Abstract
In this study, an Escherichia coli (E. coli) whole-cell biosensor for the specific detection of bioavailable arsenic was developed by placing a green fluorescent protein (GFP) reporter gene under the control of the ArsR1 (GSU2952) regulatory circuit from Geobacter sulfurreducens. E. coli cells only emitted green fluorescence in the presence of arsenite and were more sensitive to arsenite when they were grown in M9 supplemented medium compared to LB medium. Under optimal test conditions, the Geobacter arsR1 promoter had a detection limit of 0.01 μM arsenite and the GFP expression was linear within a range of 0.03–0.1 μM (2.25–7.5 μg/l). These values were well below World Health Organization’s drinking water quality standard, which is 10 μg/l. The feasibility of using this whole-cell biosensor to detect arsenic in water samples, such as arsenic polluted tap water and landfill leachate was verified. The biosensor was determined to be just as sensitive as atomic fluorescence spectrometry. This study examines the potential applications of biosensors constructed with Geobacter ArsR-Pars regulatory circuits and provides a rapid and cost-effective tool that can be used for arsenic detection in water samples.
Keywords: Arsenic detection, Whole-cell biosensor, Geobacter sulfurreducens, ars operon
Graphical abstract
Highlights
-
•
Whole-cell As biosensor was constructed with G. sulfurreducens ArsR-Pars.
-
•
Fluorescence was only detected in biosensor cells exposed to arsenite.
-
•
The biosensor could detect arsenic levels as low as 10 nM.
-
•
The As detection range was below WHO’s drinking water quality standards.
-
•
The biosensor could detect arsenic in tap water and leachate samples.
1. Introduction
Arsenic contamination in groundwater is a global problem that poses a major threat to the health of humans and wildlife. It is estimated that up to 220 million people are exposed to high arsenic concentrations in groundwater [35], and this exposure can cause skin, vascular and nervous systems disorders, and can even lead to cancer [15,43].
Landfill leachate has been identified as a major contributor to groundwater arsenic pollution [34]. There are many sources of arsenic found in municipal solid waste (MSW) landfills, including glass, metallic (e.g., metal alloys and semiconductors), and agricultural (e.g., wood preservatives, insecticides, and herbicides) products [27,34,36]. In order to minimize arsenic pollution from MSW landfills, it is important to monitor and remove arsenic from wastewater prior to release into the environment.
Arsenic in wastewater samples is usually detected by atomic fluorescence spectrometry (AFS) [26] or chemical kits [44] that require complicated chemical analysis. These methods measure total arsenic concentrations, which include both bioavailable and non-bioavailable species. Bioavailable arsenic can be defined as the form of arsenic that are taken up by cells and elicit adverse responses, and therefore poses a much greater threat to our health than non-bioavailable species [2]. Therefore, tools developed to assess environmental arsenic risks should focus more on bioavailable forms.
Bacterial cells have evolved to include a variety of mechanisms to detoxify arsenic. Some bacteria use methyltransferases (ArsM) to form methylarsine gases that are volatilized from the cells [1], while others rely on arsenic respiratory (arr) and/or arsenic-resistance (ars) transformation systems [48]. These arsenic detoxification systems are tightly regulated and are only active in the presence of certain arsenic species [40]. ArsR is a regulatory protein that binds to the promoter region of arr, ars, and certain arsM operons and inhibits their transcription unless arsenite is present [49].
This tight regulation of gene expression by the ars operon promoter (Pars) can be used for the development of whole-cell biosensors designed to produce signals such as luminescence or fluorescence. In fact, many genetically engineered whole-cell arsenic biosensors that are regulated by ArsR and Pars have already been developed (Table 1). For example, by placing an essential gene required for extracellular electron transfer and electricity production of Shewanella oneidensis under the control of Pars [47], genetically engineered a strain that produced an increased current in response to arsenic when inoculated into a bioelectrochemical system (BES). A number of E. coli whole-cell biosensors with reporter genes under the control of the ArsR-Pars circuit have also been developed (Table 1), including strains whose reporter signals have been enhanced by positive feedback loops [19] or promoter modification [5].
Table 1.
Comparison of the whole-cell biosensors constructed in this study to previously reported arsenic whole-cell biosensors.
| Host | Reporter gene | Limit of detection for As (μM) | Range of detection for As (μM) | Reference |
|---|---|---|---|---|
| Escherichia coli | luxAB | 0.09 | 0.13–1.33 | [46] |
| Escherichia coli | gfp | 0.067 | 0.13–2.67 | [20] |
| Escherichia coli | egfp | 0.13 | 0–1.33 | [3] |
| Escherichia coli | luxCDABE | 0.01 | 0.05–0.80 | [42] |
| Shewanella oneidensis | mtrB | 40 | 40–100 | [47] |
| Escherichia coli | luxCDABE | 0.10 | 0.05–5.00 | [12] |
| Escherichia coli | gfp | 0.01 | 0.013–0.67 | [22] |
| Escherichia coli | lacZ | 0.13 | 0.13–6.67 | [14] |
| Escherichia coli | egfp | 0.033 | 0.033–0.67 | [50] |
| Escherichia coli | mCherry | 0.10 | 0.067–1.87 | [19] |
| Escherichia coli | gfp | 0.01 | 0.10–4.00 | [5] |
| Escherichia coli | gfp | 0.01 | 0.03–0.1 | This study |
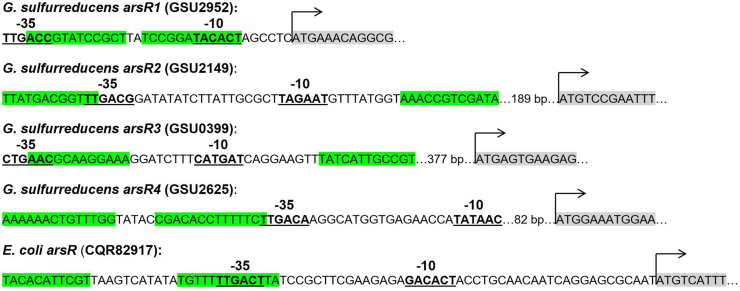
Geobacter are Fe(III) reducing bacteria that are frequently found in anoxic soils and sediments that have been contaminated with arsenic [9,11,13,17,21,31,37]. All of the Geobacter strains that have been sequenced to date have an ars detoxification system under the control of arsR [7]. There are four different genes coding for putative ArsR regulatory proteins found in the G. sulfurreducens genome (GSU2952 (arsR1), GSU2149 (arsR2), GSU0399 (arsR3), and GSU2625 (arsR4)) (Fig. S1). The promoters of these genes all contain −10 and −35 transcription binding sites that are similar to the prokaryotic consensus, and have imperfect inverted repeats located upstream from the transcriptional start sites, which is characteristic of ArsR binding sites [4,6] (Fig. 1). Among these putative ArsR regulatory proteins, only ArsR1 and ArsR2 have the metal-binding motif (ELCVCDL). The amino acid sequence of ArsR2, whose coding gene locates upstream from a gene coding for a putative CadA protein, is more closely related to those of CadC regulatory proteins [7]. This similarity suggests that arsR2 probably encodes the Cd-specific regulatory protein CadC. Our recent experimental results support this assumption (unpublished data). In addition [7], found that among the four putative ArsR regulatory proteins of G. sulfurreducens, only ArsR1 regulates transcription of the ars operon in the presence of arsenite. In order to determine whether the Geobacter ars regulatory circuit could serve as an effective regulatory unit in an arsenic biosensor, arsR1 and its promoter (Pars) from Geobacter sulfurreducens were placed upstream from a green fluorescent protein (GFP) reporter gene (gfp) so that the expression of gfp could be induced by arsenite. These whole-cell biosensors were optimized and the feasibility of using this biosensor to detect arsenite in practical water samples was assessed.
Fig. 1.
Promoters from the 4 different arsR genes found in the G. sulfurreducens PCA and E. coli K-12 genomes. The bold and underlined sequences represent the −35 and −10 transcription binding sites; the regions highlighted in green represent inverted repeats that could be putative ArsR binding sites; the arrow and grey shaded nucleotides represent the start of the ArsR coding sequence. The bacterial −10 site consensus sequence is TATAAT and the −35 consensus is TTGACA. The arsR1 -10 site has 2 mismatches with the consensus (TACACT) and 1 mismatch at the −35 site (TTGACC). The arsR2 -10 site has 1 mismatch (TAGAAT) and 1 mismatch at the −35 site (TTGACG). The arsR3 -10 site has 2 mismatches at −10 (CATGAT) and 3 at −35 (CTGAAC). The arsR4 -10 site has 1 mismatch at −10 (TATAAC) and 0 mismatches at −35 (TTGACA). The E. coli K-12 -10 site has 3 mismatches at the −10 site (GACACT) and 1 at the −35 site (TTGACT).
2. Materials and methods
2.1. Bacterial strains and culture conditions
Geobacter sulfurreducens PCA was cultured at 30 °C under strict anaerobic conditions in a mineral-based medium containing 15 mM acetate as the electron donor and 40 mM fumarate as the electron acceptor, as described by Ref. [25]. E. coli TOP10 (Tiangen, Beijing, China) was cultivated in LB (Luria Broth) medium at 37 °C. A minimum M9 supplemented medium described by Ref. [28] was used for fluorescent assay experiments.
2.2. Gene manipulation
The plasmid pPROBE-NT [29], a gift from Steven Lindow (Addgene plasmid # 37818; http://n2t.net/addgene:37818; RRID: Addgene_37818), was used to design the ArsR-Pars-GFP construct that was introduced into E. coli cells. This plasmid contained a multicloning site (MCS) upstream of a promoter-less gfp reporter gene. Four terminators located at the upstream of the MCS, which confers a low basal level of gfp expression. The DNA fragment containing Pars (regions 3,251,294–3,251,793 in the G. sulfureducens chromosome) and the arsR1 (GSU2952; regions 3,251,794–3,252,135) coding sequence was amplified from G. sulfurreducens genomic DNA via polymerase chain reaction (PCR) using ars-F and ars-R primers (Table S1). The amplified DNA fragment was then digested with restriction endonucleases Xba I and EcoR I and inserted into the pPROBE-NT plasmid in a region directly upstream from the gene coding for the GFP reporter gene (regions 4075–4791 in the plasmid). Insertion of arsR1 and Pars into the plasmid was confirmed by Sanger sequencing [41] with the oligonucleotides NT-Seq-F and NT-Seq-R (Table S1). The final ArsR1-Pars-pPROBE-NT construct (pPROBE-As) (Fig. S2) was transformed into E. coli TOP10 to make the whole-cell biosensor using the method described by Chakrabarty (1975).
2.3. Fluorescence microscopy
E. coli cells were harvested by centrifugation at 3000 rpm for 5 min and resuspended in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). After which, an aliquot of 50 μl cell suspension was mounted on a glass slide with a coverslip and observed by fluorescence microscopy using an Axio Vert. A1 microscope (Carl Zeiss, Germany) equipped with an AxioCam HRc CCD camera (Carl Zeiss, Germany).
2.4. Quantitative reverse-transcription PCR (qRT-PCR)
E. coli cells incubated together with different concentrations of arsenite (0.05, 0.1 and 0.5 μM) were defined as the experimental groups, and those incubated in the absence of arsenite were used as a control group. Four milliliters (ml) of the cell suspension was used for total RNA extraction for each sample. Total RNA was extracted using an EASYspin Plus bacteria RNA extract kit (Aidlab Biotech, Beijing, China). All RNA samples were checked for integrity using agarose gel electrophoresis and had A260/A280 ratios of between 1.8 and 2.0. The RNA samples were then used to generate the first strand of cDNA using the FastKing RT Kit (With gDNase) (Tiangen, Beijing, China). After which, the cDNA was used as qRT-PCR templates. The gene rrsA, which codes for 16S rRNA in E. coli, was used as the reference gene. Relative quantification of transcripts in comparison to rrsA transcript was performed using Talent qPCR PreMix (SYBR Green) (Tiangen, Beijing, China) on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The primers for rrsA and gfp are listed in Table S1. Each PCR mixture consisted of a total volume of 20 μl and contained 10 μl qPCR master mix, 0.6 μl of the appropriate primers (10 μM), 2 μl cDNA (<100 ng), 0.4 μl 50 × ROX reference dye, and 6.4 μl RNase-free H2O. The cycling conditions were 1 cycle of denaturation at 95 °C/15 min, followed by 40 two-segment cycles of amplification (95 °C/10 s, 60 °C/30 s) where the fluorescence was automatically measured during PCR. A melting curve was constructed for each primer pair to verify the presence of one gene-specific peak and the absence of the primer dimmer. Standard curves covering 5 orders of magnitude were constructed with serial dilutions of cDNA. These standard curves were generated using relative concentration vs. the threshold cycle (Ct). The amplification efficiency of each gene (E) was calculated based on the slopes of the standard curves:
| (1) |
The relative expression ratio of gfp in experimental group vs. control group was calculated based on the method outlined by Ref. [33]:
| (2) |
where E (gfp) and E (rrsA) are the amplification efficiencies of gfp and rrsA transcripts, respectively; Ct (gfp, control) and Ct (gfp, experimental) are Ct values for gfp in control group and experimental group, respectively; Ct (rrsA, control) and Ct (rrsA, experimental) are Ct values for rrsA in control group and experimental group, respectively. All samples were from experiments with three biological replicates. Each qPCR reaction was conducted with three technical replicates and the mean was used for analysis.
2.5. Western blotting
The synthetic efficiencies of GFP in different media were assessed by using Western blotting. E. coli cells incubated with or without arsenite in different media were collected, respectively, and subjected to protein extraction, SDS-PAGE and Western blotting with reference to a previous study [30]. β-actin was monitored as the loading control. Anti-GFP mouse monoclonal antibody (Epsilon, Beijing, China) was used as the primary antibody to monitor GFP, while anti-β-actin mouse monoclonal antibody (Epsilon, Beijing, China) was used as the primary antibody to monitor β-actin. HRP-conjugated goat anti-mouse IgG (H + L) (Epsilon, Beijing, China) was used as the secondary antibody. The relative abundance of GFP was determined by dividing the signal intensity value of GFP by that of β-actin.
2.6. Fluorescence intensity assay
Fluorescence intensity assays were conducted using the method described by Ref. [23] with some modifications. E. coli cells cultured in the LB medium containing 50 μg/ml kanamycin were collected by centrifugation for 5 min at 5000 rpm and 4 °C. Cell pellets were then resuspended in 2 × LB or 2 × M9 supplemented medium to reach a specified absorbance at 600 nm. Then the cell suspension and the sample to be tested were mixed at a volumetric ratio of 1:1. These mixtures were then incubated at 25, 30 or 37 °C with shaking at 180 rpm for 4 h. A 200 μl aliquot of these cell suspensions were used to measure fluorescence with a microplate reader (Tecan infinite 200) in a 96-well Costar plate. A water sample was prepared by diluting a stock solution of sodium arsenite or other heavy metal salts such as NaAsO2, Cu(NO3)2, Zn(NO3)2, Pb(NO3)2, Mn(NO3)2, Cr(NO3)3, Ni(NO3)2, Cd(NO3)2, Co(NO3)2 and Hg(NO3)2 to a specified concentration in distilled-deionized water. The metal concentration in each stock solution was 1000 mg/l.
2.7. Practical samples
Tap water (pH 7.8 with 3.0 mM Cl−; 0.04 mM NO3−; 0.22 mM SO42−; 0.80 mM Ca2+; 2.1 μM Fe) and different batches of treated landfill leachates from anaerobic dynamic membrane bioreactors (AnDMBR) [18] (pH 7.2–7.6 with 1220–1680 mg/l COD; 0.43–0.47 mM NH4+-N; 0.36–0.40 mM Fe) were used to assess whether the biosensor could detect arsenite in drinking water and treated wastewater samples. Sodium arsenite was added to the tap water sample to simulate arsenic pollution. The concentrations of arsenic in these samples were analyzed by the biosensor and AFS as described by Ref. [26].
2.8. Data analysis
The response of the whole-cell biosensor to arsenic or to other metals (such as Cu, Zn, Pb, Mn, Cr, Ni, Cd, Co and Hg) was expressed as the induction coefficient (IC), calculated as follows:
| (3) |
where Fx is the mean value of fluorescence intensity induced with metals at a certain cell density and F0 is the mean value of background fluorescence intensity induced with distilled-deionized water (0 M metal solutions) at the same cell density.
The limit of detection for IC (LODIC) was determined as follows [10]:
| (4) |
where F0 is the mean value of background fluorescence intensity and SD is the standard deviation of F0.
The limit of detection for arsenite concentration (LODc) was the minimum concentration of arsenite that could induce significantly stronger fluorescence (p < 0.05) than distilled-deionized water.
All statistical analyses were conducted using GraphPad Prism v6.02. Student’s t-test was used to determine statistical significance (p < 0.05). Linear regression analysis was used to assess the possible correlation between IC and arsenite concentration and to construct standard curves for qRT-PCR. Grubb’s test was used to identify statistical outliers.
3. Results and discussion
3.1. Construction of the whole-cell biosensor
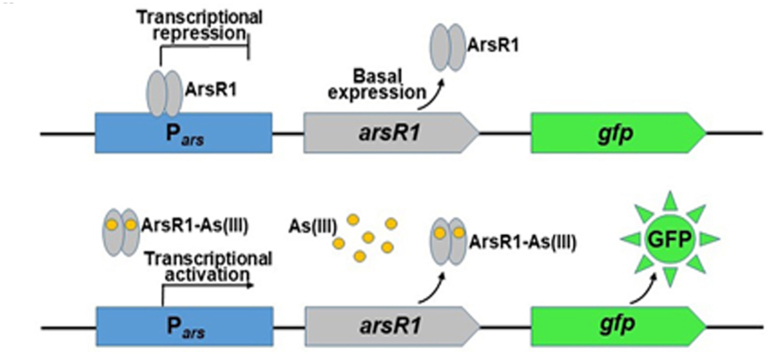
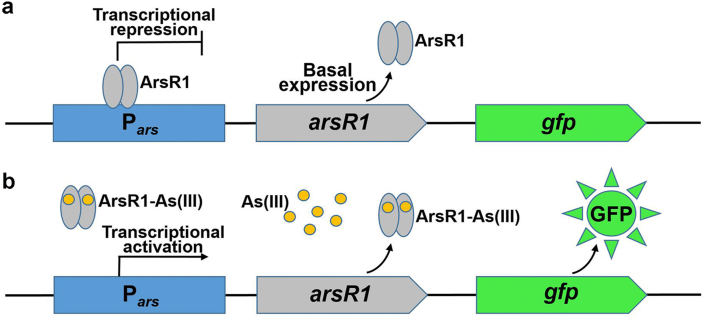
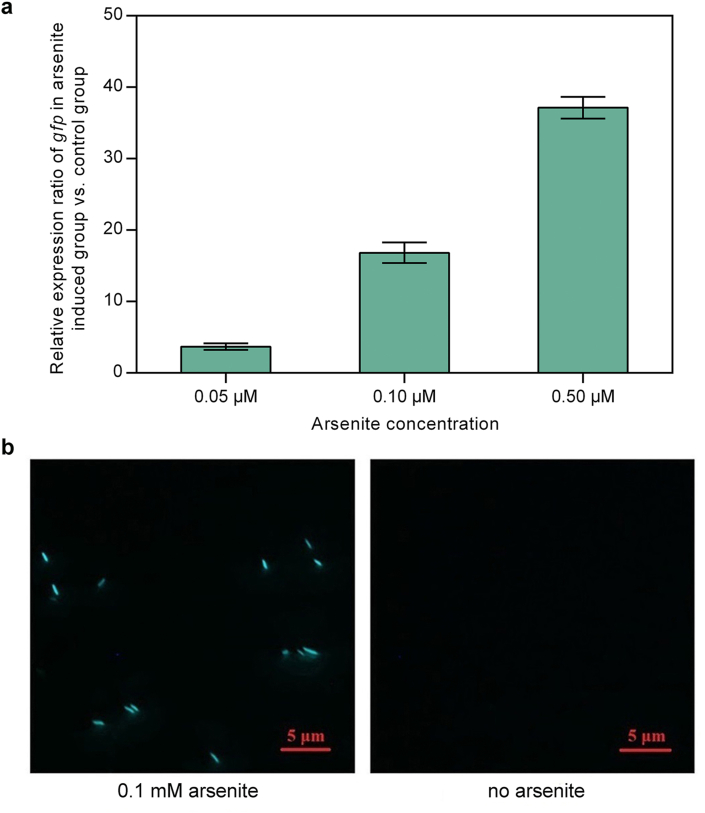
As shown in Fig. 2a, in the absence of arsenite, the transcriptional repressor ArsR1, the product of the arsR1 gene, bound Pars and repressed the transcription of gfp, which meant that no green fluorescence was observed (Fig. 3b). When arsenite was added to the cells, it bound to ArsR1 and caused it to be released from Pars, thereby inducing the transcription of arsR1 and gfp (Fig. 2b). As the concentration of arsenite increased, the relative transcription level of gfp gradually increased (Fig. 3a) and green fluorescence was observed (Fig. 3b).
Fig. 2.
Schematic of the whole-cell biosensor based on the ArsR1-Pars-gfp regulatory circuit. (a) In the absence of arsenite, (b) in the presence of arsenite.
Fig. 3.
(a) The number of gfp transcripts normalized against transcripts from the constitutively expressed rrsA gene (which codes for 16S rRNA) in cells exposed to various concentrations of arsenite. (Temperature: 30 °C; cell density: OD600 = 0.2; medium: M9 supplemented medium; induction time: 4 h. All experiments were conducted in triplicate.) (b) Fluorescence micrographs of the E. coli cells constructed in this study after induction with 0.1 mM arsenite.
3.2. Metal specificity
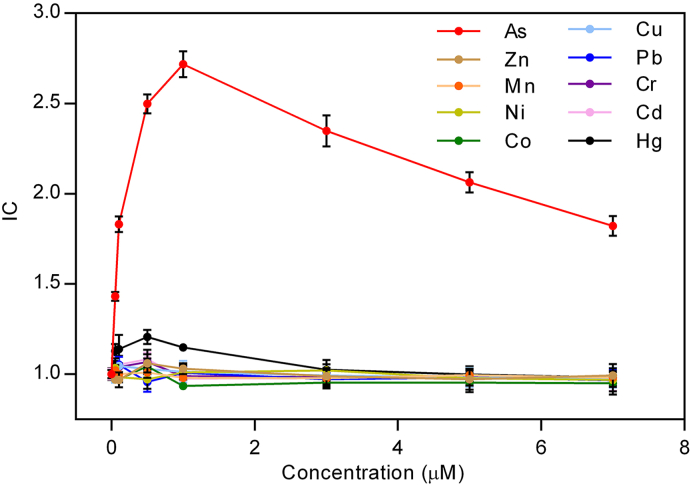
The specificity of the whole-cell biosensor to arsenic was tested with 5 different concentrations (0–7.0 μM) of 9 common heavy metal salts (Cu(NO3)2, Zn(NO3)2, Pb(NO3)2, Mn(NO3)2, Cr(NO3)3, Ni(NO3)2, Cd(NO3)2, Co(NO3)2 and Hg(NO3)2) in addition to sodium arsenite. GFP could be synthesized in response to arsenite, resulting in a statistically significant increase in the fluorescence intensity. Besides arsenite, Hg also induced an increased fluorescence, but the signal was much lower relative to arsenite. Other heavy metal salts did not induce a statistically significant increase in the fluorescence intensity (Fig. 4).
Fig. 4.
Induction coefficient (IC) of the whole-cell biosensor in response to different metal ions. Induction from 10 common metal ions was tested. (All cells were induced for 4 h at 30 °C in M9 supplemented medium. Values are means and standard deviations obtained from experiments with three biological replicates.)
3.3. Optimization of test conditions
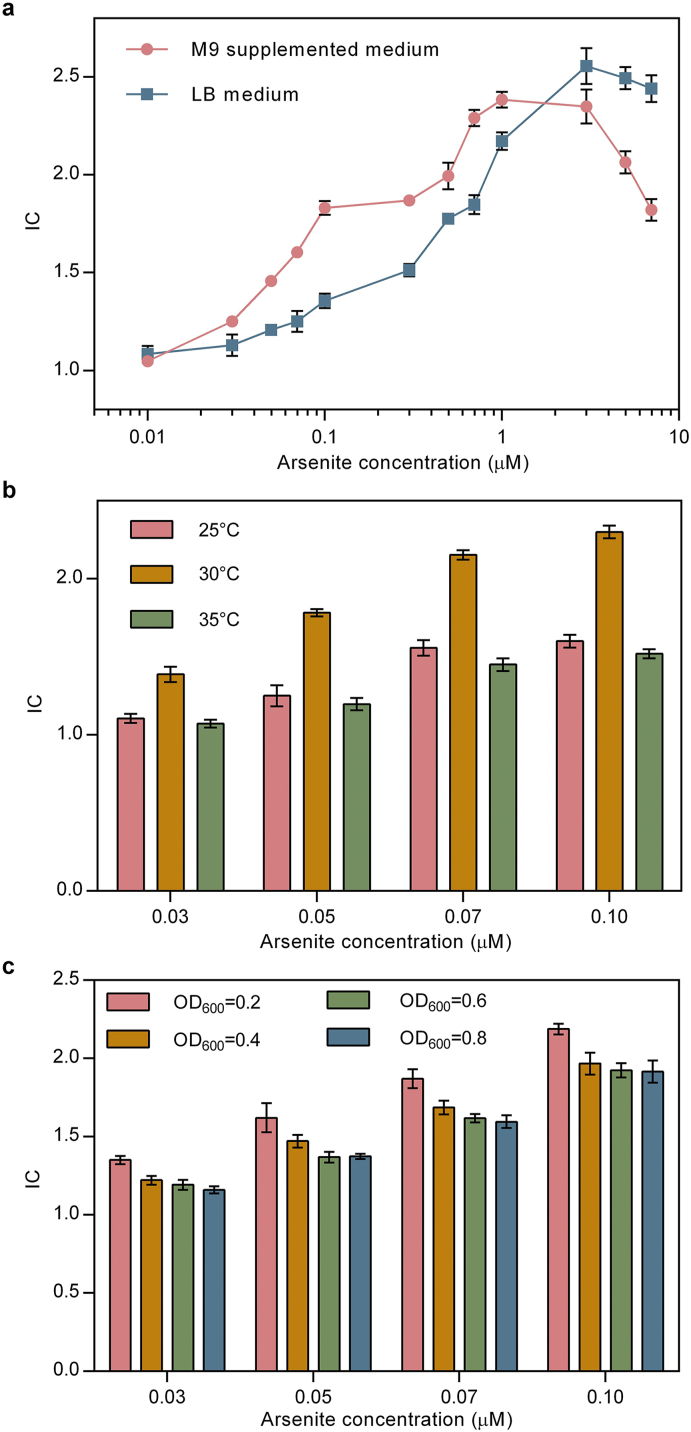
Different media, temperatures and cell densities were tested to determine the optimal conditions that should be used for whole-cell biosensor assays. Arsenite induction experiments were conducted in LB and M9 supplemented medium. Cells grown in LB medium could detect arsenite in the range of 0.03–0.3 μM, while those grown in M9 supplemented medium had a detection limit of 0.01 μM. A linear relationship (R2 = 0.998; p = 0.0005) between IC and arsenite concentration was found within the concentration range of 0.03–0.1 μM (2.25–7.5 μg/l) (Fig. 5a). These detection values are comparable to the whole-cell biosensors constructed based on E. coli strains that contained optimized arsenite-sensing elements [5,22] or those isolated from the arsenic polluted sites [42]. By altering the base composition of the −10 site and the location of the ArsR binding site (ABS) in the arsenite-regulated promoter of E. coli, the induction of the promoter could be increased by 11 fold [5]. [22] identified the arsenite-induced arsR operon for both low background and high expression through three successive rounds of fluorescence activated cell sorting (FACS) with a bidirectional strategy [42]. isolated an E. coli strain with high tolerance towards arsenic from Hooghly River (West Bengal, India) and a whole-cell biosensor with the range of detection of 0.05–0.80 μM was then developed based on this strain. It is possible that by using these aforementioned strategies, the ArsR1-Pars of G. sulfurreducens with a higher sensitivity towards arsenite can be obtained. These results show that the ArsR1-Pars of G. sulfurreducens has a greater potential for a more sensitive detection of arsenite.
Fig. 5.
Optimization of arsenic detection conditions. (a) Induction coefficient (IC) in response to arsenite in LB and M9 supplemented medium. (Temperature: 30 °C; cell density: OD600 = 0.2; induction time: 4 h) (b) IC in response to arsenite at different temperatures. (Medium: M9 supplemented medium; cell density: OD600 = 0.2; induction time: 4 h) (c) IC in response to arsenite under different cell densities. (Medium: M9 supplemented medium; temperature: 30 °C; induction time: 4 h. Values are means and standard deviations obtained from experiments with three biological replicates.)
Differences in biosensor sensitivity was observed when cells were grown on two different media types and were consistent with studies that have shown that the sensitivities of whole-cell biosensors for heavy metals such as cadmium [45] and copper [23] were higher in M9 supplemented medium than they were in LB medium. We also assessed the relative abundance of GFP in different media by using Western blotting. After arsenite induction, there was no significant difference in the relative abundance of GFP between E. coli cells cultured in LB and M9 medium. In the absence of arsenite, however, the relative abundance of GFP in E. coli cells cultured in LB medium was higher than that in M9 medium, indicating that the background synthesis of GFP was higher in LB medium (Fig. S5). This observation was consistent with the results of the fluorescence intensity (Fig. 5a).
Since the biosensor performed better in M9 supplemented medium than in LB medium, the former was used to determine the optimal temperature and cell density parameters. As shown in Fig. 5b and S6, the optimal temperature for arsenite induction was 30 °C. Biosensor assays conducted at various cell densities showed that arsenic sensitivity was greatest when the OD600 was 0.2 (Fig. 5c and S7). Previous studies also found that the response of the whole-cell biosensors decreased at higher cell densities [8,23,39]. Under optimal conditions, there was a strong positive correlation (R2 = 0.998; p = 0.0005) between IC and arsenite concentration within the concentration range of 0.03–0.1 μM (2.25–7.5 μg/l), which is below the WHO’s standard for drinking water quality [32]. Samples with higher concentrations of arsenite can be quantified after dilution.
3.4. Practical application and perspective
Tests were conducted to assess whether the biosensor could be used to detect arsenic in drinking water and treated wastewater samples. Sodium arsenite was added to tap water samples to simulate arsenic pollution. Detection of arsenite in tap water and treated leachate samples from AnDMBRs using the biosensors was just as accurate (relative differences were no more than 7%) as AFS (Table 2). Considering that AFS requires concentrated hydrochloric acid, thiourea and ascorbic acid for pretreatment and 5% hydrochloric acid and argon gas for measurement, the whole-cell biosensor only requires cell suspensions. Therefore, the latter is more cost-effective and produces considerably less liquid waste. Although some organics and bacteria exist in the treated leachate samples, they did not result in significant interferences with the testing results in this study. However, the organics and bacteria in other environmental water samples may affect the performance of the whole-cell biosensors. Given that the E. coli strain constructed in this study harbors a plasmid with a kanamycin resistance marker, other bacteria cannot grow during the testing process and their influences on the testing results can be minimized. In addition, a strain that constitutively produces a GFP signal can be used as a control to measure the signal changes caused by the organics and bacteria in the samples. Therefore, the influences of organics and other bacteria can be overcome by subtracting out the signal changes [16]. By eliminating these biotic and abiotic interferences, the feasibility of using a whole-cell biosensor for low-cost and low-emission arsenite detection in real world scenarios could be greatly improved.
Table 2.
Tap water supplemented with arsenite and treated landfill leachate from AnDMBRs were used to assess the efficacy of the whole-cell biosensor and to compare its sensitivity to AFS. The relative difference was calculated as follows: relative difference = [(result by biosensor - result by AFS)/result by AFS] × 100%.
| Source of sample | Results by AFS (μM) | Results by biosensor (μM) | Relative difference (%) |
|---|---|---|---|
| Polluted tap water 1 | 0.039 | 0.036 | −7 |
| Polluted tap water 2 | 0.058 | 0.054 | −7 |
| Polluted tap water 3 | 0.092 | 0.088 | −4 |
| Treated leachate 1 | 0.059 | 0.056 | −5 |
| Treated leachate 2 | 0.043 | 0.045 | +6 |
| Treated leachate 3 | 0.068 | 0.070 | +3 |
G. sulfurreducens is an electrogenic bacterium that can generate electric current via type IV pili-based direct electron transfer in a microbial fuel cell (MFC) [38], which makes it an ideal candidate for the construction of bioelectrochemical sensors [24]. demonstrated that the expression of high conductive pili in a pili-deficient mutant strain of P. aeruginosa could increase the bioelectricity output of P. aeruginosa in MFS systems. If the gene coding for electrically conductive pili is placed under the control of the arsR regulatory circuit in a pili-deficient strain of G. sulfurreducens or P. aeruginosa, the bioelectricity output can most likely be controlled by arsenite so that in situ arsenite monitoring without having to collect samples can be developed. Considering that the biosensor relies on genetically modified bacteria, the potential environmental risks caused by its pass through need to be assessed before field application.
4. Conclusions
In this study, an E. coli whole-cell biosensor was designed in which the gene coding for GFP was placed under the control of the ArsR1-Pars regulatory circuit from G. sulfurreducens. Optimal biosensor assay conditions were determined and resulted in a detection limit of 0.01 μM with a linear range of detection between 0.03 and 0.1 μM (2.25–7.5 μg/l). These values are significantly lower than WHO’s drinking water quality standard. The feasibility of using the whole-cell biosensor to detect arsenic in practical water samples such as drinking water and treated landfill leachate was verified. This study demonstrates that the ArsR1-Pars from G. sulfurreducens provides a sensitive detection of As and information from this study can be used to design an even more effective arsenic biosensor in the future.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the Fundamental Research Funds for the Central Universities [grant numbers BLX201934, 2019ZY19] and Beijing Municipal Education Commission through Innovative Transdisciplinary Program “Ecological Restoration Engineering”. We thank Prof. Shizhong Li, Tsinghua University, for his help with qRT-PCR analyses.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2021.100092.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bentley R., Chasteen T.G. Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol. Mol. Biol. Rev. 2002;66(2):250–271. doi: 10.1128/MMBR.66.2.250-271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradham K.D., Diamond G.L., Burgess M., Juhasz A., Klotzbach J.M., Maddaloni M., Nelson C., Scheckel K., Serda S.M., Stifelman M., Thomas D.J. In vivo and in vitro methods for evaluating soil arsenic bioavailability: relevant to human health risk assessment. J. Toxicol. Environ. Health B Crit. Rev. 2018;21(2):83–114. doi: 10.1080/10937404.2018.1440902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buffi N., Merulla D., Beutier J., Barbaud F., Beggah S., Van Lintel H., Renaud P., Van Der Meer J.R. Development of a microfluidics biosensor for agarose-bead immobilized Escherichia coli bioreporter cells for arsenite detection in aqueous samples. Lab Chip. 2011;11(14):2369–2377. doi: 10.1039/c1lc20274j. [DOI] [PubMed] [Google Scholar]
- 4.Busenlehner L.S., Pennella M.A., Giedroc D.P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2003;27(2–3):131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen S.Y., Wei W., Yin B.C., Tong Y., Lu J., Ye B.C. Development of a highly sensitive whole-cell biosensor for arsenite detection through engineered promoter modifications. ACS Synth. Biol. 2019;8(10):2295–2302. doi: 10.1021/acssynbio.9b00093. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Jiang X., Tie C., Yoo J., Wang Y., Xu M., Sun G., Guo J., Li X. Contribution of nonconsensus base pairs within ArsR binding sequences toward ArsR-DNA binding and arsenic-mediated transcriptional induction. J. Biol. Eng. 2019;13(1):53. doi: 10.1186/s13036-019-0181-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang Y., Walker D.J.F., Vautour K.E., Dixon S., Holmes D.E. Arsenic detoxification by Geobacter species. Appl. Environ. Microbiol. 2017;83(4):e02689. doi: 10.1128/AEM.02689-16. 02616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y.J., Chen W.L., Huang Q.Y. Construction of two lux-tagged Hg2+-specific biosensors and their luminescence performance. Appl. Microbiol. Biotechnol. 2008;79(3):363–370. doi: 10.1007/s00253-008-1442-1. [DOI] [PubMed] [Google Scholar]
- 9.Giloteaux L., Holmes D.E., Williams K.H., Wrighton K.C., Wilkins M.J., Montgomery A.P., Smith J.A., Orellana R., Thompson C.A., Roper T.J., Long P.E., Lovley D.R. Characterization and transcription of arsenic respiration and resistance genes during in situ uranium bioremediation. ISME J. 2013;7:370–383. doi: 10.1038/ismej.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakkila K.M., Nikander P.A., Junttila S.M., Lamminmaki U.J., Virta M.P. Cd-specific mutants of mercury-sensing regulatory protein MerR, generated by directed evolution. Appl. Environ. Microbiol. 2011;77(17):6215–6224. doi: 10.1128/AEM.00662-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hery M., Van Dongen B.E., Gill F., Mondal D., Vaughan D.J., Pancost R.D., Polya D.A., Lloyd J.R. Arsenic release and attenuation in low organic carbon aquifer sediments from West Bengal. Geobiology. 2010;8(2):155–168. doi: 10.1111/j.1472-4669.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- 12.Hou Q.-H., Ma A.-Z., Lv D., Bai Z.-H., Zhuang X.-L., Zhuang G.-Q. The impacts of different long-term fertilization regimes on the bioavailability of arsenic in soil: integrating chemical approach with Escherichia coli arsRp::luc-based biosensor. Appl. Microbiol. Biotechnol. 2014;98(13):6137–6146. doi: 10.1007/s00253-014-5656-0. [DOI] [PubMed] [Google Scholar]
- 13.Hu M., Sun W., Krumins V., Li F. Arsenic contamination influences microbial community structure and putative arsenic metabolism gene abundance in iron plaque on paddy rice root. Sci. Total Environ. 2019;649:405–412. doi: 10.1016/j.scitotenv.2018.08.388. [DOI] [PubMed] [Google Scholar]
- 14.Huang C.W., Wei C.C., Liao V.H. A low cost color-based bacterial biosensor for measuring arsenic in groundwater. Chemosphere. 2015;141:44–49. doi: 10.1016/j.chemosphere.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Hughes M.F. Arsenic toxicity and potential mechanisms of action. Toxicol. Lett. 2002;133(1):1–16. doi: 10.1016/s0378-4274(02)00084-x. [DOI] [PubMed] [Google Scholar]
- 16.Hynninen A., Virta M. In: Whole Cell Sensing Systems II. Belkin S., Gu M.B., editors. Springer; Berlin: 2009. pp. 31–63. [Google Scholar]
- 17.Islam F.S., Gault A.G., Boothman C., Polya D.A., Charnock J.M., Chatterjee D., Lloyd J.R. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature. 2004;430(6995):68–71. doi: 10.1038/nature02638. [DOI] [PubMed] [Google Scholar]
- 18.Jia R., Sun D., Dang Y., Meier D., Holmes D.E., Smith J.A. Carbon cloth enhances treatment of high-strength brewery wastewater in anaerobic dynamic membrane bioreactors. Bioresour. Technol. 2020;298:122547. doi: 10.1016/j.biortech.2019.122547. [DOI] [PubMed] [Google Scholar]
- 19.Jia X., Bu R., Zhao T., Wu K. Sensitive and specific whole-cell biosensor for arsenic detection. Appl. Environ. Microbiol. 2019;85(11):e00694. doi: 10.1128/AEM.00694-19. 00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami Y., Siddiki M.S.R., Inoue K., Otabayashi H., Yoshida K., Ueda S., Miyasaka H., Maeda I. Application of fluorescent protein-tagged trans factors and immobilized cis elements to monitoring of toxic metals based on in vitro protein-DNA interactions. Biosens. Bioelectron. 2010;26(4):1466–1473. doi: 10.1016/j.bios.2010.07.082. [DOI] [PubMed] [Google Scholar]
- 21.Lear G., Song B., Gault A.G., Polya D.A., Lloyd J.R. Molecular analysis of arsenate-reducing bacteria within Cambodian sediments following amendment with acetate. Appl. Environ. Microbiol. 2007;73(4):1041–1048. doi: 10.1128/AEM.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L., Liang J., Hong W., Zhao Y., Sun S., Yang X., Xu A., Hang H., Wu L., Chen S. Evolved bacterial biosensor for arsenite detection in environmental water. Environ. Sci. Technol. 2015;49(10):6149–6155. doi: 10.1021/acs.est.5b00832. [DOI] [PubMed] [Google Scholar]
- 23.Li P.-S., Peng Z.-W., Su J., Tao H.-C. Construction and optimization of a Pseudomonas putida whole-cell bioreporter for detection of bioavailable copper. Biotechnol. Lett. 2014;36(4):761–766. doi: 10.1007/s10529-013-1420-2. [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Wang S., Xu A., Zhang L., Liu H., Ma L.Z. Biological synthesis of high-conductive pili in aerobic bacterium Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2019;103(3):1535–1544. doi: 10.1007/s00253-018-9484-5. [DOI] [PubMed] [Google Scholar]
- 25.Lovley D.R., Phillips E.J. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 1988;54(6):1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo G. Determination of total arsenic in wastewater and sewage sludge samples by using hydride-generation atomic fluorescence spectrometry under the optimized analytical conditions. Anal. Lett. 2012;45(17):2493–2507. [Google Scholar]
- 27.Mandal B.K., Suzuki K.T. Arsenic round the world: a review. Talanta. 2002;58(1):201–235. [PubMed] [Google Scholar]
- 28.Miller J.H. Cold Spring Harbor Laboratory; New York: 1972. Experiments in Molecular Genetics. [Google Scholar]
- 29.Miller W.G., Leveau J.H.J., Lindow S.E. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 2000;13(11):1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen B.L., Willis V.C., Lin C.Y. Western blot analysis to illustrate relative control levels of the lac and ara promoters in Escherichia coli. Biochem. Mol. Biol. Educ. 2007;35(2):133–137. doi: 10.1002/bmb.25. [DOI] [PubMed] [Google Scholar]
- 31.Ohtsuka T., Yamaguchi N., Makino T., Sakurai K., Kimura K., Kudo K., Homma E., Dong D.T., Amachi S. Arsenic dissolution from Japanese paddy soil by a dissimilatory arsenate-reducing bacterium Geobacter sp. OR-1. Environ. Sci. Technol. 2013;47(12):6263–6271. doi: 10.1021/es400231x. [DOI] [PubMed] [Google Scholar]
- 32.Organization W.H. World Health Organization; 2017. Guidelines for Drinking-Water Quality. [Google Scholar]
- 33.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinel-Raffaitin P., Le Hecho I., Amouroux D., Potin-Gautier M. Distribution and fate of inorganic and organic arsenic species in landfill leachates and biogases. Environ. Sci. Technol. 2007;41(13):4536–4541. doi: 10.1021/es0628506. [DOI] [PubMed] [Google Scholar]
- 35.Podgorski J., Berg M. Global threat of arsenic in groundwater. Science. 2020;368(6493):845–850. doi: 10.1126/science.aba1510. [DOI] [PubMed] [Google Scholar]
- 36.Ponthieu M., Pinel-Raffaitin P., Le Hecho I., Mazeas L., Amouroux D., Donard O.F., Potin-Gautier M. Speciation analysis of arsenic in landfill leachate. Water Res. 2007;41(14):3177–3185. doi: 10.1016/j.watres.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 37.Qiao J., Li X., Li F., Liu T., Young L.Y., Huang W., Sun K., Tong H., Hu M. Humic substances facilitate arsenic reduction and release in flooded paddy soil. Environ. Sci. Technol. 2019;53(9):5034–5042. doi: 10.1021/acs.est.8b06333. [DOI] [PubMed] [Google Scholar]
- 38.Reguera G., Nevin K.P., Nicoll J.S., Covalla S.F., Woodard T.L., Lovley D.R. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 2006;72(11):7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riether K., Dollard M.A., Billard P. Assessment of heavy metal bioavailability using Escherichia coli zntAp::lux and copAp::lux-based biosensors. Appl. Microbiol. Biotechnol. 2001;57(5–6):712–716. doi: 10.1007/s00253-001-0852-0. [DOI] [PubMed] [Google Scholar]
- 40.Rosen B.P. Biochemistry of arsenic detoxification. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2002;529(1):86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 41.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma P., Asad S., Ali A. Bioluminescent bioreporter for assessment of arsenic contamination in water samples of India. J. Biosci. 2013;38(2):251–258. doi: 10.1007/s12038-013-9305-z. [DOI] [PubMed] [Google Scholar]
- 43.Smith A.H., Lingas E.O., Rahman M. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull. World Health Organ. 2000;78(9):1093–1103. [PMC free article] [PubMed] [Google Scholar]
- 44.Steinmaus C.M., George C.M., Kalman D.A., Smith A.H. Evaluation of two new arsenic field test kits capable of detecting arsenic water concentrations close to 10 microg/L. Environ. Sci. Technol. 2006;40(10):3362–3366. doi: 10.1021/es060015i. [DOI] [PubMed] [Google Scholar]
- 45.Tao H.-C., Peng Z.-W., Li P.-S., Yu T.-A., Su J. Optimizing cadmium and mercury specificity of CadR-based E. coli biosensors by redesign of CadR. Biotechnol. Lett. 2013;35(8):1253–1258. doi: 10.1007/s10529-013-1216-4. [DOI] [PubMed] [Google Scholar]
- 46.Trang P.T., Berg M., Viet P.H., Van Mui N., Van Der Meer J.R. Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ. Sci. Technol. 2005;39(19):7625–7630. doi: 10.1021/es050992e. [DOI] [PubMed] [Google Scholar]
- 47.Webster D.P., Teravest M.A., Doud D.F., Chakravorty A., Holmes E.C., Radens C.M., Sureka S., Gralnick J.A., Angenent L.T. An arsenic-specific biosensor with genetically engineered Shewanella oneidensis in a bioelectrochemical system. Biosens. Bioelectron. 2014;62:320–324. doi: 10.1016/j.bios.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Xiao K.Q., Li L.G., Ma L.P., Zhang S.Y., Bao P., Zhang T., Zhu Y.G. Metagenomic analysis revealed highly diverse microbial arsenic metabolism genes in paddy soils with low-arsenic contents. Environ. Pollut. 2016;211:1–8. doi: 10.1016/j.envpol.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 49.Ye J., Rensing C., Rosen B.P., Zhu Y.G. Arsenic biomethylation by photosynthetic organisms. Trends Plant Sci. 2012;17(3):155–162. doi: 10.1016/j.tplants.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon Y., Kim S., Chae Y., Jeong S.-W., An Y.-J. Evaluation of bioavailable arsenic and remediation performance using a whole-cell bioreporter. Sci. Total Environ. 2016;547:125–131. doi: 10.1016/j.scitotenv.2015.12.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.