Abstract
Certain strains of root-colonizing fluorescent Pseudomonas spp. produce phenazines, a class of antifungal metabolites that can provide protection against various soilborne root pathogens. Despite the fact that the phenazine biosynthetic locus is highly conserved among fluorescent Pseudomonas spp., individual strains differ in the range of phenazine compounds they produce. This study focuses on the ability of Pseudomonas aureofaciens 30-84 to produce 2-hydroxyphenazine-1-carboxylic acid (2-OH-PCA) and 2-hydroxyphenazine from the common phenazine metabolite phenazine-1-carboxylic acid (PCA). P. aureofaciens 30-84 contains a novel gene located downstream from the core phenazine operon that encodes a 55-kDa aromatic monooxygenase responsible for the hydroxylation of PCA to produce 2-OH-PCA. Knowledge of the genes responsible for phenazine product specificity could ultimately reveal ways to manipulate organisms to produce multiple phenazines or novel phenazines not previously described.
Certain strains of root-colonizing fluorescent Pseudomonas spp. have gained attention in recent years because they produce broad-spectrum antibiotic metabolites that can provide protection against soilborne root diseases (46). One such class of antibiotics, the phenazines, encompasses a large family of heterocyclic nitrogen-containing compounds produced in late exponential and stationary phase. The ability to produce phenazines is limited almost exclusively to bacteria and has been reported in members of the genera Pseudomonas, Streptomyces, Nocardia, Sorangium, Brevibacterium, and Burkholderia (48). There are currently over 50 known phenazine compounds with the same basic structure, differing only in the derivatization of the heterocyclic core. These modifications largely determine the physical properties of phenazines and influence their biological activity against plant and animal pathogens.
The broad-spectrum activity exhibited by phenazine compounds against fungi and other bacteria is not understood. It is thought that they diffuse across the membrane and, once inside the cell, accept a single electron, disrupting respiration by interfering with the normal process of electron transport. This results in the overproduction of O2− and H2O2, which overwhelm cellular superoxide dismutases and ultimately cause cell death. The cellular superoxide dismutases of Pseudomonas aeruginosa, a bacterium which produces the phenazine compound pyocyanin, are more active than those of phenazine-nonproducing bacteria such as Escherichia coli, and they provide protection against phenazines (18, 19).
Several studies conducted in the early 1970s revealed tight links between phenazine biosynthesis and the shikimic acid pathway (48), but the biochemistry and genetic control of phenazine synthesis are still not fully understood. Chorismic acid has long been recognized as the branch point from the shikimic acid pathway to phenazine synthesis (26). Studies with radiolabeled precursors suggest that the phenazine core is formed by the symmetrical condensation of two molecules of chorismic acid (7, 20, 22, 26), while the amide nitrogen of glutamine serves as the immediate source of nitrogen in the heterocyclic nucleus of phenazine compounds (36). Phenazine-1,6-dicarboxylic acid is the first phenazine formed, and it is thought to be converted to phenazine-1-carboxylic acid (PCA), a key intermediate in the synthesis of other phenazines by fluorescent pseudomonads (6, 20, 22, 28).
Genetic studies in fluorescent Pseudomonas spp., the only microorganisms for which the genes responsible for the assembly of the heterocyclic phenazine nucleus have been cloned and sequenced, support this model. The phenazine biosynthetic loci from P. fluorescens 2-79 (27), P. aureofaciens 30-84 (27, 33), P. aeruginosa PA01 (D. V. Mavrodi and L. S. Thomashow, unpublished data), and P. chlororaphis PCL1391 (T. F. C. Chin-A-Woeng, D. van den Broek, G. de Voer, K. M. G. M. van der Drift, J. E. Thomas-Oates, B. J. J. Lugtenberg, and G. V. Bloemberg, Pseudomonas '99: Biotechnology and Pathogenesis, abstr. S48, 1999) are highly conserved. Each contains a seven-gene core operon regulated in a cell density-dependent manner by homologues of LuxI and LuxR (25, 52; D. V. Mavrodi and S. K. Farrand, unpublished data). In P. fluorescens 2-79, P. aureofaciens 30-84, and P. chlororaphis PCL1391, the phzI/R genes are found directly upstream from the phenazine core. Phenazine production in P. aeruginosa is controlled by two sets of regulatory proteins, rhlI/R and lasI/R, located elsewhere in the genome. The core gene products PhzC, PhzD, and PhzE, which are homologous with PhzF, PhzA, and PhzB in strain 30-84, are similar to enzymes of shikimic and chorismic acid metabolism. Sequence comparisons of PhzD and PhzE with other chorismate-modifying enzymes have shed new light on probable intermediates in the PCA pathway, suggesting that phenazine synthesis proceeds via the intermediates aminodeoxyisochorismic acid and 3-hydroxyanthranilate (27) rather than anthranilate, as suggested previously (12).
Although the phenazine biosynthetic loci of fluorescent pseudomonads are highly homologous, individual species typically differ in the range of compounds they produce. Previous work by Pierson et al. (33) suggested that the phzC gene of P. aureofaciens 30-84, and in particular the last 28 amino acids of the PhzC protein, are essential for the production of 2-hydroxyphenazine-1-carboxylic acid (2-OH-PCA) and 2-hydroxyphenazine (2-OH-PHZ), derivatives that are characteristic of strains previously designated P. aureofaciens but now classified as P. chlororaphis (24). The purpose of the present study was to determine the genetic basis for the production of these hydroxyphenazines by P. aureofaciens 30-84. Two possibilities were considered: first, that product specificity is determined by amino acid substitutions within the core biosynthetic genes, as suggested previously (33); or second, that a core pathway conserved among fluorescent pseudomonads is responsible for the synthesis of PCA, which then can be modified in a strain- or species-specific manner to yield a variety of different phenazine products. Knowledge of the mechanisms responsible for phenazine product specificity ultimately could reveal ways to manipulate organisms to produce multiple phenazines or hybrid phenazine products not previously described. Such compounds may have improved activity against soilborne plant pathogens or may lead to the development of novel pharmaceutical products.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are described in Table 1. Rifampin-resistant derivatives of P. fluorescens strains 2-79, M4-80R, and Q8r1-96 and P. chlororaphis 30-84 (referred to here by its original designation, P. aureofaciens 30-84) were used. Pseudomonas strains were grown at 28°C in Luria-Bertani (LB) broth, 2× YT broth (37), or M9 minimal medium (2) supplemented with sodium citrate to a final concentration of 40 mM as a carbon source. E. coli strains were grown in LB broth or 2× YT broth at 28 or 37°C. To enhance phenazine production, Pseudomonas strains were grown in LB broth supplemented with 1.5% glucose. When appropriate, antibiotic supplements were used at the following concentrations: tetracycline, 12.5 μg/ml (E. coli) or 25 μg/ml (Pseudomonas strains); rifampin, 100 μg/ml; kanamycin, 100 μg/ml; neomycin, 100 μg/ml (P. fluorescens 2-79); chloramphenicol, 35 μg/ml; and ampicillin, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Phz+, produces pyocyanin | 12 |
| ATCC 25007 | Phz+, produces pyocyanin, aeruginosins A and B | 40 |
| ATCC 25011 | Phz+, produces aeruginosins A and B | 53 |
| KN-1 | Phz+, produces pyocyanin | Laboratory collection |
| KNP-1 | Phz+, produces pyocyanin | Laboratory collection |
| KNP-2 | Phz+, produces pyocyanin | Laboratory collection |
| P. aureofaciens | ||
| 30-84 | Phz+, produces PCA, 2-OH-PCA, and 2-OH-PHZ | 32 |
| 30-84mxO | Phz+, produces PCA | This study |
| ATCC 13985 | Phz+, produces PCA, 2-OH-PCA, and 2-OH-PHZ | 40 |
| BS 1391 | Phz+, produces PCA, 2-OH-PCA, and 2-OH-PHZ | V. Kotchetkov |
| BS 1393 | Phz+, produces PCA, 2-OH-PCA, and 2-OH-PHZ | V. Kotchetkov |
| PGS12 | Phz+, produces PCA, 2-OH-PCA, and 2-OH-PHZ | 15 |
| AP-9 | Phz+, produces PCA, 2-OH-PCA, and 2-OH-PHZ | 1 |
| TX-1 | Phz+, produces PCA, 2-OH-PCA, and 2-OH-PHZ | EcoSoil Systems |
| P. chlororaphis | ||
| ATCC 17411 | Phz+, produces chlororaphin | 48 |
| ATCC 17809 | Phz+, produces chlororaphin | ATCC |
| ATCC 9446 | Phz+, produces chlororaphin | 40 |
| P. fluorescens | ||
| 2-79 | Phz+ Rifr, produces PCA | 50 |
| M4-80R | Phz− Rifr | 17 |
| Q8r1-96 | Phl+ Rifr | 35 |
| UQ 112 | Phz+, produces PCA | G. Botelho |
| UN 15 | Phz+, produces PCA | G. Botelho |
| UN 4127 | Phz+, produces PCA | G. Botelho |
| E. coli | ||
| SM17 (λ-pir) | thi pro hsdR hsdM recA rpsL RP4-2 Tetr::Mu Kanr::Tn7 | 39 |
| JM109 | F′ traD36 proA+ proB+ lacI lacZΔM15/recA1 | Promega Corp. |
| Plasmids | ||
| pALTER-Ex1 | ColE1 Tetr SP6, tac, and T7 promoters | Promega Corp. |
| pNOT19 | ColE1 bla accessory plasmid | 38 |
| pUCP26 | pUCP18-derived broad-host-range vector Tetr | 51 |
| pMOB3 | Kanrcat sacBR | 38 |
| pUCP4.5 | pUCP26 containing 4.5 kb-XbaI-EcoRI DNA fragment from P. aureofaciens 30-84; Tetr | This study |
| pUCP2.9XP | pUCP26 containing 2.9-kb XbaI-PstI DNA fragment from P. aureofaciens 30-84; contains phzO, Tetr | This study |
| pGEM-T Easy | Ampr, pUC18-derived SP6 and T7 promoters, f1 ori, lacZα | Promega Corp. |
| pGEM-PHZO | pGEM-T Easy containing 1.8-kb phzO fragment amplified from P. aureofaciens 30-84 by PCR | This study |
| p18Sfi | Ampr, pUCP18-derived vector with SfiI-EcoRI-SalI-HindIII-SfiI as multiple cloning site | 21 |
| pUT-Km-Tn5 | Ampr, Tn5-based delivery plasmid with Kanr | 21 |
| pUT-Km/30-84 | pUT-Km-Tn5 with phzXYFABCD from P. aureofaciens 30-84 cloned between SfiI sites | This study |
| pLAFR3 | IncP Tetrcos+ rlx+ | 41 |
| pLSP259 | pLAFR3 containing 20.9-kb DNA fragment from P. aureofaciens 30-84; Tetr | 32 |
| pLSP282 | pLAFR3 containing 24.7-kb DNA fragment from P. aureofaciens 30-84; Tetr | 32 |
| pLSP282Δ20-9 | pLAFR3 containing 15.0-kb DNA fragment from P. aureofaciens 30-84; Tetr | 32 |
| pLSP282Δ30-8 | pLAFR3 containing 11.2-kb DNA fragment from P. aureofaciens 30-84; Tetr | 32 |
| pNOT2.9T-1 | pNOT19 containing 2.9-kb XbaI-PstI DNA fragment from P. aureofaciens 30-84; Tet resistance gene inserted at NcoI in same orientation as phzO; Tetr Ampr | This study |
| pOT-1 | 2.5-kb EcoRI-NotI DNA fragment from pNOT2.9T-1 in pNOT19; phzO+ Tetr Ampr | This study |
| pOT1-1 | pOT-1 containing 5.6-kb sac genes from pMOB3; phzO+ Tetr Ampr Chlr | This study |
Phz+/−, the strain does (+) or does not (−) produce phenazines; Phl+, production of 2,4-diacetylphloroglucinol; bla, β-lactamase; cat, chloramphenicol acetyltransferase; Ampr, ampicillin resistance; Chlr, chloramphenicol resistance; Kanr, kanamycin resistance; Rifr, rifampin resistance; Tetr, tetracycline resistance.
DNA manipulations.
Standard methods were used for DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, and ligation (2). Pseudomonas and E. coli cells were transformed by electroporation in a Gene Pulser II system (Bio-Rad, Hercules, Calif.) according to the method of Enderle and Farwell (11) at settings of 25 μF for the capacitor, 200 Ω resistance, and an electric field of 1.8 kV/cm. Genomic DNA was isolated and purified by a cetyltrimethylammonium bromide (CTAB) miniprep procedure (2). For Southern blotting and hybridization, 500 ng of genomic DNA was digested with EcoRI and PstI, separated by electrophoresis in an 0.8% agarose gel, and transferred onto a BrightStar-Plus nylon membrane (Ambion, Inc., Austin, Tex.) in 0.4 M NaOH with subsequent cross-linking by exposure to UV irradiation (2). Membranes were prehybridized for 3 h at 60°C in a solution containing 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 4× Denhardt's solution, 0.1% sodium dodecyl sulfate (SDS), and 250 mg of denatured salmon sperm DNA/ml. After prehybridization, the membranes were incubated with specific probes overnight under the same conditions and washed with 2× SSC and 0.1% SDS at room temperature, 0.2× SSC and 0.1% SDS at room temperature, 0.2× SSC and 0.1% SDS at 60°C, and 0.1× SSC and 0.1% SDS at 60°C. DNA-DNA hybrids were detected with the BrightStar non-isotopic detection kit (Ambion Inc.) according to the manufacturer's protocol. The 2.1-kb phzO probe was amplified by PCR from P. aureofaciens 30-84 genomic DNA with the oligonucleotide primers 30-84XBA (5′-AAG TCC AGA TGC GAA AGA ACG-3′) and PHZO10 (5′-AAG TGG CAT GGC TCG AAC AAA G-3′). Amplification was carried out in a 25-μl reaction mixture containing 1× thermophilic DNA polymerase buffer (Promega Corp., Madison, Wis.), 1.5 mM MgCl2, 5.0% (final concentration) dimethyl sulfoxide (Sigma Chemical Co., St. Louis, Mo.), 200-μM concentrations of dGTP, dATP, dCTP, and dTTP (Perkin-Elmer, Norwalk, Conn.), 20 pM of each primer, and 1.2 U of Taq DNA polymerase (Promega Corp.). Amplifications were performed with a PTC-200 thermal cycler (MJ Research Inc., Watertown, Mass.). The cycling program included a 45-s initial denaturation at 94°C followed by 30 cycles of 94°C for 45 s, 51°C for 45 s, and 72°C for 1.5 min. Amplified DNA was labeled with a random primer biotin labeling kit (NEN Life Science Products Inc., Boston, Mass.).
DNA sequencing and analysis.
DNA was sequenced by using the ABI Prism Dye Terminator Cycle sequencing kit (Perkin-Elmer), according to the manufacturer's instructions. All custom-designed oligonucleotides came from Operon Technologies Inc. (Alameda, Calif.). Sequence data were compiled and analyzed for open reading frames and codon usage with the Omiga version 1.1.3 software package (Oxford Molecular Ltd., Oxford, United Kingdom). A database search for similar protein sequences was carried out with the BLAST (44) and FASTA network servers at the National Center for Biotechnology Information and the European Molecular Biology Laboratory (EMBL), respectively. The probable domain homologies search was performed with PROSITE (EMBL, Heidelberg, Germany) (3) and ISREC ProfileScan (Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland [www.isrec.isb-sib.ch/software/PFSCAN_form.html]) computer services. The significance of the similarity of a predicted protein to known proteins was determined by calculating the binary comparison score (Z score). Pairwise alignments were obtained by using the BESTFIT program from the Wisconsin Package (Genetics Computer Group, Madison, Wis.), and the resulting percent identities, percent similarities, alignment scores (A), mean random alignment scores (R), and standard deviations (SD) (n = 100) were noted. Z scores were then calculated by the equation Z = (A − R)/SD. Multiple sequence alignments were built with Omiga's ClustalW and analyzed with the TreeView version 1.5.0 software package (31).
Mating and screening of transconjugants.
Plasmids were mobilized from the donor strain E. coli S-17 (λ-pir) into Pseudomonas recipients by using the filter mating technique described by van Overbeek (49). To counterselect E. coli donor cells, mating mixtures were plated on M9 agar supplemented with appropriate antibiotics and sodium citrate as a carbon source. Positive isolates were replated and screened for the presence of phenazine genes by PCR with primers PHZ1 and PHZ2. The oligonucleotide primers PHZ1 (5′-GGC GAC ATG GTC AAC GG-3′) and PHZ2 (5′-CGG CTG GCG GCG TAT TC-3′) were used as universal phenazine primers to amplify a 1.4-kb fragment containing parts of phzF and phzA in P. aureofaciens 30-84, which correspond to phzC and phzD in P. fluorescens 2-79. The amplification was carried out in a 15-μl reaction mixture. The cycling program included an initial denaturation for 2 min at 94°C followed by 25 cycles of 94°C for 1 min, 56°C for 45 s, 72°C for 1.75 min, and a final extension at 75°C for 1 min. The oligonucleotide primers PHZX (5′-TTT TTT CAT ATG CCT GCT TCG CTT TC-3′) and PHZY (5′-TTT GGA TCC TTA AGT TGG AAT GCC TCC G-3′) were used to distinguish between the phenazine operons of P. aureofaciens 30-84 and P. fluorescens 2-79. These primers amplify a 1.1-kb DNA fragment containing parts of phzX and phzY from strain 30-84 but not from the corresponding, homologous phzA and phzB sequences of strain 2-79. The program included an initial denaturation at 94°C for 1.5 min followed by 30 cycles of 94°C for 45 s, 58°C for 30 s, 72°C for 1.75 min, and a final extension at 72°C for 1 min.
Protein expression.
The phzO gene was cloned from P. aureofaciens 30-84 and expressed under the control of the lac promoter in the plasmid vector pUCP26. E. coli JM109 harboring pUCP26, pUCP2.9XP, or pUCP4.5 was grown in LB broth to an optical density at 600 nm of 0.6 and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested 3 h later and total cellular protein was analyzed by electrophoresis in an SDS–10% polyacrylamide gel as described by Copeland (9). Alternatively, phzO was amplified by PCR from P. aureofaciens 30-84 genomic DNA with the primers PHZOstart (5′-CGA CTC TAG AAC GTT GTC CTT GAC C-3′) and PHZO10 in a 30-μl reaction mixture with a cycling program that included a 45-s initial denaturation at 94°C and 29 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 3.25 min. The 1.8-kb reaction product was ligated into pGEM-T Easy (Promega) to give pGEM-PHZO, which was transformed into E. coli JM109. The resulting plasmid, pGEM-PHZO, contained the entire phzO gene preceded by 88 bp upstream of the start codon and 265 bp downstream of the coding sequence. Expression was induced as described above.
PCA transformation assay.
E. coli JM109 bearing pUCP26, pUCP2.9XP, pGEM-T Easy, or pGEM-PHZO was grown at 37°C in 2× YT supplemented with tetracycline. The cells were harvested, suspended in fresh medium, and induced with 0.5 mM IPTG. PCA was added to a final concentration of 0.3 or 0.5 mg/ml from a 25 mM stock solution in 5% (wt/vol) NaHCO3. Samples were taken at 3-h intervals, extracted, and analyzed for phenazine composition by reverse-phase high-performance liquid chromatography (RP-HPLC).
Gene replacement mutagenesis of phzO.
A phzO knockout mutant of P. aureofaciens 30-84 was generated by gene replacement as described by Schweizer (38). Briefly, a 2.5-kb PvuII fragment bearing a tetracycline resistance gene from pALTER-Ex1 (Promega) was inserted into phzO at the NcoI site. The interrupted gene was subcloned into pNOT19, yielding pOT1, which was digested with NotI and ligated with a 5.3-kb pMOB3 sacB cassette. The resulting plasmid, pOT1-1, was mobilized into P. aureofaciens 30-84 from E. coli S-17 (λ-pir), and double crossover progeny were selected as described previously (38).
Analysis of phenazine compounds.
Phenazine compounds were extracted according to the method of Bonsall et al. (5). Bacterial strains were cultivated for 72 h in LB broth supplemented with 1.5% glucose. The cultures were acidified with 10% trifluoroacetic acid (TFA) and then extracted twice with ethyl acetate. The organic phase containing the phenazines was evaporated to dryness and suspended in 35% acetonitrile (ACN)–0.1% TFA.
Since phenazine-producing Pseudomonas spp. often produce mixtures of phenazine compounds, a generalized HPLC protocol for detection of these metabolites was developed. The protocol utilized a NOVA-PAK C18 reverse-phase Radial-PAK cartridge (4 μm, 8 by 100 mm) (Waters Corp., Milford, Mass.) and solvent conditions consisting of a 2-min initial wash with 35% ACN–0.1% TFA in H2O followed by a 25-min linear gradient to 100% ACN–0.1% TFA at a flow rate of 1.0 ml/min. The Waters HPLC system included a 710B WISP, 510 pumps, and a 680 automated gradient controller with a 990 photodiode array detector (Waters Corp.). Phenazine compounds were identified by retention time and UV spectrum. Standards included compounds purified from well-characterized strains (27, 32) and chemically synthesized compounds (PCA, 2-OH-PCA, and 2-OH-PHZ) obtained from Colour Your Enzyme (Bath, Ontario, Canada). Although the protocol allowed simultaneous identification of phenazine compounds including unsubstituted phenazine, PCA, 2-OH-PCA, 2-OH-PHZ, chlororaphin, and 1-OH-PCA, it failed to clearly separate PCA and 2-OH-PCA. 2-OH-PHZ, which is formed by spontaneous decarboxylation of 2-OH-PCA (see below), was therefore used as an indicator of 2-hydroxyphenazine synthesis, and when necessary, the presence of 2-OH-PCA in samples containing PCA was determined by peak purity and spectral analyses using the Waters 991 photodiode array (Waters Corp.).
Fungal inhibition assay.
The inhibition of hyphal growth of Gaeumannomyces graminis var. tritici by Pseudomonas spp. strains 30-84, 30-84mxO, 2-79, and 2-79 harboring pUCP2.9XP was assayed as described by Ownley et al. (30) using Kanner agar supplemented with potato extract (KMPE), which supports the production of phenazine compounds. Plates were incubated at room temperature in the dark and radial growth of the fungus was measured after 5 days. The experiment was repeated twice with 7 or 8 replicates each time. Inhibition of mean fungal radial growth by each bacterial strain was analyzed for significance by the Student t test at a P level of 0.05.
Nucleotide sequence accession number.
The nucleotide sequence for the phzO gene has been deposited in GenBank under accession number AF230879.
RESULTS
Localization of the 2-hydroxyphenazine gene.
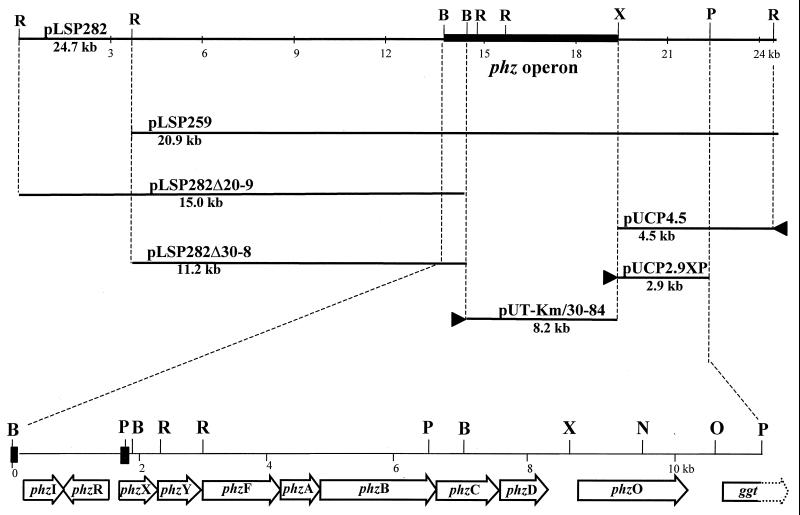
Previous studies by Pierson and Thomashow (32) identified two cosmids, pLSP259 and pLSP282, from a genomic library of P. aureofaciens 30-84 that were able to restore Phz− mutants of 30-84 to production of PCA, 2-OH-PCA, and 2-OH-PHZ. These cosmids contain identical 11.2- and 9.2-kb EcoRI fragments, and an additional 3.8-kb EcoRI fragment is present in pLSP282 (Fig. 1). To determine whether pLSP259 and pLSP282 are sufficient to enable the synthesis of PCA, the two hydroxyphenazines, or all three products, each cosmid was introduced into P. fluorescens strains 2-79, M4-80R, and Q8r1-96 and the cosmid's presence was confirmed by PCR with phzXY-specific primers. Phenazine compounds produced by the transformed strains were extracted and analyzed by RP-HPLC. Transformants of all three strains harboring either cosmid produced both PCA and the 2-hydroxyphenazines (Table 2), indicating that pLSP259 and pLSP282 contain the necessary information required for the synthesis of all three compounds.
FIG. 1.
Physical map of the constructs used in this study and description of the phz operon (phzIRXYFABCD) and phzO region in P. aureofaciens 30-84. The phenazine operon has been described previously (33). The lux boxes upstream of phzI and phzX are represented by ■. ▸ indicates the orientation of the lac promoters in pUT-Km and pUCP26. ggt indicates the position of an open reading frame with similarity to gamma-glutamyl transpeptidase. The restriction enzymes indicated on the map are B, BglII; N, NcoI; O, NotI; P, PstI; R, EcoRI; and X, XbaI.
TABLE 2.
Phenazine production as the result of introduction of plasmids into P. fluorescens strains
| Plasmid | Phenazine compound | Result in
recipient P. fluorescens strain:
|
||
|---|---|---|---|---|
| 2-79 | Q8r1-96 | M4-80R | ||
| pLSP282 | PCA | + | + | + |
| 2-OH-PHZ | + | + | + | |
| pLSP259 | PCA | + | + | + |
| 2-OH-PHZ | + | + | + | |
| pUT-Km30-84 | PCA | + | + | + |
| 2-OH-PHZ | − | − | − | |
| pLSP282Δ20-9 | PCA | + | N/Aa | N/A |
| 2-OH-PHZ | − | N/A | N/A | |
| pLSP282Δ30-8 | PCA | + | N/A | N/A |
| 2-OH-PHZ | − | N/A | N/A | |
| pUCP4.5 | PCA | + | N/A | N/A |
| 2-OH-PHZ | − | N/A | N/A | |
| pUCP2.9 | PCA | + | N/A | N/A |
| 2-OH-PHZ | + | N/A | N/A | |
N/A, not analyzed.
We next determined whether the core phenazine operon, phzXYFABCD, which was present in both cosmids, was sufficient for the synthesis of the three phenazine products. Previous studies (33) had suggested that in P. aureofaciens 30-84, the C-terminal 28 amino acids of PhzC were necessary for the synthesis of 2-OH-PCA and 2-OH-PHZ. PhzC is a 278-amino acid, 30.3-kDa protein with 94% amino acid sequence identity to PhzF from P. fluorescens 2-79. These proteins have no common motifs or other similarities with other proteins of known function, but PhzF is absolutely required for the synthesis of PCA in strain 2-79 (27). A pairwise alignment of the terminal 28 amino acids of the two proteins revealed three conservative substitutions: lysine at position 251 in strain 30-84 instead of arginine in strain 2-79; glutamic acid at position 257 instead of aspartic acid; and valine at position 269 instead of isoleucine. To determine the biosynthetic potential of phzXYFABCD, the core operon was cloned downstream of a tac promoter and transposed from pUT-Km/30-84 into the genomes of P. fluorescens 2-79, Q8r1-96, and M4-80R. Transposition in each recipient was confirmed by PCR with phzXY-specific primers. RP-HPLC revealed that all three transformed strains produced PCA but not 2-OH-PCA or 2-OH-PHZ (Table 2), indicating that the core genes from strain 30-84 do not contain the information necessary for the synthesis of the 2-hydroxyphenazines.
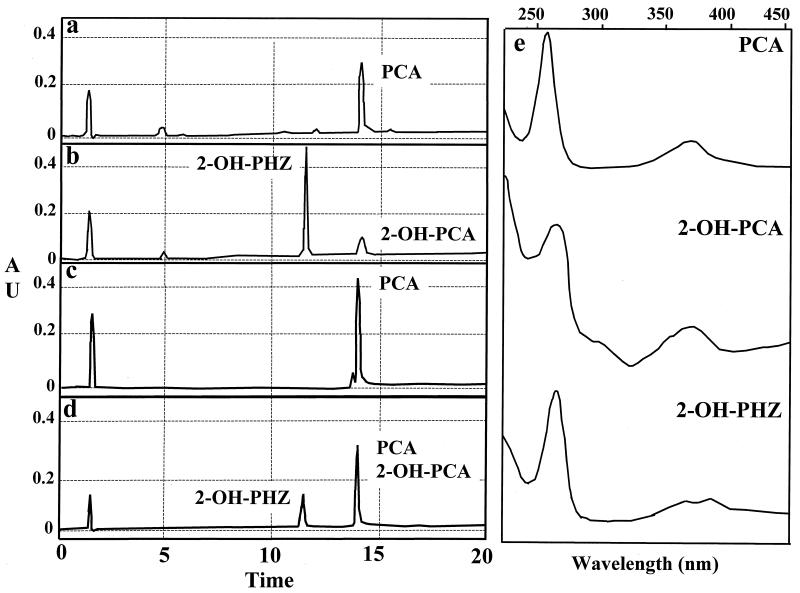
The regions upstream and downstream of phzXYFABCD were next analyzed for genes enabling the conversion of PCA to 2-hydroxyphenazine derivatives. P. fluorescens 2-79 harboring either pLSP282Δ20-9, containing the 3.8- and 11.2-kb EcoRI fragments 5′ to the phz operon, or pLSP282Δ30-8, containing the 11.2-kb fragment (32) (Fig. 1), produced only PCA (Table 2), suggesting that the genes required for 2-hydroxyphenazine synthesis do not reside upstream of the core locus. The remaining 4.5-kb fragment downstream of the phenazine operon in pLSP282 was cloned into the broad-host-range vector pUCP26. A smaller 2.9-kb XbaI-PstI fragment also was cloned into pUCP26 in the opposite orientation. Plasmid pUCP2.9XP contained the C-terminal region of phzD and downstream sequences under the control of the vector's lac promoter. Both plasmids were introduced into P. fluorescens 2-79 and the phenazines were extracted for RP-HPLC analysis. Strain 2-79 containing pUCP2.9XP, but not pUCP4.5, produced 2-OH-PHZ in addition to the PCA (Fig. 2), indicating that the 2.9-kb DNA fragment lacked a promoter, was colinear with the phenazine biosynthetic locus, and contained the gene(s) required for the conversion of PCA.
FIG. 2.
HPLC analyses of phenazine compounds produced by P. fluorescens 2-79 harboring the pUCP26 vector (a), P. fluorescens 2-79 harboring pUCP2.9XP and containing phzO (b), E. coli JM109 harboring pUCP26 (c), E. coli JM109 harboring pUCP2.9XP and containing phzO (d), and peak identity of PCA and 2-OH-PHZ confirmed by spectral analysis (e). Retention times for PCA and 2-OH-PHZ are 14.1 and 11.4 min, respectively. Absorption maxima for PCA are 248 and 371 nm. Absorption maxima for 2-OH-PCA are 257 and 369 nm. Absorption maxima for 2-OH-PHZ are 257, 368, and 387 nm.
DNA sequence analysis.
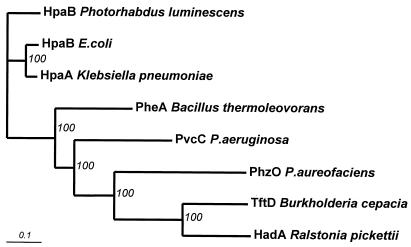
The 2.9-kb XbaI-PstI fragment from P. aureofaciens 30-84 was sequenced in both directions and compiled with Omiga. Computer analysis revealed a large open reading frame, designated phzO, located 271 nucleotides downstream from phzD and preceded by a well-conserved ribosome binding site, GAGG. phzO encoded a 491-amino acid protein with a calculated molecular mass of 55.1 kDa. Homology searches with the deduced amino acid sequence revealed similarity to bacterial aromatic hydroxylases and monooxygenases (Table 3). Phylogenetic analysis of these aligned protein sequences resulted in the tree shown in Fig. 3. The high bootstrap values (from 1,000 resamplings) showed the robustness of these groups. A second open reading frame with the initiation codon GTG, preceded by the ribosome binding site GGAG and encoding a putative polypeptide with significant similarity (BLAST values of 6.6e−43) to gamma-glutamyl transpeptidase enzyme precursor proteins, was identified 656 bp downstream of the phzO termination codon.
TABLE 3.
Proteins displaying similarity to PhzO
| Organism | Protein name | Enzyme | NCBI accession no. | Identity (%)a | Similarity (%)a | Z scorea |
|---|---|---|---|---|---|---|
| Bacillus thermoleovorans A2 | PheA | Phenol hydroxylase | AAC38324 | 28.1 | 37.9 | 42.1 |
| Klebsiella pneumoniae | HpaA | 4-hydroxyphenylacetate-3-hydroxylase | AAC37120 | 24.8 | 36.5 | 44.0 |
| Escherichia coli ATCC 11105 | HpaB | 4-hydroxyphenylacetate hydroxylase | CAA82321 | 25.2 | 36.5 | 35.8 |
| Ralstonia pickettii DTP0602 | HadA | Chlorophenol-4-hydroxylase | BAA13105 | 26.7 | 39.9 | 64.4 |
| Pseudomonas aeruginosa PAO1 | PvcC | 4-hydroxylphenylacetate hydroxylase | AAC21673 | 24.5 | 35.5 | 61.8 |
| Burkholderia cepacia AC1100 | TftD | Chlorophenol-4-monooxygenase | AAC23548 | 25.1 | 37.1 | 65.0 |
| Photorhabdus luminescens | HpaB | 4-hydroxylphenylacetate hydroxylase | AAC08739 | 24.0 | 35.9 | 37.0 |
Identities, similarities, and Z scores were determined by the program BESTFIT from the Genetics Computer Group package.
FIG. 3.
Phylogenetic relationship between PhzO and various bacterial aromatic monooxygenases. The neighbor-joining tree with bootstrap support was constructed and visualized by using the CLUSTAL W and TreeView version 1.5.0 programs (31), respectively.
Expression and functional analysis of PhzO.
The phzO gene from P. aureofaciens 30-84 was cloned in pUCP26 under the control of the lac promoter and expressed in E. coli JM109. Cells from induced cultures expressing PhzO produced a unique band of approximately 55 kDa on SDS-polyacrylamide gels, in good agreement with the size predicted by nucleotide sequence analysis. Induced cultures of E. coli expressing PhzO, either in pUCP26 or in pGEM-T Easy, converted PCA (0.3 or 0.5 mg/ml in 5% NaHCO3) to 2-OH-PCA and 2-OH-PHZ within 3 h, whereas no such conversion occurred in control cultures harboring only the respective vectors (Fig. 2). These results indicate that PhzO, independent of up- and downstream sequences, is sufficient to hydroxylate PCA. To determine whether PhzO is responsible for this reaction in P. aureofaciens 30-84, a tetracycline resistance gene was inserted into phzO and introduced in the genome by homologous recombination. P. aureofaciens 30-84mxO produced PCA but not 2-OH-PCA and 2-OH-PHZ. Finally, to test the hypothesis that the conversion of 2-OH-PCA to 2-OH-PZ occurs spontaneously in the absence of enzymatic activity, as suggested previously (13), solutions of synthetic 2-OH-PCA were incubated for 18 h in 0.1 M sodium phosphate buffer at pHs of 4.0, 6.0, 7.0, and 8.0, extracted, and analyzed by RP-HPLC. At pH 4.0, 2-OH-PZ accounted for only 0.2% of the total phenazine present after 18 h, but at pHs of 6.0, 7.0, and 8.0, 33.3, 74, and 64% of the 2-OH-PCA initially present was converted to 2-OH-PZ.
Conservation of PhzO among phenazine-producing fluorescent Pseudomonas spp.
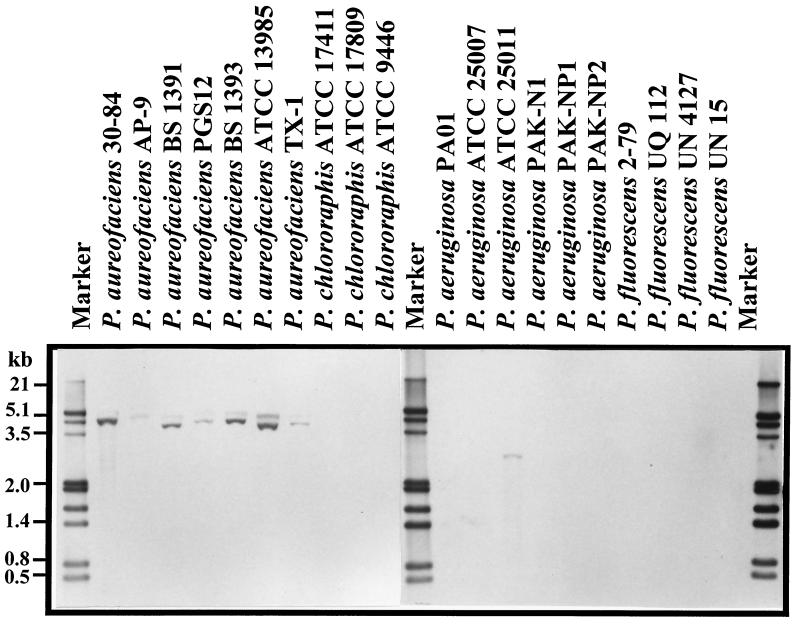
A 2.1-kb probe containing the 1.5-kb phzO gene and flanking regions was hybridized to total genomic DNA from 20 known phenazine-producing fluorescent pseudomonads to determine whether the gene is unique to producers of 2-hydroxyphenazines or if it also is conserved in other phenazine-producing strains. All seven strains of P. aureofaciens contained sequences that hybridized to the probe (Fig. 4), and each produced 2-OH-PCA and 2-OH-PHZ in addition to PCA, as determined by RP-HPLC. Two additional strains, P. chlororaphis 9446 and P. aeruginosa 25011, contained a faintly hybridizing band. However, no 2-hydroxylated phenazines were found in the extracts from cultures of these strains (data not shown), which previously were reported to produce chlororaphin and aeruginosins A and B, respectively (24, 54). No hybridization was detected between the phzO probe and DNA from P. chlororaphis strains ATCC 17411 and ATCC 17809, P. aeruginosa strains PA01, PAK-N1, PAK-NP1, PAK-NP2, ATCC 25007, and ATCC 25011, or the PCA-producing P. fluorescens strains 2-79, UQ 112, UN 4127, and UN 15 (Fig. 4), even after very heavy overexposure of the films (data not shown).
FIG. 4.
Southern hybridization of phzO probe to total genomic DNA from 20 phenazine-producing Pseudomonas strains. Total DNA samples were digested with endonucleases PstI and EcoRI.
Fungal inhibition assays.
Assays were conducted in vitro to determine if strains producing hydroxyphenazine compounds inhibited the hyphal growth of G. graminis var. tritici more than those producing only PCA. The radial growth of the fungus on plates in the presence of strain 30-84 was significantly less than that in the presence of the mutant 30-84mxO, which produced only PCA (18 versus 22 mm, P ≤ 0.05). Similarly, P. fluorescens 2-79(pUCP2.9XP), transformed to hydroxyphenazine production, was more inhibitory than wild-type 2-79 (14 versus 17 mm, P ≤ 0.05).
DISCUSSION
Results of the current study show clearly that phzXYFABCD, the core phenazine biosynthetic operon of strain 30-84, is responsible only for the synthesis of PCA (Fig. 5). When transformed with these genes, the sole phenazine product synthesized by P. fluorescens strains Q8r1-96 and M4-80R (which themselves do not produce phenazines) was PCA. A novel gene designated phzO was identified immediately downstream of the core biosynthetic operon of strain 30-84. Phenazine-nonproducing strains transformed with phzXYFABCD and phzO, or P. fluorescens 2-79 transformed with phzO, synthesized hydroxyphenazine compounds in addition to PCA, and E. coli expressing phzO rapidly converted exogenously supplied PCA to hydroxyphenazine products. Finally, a mutant of 30-84 inactivated in phzO produced only PCA. These results are consistent with those of an earlier study (27) suggesting that minor sequence differences between PhzF of strain 2-79 and PhzC of strain 30-84 are insufficient to account for the differences in the products synthesized by the two strains and implicating additional determinants of phenazine product specificity. This hypothesis is further supported by data reported by Chin-A-Woeng et al. (8), who recently described an aminotransferase gene designated phzH located downstream of the phenazine operon in P. chlororaphis PCL1391. PhzH was found to be responsible for the conversion of PCA to phenazine-1-carboxamide (chlororaphin), the green phenazine compound characteristic of P. chlororaphis. It thus appears that the presence of species-specific phenazine-modifying genes adjacent to the core biosynthetic locus may be a common feature among fluorescent Pseudomonas spp.
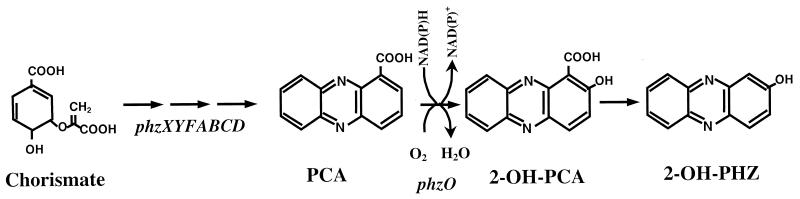
FIG. 5.
Proposed mechanism for the production of 2-hydroxyphenazine-1-carboxylic acid and 2-hydroxyphenazine in P. aureofaciens 30-84.
That PhzO belongs to a recently defined (14) family of two-component nonheme flavin-diffusible bacterial aromatic monooxygenases (TC-FDMs) is supported by results of both pairwise comparisons (high Z scores) and multiple sequence alignments (high bootstrap values). These enzymes are NAD(P)H-dependent flavoproteins that lack the defined GXGXXG FAD/NADH binding site typical of aromatic monooxygenases. Instead, they function in concert with a reductase component that uses NAD(P)H to generate a reduced flavin. The flavin then diffuses to the oxygenase, where it serves as a cosubstrate in the oxidation of aromatic compounds by molecular oxygen (14, 53). TC-FDMs hydroxylate aromatic substrates in either the ortho or the para position and include both dehalogenating (HadA from Ralstonia pickettii, PheA from Bacillus thermoleovorans, and TftD from Burkholderia cepacia) and nondehalogenating (HpaB from E. coli, HpaB from Photorhabdus luminescens, HpaA from Klebsiella pneumoniae, and PvcC from P. aeruginosa) enzymes (10, 14, 16, 23, 34, 42, 43). Many require the presence of an additional 19- to 21-kDa “coupling” subunit (16, 23, 34, 43) to provide reduced flavin, but at least for HpaB, the archetype of this family, this requirement can be satisfied with reduced flavin adenine dinucleotide provided exogenously or generated by an alternative flavin reductase (14, 53). This apparently is also the case for PhzO, since the cloned gene in either pUCP2.9XP or pGEM-PHZO (which contained very little flanking sequence from P. aureofaciens 30-84) was sufficient to catalyze the conversion of PCA to 2-hydroxyphenazines in E. coli. Whereas the genes encoding the oxidase and reductase components of most known TC-FDM enzymes are situated near one other on the chromosome (14), we found no detectable similarity with known flavin reductases in the 1.3-kb DNA segment downstream of phzO. Although a functionally “dedicated” reductase may be encoded elsewhere in the genome of strain 30-84, such an enzyme clearly is not required for phenazine 2-hydroxylation in E. coli. The apparent absence of a linked reductase gene and the relatively low level of overall homology between PhzO and other members of the TC-FDM family distinguish this phenazine-modifying enzyme from other oxygenases.
Earlier, Flood et al. (13) in a study with deuterated precursors revealed that hydroxylated phenazines are synthesized in P. aureofaciens through the formation of a hypothetical arene intermediate in the following order: PCA→2-OH-PCA→2-OH-PHZ (13). The authors also concluded that the hydroxylation of PCA occurred inefficiently, since PCA was more abundant in the extracts than were the hydroxylated derivatives. Based on the results of our study, we speculate that 2-hydroxylation of PCA is carried out in P. aureofaciens by a nonheme, flavin-diffusible monooxygenase, PhzO, which adds a hydroxyl group to PCA at the ortho position relative to the carboxyl group, resulting in the synthesis of 2-OH-PCA (Fig. 5). The reaction presumably also requires a yet-unidentified, highly active reductase, NAD(P)H, flavin, and O2. As speculated previously (13), the subsequent decarboxylation of 2-OH-PCA to 2-OH-PHZ occurs spontaneously in the absence of enzymes. Up to 74% of 2-OH-PCA in phosphate buffer at pH 7 was converted to 2-OH-PHZ after 18 h, whereas lesser amounts (33 and 62%) were converted in buffers at pH values of 6 and 8, respectively (G. Phillips and L. S. Thomashow, unpublished data).
We screened a collection of phenazine-producing Pseudomonas spp. for the presence of phzO by Southern hybridization (Fig. 4). Our results indicate that this gene is found almost exclusively in isolates of P. aureofaciens. The only two non-P. aureofaciens strains that hybridized with the phzO probe were P. chlororaphis 9446 and P. aeruginosa 25011 (Fig. 4). However, it is possible that these strains do not have the phzO homologue, since in both cases the hybridization signal was very weak, the size of the hybridizing fragment was different from that in P. aureofaciens strains, and no hydroxylated phenazines were detected in the culture extracts. Based on these findings, we speculate that phzO (and probably phzH) is a species-specific gene in fluorescent Pseudomonas spp. Moreover, the fact that all the tested strains possess a well-conserved core phenazine locus (D. M. Mavrodi and L. S. Thomashow, unpublished observation) may indicate that the acquisition of phenazine-modifying genes by phenazine-producing pseudomonads is a fairly recent event.
Interest in strains of P. aureofaciens frequently has centered on their ability to suppress soilborne plant pathogens (4, 32, 40, 45). We used derivatives of strain 30-84 mutated in phzO and strain 2-79 transformed with phzO to evaluate the importance of hydroxylated phenazines in biological control activity against G. graminis var. tritici in vitro. For both strains, the ability to produce hydroxyphenazine compounds was correlated with greater antifungal activity than was production of PCA alone. These results are consistent with the findings of Smirnov and Kiprianova (40), who compared the inhibitory effects of PCA, 2-OH-PCA, and 2-OH-PHZ against a variety of bacterial and fungal animal and plant pathogens and found that in all cases the 2-hydroxyphenazines exhibited stronger bacteriostatic and fungistatic activity. We have recently demonstrated that the introduction of the core biosynthetic genes in other biocontrol microorganisms resulted in increased suppression of certain phytopathogenic fungi (46a; Huang et al., unpublished data). The phzO gene from P. aureofaciens 30-84 is an attractive target for such genetic manipulations because of the wide antimicrobial and antifungal activity of 2-hydroxyphenazines, which, on the other hand, exhibit little or no toxicity to fish, insects, or mammals (29, 47).
ACKNOWLEDGMENTS
We are grateful to Michael Konkel, Luying Xun, and David Weller for their interest and insightful comments on this work, to Tracey Timms-Wilson and Mark Bailey for making available their unpublished data, to Greg Phillips for the 2-hydroxyphenazine-1-carboxylic acid conversion assays, and to David Odelson and Glória Botelho for providing phenazine-producing strains.
REFERENCES
- 1.Aprill W A. Effect of Pseudomonas rhizobacteria on the growth of barley. M. S. thesis. Pullman: Washington State University; 1986. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingstons R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Bairoch A, Bucher P, Hofman K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1995;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker J O, Hepter C A, Yuen G Y, Van Gundy S D, Schroth M N, Hancock J G, Weinhold A R, Bowman T. Effect of rhizobacteria and metham-sodium on growth and root microflora of celery cultivars. Phytopathology. 1990;80:206–211. [Google Scholar]
- 5.Bonsall R F, Weller D M, Thomashow L S. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonasspp. in vitro and in the rhizosphere of wheat. Appl Environ Microbiol. 1997;63:951–955. doi: 10.1128/aem.63.3.951-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byng G S, Turner J M. Isolation of pigmentation mutants of Pseudomonas phenazinium. J Gen Microbiol. 1976;97:57–62. doi: 10.1099/00221287-97-1-57. [DOI] [PubMed] [Google Scholar]
- 7.Chang P C, Blackwood A C. Simultaneous production of three phenazine pigments by Pseudomonas aeruginosaMac 436. Can J Microbiol. 1969;72:581–583. doi: 10.1139/m69-077. [DOI] [PubMed] [Google Scholar]
- 8.Chin-A-Woeng T F C, Bloemberg G V, van der Bij A J, van der Drift K M G M, Schripsema J, Kroon B, Scheffer R J, Keel C, Bakker P A H M, Tichy H, de Bruijn F J, Thomas-Oates J E, Lugtenberg B J J. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact. 1998;11:1069–1077. [Google Scholar]
- 9.Copeland R A. Methods for protein analysis. New York, N.Y: Chapman & Hall; 1994. pp. 62–71. [Google Scholar]
- 10.Duffner F M, Müller R. A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovoransstrain A2: nucleotide sequence and analysis of the genes. FEMS Microbiol Lett. 1998;161:37–45. doi: 10.1111/j.1574-6968.1998.tb12926.x. [DOI] [PubMed] [Google Scholar]
- 11.Enderle P J, Farwell M A. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosacells. BioTechniques. 1998;25:954–958. doi: 10.2144/98256bm05. [DOI] [PubMed] [Google Scholar]
- 12.Essar D W, Eberly L, Hadero A, Crawford I P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flood M E, Herbert R B, Holliman F G. Pigments of Pseudomonas species. Part V; Biosynthesis of pyocyanin and the pigments of Ps. aureofaciens. J C S Perkins Transactions. 1972;1:622–627. doi: 10.1039/p19720000622. [DOI] [PubMed] [Google Scholar]
- 14.Galán B, Díaz E, Prieto M A, García J L. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coliW: a prototype of a new flavin:NAD(P)H reductase subfamily. J Bacteriol. 2000;182:627–636. doi: 10.1128/jb.182.3.627-636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgakopoulos D, Hendson M, Panopoulos N J, Schroth M N. Cloning of a phenazine biosynthetic locus of Pseudomonas aureofaciensPGS12 and analysis of its expression in vitro with the ice nucleation reporter gene. Appl Environ Microbiol. 1994;60:2931–2938. doi: 10.1128/aem.60.8.2931-2938.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibello A, Suárez M, Allende J L. Molecular cloning and analysis of the genes encoding the 4-hydroxyphenylacetate hydroxylase from Klebsiella pneumoniae. Arch Microbiol. 1997;167:160–166. [PubMed] [Google Scholar]
- 17.Hamdan H, Weller D M, Thomashow L S. Relative importance of fluorescent siderophores and other factors in biological control of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens2-79 and M4–80R. Appl Environ Microbiol. 1991;57:3270–3277. doi: 10.1128/aem.57.11.3270-3277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan H M, Fridovich I. Mechanism of the antibiotic action of pyocyanine. J Bacteriol. 1980;141:156–163. doi: 10.1128/jb.141.1.156-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodBmutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herbert R B, Holliman J G, Sheridan J B. Biosynthesis of microbial phenazines: incorporation of shikimic acid. Tetrahedron Lett. 1976;8:639–642. [Google Scholar]
- 21.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollstein U, McCamey D A. Biosynthesis of phenazines II: incorporation of (6-14C)-d-shikimic acid into phenazines—carboxylic acid and iodinin. J Org Chem. 1973;38:3415–3417. doi: 10.1021/jo00959a041. [DOI] [PubMed] [Google Scholar]
- 23.Hübner A, Danganan C E, Xun L, Chakrabarty A M, Hendrickson W. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia AC1100: characterization of the tftC and tftD genes and locations of the tftoperons on multiple replicons. Appl Environ Microbiol. 1998;64:2086–2093. doi: 10.1128/aem.64.6.2086-2093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson J L, Palleroni N J. Deoxyribonucleic acid similarities among Pseudomonasspecies. Int J Syst Bacteriol. 1989;39:230–235. [Google Scholar]
- 25.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosaPA01. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 26.Longley R P, Halliwell J E, Campbell J J R, Ingledew W M. The branchpoint of pyocyanine biosynthesis. Can J Microbiol. 1972;18:1357–1368. doi: 10.1139/m72-210. [DOI] [PubMed] [Google Scholar]
- 27.Mavrodi D V, Ksenzenko V N, Bonsall R F, Cook R J, Boronin A M, Thomashow L S. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens2-79. J Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messenger A J M, Turner J M. Phenazine-1,6-dicarboxylate and its dimethyl ester as precursors of other phenazines in bacteria. FEMS Microbiol Lett. 1983;18:65–68. [Google Scholar]
- 29.Nelson, C. D., and J. I. Toohey. 1968. Controlling the growth of algae and noxious plants. U.S. Patent 3,367,765.
- 30.Ownley B H, Weller D M, Thomashow L S. Influence of in situ and in vitro pH on suppression of Gaemannomyces graminis var. tritici by Pseudomonas fluorescens2-79. Phytopathology. 1992;82:178–184. [Google Scholar]
- 31.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Applic Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 32.Pierson L S, III, Thomashow L S. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens30-84. Mol Plant-Microbe Interact. 1992;5:330–339. doi: 10.1094/mpmi-5-330. [DOI] [PubMed] [Google Scholar]
- 33.Pierson L S, III, Gaffney T, Lam S, Gong F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens30-84. FEMS Microbiol Lett. 1995;134:299–307. doi: 10.1111/j.1574-6968.1995.tb07954.x. [DOI] [PubMed] [Google Scholar]
- 34.Prieto M A, García J L. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. J Biol Chem. 1994;269:22823–22829. [PubMed] [Google Scholar]
- 35.Raaijmakers J M, Weller D M. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonasspp. in take-all decline soils. Mol Plant-Microbe Interact. 1998;11:144–152. [Google Scholar]
- 36.Römer A, Herbert R B. Further observations on the source of nitrogen in phenazine biosynthesis. Z Naturforsch C. 1982;37:1070–1074. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 40.Smirnov V V, Kiprianova E A. Bacteria of Pseudomonas genus. Kiev, Ukraine: Naukova Dumka; 1990. pp. 100–111. . [Translation by D. V. Mavrodi.] [Google Scholar]
- 41.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stintzi A, Johnson Z, Stonehouse M, Ochsner U, Meyer J, Vasil M L, Poole K. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS. J Bacteriol. 1999;181:4118–4124. doi: 10.1128/jb.181.13.4118-4124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takizawa N, Yokoyama H, Yanagihara K, Hatta T, Kiyohara H. A locus of Pseudomonas pickettii DTP0602, had, that encodes 2,4,6-trichlorophenol-4-dechlorinase with hydroxylase activity, and hydroxylation of various chlorophenols by the enzyme. J Ferment Bioeng. 1995;80:318–327. [Google Scholar]
- 44.Tatusova T A, Madden T L. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 45.Thomashow L S, Weller D M. Role of phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomashow L S, Weller D M. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites. In: Stacey G, Keen N T, editors. Plant-microbe interactions. New York, N.Y: Chapman and Hall; 1996. pp. 187–235. [Google Scholar]
- 46a.Timms-Wilson T M, Ellis R J, Renwick A, Rhodes D J, Mavrodi D V, Weller D M, Thomashow L S, Bailey M J. Chromosomal insertion of phenazine-1-carboxylic acid biosynthetic pathway enhances efficacy of damping-off disease control by Pseudomonas fluorescens. Mol Plant-Microbe Interact. 2000;13:1293–1300. doi: 10.1094/MPMI.2000.13.12.1293. [DOI] [PubMed] [Google Scholar]
- 47.Toohey J L, Nelson C D, Kritkov G. Toxicity of phenazine carboxylic acids to some bacteria, algae, higher plants, and animals. Can J Bot. 1965;43:1151–1155. [Google Scholar]
- 48.Turner J M, Messenger A J. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/s0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 49.van Overbeek L S. Responses of bacterial inoculants to soil conditions. Ph.D. dissertation. Leiden, The Netherlands: University of Leiden; 1998. [Google Scholar]
- 50.Weller D M. Colonization of wheat roots by a fluorescent pseudomonad suppressive to take-all. Phytopathology. 1983;73:1548–1553. [Google Scholar]
- 51.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 52.Wood D W, Pierson L S., III The phzI gene of Pseudomonas aureofaciens30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene. 1996;108:49–53. doi: 10.1016/0378-1119(95)00754-7. [DOI] [PubMed] [Google Scholar]
- 53.Xun L, Sandvik E R. Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia colias a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl Environ Microbiol. 2000;66:481–486. doi: 10.1128/aem.66.2.481-486.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yabuuchi E A, Ohyama A. Characterization of “pyomelanin”-producing strains of Pseudomonas aeruginosa. Int J Syst Bacteriol. 1972;22:53–64. [Google Scholar]