Abstract
Bioremediation of groundwater contaminated by a mixture of aromatic hydrocarbons and chlorinated solvents is typically challenged because these contaminants are degraded via distinctive oxidative and reductive pathways, thus requiring different amendments and redox conditions. Here, we provided the proof-of-concept of a single-stage treatment of synthetic groundwater containing toluene and trichloroethene (TCE) in a tubular bioelectrochemical reactor, known as a “bioelectric well”. Toluene was degraded by a microbial bioanode (up to 150 μmol L−1 d−1) with a polarized graphite anode (+0.2 V vs. SHE) serving as the terminal electron acceptor. The electric current deriving from microbially-driven toluene oxidation resulted in (abiotic) hydrogen production (at a stainless-steel cathode), which sustained the reductive dechlorination of TCE to less-chlorinated intermediates (i.e., cis-DCE, VC, and ETH), at a maximum rate of 500 μeq L−1 d−1, in the bulk of the reactor. A phylogenetic and functional gene-based analysis of the “bioelectric well” confirmed the establishment of a microbiome harboring the metabolic potential for anaerobic toluene oxidation and TCE reductive dechlorination. However, Toluene degradation and current generation were found to be rate-limited by external mass transport phenomena, thus indicating the existing potential for further process optimization.
Keywords: Bioelectric well, Dehalococcoides mccartyi, Electrobioremediation, Groundwater remediation, Toluene, Trichloroethene
Graphical abstract
Highlights
-
•
Toluene and TCE are biodegraded in a single-stage bioelectrochemical reactor.
-
•
Toluene is oxidazied at the anode while TCE is dechlorinated at the cathode.
-
•
Mass-transport limitations affects toluene degradation and current generation.
-
•
Phylogenetic and functional gene based analysis reveal a competent microbiome.
1. Introduction
Petroleum hydrocarbons (PH) and chlorinated aliphatic hydrocarbons (CAHs) are among the most frequent and harmful soil and groundwater contaminants [1]. Their occurrence in subsurface environments, typically caused by accidental spills, leakage from underground storage tanks, and improper manufacturing or disposal practices, poses severe environmental and health concerns due to the relevant toxicity and recalcitrance of such compounds [2]. In the last decades, the ever-increasing knowledge gathered on the ability of microorganisms to degrade or transform pollutants into harmless end-products and on their degradative metabolic pathways has spurred the interest towards the application of bioremediation approaches for the cleanup of contaminated sites [[3], [4], [5], [6], [7]]. These are typically based on manipulating environmental conditions through, for instance, the control of the redox potential and/or the supplementation of electron donors or acceptors [8]. A challenging problem related to the bioremediation of sites containing a mixture of PH and CAHs is, however, the fact that these contaminants are degraded via distinctive oxidative and reductive pathways, thus requiring different amendments and redox conditions [[9], [10], [11], [12]]. In particular, the (aerobic or anaerobic) oxidative biodegradation of PH can be typically stimulated by providing naturally occurring microbial communities with an otherwise limiting electron acceptor (e.g., oxygen, nitrate, sulfate). By contrast, CAHs are preferably biodegraded via a reductive pathway, often referred to as reductive dechlorination (RD) in which the chlorinated contaminant serves as a respiratory electron acceptor in the energy metabolism of so-called organohalide-respiring bacteria (OHRB) [13,14]. Among them, Dehalococcoides mccartyi is the only one capable of dechlorinating CAHs to harmless ethene through the catalytic activity of the reductive dehalogenases (i.e., tceA, bvcA, vcrA) directly responsible for the RD process [15]. Thus, the RD of CAHs can be stimulated by providing autochthonous OHRB with suitable electron donors (i.e., H2 or fermentable substrates).
Clearly, to minimize the establishment of competitive reactions which may adversely affect the rate and efficiency of the bioremediation process, the supply of the electron acceptor (to drive the oxidation of PH) and the electron donor (to drive the RD of CAH) need to be kept separated in space or time, thereby complicating the design, operation, and control of the whole bioremediation process.
In principle, a possibility exists that PH may serve as electron donors in the RD of CAH, thus simplifying the overall treatment of PH and CAH mixtures. However, this chance is greatly limited because the majority of OHRB (e.g., Dehalococcoides mccartyi, the only one capable of dechlorinating chloroethenes to harmless ethane) are restricted to using H2 as the sole electron donor [16]. Hence, a single-stage biotreatment of PH and CAH, although of potentially great practical and economical value, would require the establishment of close syntrophic cooperation among PH- and CAH-degrading microorganisms which is seldom observed both in the field and in laboratory experiments.
In recent years, microbial electrochemical technologies (METs) have emerged as a novel and highly versatile platform for treating soils and groundwater contaminated by either PH or CAH [17,18]. METs employ electro-active microorganisms to electro-catalyze oxidation or reduction reactions using solid-state electrodes as virtually inexhaustible electron acceptors or donors. In previous studies, METs have been successfully employed to stimulate the oxidative treatment of groundwater containing PH such as benzene, toluene, xylenes, and ethyl-benzene (BTEX) [[19], [20], [21], [22]], as well as the reductive dechlorination of a variety of CAHs, including perchloroethene (PCE), trichloroethene (TCE), and 1,2,-dichloroethane (1,2-DCA) [[23], [24], [25], [26]]. However, none of these studies has attempted to treat PH and CAH simultaneously at the anode and cathode of the same MET to achieve a single-stage treatment of commingled PH/CAH groundwater.
Here, we explored for the first time the possibility of using a “bioelectric well”, a previously developed MET specifically designed for in situ treatment of contaminated groundwater [27], for the bioremediation of a synthetic groundwater containing a mixture of toluene (model PH) and TCE (model CAH). Results demonstrated that the electric current deriving from the microbially-driven oxidation of toluene at the anode generates H2 at the cathode, which, in turn, can sustain the RD of TCE to less-chlorinated or eventually non-chlorinated end-products.
2. Materials and methods
2.1. Reactor setup and operation
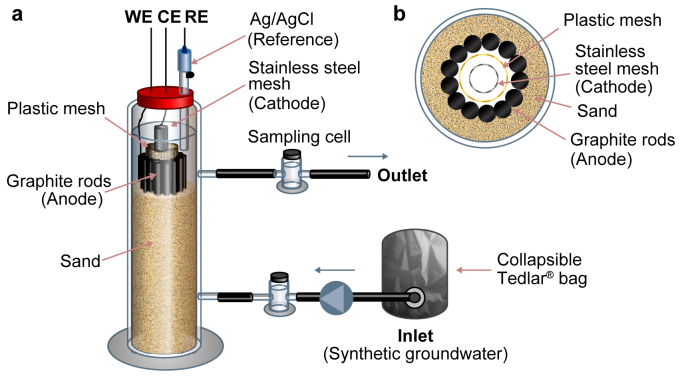
The bioelectrochemical reactor used in the present study consisted of a 250 mL-glass cylinder filled with river sand and housing eight contiguous graphite rods (purity: 99.995%, length: 30 cm, ø: 0.6 cm; Sigma-Aldrich, Italy) and a concentric stainless-steel mesh cathode (dimensions: 3 × 30 cm; type 304, Alpha Aesar, USA) (Fig. 1a). Anode and cathode were kept physically separated by a polyethylene mesh (ø: 1 cm, length: 30 cm; Fig. 1b), yet allowing the hydraulic connection between the anodic and cathodic zones. An Ag/AgCl reference electrode (+198 mV versus the standard hydrogen electrode, SHE) was placed on top of the cylinder to control, using an IVIUMnSTAT potentiostat (IVIUM Technologies, The Netherlands), the potential of the anode at the desired value (i.e., +200 mV vs. SHE). Titanium wires (ø: 0.81 mm, Alfa Aesar, USA) connected anode and cathode to the potentiostat. At the start of the study, the reactor was inoculated with 0.2 L of groundwater from a toluene-contaminated site in Italy [28], and 50 mL of a TCE-to ethene dechlorinating enrichment culture [29].
Fig. 1.
a, Schematic drawing of the continuous flow bioelectrochemical reactor. b, Cross-sectional view of the bioelectrochemical reactor displaying the relative position of electrodes.
Throughout the study, the reactor was continuously fed with synthetic groundwater consisting of an anaerobic mineral medium spiked with toluene and TCE (Sigma-Aldrich, Italy) at the desired concentrations (Table 1). The medium contained the following components: NH4Cl (0.5 g L−1), MgCl2·6H2O (0.1 g L−1), K2HPO4 (0.4 g L−1), CaCl2·2H2O (0.05 g L−1), 2 mL L−1 of a trace metal solution [30], and 2 mL L−1 of vitamin solution [31]. The electrical conductivity of the medium was 4.8 mS cm−1, hence within the range of values typically reported for highly contaminated groundwater (i.e., 0.67–7.98 mS cm−1) [32].
Table 1.
Main operating conditions applied during the different experimental runs.
| Run I | Run II | Run III | Run IV | |
|---|---|---|---|---|
| Operational period (days) | 0–10 | 11–24 | 25–38 | 39–53 |
| Anode polarization (V vs. SHE) | +0.2 | +0.2 | +0.2 | OCP |
| HRT (h) | 9.3 | 9.3 | 9.3 | 9.3 |
| Average influent Toluene conc. (μmol L−1) | 170 ± 6 | 161 ± 7 | 82 ± 8 | 119 ± 3 |
| Average influent TCE conc. (μmol L−1) | 170 ± 15 | – | 110 ± 9 | 157 ± 6 |
During operation, the synthetic groundwater was maintained in a 5 L collapsible Tedlar® bag and entered the reactor through a port situated at the bottom of the cylinder (flow rate: 0.75 L d−1, HRT: 9.3 h), while the treated effluent exited from a port positioned near the upper end (Fig. 1). The inlet and outlet of the reactor were equipped with flow-through, vigorously stirred sampling cells (volume: 25 mL). All the tubings were made of Viton® (Sigma-Aldrich, Italy), which keeps volatilization losses and organic contaminant adsorption to a minimum. Throughout the whole study, the system was kept at room temperature (i.e., 24 ± 3 °C).
2.2. Gas analyses
Gaseous samples, taken from the sampling cells using gastight syringes, were analyzed in O2, H2 and CH4 using a gas-chromatograph (Agilent 8860, GC system, USA) equipped with a thermal conductivity detector (TCD). The concentration of toluene, TCE, cis-dichloroethene (cis-DCE), vinyl chloride (VC) and ethene (ETH), were measured by injecting gaseous samples into a gas-chromatograph (Agilent 8860, GC system, USA) equipped with a flame ionization detector (FID). Gas-phase concentrations were converted into liquid-phase concentrations using tabulated Henry's Law constants [33]. The GC methods, calibration ranges and LOD of analytical methods are reported elsewhere [28,29].
2.3. High-throughput bacterial and archaeal 16S rRNA gene sequencing
Sample of effluent at the beginning of the experiment T0: 1 L) and at the end of Run III (15 mL) were filtered through polycarbonate membranes (pore size: 0.2 mm, diameter: 47 mm, Nuclepore) and immediately stored at −20 °C. Genomic DNA was extracted with the DNeasy PowerSoil Pro Kit (QIAGEN, Germantown, MD) and utilized as the template for the amplification of the V1–V3 region of 16S rRNA gene of Bacteria (27F 5′-AGAGTTTGATCCTGGCTCAG-3’; 534R 5′-ATTACCGCGGCTGCTGG-3′) and the region V3–V5 of 16S rRNA gene of Archaea (340F 5′-CCCTAHGGGGYGCASCA-3’; 915R 5′-GWGCYCCCCCGYCAATTC-3′) following the procedure for library preparation and sequencing described in Ref. [34]. The paired-end sequencing (2 × 301bp) was performed on a MiSeq platform (Illumina) using a MiSeq Reagent kit v3, 600 cycles (Illumina, USA) following the standard guidelines for preparing and loading samples. Phix control library was spiked at a concentration of 20%.
Bioinformatics analyses were carried out using QIIME2 v. 2018.2 [35], following the procedure previously reported [36]. High-throughput sequencing of the V1–V3 and V3–V5 regions of the bacterial and archaeal 16S rRNA gene yielded a total of 20,438 and 30,988 sequence reads after quality control and bioinformatic processing that resolved into 318 and 15 ASVs, respectively. Datasets are available through the Sequence Read Archive (SRA) under accession PRJNA799244.
2.4. Droplet Digital PCR quantification of key-functional genes
The QX200™ Droplet Digital™ PCR System (ddPCR™, Bio-Rad, USA) was used to perform absolute quantification of the functional genes involved in the anaerobic degradation of PH and CAH.
For the estimation of PH related genes, such as benzylsuccinate synthase (bssA gene) and benzoyl CoA reductases class I (bcrC, bzdN) and class II (bamB), the ddPCR reaction mixture consisted of 11 μL of 2 × ddPCR EvaGreen supermix (Bio-Rad, USA), 1 μL of each primer (at 7.5 μM concentration), 6 μL of nuclease-free water, and 3 μL of sample DNA.
For the quantification of the reductive dehalogenase genes tceA, bvcA and vcrA, the PCR reaction mixtures were prepared in a 22 μL total volume for each sample, including ddPCR Supermix for Probes® (Bio-Rad, USA), 3 μL of DNA as a template, 900 nM of each primer and 300 nM of TaqMan probe. The set of primer and probes used are summarized in Table S1.
Droplets were generated using an eight-channel DG8 cartridge and cartridge holder (Bio-Rad, USA). 20 μL of PCR reaction mixture were combined with 70 μL of droplet generation oil and placed in QX200 Droplet Generator (Bio-Rad, USA). Following droplet generation, 40 μL of water-in-oil droplets were transferred to a standard 96-well PCR plate, which was heat-sealed with foil plate using the PX1™ PCR plate sealer (Bio-Rad, USA) and amplified with the T100 thermal cycler (Bio-Rad, USA).
PCR cycle parameters for PH-related genes were as follows: 5 min at 95 °C, followed by 39 cycles of 30 s at 95 °C and 1 min at 55–60 °C according to the primer pair (ramping speed: 2 °C s−1), followed by 5 min hold at 4 °C and 5 min at 95 °C. Whereas the PCR cycling conditions for the tceA, bvcA and vcrA genes were: 10 min at 95 °C, 39 cycles for 30 s at 94 °C, and 1 min at 60 °C (ramping rate: 2 °C s−1), 10 min at 98 °C, ending at 4 °C.
Upon completion of PCR, the plate was transferred to QX200 Droplet Reader (Bio-Rad, USA) to detect positive and negative fluorescent droplets to calculate the targeted gene concentrations. Data were analyzed using QuantaSoft Software® (Bio-Rad, USA), and quantitative data were reported as gene copy numbers per volume of sample (95% confidence intervals).
2.5. Calculations
The removal rate of toluene and TCE q (μmol L−1 d−1) were calculated using the following equation:
| (1) |
where Cin and Cout (μmol L−1 d−1) are the toluene or TCE liquid phase concentrations measured in the influent and the effluent, Vr (L) is the empty volume of the reactor and Q (L d−1) is the volumetric flow rate.
Similarly, the formation rate of TCE reductive dechlorination products qRD (μeq L−1 d−1) was calculated as:
| (2) |
where Cout,DCE, Cout,VC, and Cout,ETH (μmol L−1 d−1) are the measured liquid phase concentration of dechlorination products and 2, 4, or 6 are the number of moles of electrons required for the formation of 1 mol of cis-DCE, VC, ethene from TCE, respectively [37].
The toluene or TCE removal efficiencies (η%) were calculated as follows:
| (3) |
The columbic efficiency (CE) was calculated as the ratio between charge, that is the integral of the electric current over time, and the theoretical charge deriving from the oxidation of removed toluene, according to the following equation:
| (4) |
where i is the measured current (mA), F is the Faraday's constant and ΔTol is the amounts of removed toluene per day (mmol d−1), fTol represents the number of mmol of electrons released from the complete oxidation of 1 mmol of toluene.
The apparent activation energy of the current-producing toluene degradation rate was estimated as reported elsewhere [38]. In brief, as electric current is proportional to toluene degradation rate, which is, in turn, proportional to the rate constant, then in a narrow temperature range (e.g., 15–30 °C), it is possible to represent the data by the following Arrhenius equation:
| (5) |
where k is the rate constant, Ea (J mol−1) the apparent activation energy, R the gas constant (J mol−1 K−1), T (K) the temperature, and A the preexponential factor.
3. Results and discussion
3.1. Performance of the continuous flow bioelectrochemical reactor
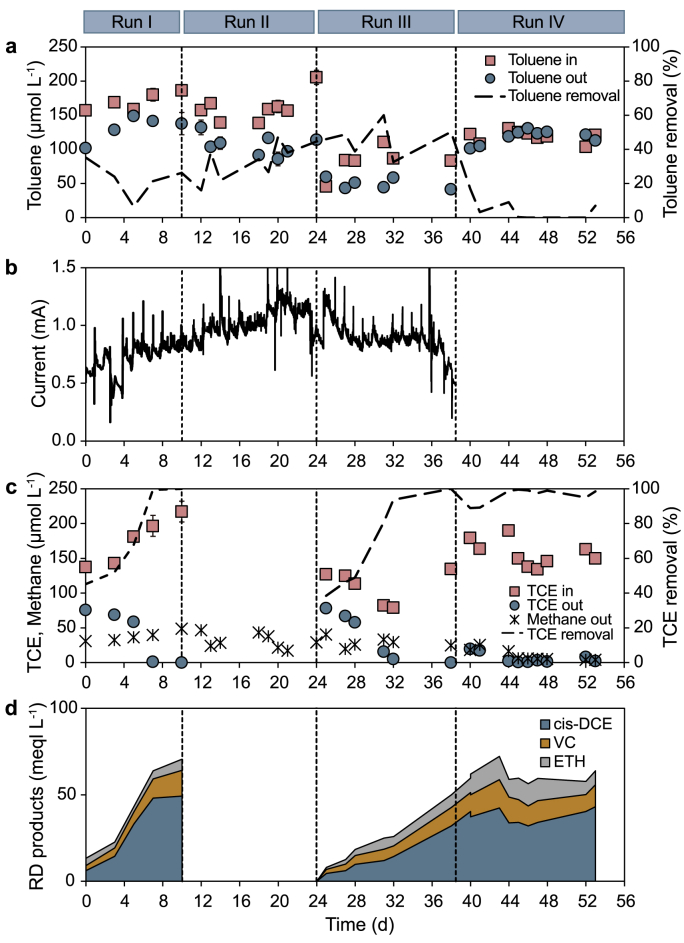
The continuous flow bioelectrochemical reactor was operated for 53 days (i.e., corresponding to nearly 140 HRT) under different operating conditions, as summarized in Table 1. During Run I, the feed solution contained a mixture of toluene (170 ± 6 μmol L−1) and TCE (170 ± 15 μmol L−1) as co-contaminants, while the anode of the reactor was poised at +0.2 V vs. SHE. On average, during this run, 23 ± 5% of the influent toluene was removed (Fig. 2a). Toluene removal was accompanied by electric current generation, which gradually increased from around 0.55 mA to around 0.85 mA throughout the run (Fig. 2b). The resulting average coulombic efficiency was 40 ± 4%, thus likely indicating that other biotic (e.g., methanogenic biodegradation) or abiotic (e.g., adsorption and/or volatilization) processes also contributed to the observed toluene removal.
Fig. 2.
Performance parameters of the continuous flow bioelectrochemical reactor under the different experimental runs. a, Time-course of influent and effluent toluene concentration and toluene removal efficiency. b, Electric current generated from toluene oxidation. c, Influent and effluent TCE concentration and effluent methane concentration. d, Concentration of TCE reductive dechlorination products.
Interestingly, TCE removal (Fig. 2c) also increased over time (up to nearly 100% by day 7), as well as the sum of reductive dechlorination products (Fig. 2d), mainly consisting of cis-DCE (50%, on an electron equivalent basis), VC (30%), and ETH (20%), thus mirroring the observed trend of the electric current.
Methane was also detected in the effluent of the reactor at an average concentration of 38 ± 3 μmol L−1. In principle, methane production could derive both from the syntrophic conversion of toluene [39], a metabolic process in competition with the electric current generation, or from the biological reduction of carbon dioxide fueled by cathodic H2.
Throughout the whole experimental period, the pH of the influent and effluent of the reactor were typically in the range 6.5–7 and were statistically indistinguishable. This reflects that the (proton-releasing) anodic reaction and the (proton-consuming) cathodic reaction occurred in the same reaction environment, hence preventing the establishment of pH gradients.
During Run II, TCE was omitted from the synthetic groundwater, and the reactor was fed with toluene as the only organic contaminant. This change resulted in a slight increase in toluene removal, which averaged 33 ± 4% (Fig. 3a), in electric current generation, which steadily increased up to nearly 1.2 mA (Fig. 3b), and in the average coulombic efficiency, which accounted for 58 ± 7%. This latter value clearly indicates that electric current generation was the primary biological mechanism underlying toluene removal.
Fig. 3.
Average toluene removal rate (a), electric current generation (b), and cumulative reductive dechlorination rate (c) during the different experimental runs. As for the reductive dechlorination rate, reported values refer to the last data points of each run when a nearly stable activity was achieved.
Taken as a whole, these findings point to a slight inhibitory effect of TCE on the conversion of toluene into electric current, which can at least partially explain the lower toluene degradation rates observed in the present study compared to previous investigations in which, however, toluene was supplied as the only organic contaminant [21,28].
During Run III, the influent toluene concentration was halved (82 ± 8 μmol L−1), and the influent TCE concentration was reduced by 35%. (Fig. 2). This resulted in a decrease of the average toluene removal rate (Fig. 3a) and of the produced electric current (Fig. 3b), hence confirming the existing correlation between these two parameters. Analogously to what was observed during Run I, also during Run III the removal of TCE (and the corresponding formation of RD products) steadily increased over time until reaching, by the end of the run, values comparable to those observed during Run I (Fig. 3c). Collectively, this finding suggests that TCE dechlorination was not limited by electron donor (H2 or electrons) availability, in agreement with the stoichiometry of the involved reactions which indicates that the complete electrogenic biodegradation of 1 mol of toluene (equation (6)) would provide sufficient H2 (equation (7)) to drive the complete reduction of 6 mol of TCE to ethene (equation (8)).
| (6) |
| (7) |
| (8) |
During the last operational run, toluene and TCE were simultaneously fed to the bioelectrochemical reactor, which was maintained at open circuit potential (OCP). Notably, upon removal of anode polarization, toluene biodegradation ceased almost immediately, and the effluent concentration rapidly equaled the influent concentration (Fig. 2a). Upon interruption of the anodic polarization, the RD of TCE ceased to increase and stabilized at values similar to those observed during the previous runs. Most probably, during the whole Run III, the reductive dechlorination of TCE was fueled by electrons deriving from the endogenous decay of the biomass present within the bioreactor (e.g., the anodic biofilm grown on toluene). Although such a metabolic activity is meant to decline over time and therefore cannot be sustained over a long term, it may still be important in buffering TCE dechlorination during periods in which electrode polarization is temporarily dismissed.
3.2. Mass-transport limitations
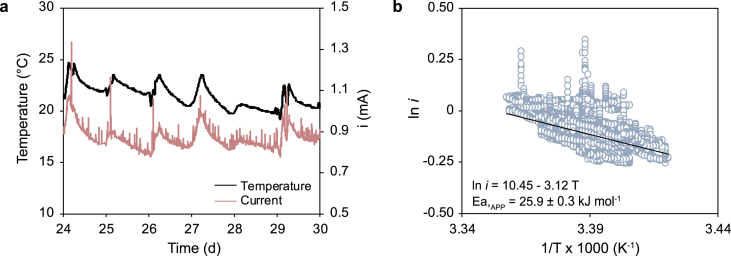
The observed inhibitory effect of TCE on the electrogenic toluene degradation was not sufficient to explain the substantially lower (around 3 fold) performance of the bioelectrochemical reactor, particularly in terms of toluene removal and electric current generation, relative to previous studies, although the same inoculum and identical anodes were employed [28]. In that previous case, however, the bioelectrochemical reactor was equipped with an internal recycle of the liquid phase to minimize the establishment of substrate/products concentration gradients and was not filled with sand, as in the present case. Hence, to verify whether the performance of the reactor was controlled by mass-transport limitations triggered by the adopted changes in the reactor configuration, we analyzed the existing correlations between the electric current and the ambient temperature (Fig. 4a). In brief, as the electric current is proportional to the rate of toluene oxidation which is, in turn, proportional to the rate constant of the reaction, experimental data collected during Run III were plotted in Arrhenius form (ln i vs. 1/T) to determine the “apparent” activation energy of the reaction (i.e., the current response of the system to temperature variations; Fig. 4b). Although to the best of our knowledge, no other values of the activation energy are available in the literature for the electrogenic toluene oxidation reaction, the herein obtained value of 25.9 ± 0.3 kJ mol−1 is substantially lower concerning values reported for other bioelectrocatalytic reactions, or for (bio)chemical reactions in general, which typically fall within the range (40–80 kJ mol−1) [38,40,41]. Collectively, this result provides a strong line of evidence that the electric current deriving from the microbially-driven toluene oxidation was in turn rate-limited by mass-transport of the substrate (or products) rather than by the intrinsic kinetics of the bioelectrocatalytic reaction.
Fig. 4.
a, Fluctuations of electric current and ambient temperature relative to the period of Run 3 from day 24 to day 30. b, Dependence of electric current on temperature: Arrhenius plot.
3.3. Microbial community characterization
The biomolecular characterization revealed slight differences among microbial communities at the beginning of the experiment and at the end of Run III within the bioelectrochemical reactor. The results obtained with the high-throughput sequencing of bacterial and archaeal 16S rRNA gene suggested an initial high potential in PH degradation and TCE-RD. Indeed, the microbiome at T0 is mainly composed by genera Pseudomonas (24.2% of total reads), Acinetobacter (13.0%), Dechloromonas (5.7%), Dehalococcoides (4.4%), Dechlorobacter (1.0%), and unidentified members of Burkholderiaceae family (19.0%) (Fig. 5a). The presence of these genera was often reported in previous studies for their capability to degrade petroleum hydrocarbon and to reduce chlorinated compounds [42,43]. The archaeal microbiome was mostly composed of genus Methanobrevibacter (91.0%), followed to a minor extent by Methanospirillum (5.2%) and Methanobacterium (2.4%) (Fig. 5a).
Fig. 5.
Microbial characterization at genus-level of bacterial (a) (only genera >1% of total reads in at least one sample are depicted) and archaeal (b) communities within the bioelectrochemical reactor estimated by high-throughput sequencing.
The characterization at the end of Run III revealed a bacterial community mainly represented by genera Azospira (30.9%), Thermincola (7.0%), Zooglea (6.7%), and Azovibrio (6.5%). Overall, AVSs affiliated with Rhodocyclaceae family represented 45.5% of the total sequences. Notably, its members are commonly found in polluted environments and are typically associated with the anaerobic biodegradation of a wide range of aromatic hydrocarbons (Oren, 2014). The presence of genera Thermincola and Zooglea can be related to the toluene degradation observed in the bioelectrochemical reactor. These genera are, indeed, reported to be active degraders of petroleum hydrocarbons [44,45].
The presence of Azovibrio, a microaerophilic, N2 fixing bacterium, capable of using oxygen, nitrate or even perchlorate as the terminal electron acceptor, have been previously demonstrated to boost bioremediation processes in oil-contaminated soils [46,47]. Members of genus Dechloromonas, together with the closely related Azospira, represent the predominant perchlorate-reducing bacteria in the environment and have been found to be ubiquitous [[48], [49], [50]]. Several strains of Dechloromonas have been studied for their capability to degrade benzene, toluene, ethylbenzene, and xylene compounds both aerobically and anaerobically with perchlorate or chlorate as a suitable electron acceptor [48,51].
Furthermore, in line with low CH4 production herein measured and with previous evidence in methanogenic bioelectrochemical reactors [[52], [53], [54], [55]], the archaeal microbiome was mostly represented by hydrogenotrophic methanogens affiliated with genera Methanobacterium (79.9%), Methanospirillum (10.4%), and Methanobrevibacter (9.2%) (Fig. 5b).
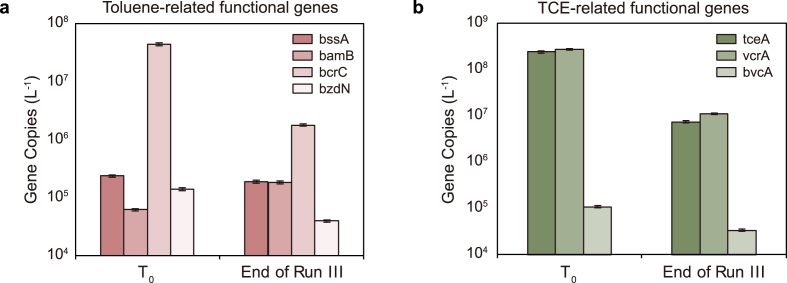
In line with sequencing output and process data, the abundance of key-functional genes involved in PH-degradation (i.e., bssA, brcC, bzdN, bamB) and TCE-reductive dechlorination (i.e., tceA, vcrA, bvcA) were quantified in the samples at T0 and the end of run III (Fig. 6). In detail, the abundance of benzylsuccinate synthase (bssA), the biomarker gene of anaerobic toluene degrading bacteria that use fumarate addition pathway [56], showed similar values between the beginning of the experiment and the end of run III (2.1 × 105 gene copies L−1 on average; Fig. 6a), consistently with the toluene degradation rate observed in the reactor. Furthermore, the abundance of bcrC and bzdN genes, encoding for the ATP-dependent class I benzoyl CoA reductases [57,58], decreased from 4.4 × 107 to 1.8 × 106 and from 1.4 × 105 to 4.0 × 104 gene copies L−1, respectively (Fig. 6a). A slightly increasing trend was observed for the ATP-independent class II (bamB) benzoyl. Instead, CoA reductases counting 6.2 × 104 gene copies L−1 in the sample at T0 and 1.8 × 105 gene copies L−1 at the end of run III (Fig. 6a).
Fig. 6.
Abundance of key-functional genes involved in anaerobic toluene degradation (a) and TCE reductive dechlorination (b) estimated by ddPCR in the effluent of the reactor at the beginning of the experiment (T0) and at the end of run III. Data are reported in Log scale.
In addition, at the beginning of the experiment, tceA (2.4 × 108 gene copies L−1) and vcrA (2.7 × 108 gene copies L−1) were the most abundant reductive dehalogenase genes found, while bvcA (1.1 × 105 gene copies L−1) was detected at minor extent (Fig. 6b). In line with the decrement of the cumulative RD rate observed at the end of the run III, all the reductive dehalogenase genes analyzed decreased at least by one order of magnitude and accounted for 7.4× 106, 1.1 × 107, and 3.3 × 104 gene copies L−1 of tceA, vcrA, and bvcA, respectively (Fig. 6b).
Overall, differences in microbial composition and abundance of key functional genes were observed at the end of run III compared to T0, most likely due to the various experimental conditions tested. However, even though these variations affected the microbial communities, a microbiome highly involved in the simultaneous toluene degradation and TCE-reductive dechlorination was strongly established. In fact, the microbiological results fully supported the PH-degradation and TCE-reductive dechlorination rates observed in this study.
4. Conclusions
This study demonstrated, for the first time, the possibility to treat (synthetic) groundwater containing a mixture of toluene and TCE using a single-stage bioelectrochemical system, which exploited a graphite anode (for toluene oxidation) and a stainless-steel cathode (for TCE reduction) positioned in the same reaction environment. The electric current (up to nearly 1 mA) resulting from the microbially-catalyzed oxidation of toluene (with a maximum observed removal rate of 150 μmol L−1 d−1), with a polarized anode (+0.2 V vs. SHE) serving as the terminal electron acceptor. In the bulk of the reactor, the hydrogen produced (abiotically) at the cathode sustained the partial dechlorination of TCE to less-chlorinated intermediates (i.e., cis-DCE, VC, and ETH), at a maximum rate of 500 μeq L−1 d−1. However, toluene degradation and current generation were found to be rate-limited by external mass transport phenomena. Further studies focusing on the identification of alternative packing materials and different hydrodynamic regimes are thus needed to improve the catalytic efficiency of the treatment system. Furthermore, despite the abundance of Dehalococcoides mccartyi and the related dechlorination functional genes (up to nearly 107 copies L−1 for tceA and vcrA), TCE degradation appeared to be relatively slow and incomplete, likely due to ineffective utilization of the generated hydrogen. Thus, in this case, further research efforts are certainly warranted to increase the rate and yield of TCE conversion, possibly into harmless non-chlorinated end products.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Camilla de Laurentiis is acknowledged for her skillful support with the gas-chromatographic analyses. This study was supported by the European Union’s Horizon 2020 project ELECTRA (www.electra.site) under grant agreement No. 826244.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2022.100171.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Majone M., Verdini R., Aulenta F., Rossetti S., Tandoi V., Kalogerakis N., Agathos S., Puig S., Zanaroli G., Fava F. In situ groundwater and sediment bioremediation: barriers and perspectives at European contaminated sites. N. Biotechnol. 2015;32:133–146. doi: 10.1016/j.nbt.2014.02.011. https://linkinghub.elsevier.com/retrieve/pii/S1871678414000247 [DOI] [PubMed] [Google Scholar]
- 2.Lhotský O., Krákorová E., Linhartová L., Křesinová Z., Steinová J., Dvořák L., Rodsand T., Filipová A., Kroupová K., Wimmerová L., Kukačka J., Cajthaml T. Assessment of biodegradation potential at a site contaminated by a mixture of BTEX, chlorinated pollutants and pharmaceuticals using passive sampling methods – case study. Sci. Total Environ. 2017;607–608:1451–1465. doi: 10.1016/j.scitotenv.2017.06.193. https://www.sciencedirect.com/science/article/pii/S0048969717316121 [DOI] [PubMed] [Google Scholar]
- 3.Alvarez P.J.J., Illman W.A. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2005. Bioremediation and Natural Attenuation. Bioremediation Nat. Attenuation. [DOI] [Google Scholar]
- 4.Jørgensen K.S. Compr. Biotechnol. Elsevier; 2011. In situ bioremediation; pp. 59–67.https://linkinghub.elsevier.com/retrieve/pii/B978008088504900372X [Google Scholar]
- 5.Megharaj M., Ramakrishnan B., Venkateswarlu K., Sethunathan N., Naidu R. Bioremediation approaches for organic pollutants: a critical perspective. Environ. Int. 2011;37:1362–1375. doi: 10.1016/j.envint.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Sadañoski M.A., Tatarin A.S., Barchuk M.L., Gonzalez M., Pegoraro C.N., Fonseca M.I., Levin L.N., Villalba L.L. Evaluation of bioremediation strategies for treating recalcitrant halo-organic pollutants in soil environments. Ecotoxicol. Environ. Saf. 2020;202 doi: 10.1016/j.ecoenv.2020.110929. https://linkinghub.elsevier.com/retrieve/pii/S0147651320307685 [DOI] [PubMed] [Google Scholar]
- 7.Wu M., Li W., Dick W.A., Ye X., Chen K., Kost D., Chen L. Bioremediation of hydrocarbon degradation in a petroleum-contaminated soil and microbial population and activity determination. Chemosphere. 2017;169:124–130. doi: 10.1016/j.chemosphere.2016.11.059. https://linkinghub.elsevier.com/retrieve/pii/S0045653516315946 [DOI] [PubMed] [Google Scholar]
- 8.Davoodi S.M., Miri S., Taheran M., Brar S.K., Galvez-Cloutier R., Martel R. Bioremediation of unconventional oil contaminated ecosystems under natural and assisted conditions: a review. Environ. Sci. Technol. 2020;54:2054–2067. doi: 10.1021/acs.est.9b00906. [DOI] [PubMed] [Google Scholar]
- 9.Aulenta F., Bianchi A., Majone M., Petrangeli Papini M., Potalivo M., Tandoi V. Assessment of natural or enhanced in situ bioremediation at a chlorinated solvent-contaminated aquifer in Italy: a microcosm study. Environ. Int. 2005;31:185–190. doi: 10.1016/j.envint.2004.09.014. https://linkinghub.elsevier.com/retrieve/pii/S0160412004001643 [DOI] [PubMed] [Google Scholar]
- 10.Aulenta F., Majone M., Tandoi V. Enhanced anaerobic bioremediation of chlorinated solvents: environmental factors influencing microbial activity and their relevance under field conditions. J. Chem. Technol. Biotechnol. 2006;81 [Google Scholar]
- 11.Rabus R., Boll M., Heider J., Meckenstock R.U., Buckel W., Einsle O., Ermler U., Golding B.T., Gunsalus R.P., Kroneck P.M.H., Krüger M., Lueders T., Martins B.M., Musat F., Richnow H.H., Schink B., Seifert J., Szaleniec M., Treude T., Ullmann G.M., Vogt C., von Bergen M., Wilkes H. Anaerobic microbial degradation of hydrocarbons: from enzymatic reactions to the environment. J. Mol. Microbiol. Biotechnol. 2016;26:5–28. doi: 10.1159/000443997. https://www.karger.com/Article/FullText/443997 [DOI] [PubMed] [Google Scholar]
- 12.Vogt C., Dorer C., Musat F., Richnow H.-H. Multi-element isotope fractionation concepts to characterize the biodegradation of hydrocarbons — from enzymes to the environment. Curr. Opin. Biotechnol. 2016;41:90–98. doi: 10.1016/j.copbio.2016.04.027. https://linkinghub.elsevier.com/retrieve/pii/S0958166916301355 [DOI] [PubMed] [Google Scholar]
- 13.Atashgahi S., Lu Y., Smidt H. Organohalide-Respiring Bact. Springer Berlin Heidelberg; Berlin, Heidelberg: 2016. Overview of known organohalide-respiring bacteria—phylogenetic diversity and environmental distribution; pp. 63–105.http://link.springer.com/10.1007/978-3-662-49875-0_5 [Google Scholar]
- 14.Aulenta F., Rossetti S., Matturro B., Tandoi V., Verdini R., Majone M. Organohalide-Respiring Bact. Springer Berlin Heidelberg; Berlin, Heidelberg: 2016. Redox interactions of organohalide-respiring bacteria (OHRB) with solid-state electrodes: principles and perspectives of microbial electrochemical remediation; pp. 499–516.http://link.springer.com/10.1007/978-3-662-49875-0_21 [Google Scholar]
- 15.Ritalahti K.M., Amos B.K., Sung Y., Wu Q., Koenigsberg S.S., Löffler F.E. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 2006;72:2765–2774. doi: 10.1128/AEM.72.4.2765-2774.2006. https://aem.asm.org/content/72/4/2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Löffler F.E., Yan J., Ritalahti K.M., Adrian L., Edwards E.A., Konstantinidis K.T., Müller J.A., Fullerton H., Zinder S.H., Spormann A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013;63 doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 17.Daghio M., Aulenta F., Vaiopoulou E., Franzetti A., Arends J.B.A., Sherry A., Suárez-Suárez A., Head I.M., Bestetti G., Rabaey K. Electrobioremediation of oil spills. Water Res. 2017;114:351–370. doi: 10.1016/j.watres.2017.02.030. https://linkinghub.elsevier.com/retrieve/pii/S0043135417301185 [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Luo H., Fallgren P.H., Jin S., Ren Z.J. Bioelectrochemical system platform for sustainable environmental remediation and energy generation. Biotechnol. Adv. 2015;33:317–334. doi: 10.1016/j.biotechadv.2015.04.003. http://www.ncbi.nlm.nih.gov/pubmed/25886880 [DOI] [PubMed] [Google Scholar]
- 19.Espinoza-Tofalos A., Daghio M., Palma E., Aulenta F., Franzetti A. Structure and functions of hydrocarbon-degrading microbial communities in bioelectrochemical systems. Water. 2020;12:343. https://www.mdpi.com/2073-4441/12/2/343 [Google Scholar]
- 20.Marzocchi U., Palma E., Rossetti S., Aulenta F., Scoma A. Parallel artificial and biological electric circuits power petroleum decontamination: the case of snorkel and cable bacteria. Water Res. 2020;173:115520. doi: 10.1016/j.watres.2020.115520. https://linkinghub.elsevier.com/retrieve/pii/S0043135420300567 [DOI] [PubMed] [Google Scholar]
- 21.Palma E., Daghio M., Espinoza Tofalos A., Franzetti A., Cruz Viggi C., Fazi S., Petrangeli Papini M., Aulenta F. Anaerobic electrogenic oxidation of toluene in a continuous-flow bioelectrochemical reactor: process performance, microbial community analysis, and biodegradation pathways. Environ. Sci. Water Res. Technol. 2018;4:2136–2145. [Google Scholar]
- 22.Palma E., Espinoza Tofalos A., Daghio M., Franzetti A., Tsiota P., Cruz Viggi C., Papini M.P., Aulenta F. Bioelectrochemical treatment of groundwater containing BTEX in a continuous-flow system: substrate interactions, microbial community analysis, and impact of sulfate as a co-contaminant. N. Biotechnol. 2019;53:41–48. doi: 10.1016/j.nbt.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Aulenta F., Canosa A., Majone M., Panero S., Reale P., Rossetti S. Trichloroethene dechlorination and H 2 evolution are alternative biological pathways of electric charge utilization by a dechlorinating culture in a bioelectrochemical system. Environ. Sci. Technol. 2008;42:6185–6190. doi: 10.1021/es800265b. https://pubs.acs.org/doi/10.1021/es800265b [DOI] [PubMed] [Google Scholar]
- 24.Lai A., Aulenta F., Mingazzini M., Palumbo M.T., Papini M.P., Verdini R., Majone M. Bioelectrochemical approach for reductive and oxidative dechlorination of chlorinated aliphatic hydrocarbons (CAHs) Chemosphere. 2017;169 doi: 10.1016/j.chemosphere.2016.11.072. [DOI] [PubMed] [Google Scholar]
- 25.Leitão P., Rossetti S., Nouws H.P.A., Danko A.S., Majone M., Aulenta F. Bioelectrochemically-assisted reductive dechlorination of 1,2-dichloroethane by a Dehalococcoides- enriched microbial culture. Bioresour. Technol. 2015;195:78–82. doi: 10.1016/j.biortech.2015.06.027. https://linkinghub.elsevier.com/retrieve/pii/S0960852415008135 [DOI] [PubMed] [Google Scholar]
- 26.Verdini R., Aulenta F., De Tora F., Lai A., Majone M. Relative contribution of set cathode potential and external mass transport on TCE dechlorination in a continuous-flow bioelectrochemical reactor. Chemosphere. 2015;136 doi: 10.1016/j.chemosphere.2015.03.092. [DOI] [PubMed] [Google Scholar]
- 27.Palma E., Daghio M., Franzetti A., Petrangeli Papini M., Aulenta F. The bioelectric well: a novel approach for in situ treatment of hydrocarbon-contaminated groundwater. Microb. Biotechnol. 2018;11:112–118. doi: 10.1111/1751-7915.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucci M., Cruz Viggi C., Resitano M., Matturro B., Crognale S., Pietrini I., Rossetti S., Harnisch F., Aulenta F. Simultaneous removal of hydrocarbons and sulfate from groundwater using a “bioelectric well. Electrochim. Acta. 2021;388 https://linkinghub.elsevier.com/retrieve/pii/S0013468621009269 [Google Scholar]
- 29.Masut E., Battaglia A., Ferioli L., Legnani A., Cruz Viggi C., Tucci M., Resitano M., Milani A., de Laurentiis C., Matturro B., Di Franca M.L., Rossetti S., Aulenta F. A microcosm treatability study for evaluating wood mulch-based amendments as electron donors for trichloroethene (TCE) reductive dechlorination. Water. 2021;13:1949. https://www.mdpi.com/2073-4441/13/14/1949 [Google Scholar]
- 30.Balch W.E., Fox G.E., Magrum L.J., Woese C.R., Wolfe R.S. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. https://mmbr.asm.org/content/43/2/260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeikus J.G. The biology of methanogenic bacteria. Bacteriol. Rev. 1977;41:514–541. doi: 10.1128/br.41.2.514-541.1977. https://mmbr.asm.org/content/41/2/514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naudet V., Revil A., Rizzo E., Bottero J.-Y., Bégassat P. Groundwater redox conditions and conductivity in a contaminant plume from geoelectrical investigations. Hydrol. Earth Syst. Sci. 2004;8:8–22. https://hess.copernicus.org/articles/8/8/2004/ [Google Scholar]
- 33.Sander R. Compilation of Henry's law constants (version 4.0) for water as solvent. Atmos. Chem. Phys. 2015;15 4399–4981. [Google Scholar]
- 34.Crognale S., Casentini B., Amalfitano S., Fazi S., Petruccioli M., Rossetti S. Biological As(III) oxidation in biofilters by using native groundwater microorganisms. Sci. Total Environ. 2019;651 doi: 10.1016/j.scitotenv.2018.09.176. [DOI] [PubMed] [Google Scholar]
- 35.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L., Kaehler B.D., Kang K Bin, Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y.X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft JJJ, Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crognale S., Braguglia C.M., Gallipoli A., Gianico A., Rossetti S., Montecchio D. Direct conversion of food waste extract into caproate: metagenomics assessment of chain elongation process. Microorganisms. 2021;9 doi: 10.3390/microorganisms9020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aulenta F., Tocca L., Verdini R., Reale P., Majone M. Dechlorination of trichloroethene in a continuous-flow bioelectrochemical reactor: effect of cathode potential on rate, selectivity, and electron transfer mechanisms. Environ. Sci. Technol. 2011;45:8444–8451. doi: 10.1021/es202262y. https://pubs.acs.org/doi/10.1021/es202262y [DOI] [PubMed] [Google Scholar]
- 38.Liu Y., Climent V., Berná A., Feliu J.M. Effect of temperature on the catalytic ability of electrochemically active biofilm as anode catalyst in microbial fuel cells. Electroanalysis. 2011;23:387–394. https://onlinelibrary.wiley.com/doi/10.1002/elan.201000499 [Google Scholar]
- 39.Edwards E.A., Grbić-Galić D. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 1994;60:313–322. doi: 10.1128/aem.60.1.313-322.1994. https://aem.asm.org/content/60/1/313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey J.E. Biochemical reaction engineering and biochemical reactors. Chem. Eng. Sci. 1980;35:1854–1886. https://linkinghub.elsevier.com/retrieve/pii/0009250980801345 [Google Scholar]
- 41.Villano M., Monaco G., Aulenta F., Majone M. Electrochemically assisted methane production in a biofilm reactor. J. Power Sources. 2011;196:9467–9472. [Google Scholar]
- 42.Crampon M., Bodilis J., Portet-Koltalo F., Crampon M., Portet-Koltalo F. Linking initial soil bacterial diversity and polycyclic aromatic hydrocarbons (PAHs) degradation potential. J. Hazard Mater. 2018;359:500–509. doi: 10.1016/j.jhazmat.2018.07.088. [DOI] [PubMed] [Google Scholar]
- 43.Saiyari D.M., Chuang H.P., Senoro D.B., Lin T.F., Whang L.M., Chiu Y.T., Chen Y.H. A review in the current developments of genus Dehalococcoides, its consortia and kinetics for bioremediation options of contaminated groundwater. Sustain. Environ. Res. 2018;28:149–157. [Google Scholar]
- 44.Táncsics A., Farkas M., Horváth B., Maróti G., Bradford L.M., Lueders T., Kriszt B. Genome analysis provides insights into microaerobic toluene-degradation pathway of Zoogloea oleivorans BucT. Arch. Microbiol. 2020;202:421–426. doi: 10.1007/s00203-019-01743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toth C.R.A., Luo F., Bawa N., Webb J., Guo S., Dworatzek S., Edwards E.A. Anaerobic benzene biodegradation linked to the growth of highly specific bacterial clades. Environ. Sci. Technol. 2021;55:7970–7980. doi: 10.1021/acs.est.1c00508. [DOI] [PubMed] [Google Scholar]
- 46.Sarkar J., Kazy S.K., Gupta A., Dutta A., Mohapatra B., Roy A., Bera P., Mitra A., Sar P. Biostimulation of indigenous microbial community for bioremediation of petroleum refinery sludge. Front. Microbiol. 2016;7:1–20. doi: 10.3389/fmicb.2016.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang S., Wen X., Zhao L., Shi Y., Jin H. Crude oil treatment leads to shift of bacterial communities in soils from the deep active layer and upper permafrost along the China-Russia Crude Oil Pipeline route. PLoS One. 2014;9:12–14. doi: 10.1371/journal.pone.0096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakraborty R., O'Connor S.M., Chan E., Coates J.D. Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene compounds by Dechloromonas strain RCB. Appl. Environ. Microbiol. 2005;71:8649–8655. doi: 10.1128/AEM.71.12.8649-8655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coates J.D., Michaelidou U., Bruce R.A., O'Connor S.M., Crespi J.N., Achenbach L.A. The ubiquity and diversity of dissimilatory (per-)chlorate-reducing bacteria. Appl. Environ. Microbiol. 1999;65:5234–5241. doi: 10.1128/aem.65.12.5234-5241.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weelink S.A.B., van Eekert M.H.A., Stams A.J.M. Degradation of BTEX by anaerobic bacteria: physiology and application. Rev. Environ. Sci. Bio/Technology. 2010:359–385. 2010 94 9. [Google Scholar]
- 51.Chakraborty R., Coates J.D. Anaerobic degradation of monoaromatic hydrocarbons. Appl. Microbiol. Biotechnol. 2004:437–446. doi: 10.1007/s00253-003-1526-x. 2004 644 64. [DOI] [PubMed] [Google Scholar]
- 52.Cheng S., Xing D., Call D., Logan B. Direct biological conversion of electrical current into methane by electromethanogenesis. Environ. Sci. Technol. 2009;43:3953–3958. doi: 10.1021/es803531g. [DOI] [PubMed] [Google Scholar]
- 53.Van Eerten-Jansen M.C.A.A., Veldhoen A.B., Plugge C.M., Stams A.J.M., Buisman C.J.N., Ter Heijne A. Microbial community analysis of a methane-producing biocathode in a bioelectrochemical system. Archaea. 2013:481784. doi: 10.1155/2013/481784. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasaki K., Morita M., Sasaki D., Hirano S ichi, Matsumoto N., Ohmura N., Igarashi Y. Methanogenic communities on the electrodes of bioelectrochemical reactors without membranes. J. Biosci. Bioeng. 2011;111:47–49. doi: 10.1016/j.jbiosc.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 55.Siegert M., Li X.F., Yates M.D., Logan B.E. The presence of hydrogenotrophic methanogens in the inoculum improves methane gas production in microbial electrolysis cells. Front. Microbiol. 2014;5:1–12. doi: 10.3389/fmicb.2014.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Von Netzer F., Kuntze K., Vogt C., Richnow H.H., Boll M., Lueders T. Functional gene markers for fumarate-adding and dearomatizing key enzymes in anaerobic aromatic hydrocarbon degradation in terrestrial environments. J. Mol. Microbiol. Biotechnol. 2016;26:180–194. doi: 10.1159/000441946. [DOI] [PubMed] [Google Scholar]
- 57.Boll M., Löffler C., Morris B.E.L., Kung J.W. Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme A esters: organisms, strategies and key enzymes. Environ. Microbiol. 2014;16:612–627. doi: 10.1111/1462-2920.12328. [DOI] [PubMed] [Google Scholar]
- 58.Fuchs G., Boll M., Heider J. Microbial degradation of aromatic compounds- from one strategy to four. Nat. Rev. Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.