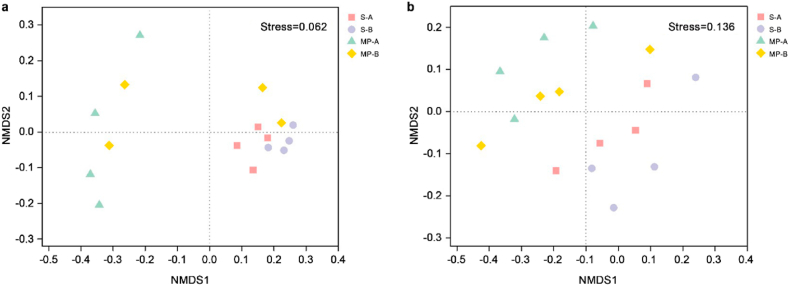
We performed non-metric multidimensional scaling (NMDS) of weighted UniFrac distances to analyze the patterns of separation between microbial communities (Fig. 5). The NMDS stress coefficients of bacterial and fungal communities were 0.062 and 0.136, respectively. The result based on NMDS analysis better reveals the differences in community structures of bacteria and fungi on microplastic surfaces and in surrounding “rhizosphere-like” soil. NMDS ordination clearly distinguished between communities on microplastics and soil samples (Fig. 5). The bacterial communities on microplastic surfaces and in “rhizosphere-like” soil were significantly different (Fig. 5a), showing that microplastics significantly impact the bacterial community structure. The fungal communities on microplastic surfaces and in “rhizosphere-like” soil were more clearly separated compared to the bacterial communities (Fig. 5b), indicating that fungal communities may respond more strongly to microplastics. This result supported the results of LefSe. However, the fungal community structures in the different soil horizons did not differ significantly.
Fig. 5.
NMDS analysis of bacterial (a) and fungal (b) communities in soil surrounding microplastics and on microplastic surfaces. S-A and MP-A represent soil and microplastic samples, respectively, in the humus horizon; S-B and MP-B represent soil and microplastic samples, respectively, in eluvial horizon.
3.3. Effects of microplastics on microbial function
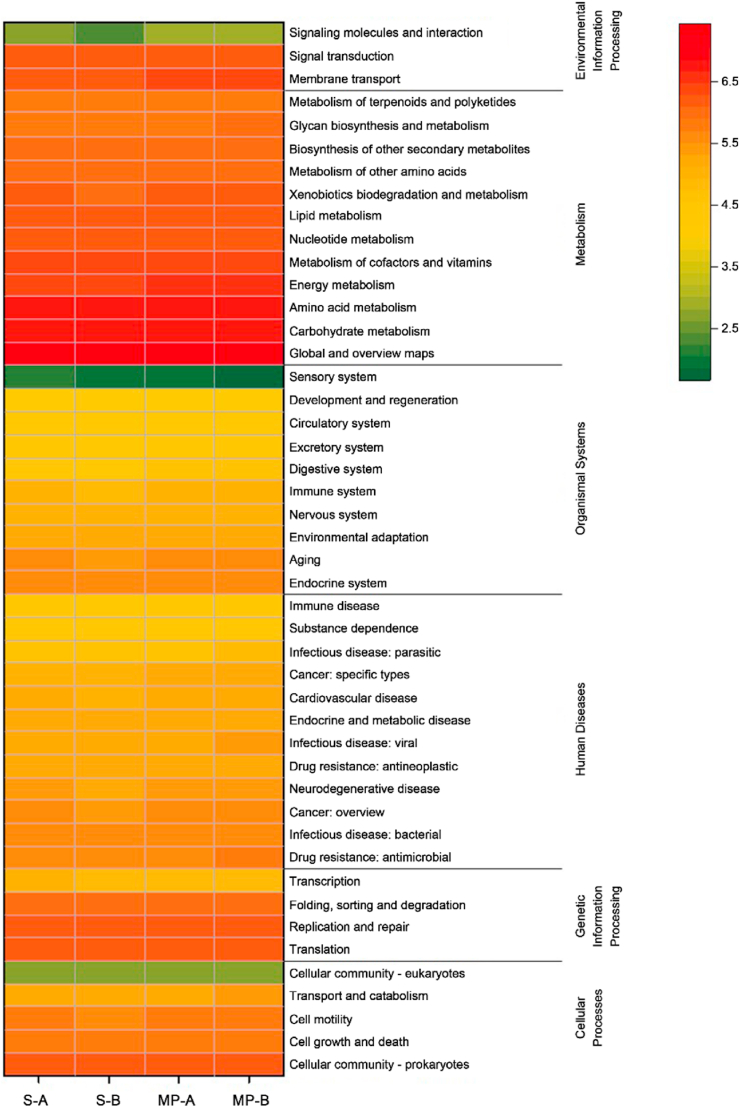
Changes in microbial communities may potentially impact metabolic functional diversity [39]. Therefore, PICRUSt was used to predict bacterial functional content based on the 16S rRNA gene. The gene profiles of microplastics and soil samples were annotated using the KEGG database to evaluate the effect of microplastics on bacterial functions (Fig. 6). Pathways related to human diseases were present in significantly higher levels on microplastic surfaces than in “rhizosphere-like” soil. For example, the relative abundance of pathways related to bacterial and parasitic infectious diseases and substance dependence was higher by more than 20% on microplastic surfaces. This finding may have implications for human health. This finding may be attributed to microplastics can not only adsorb chemical pollutants, including heavy metals, dioxins, and persistent organic pollutants [40,41], but also release flame retardants, plasticizers, heat stabilizers, and antioxidants [42], and these pollutants have great biological toxicity. In addition, microplastic surfaces can also carry pathogens, which further threatens human health [43]. However, PICRUSt analysis provides only give predictive functional profiles of bacterial communities. Therefore, a metagenomic analysis will be required to evaluate the effects of microplastics on soil bacterial function.
Fig. 6.
Relative abundance of bacterial functions in soil surrounding microplastics and on microplastic surfaces.
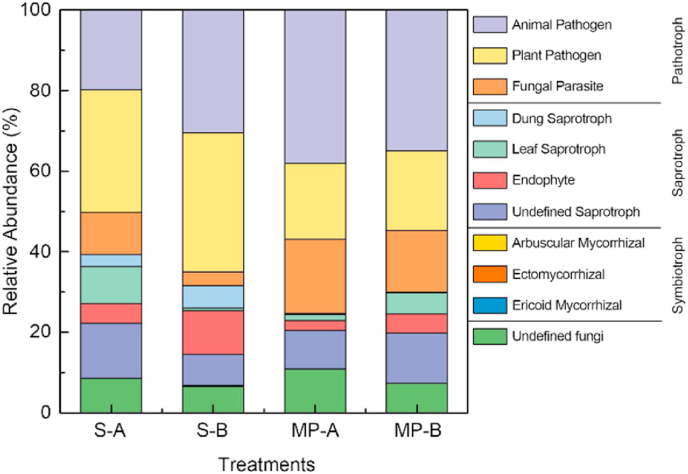
To study the effects of microplastics on fungal functions, we used FunGuild to assign fungal taxa to three ecologically relevant trophic modes– saprotrophy, symbiotrophy, and pathotrophy (Fig. 7). Pathotrophs and saprotrophs accounted for approximately 90% of all fungal OTUs, with symbiotrophs accounting for less than 0.5% of OTUs. In the pathotroph group, the relative abundance levels of animal pathogen and fungal parasites on microplastic surfaces were significantly higher than those in “rhizosphere-like” soil, while the relative abundance of plant pathogen in “rhizosphere-like” soil was higher. In the saprotroph group, the relative abundance levels of dung saprotrophs and endophytes on microplastic surfaces were significantly lower than those in “rhizosphere-like” soil. The relative abundance levels of leaf saprotrophs and undefined saprotrophs on microplastic surfaces in the humus horizon were lower than those in “rhizosphere-like” soil, while in the eluvial horizon, these saprotrophs were more abundant in “rhizosphere-like” soil. Microplastics alter soil microbial communities and function, and thus shift plant mycorrhizal symbiosis as well as causing an alteration in C, N and P-related enzymes, as a consequence, is likely to affect the cycling of key nutrients and greenhouse gases emissions.
Fig. 7.
Relative abundance of fungal functional guilds in soil surrounding microplastics and on microplastic surfaces.
4. Conclusions
In this study, we proved for the first time that there is a “rhizosphere-like neighboring effect” between microplastics and the surrounding soil. The Sobs, Chao1, Ace, and Shannon index values of microbial communities on microplastic surfaces were lower than in surrounding “rhizosphere-like” soil, indicating that microplastics reduced soil microbial richness and diversity. In addition, the values for these indexes were reduced to a greater degree in the humus horizon. Microplastics changed the compositions of the bacterial and fungal communities, enriching for community members involved in microplastic biodegradation. The relative abundance levels of Cyanobacteria and Basidiomycota on microplastic surfaces were significantly higher than those in surrounding “rhizosphere-like” soil. Furthermore, microplastics changed the metabolic functional diversity of microorganisms. The relative abundance levels of pathways related to human diseases, animal pathogen, and fungal parasites were significantly higher on microplastic surfaces than in “rhizosphere-like” soil. These results show that the microbial diversity, richness, community structure and function between microplastic surfaces and surrounding “rhizosphere-like” soil are significantly different, leading to a “rhizosphere-like neighbor avoidance effect” between microplastic surfaces and soil surrounding microplastics. Future research needs to implement a comprehensive sampling within cropping systems to better clarify the effect of microplastics on microorganisms in agricultural settings, which is more meaningful for the assessment of the ecological consequences of microplastics in soil.
Author contributions
Hong Yu: Conceptualization, Investigation, Data curation, Funding acquisition, Writing - original draft. Ying Zhang: Data curation, Methodology, Formal analysis. Wenbing Tan: Conceptualization, Writing - review & editing. Beidou Xi: Conceptualization, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (No. 2020YFC1909502), the National Natural Science Foundation of China (No. 41977030), the Joint Research Project for the Yangtze River Conservation (Phase I) (No. 2019-LHYJ-01-0206), and the Departmental Budget Project of Ministry of Ecology and Environment (No. 144026000000200026).
Contributor Information
Hong Yu, Email: yuhong1018@126.com.
Ying Zhang, Email: zhying010@126.com.
References
- 1.Collignon A., Hecq J.H., Galgani F., Collard F., Goffart A. Annual variation in neustonic micro- and meso-plastic particles and zooplankton in the Bay of Calvi (Mediterranean–Corsica) Mar. Pollut. Bull. 2014;79(1):293–298. doi: 10.1016/j.marpolbul.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y., Liu Q., Jia W., Yan C., Wang J. Agricultural plastic mulching as a source of microplastics in the terrestrial environment. Environ. Pollut. 2020;260 doi: 10.1016/j.envpol.2020.114096. [DOI] [PubMed] [Google Scholar]
- 3.Weithmann N., Möller J.N., Löder M.G.J., Piehl S., Laforsch C., Freitag R. Organic fertilizer as a vehicle for the entry of microplastic into the environment. Sci. Adv. 2018;4(4) doi: 10.1126/sciadv.aap8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nizzetto L., Futter M., Langaas S. Are agricultural soils dumps for microplastics of urban origin? Environ. Sci. Technol. 2016;50(20):10777–10779. doi: 10.1021/acs.est.6b04140. [DOI] [PubMed] [Google Scholar]
- 5.China Agriculture Yearbook . China Agriculture Press; Beijing: 2019. China Agriculture Yearbook. [Google Scholar]
- 6.Zhu Y., Cao M., Luo J., Zhang Q., Cao J. Distribution and potential risks of microplastics in China: a review. Res. Environ. Sci. 2019;32:1437–1447. [Google Scholar]
- 7.Zhou B., Wang J., Zhang H., Shi H., Fei Y., Huang S., Tong Y., Wen D., Luo Y., Barcelo D. Microplastics in agricultural soils on the coastal plain of Hangzhou Bay, east China: multiple sources other than plastic mulching film. J. Hazard Mater. 2020;388:121814. doi: 10.1016/j.jhazmat.2019.121814. [DOI] [PubMed] [Google Scholar]
- 8.Zhang D., Liu H., Hu W., Qin X., Ma X., Yan C., Wang H. The status and distribution characteristics of residual mulching film in Xinjiang, China. J. Integr. Agr. 2016;15(11):2639–2646. [Google Scholar]
- 9.Zhang G., Liu Y. The distribution of microplastics in soil aggregate fractions in southwestern China. Sci. Total Environ. 2018;642:12–20. doi: 10.1016/j.scitotenv.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S., Liu X., Hao X., Wang J., Zhang Y. Distribution of low-density microplastics in the Mollisol farmlands of northeast China. Sci. Total Environ. 2019;708 doi: 10.1016/j.scitotenv.2019.135091. [DOI] [PubMed] [Google Scholar]
- 11.Ding L., Zhang S., Wang X., Yang X., Zhang C., Qi Y., Guo X. The occurrence and distribution characteristics of microplastics in the agricultural soils of Shaanxi Province, in north-western China, Sci. Total Environ. 2020;720 doi: 10.1016/j.scitotenv.2020.137525. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y., Liu X., Wang J. Characterization of microplastics and the association of heavy metals with microplastics in suburban soil of central China. Sci. Total Environ. 2019;694 doi: 10.1016/j.scitotenv.2019.133798. [DOI] [PubMed] [Google Scholar]
- 13.de Souza Machado A.A., Lau C.W., Till J., Kloas W., Lehmann A., Becker R., Rillig M.C. Impacts of microplastics on the soil biophysical environment. Environ. Sci. Technol. 2018;52(17):9656–9665. doi: 10.1021/acs.est.8b02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Souza Machado A.A., Lau C.W., Kloas W., Bergmann J., Bachelier J.B., Faltin E., Becker R., Gorlich A.S., Rillig M.C. Microplastics can change soil properties and affect plant performance. Environ. Sci. Technol. 2019;53(10):6044–6052. doi: 10.1021/acs.est.9b01339. [DOI] [PubMed] [Google Scholar]
- 15.Zhang D., Ng E.L., Hu W., Wang H., Galaviz P., Yang H., Sun W., Li C., Ma X., Fu B., Zhao P., Zhang F., Jin S., Zhou M., Du L., Peng C., Zhang X., Xu Z., Xi B., Liu X., Sun S., Cheng Z., Jiang L., Wang Y., Gong L., Kou C., Li Y., Ma Y., Huang D., Zhu J., Yao J., Lin C., Qin S., Zhou L., He B., Chen D., Li H., Zhai L., Lei Q., Wu S., Zhang Y., Pan J., Gu B., Liu H. Plastic pollution in croplands threatens long-term food security. Global Change Biology Bioenergy. 2020;26(6):3356–3367. doi: 10.1111/gcb.15043. [DOI] [PubMed] [Google Scholar]
- 16.Huerta Lwanga E., Gertsen H., Gooren H., Peters P., Salanki T., van der Ploeg M., Besseling E., Koelmans A.A., Geissen V. Incorporation of microplastics from litter into burrows of Lumbricus terrestris. Environ. Pollut. 2017;220(Pt A):523–531. doi: 10.1016/j.envpol.2016.09.096. [DOI] [PubMed] [Google Scholar]
- 17.Wan Y., Wu C., Xue Q., Hui X. Effects of plastic contamination on water evaporation and desiccation cracking in soil. Sci. Total Environ. 2019;654:576–582. doi: 10.1016/j.scitotenv.2018.11.123. [DOI] [PubMed] [Google Scholar]
- 18.Qi Y., Ossowicki A., Yang X., Huerta Lwanga E., Dini-Andreote F., Geissen V., Garbeva P. Effects of plastic mulch film residues on wheat rhizosphere and soil properties. J. Hazard Mater. 2020;387 doi: 10.1016/j.jhazmat.2019.121711. [DOI] [PubMed] [Google Scholar]
- 19.Liu H., Yang X., Liu G., Liang C., Xue S., Chen H., Ritsema C.J., Geissen V. Response of soil dissolved organic matter to microplastic addition in Chinese loess soil. Chemosphere. 2017;185:907–917. doi: 10.1016/j.chemosphere.2017.07.064. [DOI] [PubMed] [Google Scholar]
- 20.Boots B., Russell C.W., Green D.S. Effects of Microplastics in soil ecosystems: above and below ground. Environ. Sci. Technol. 2019;53(19):11496–11506. doi: 10.1021/acs.est.9b03304. [DOI] [PubMed] [Google Scholar]
- 21.Yu H., Fan P., Hou J., Dang Q., Cui D., Xi B., Tan W. Inhibitory effect of microplastics on soil extracellular enzymatic activities by changing soil properties and direct adsorption: an investigation at the aggregate-fraction level. Environ. Pollut. 2020;267 doi: 10.1016/j.envpol.2020.115544. [DOI] [PubMed] [Google Scholar]
- 22.Fei Y., Huang S., Zhang H., Tong Y., Wen D., Xia X., Wang H., Luo Y., Barcelo D. Response of soil enzyme activities and bacterial communities to the accumulation of microplastics in an acid cropped soil. Sci. Total Environ. 2020;707 doi: 10.1016/j.scitotenv.2019.135634. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y., Zhao Y., Wang J., Zhang M., Jia W., Qin X. LDPE microplastic films alter microbial community composition and enzymatic activities in soil. Environ. Pollut. 2019;254(Pt A) doi: 10.1016/j.envpol.2019.112983. [DOI] [PubMed] [Google Scholar]
- 24.Qian H., Zhang M., Liu G., Lu T., Qu Q., Du B., Pan X. Effects of soil residual plastic film on soil microbial community structure and fertility. Water, Air Soil Pollution. 2018;229(8):261. [Google Scholar]
- 25.Wang J., Huang M., Wang Q., Sun Y., Zhao Y., Huang Y. LDPE microplastics significantly alter the temporal turnover of soil microbial communities. Sci. Total Environ. 2020;726 doi: 10.1016/j.scitotenv.2020.138682. [DOI] [PubMed] [Google Scholar]
- 26.Miao L., Wang P., Hou J., Yao Y., Liu Z., Liu S., Li T. Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci. Total Environ. 2019;650(Pt 2):2395–2402. doi: 10.1016/j.scitotenv.2018.09.378. [DOI] [PubMed] [Google Scholar]
- 27.Hou J., Xu X., Yu H., Xi B., Tan W. Comparing the long-term responses of soil microbial structures and diversities to polyethylene microplastics in different aggregate fractions. Environ. Int. 2021;149 doi: 10.1016/j.envint.2021.106398. [DOI] [PubMed] [Google Scholar]
- 28.Ren X., Tang J., Liu X., Liu Q. Effects of microplastics on greenhouse gas emissions and the microbial community in fertilized soil. Environ. Pollut. 2020;256:113347. doi: 10.1016/j.envpol.2019.113347. [DOI] [PubMed] [Google Scholar]
- 29.Doran J.W., Zeiss M.R. Soil health and sustainability: managing the biotic component of soil quality. Appl. Soil Ecol. 2000;15(1):3–11. [Google Scholar]
- 30.Kuzyakov Y., Razavi B.S. Rhizosphere size and shape: temporal dynamics and spatial stationarity. Soil Biol. Biochem. 2019;135:343–360. [Google Scholar]
- 31.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 33.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huhe X. Chen, Hou F., Wu Y., Cheng Y.X. Bacterial and fungal community structures in loess plateau grasslands with different grazing intensities. Front. Microbiol. 2017;8:606. doi: 10.3389/fmicb.2017.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Q., Xiao M., Ma Y., Niu H., Zhang G. Polyester microfiber and natural organic matter impact microbial communities, carbon-degraded enzymes, and carbon accumulation in a clayey soil. J. Hazard Mater. 2021;405 doi: 10.1016/j.jhazmat.2020.124701. [DOI] [PubMed] [Google Scholar]
- 36.Totsche K.U., Amelung W., Gerzabek M.H., Guggenberger G., Klumpp E., Knief C., Lehndorff E., Mikutta R., Peth S., Prechtel A., Ray N., Kögel-Knabner I. Microaggregates in soils. J. Plant Nutr. Soil Sci. 2018;181(1):104–136. [Google Scholar]
- 37.Joergensen R.G., Wichern F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol. Biochem. 2008;40(12):2977–2991. [Google Scholar]
- 38.Zhang M., Zhao Y., Qin X., Jia W., Chai L., Huang M., Huang Y. Microplastics from mulching film is a distinct habitat for bacteria in farmland soil. Sci. Total Environ. 2019;688:470–478. doi: 10.1016/j.scitotenv.2019.06.108. [DOI] [PubMed] [Google Scholar]
- 39.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar M., Xiong X., He M., Tsang D.C.W., Gupta J., Khan E., Harrad S., Hou D., Ok Y.S., Bolan N.S. Microplastics as pollutants in agricultural soils. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.114980. [DOI] [PubMed] [Google Scholar]
- 41.Wang W., Ge J., Yu X., Li H. Environmental fate and impacts of microplastics in soil ecosystems: progress and perspective. Sci. Total Environ. 2020;708 doi: 10.1016/j.scitotenv.2019.134841. [DOI] [PubMed] [Google Scholar]
- 42.Yang L., Zhang Y., Kang S., Wang Z., Wu C. Microplastics in soil: a review on methods, occurrence, sources, and potential risk. Sci. Total Environ. 2021;780 doi: 10.1016/j.scitotenv.2021.146546. [DOI] [PubMed] [Google Scholar]
- 43.Amaral-Zettler L.A., Zettler E.R., Mincer T.J. Ecology of the plastisphere, nat. Rev. Microbiol. 2020;18(3):139–151. doi: 10.1038/s41579-019-0308-0. [DOI] [PubMed] [Google Scholar]