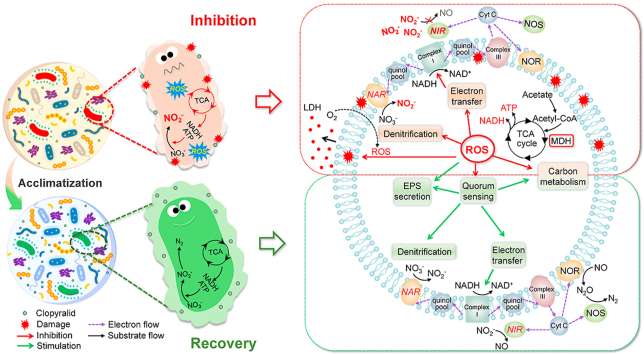
Abstract
The contaminant of herbicide clopyralid (3,6-dichloro-2- pyridine-carboxylic acid, CLP) poses a potential threat to the ecological system. However, there is a general lack of research devoted to the perturbation of CLP to the bio-denitrification process, and its biological response mechanism remains unclear. Herein, long-term exposure to CLP was systematically investigated to explore its influences on denitrification performance and dynamic microbial responses. Results showed that low-concentration of CLP (<15 mg/L) caused severe nitrite accumulation initially, while higher concentrations (35–60 mg/L) of CLP had no further effect after long-term acclimation. The mechanistic study demonstrated that CLP reduced nitrite reductase (NIR) activity and inhibited metabolic activity (carbon metabolism and nitrogen metabolism) by causing oxidative stress and membrane damage, resulting in nitrite accumulation. However, after more than 80 days of acclimation, almost no nitrite accumulation was found at 60 mg/L CLP. It was proposed that the secretion of extracellular polymeric substances (EPS) increased from 75.03 mg/g VSS at 15 mg/L CLP to 109.97 mg/g VSS at 60 mg/L CLP, which strengthened the protection of microbial cells and improved NIR activity and metabolic activities. Additionally, the biodiversity and richness of the microbial community experienced a U-shaped process. The relative abundance of denitrification- and carbon metabolism-associated microorganisms decreased initially and then recovered with the enrichment of microorganisms related to the secretion of EPS and N-acyl-homoserine lactones (AHLs). These microorganisms protected microbe from toxic substances and regulated their interactions among inter- and intra-species. This study revealed the biological response mechanism of denitrification after successive exposure to CLP and provided proper guidance for analyzing and treating herbicide-containing wastewater.
Keywords: Denitrification, Nitrite accumulation, Clopyralid, Membrane damage, Metabolism activity, Microbial community
Graphical abstract
Highlights
-
•
NO2− accumulation was significantly enhanced by initial CLP exposure.
-
•
CLP could inhibit microbial viability by causing ROS production and membrane damage.
-
•
The generation of ROS reduced NIR, metabolism and electron transport efficiency.
-
•
Long-term CLP exposure stimulated the increase of EPS secretion.
-
•
The protection of EPS and succession of microbial community restored denitrification.
Abbreviations
- CLP
clopyralid
- NIR
nitrite reductase
- ROS
reactive oxygen species
- EPS
extracellular polymeric substances
- AHLs
N-acyl-homoserine lactones
- NAR
nitrate reductase
- NOR
nitric oxide reductase
- NOS
nitrous oxide reductase
- DNB
denitrifying bacteria
- HRT
hydraulic retention time
- CTN
chlorothalonil
- DPV
differential pulse voltammetry
- ETSA
electron transfer system activity
- PN
protein
- PS
polysaccharide
- MDH
dehydrogenase
- PCR
polymerase chain reaction
- OTUs
operational taxonomic units
- HO•
hydroxyl radical
- O2−
superoxide anion
- H2O2
hydrogen peroxide
- LDH
lactate dehydrogenase
- TCA
tricarboxylic acid
- SOD
superoxide dismutase
- QS
Quorum sensing
- NADH
nicotinamide adenine dinucleotide
- ATP
adenosine triphosphate
1. Introduction
Due to the extensive use of pesticides in modern agriculture, water pollution related to organochlorine compounds released by industrial and agricultural activities has attracted significant research attention [[1], [2], [3], [4]]. Recent studies have revealed that more than 99% of pesticides were released into the environment after application, contaminating the water notably, while less than 1% of all pesticides reached the targets [5,6]. The herbicide CLP belongs to the picolinic acid family and is usually used to remove broadleaf weeds in certain crops and woody plants [7]. Its chemical stability and high water solubility (1000 mg/L) cause consistent pollution to the water environment, and it has been frequently detected in aquatic ecosystems [8,9]. Such chemicals could be inevitably transported into wastewater treatment plants as a consequence of their widespread use as well as being discharged from urban and industrial effluents. Like many other herbicides, CLP is considered to have broad-spectrum antibacterial properties and resistance to biodegradation [10]. The antibacterial property of CLP significantly influences the efficiency of microbial-based conventional wastewater treatment and has attracted much attention.
Microbial denitrification is a popular and eco-friendly way to remove nitrate-nitrogen in wastewater. It is a stepwise reduction process involving metabolic activities commonly associated with denitrifying bacteria (DNB), which reduce nitrate to nitrite and nitrogen [11,12]. However, this process is generally sensitive to toxic pollutants [13,14]. Extensive studies have been conducted on the inhibitory impacts of pesticides on denitrification performance, denitrifying enzymes, and microbial communities. He et al. proposed that the chlorothalonil (CTN) accumulation disturbed the natural microbial processes and influenced its extracellular enzyme activity and nitrogen transformation in riparian sediments [15]. High concentrations of CTN (10 and 25 mg/kg) significantly decrease the denitrification rates up to 37–62%. In addition, pesticides such as dazomet could change the microbial community structure to adapt to toxin-induced stress [16]. Previous studies have shown that Proteobacteria, the most frequently observed phylum involved in denitrification, increased after being exposed to dazomet initially and then significantly decreased, while another denitrifying phylum Chloroflexi was contrary developed Proteobacteria [17]. The adverse effects of pesticides on microbial respiration and the bacterial community may result in changes in denitrification. Therefore, it is of ecological significance to understand the response mechanism of microorganisms and denitrifying communities to CLP stress in denitrification.
Denitrification is carried out by DNB, and its efficiency depends on the microbial viability and metabolic activity of denitrifiers [18]. Oxide stress could damage microbial cells by accumulating ROS and damaging cell membrane integrity [19,20]. When the cytomembrane is severely damaged, lactate dehydrogenase (LDH) can be released to the extracellular region [20]. The amount of LDH released can usually be used as an indicator of membrane damage. Moreover, microbial metabolism plays an essential role in the process of denitrification. The nicotinamide adenine dinucleotide (NADH) and adenosine triphosphate (ATP) produced during microbial metabolism are direct electron donors and energy sources for denitrification [19]. The generated NADH could be catalyzed by NADH dehydrogenase to produce electrons. Then the electrons are transferred through the quinone pool, electron transport protein, and finally utilized by the critical denitrifying enzymes (nitrate reductase (NAR), NIR, nitric oxide reductase (NOR) and nitrous oxide reductase (NOS)) [21]. Exposure to pesticide CTN could deteriorate the microbial metabolic activity and key denitrifying enzyme activities, which explained the decline of denitrification performance [15]. Additionally, the denitrification performance is closely related to the bacterial community structure, and the abundance of dominant bacteria is significantly affected by pesticides. According to Hura et al. CLP could change the microbial community structure in the activated sludge and inhibit bacterial growth, such as Chryseobacterium, Enterobacter and Pseudomonas [22]. Revealing the connection between microbial activities, bacterial species, and denitrification performance is of great importance to better understand the denitrification process. However, there is a general lack of research devoted to the perturbation of CLP to the denitrification process at the level of microbial mechanism and microbial community, and its biological response mechanism remains unclear.
Thus, the specific aims of the study are to (a) explore the effects of increasing CLP concentration on the denitrification process and investigate its impacts on microbial membrane damage via measurements of LDH, ROS and superoxide dismutase (SOD); (b) reveal the microbial metabolic responses of denitrifiers to increasing CLP from carbon source metabolism, electron transport activity, denitrifying enzymes and EPS secretion; (c) investigate the transformation of the denitrifying microbial community through high-throughput sequencing as well as the microbial response mechanism under CLP exposure. This work conducted a comprehensive investigation and proposed the possible mechanism for the perturbation of CLP on denitrification. In light of this, in-depth insight into the mechanism of microbial response to CLP stress provides guidance for analyzing and treating herbicide-containing wastewater.
2. Materials and methods
2.1. Experimental set-up, inoculation and reactor operation
Two plexiglass packed bed reactors were constructed, with an inner diameter of 7 cm, 74 cm height and total volume of 1 L. Two peristaltic pumps were used to load influent into the reactors. The bottom ports were used for the collection of sludge samples from reactors. The whole experimental device was in an insulated room, and the operating temperature was maintained at 30–35 °C. Synthetic influent wastewater was prepared to simulate nitrate wastewater. Influent was composed of 0.10 g/L MgSO4, 0.20 g/L CaCl2, 0.02 g/L KH2PO4, 4:1 C:N molar ratio maintained by NaNO3 and CH3COONa, and 1 mL/L trace element solution (details were provided in Supplementary Materials), and CLP was added at the specified concentration. The pH of the synthetic water was adjusted to 7.3–7.5 by HCl.
The two reactors were inoculated with activated sludge from a secondary sedimentation tank in municipal wastewater treatment plant (Tianjin city, China). Both reactors started with 100 mg/L NO3−-N and 4-h hydraulic retention time (HRT). The molar ratio of C:N was maintained at 4:1 by adding CH3COONa and NaNO3. Considering that the increasing trend of CLP production and contributing to better understand the biological mechanism of the long-term perturbation of CLP on bio-denitrification, the concentration range selected in this study (0.25 mg/L to 60 mg/L) is slightly higher than the previous study [23,24]. On day 16, one reactor (R1) was operated in the addition of 0.25 mg/L CLP to the synthetic water, whereas the second reactor (control, R2) was operated continuously without changing any parameters or conditions. Except for sludge exposure to CLP in R1, all other parameters and conditions were identical in both reactors (Table 1).
Table 1.
Operational conditions from phase I to phase VII.
| Phase | Time (d) | CLP (mg/L) | Acetate COD (mg/L) | NO3−-N (mg/L) | HRT (h) |
|---|---|---|---|---|---|
| I | 0–15 | 0 | 400 | 100 | 4 |
| II | 16–29 | 0.25 | 400 | 100 | 4 |
| III | 30–41 | 1.00 | 400 | 100 | 4 |
| IV | 42–56 | 5.00 | 400 | 100 | 4 |
| V | 57–74 | 15.00 | 400 | 100 | 4 |
| VI | 75–85 | 35.00 | 400 | 100 | 4 |
| VII | 86–94 | 60.00 | 400 | 100 | 4 |
2.2. Chemical analysis
Water samples were collected and filtered with 0.22-μm filters. An ultraviolet spectrophotometer (UV-2700, Shimadzu, Japan) was used to measure nitrate (at 220 nm and 275 nm) and nitrite (at 540 nm). CLP was measured by high-performance liquid chromatography (HPLC, LC-1260, Agilent, USA). Detail measurements were in Supplementary Materials.
Differential pulse voltammetry (DPV) and Tafel were carried out to reveal the electron transfer ability of the denitrification system. They were all detected using an electrochemical workstation (CHI660E, Chehua, Shanghai, China) with a traditional three-electrode system composed of the reference electrode (saturated calomel electrode), working electrode (glassy carbon electrode) and counter electrode (platinum wire). The working electrode was firstly polished with Al2O3, diameters of 1 mm, 0.3 mm and 0.05 mm, then cleaned with deionized water, alcohol and deionized water for 30 s sequence. Finally, 5 mM potassium ferricyanide was used as the solute, and 0.1 M potassium chloride was used as the solvent to test the electrode polishing degree. The testing parameters were as follows, for DPV: amplitude = 50 mV, pulse width = 0.20 s; for Tafel: scan rate = 5 mV/s.
2.3. Half inhibitory concentration (IC50) of CLP analysis
100 mL basal medium containing a bacterial suspension of denitrifying microorganisms (OD = 0.08) were transferred into serum bottles with 100 mL effective volume. Then CLP solution was added to each serum bottle, making the final concentration to 0, 0.25, 5, 15, 35 and 60 mg/L, respectively. The influent NO3−-N in short-term exposure experiment were about 100 mg/L, and the basal medium consisted of: CH3COONa (0.51 g/L), KH2PO4 (0.01 g/L), MgSO4 (0.12 g/L), yeast extract (0.15 g/L), tryptone (0.50 g/L) and trace elements (2 mL/L) [25]. All experiments were conducted at 35 °C in the stationary constant temperature shaker. The water sample was taken out at 0, 2, 4, 6.5, 9.5 and 13 h to analyze the nitrate concentration, respectively.
The half inhibitory concentration (IC50) of CLP on the nitrate reduction was calculated according to the non-competitive inhibition model (Eq. (1)) [26]:
| (1) |
where I (%) is the inhibition response; a is the IC50 value (mg/L); b is the fitting parameter; c is the concentration of CLP.
2.4. Molecular biological analysis
Electron transfer system activity (ETSA) was used to evaluate electron transport and reception ability during the denitrification process [19], which was measured by reducing 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride to formazan [27]. The absorbance of the orange-like formazan was recorded at 590 nm against a methanol solvent blank. ATP was measured according to the previous report [28]. Briefly, 6 mL Tris-EDTA buffer solution was added to samples to extract ATP and then immediately boiled for 3 min. ATP was measured by high-performance liquid chromatography (HPLC, LC-1260, Agilent, USA). Detail procedures were provided in Supplementary Materials.
EPS, an indispensable part of microorganisms, has been proven to play a significant role in defending against external toxic substances to maintain the denitrification process [29]. EPS extraction was carried out by the thermal treatment method, and EPS was then filtered by a 0.22-μm filter, used for protein (PN) and polysaccharide (PS) analysis. The sum of these fractions was calculated as total EPSs. PS measurement was performed according to the anthrone method, and PN measurement was conducted using the coomassie brilliant blue colorimetric method.
Quorum sensing (QS) is a way of cell-to-cell communication used by microorganisms to collectively change biological behaviors in response to variations of cell density and the surrounding environment [30]. Previous studies found that AHLs-mediated QS was essential for the environmental stress resistance of microorganisms [31]. In this study, AHLs were detected according to Wang et al. [32]. An ultrasonic cell crusher crushed the samples. They were extracted with an equivalent volume of ethyl acetate after being filtered through a 0.22-μm filter and then evaporated at 40 °C with a rotary evaporator (Yarong RE-5220, China). The extracted AHLs were dissolved in methanol, and then they were detected by the Ultra-Performance Liquid Chromatography-Mass Spectrometry (UPLC-MS, 6410B, Agilent, USA). Detail procedures were provided in Supplementary Materials.
The corresponding enzyme-linked immunoassay kits determined ROS, LDH, SOD, dehydrogenase (MDH), NADH, NAR and NIR kits according to the manufacturer's procedure (Enzyme-linked Biotechnology Co., Ltd, Shanghai, China).
2.5. Evaluation of the microbial community
Illumina MiSeq sequencing based on 16S rRNA gene was performed to analyze the microbial community structures and compositions. According to the manufacturer's protocol, genomic DNA was extracted from samples using the Qubit ssDNA Assay Kit. The V3–V4 region of the bacterial 16S ribosomal RNA gene was amplified by polymerase chain reaction (PCR) using the bacterial primers 341F (CCCTACACGACGCTCTTCCGATCTGCCTACGGGNGGCWGCAG) and 805R (GACTGGAGTTCCTTGGCACCCGAGAATTCCAGACTACHVGGGTATCTAAT) [25]. The amplicons were sequenced in MiSeq sequencing platforms (Illumina, USA) using PE 300 strategy (Majorbio Bio-Pharm Technology Co., Ltd, Shanghai, China). The statistical analysis of the biological information was performed using operational taxonomic units (OTUs) definition of ≥97% sequence similarity.
2.6. Other analytical procedures
All experiments were performed in triplicate, and the results were expressed as means ± standard error. The significance was performed by a t-test using the Statistical Package for the Social Sciences (SPSS) 20.0 from the USA.
3. Results and discussion
3.1. Performance of denitrification during long-term exposure to CLP
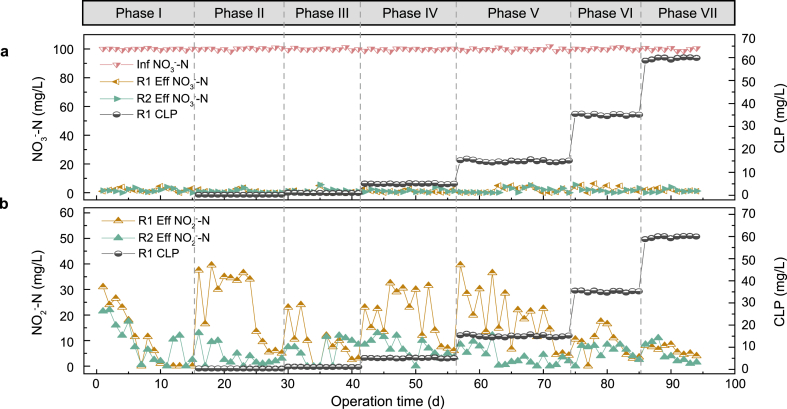
The packed bed reactor was operated for almost 100 days and divided into seven phases according to the influent concentration of CLP increasing from 0 mg/L to 60 mg/L to explore the response of the denitrification performance to CLP perturbation. The variation of nitrogen (NO3−-N and NO2−-N) and CLP concentrations during the operation period are shown in Fig. 1.
Fig. 1.
Denitrification performance along a gradient concentration of CLP: (a) NO3−-N concentration; (b) NO2−-N concentration (phase I-VII indicate the different CLP concentrations, i.e., 0.25, 1, 5, 15, 35 and 60 mg/L CLP).
In phase I, the reactors were operated for 15 days without the addition of CLP. The effluent nitrate concentration stabilized lower than 5 mg/L, and the nitrite accumulation was below 10 mg/L after a period of operation, indicating denitrification performance became stable. After the startup period, R1 was exposed to CLP with the increasing concentration ranging from 0.25 mg/L to 60 mg/L to evaluate the denitrification performance under CLP stress. The results demonstrated that there were almost no differences in NO3−-N concentration between the control (R2) and CLP group (R1). However, the nitrite accumulation in R1 showed a noticeable increase whenever the CLP influent concentration was switched to a new level during the phase of I–V. As shown in Fig. 1, in the early stages from phase II to V, the nitrite accumulation soared steeply to 37.50 mg/L, 23 mg/L, 32.50 mg/L and 39.50 mg/L, respectively. The fluctuation of nitrite indicated that the CLP inhibited denitrification, and the partially removed nitrate was converted into nitrite without further transformation. The inhibition of denitrification caused by pesticides has been reported in many previous studies [21,29], such as 40 mM 1-hydroxyethane-(1,1-bisphosphonic acid), an environmental toxicant that is toxic to aquatic organisms, can also inhibit nitrite reduction. However, the denitrification system showed good stability in overcoming the CLP shock, that was, after continuous operation for more than 10 days in each phase, the nitrite removal efficiency of R1 recovered.
More importantly, the denitrification performance of R1 remained almost stable, although the concentration of CLP continued to increase to 35 mg/L even 60 mg/L. Especially in phase VII, with the influent CLP concentration of 60 mg/L, there was almost no accumulation of nitrite in R1, indicating that after a period of operation, the reactor adapted to the toxicity of CLP. It was reported that the nitrogen removal performance of the anammox system was stable under long-term stress of gradually increasing the norfloxacin concentration to 50 mg/L [33]. Such tolerance may benefit from: (1) the metabolic activity responses of microorganisms, such as adjustment of EPS and AHLs secretion [34,35]; (2) changes in the abundance of denitrifying bacteria and functional succession of microbial community structure [36]. In addition, the low concentration of CLP could act as a necessary signaling molecule that activated the downstream pathway and promoted its adaptive response to the higher concentrations of CLP.
3.2. Responses of antioxidant systems and membrane damage after CLP exposure
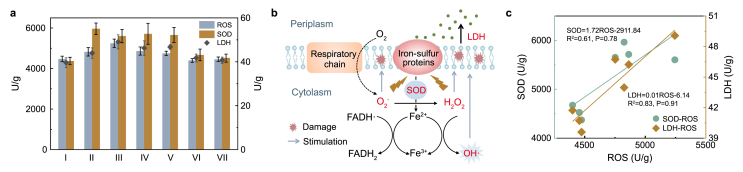
The damaged membrane allows the release of LDH [20]. As illustrated in Fig. 2a, after exposure of R1 to CLP, LDH in R1 increased significantly. In detail, the LDH of R1 without CLP was 39.60 U/g, and then increased to 42.92, 49.08, 46.23, 46.74 U/g with 0.25, 1, 5 and 15 mg/L of influent CLP, respectively. It implied that CLP might cause damage to the membrane integrity of microorganisms. Rohini et al. proposed that chlorinated organics such as diclofenac could inhibit cell viability and bacteria growth by disrupting membrane activity [37]. The damage may be caused by the Cl atom, a strong electrophile, which tends to bind to the C=C double bonds of lipid bilayers as well as peptide bond and amino groups of proteins that microbial cell membrane is mainly comprised [38].
Fig. 2.
(a) Effects of CLP concentration on the level of ROS production, SOD activity and LDH release; (b) Schematic diagram of the generation of LDH and ROS and SOD function; (c) Linear relationships between ROS with SOD and LDH. Error bars represent standard deviations of triplicate measurements.
Additionally, membrane damage might lead to abnormalities in the respiratory electron transfer and allow undesired exposure of O2 to iron or quinone moieties part of the electron transport chain, resulting in excessive production of ROS [20]. In the present study, the responses of ROS to CLP were investigated. As shown in Fig. 2a, the intracellular ROS content remarkably increased from 4474.11 U/g at 0 mg/L CLP to 4825.57, 5244.30, 4863.52, 4752.15 U/g at 0.25, 1, 5 and 15 mg/L CLP, respectively. It indicated that CLP led to the excessive accumulation of ROS, causing an intracellular imbalance between oxidants and antioxidants in cells, which might be another trigger for the destruction of cell membrane integrity [38]. In the anaerobic environment, submicromolar oxygen remains in the medium. It can react with electrons unwontedly transferring from the electron carriers, leading to the generation of superoxide anion (O2−) and hydrogen peroxide (H2O2) [39]. The abundant reduced iron (e.g., iron-sulfur clusters or semiquinones) catalyzes self-sustaining Fenton reactions with the H2O2 formed by substrate oxidation to produce hydroxyl radical (HO•), which is the major killing mechanism of the ROS-generating agents (Fig. 2b) [40]. Toxic pollutants (e.g., ampicillin) exposure have also been reported to cause excessive ROS accumulation [41]. According to the results observed in the current study, it was proposed that the CLP-induced accumulation of ROS could result in direct or indirect ROS-mediated damage of cellular macromolecules, such as nucleic acids, proteins and lipids, leading to cytotoxicity and damage [42]. Therefore, the attack of the membrane components could destroy the microenvironment of enzymes, causing the deterioration of enzymes activities related to electron behaviors and denitrification in the cell membrane [43]. Moreover, ROS could react with enzymes directly, and its primary targets are in the cytoplasm [44], leading to cytoplasmic enzyme dysfunction related to denitrification, carbon metabolism and electron transfer. This might be one of the reasons why the denitrification process was inhibited when R1 was exposed to CLP.
Meanwhile, in response to oxidative stress, microbial cells have evolved a variety of antioxidant enzymes, such as SOD, to mitigate ROS to maintain a balance between ROS production and the ability to detoxify the reactive intermediates [45]. As the activities of SOD are usually related to the oxidative damage effect, changes in activities of SOD under various dosages of CLP have been investigated. The data showed that the activities of SOD displayed similar change trends as the ROS. It increased from 4372.71 U/g at 0 mg/L CLP to 5964.26, 5603.27, 5713.54, 5653.90 U/g at 0.25, 1, 5 and 15 mg/L CLP, respectively, suggesting that CLP might stimulate SOD activity in response to exposure to oxidative stress (Fig. 2a). The LDH release and SOD activity were positively correlated with ROS production (R2 = 0.61 and 0.83, respectively) (Fig. 2c), confirming CLP inhibited microbial viability. Finally, the level of ROS, SOD, and LDH decreased and tended to stabilize with the continuous increase of CLP from 35 mg/L to 60 mg/L through negative feedback regulation. No significant difference was measured between the control and R1 (from phase V to VII). It could be found that the accumulation of nitrite decreased significantly in the early stage of phase V and the denitrification performance of R1 recovered gradually, which was in correspondence with the results of low oxidative stress level and no obvious decrease in microbial viability. Accordingly, the IC50 value was calculated to be 498.44 mg/L through a regression equation (Eq. (1)) (Fig. S1), indicating that denitrifying microorganisms had strong tolerance to CLP. Because CLP concentration in R1 (≤60 mg/L) was much lower than IC50, the inhibitory effect of CLP on denitrification performance and enzyme activities was not significant. Also, the phenomenon indicated that the microorganisms have almost adapted to the stress of CLP, which could also be explained by the succession of the microbial community and the more secretion of EPS by microorganisms to resist external toxic interference. Detailed analysis and verification are carried out in sections 3.3, 3.4.
3.3. Responses of microbial metabolism after CLP exposure in denitrifying microorganism
3.3.1. Effects of CLP on key enzymes activity of denitrification
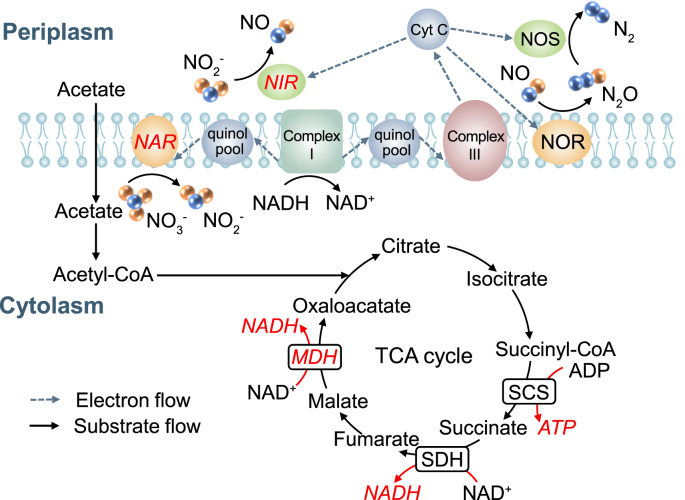
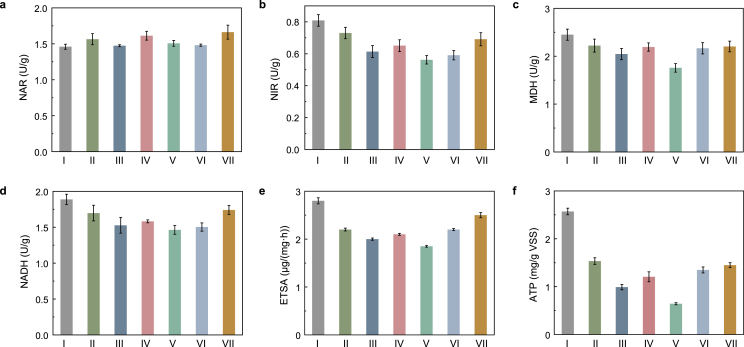
NAR and NIR are key denitrification enzymes that catalyze the reduction of nitrate to nitrite, nitrite to nitric oxide, respectively (Fig. 3). As shown in Fig. 4a, CLP almost had no detectable influence on NAR activity, which was consistent with no measurable effects on the process of nitrate reduction. However, the NIR activity was remarkably inhibited by CLP, and its activity decreased from 0.81 U/g at 0 mg/L CLP to 0.73, 0.61, 0.65, 0.56 U/g at 0.25, 1, 5, 15 mg/L CLP, respectively (Fig. 4b). The declined NIR activity could be the direct reason for nitrite accumulation. Subsequently, the NIR activity increased gradually when the CLP concentration increased to 35 and 60 mg/L. This observation was consistent with the result that the nitrite accumulation in the denitrification system showed a trend of increasing first and then decreasing. The phenomenon of no measurable effect on NAR activity can be explained by the fact that NAR is stably embedded in the cell membrane, whereas NIR is freely distributed in the periplasm of denitrifiers, suggesting that the activity of NIR is more susceptible to CLP than NAR [19]. It was also reported that periplasmic enzymes were easily inhibited by some external toxicants, such as chlorothalonil [15].
Fig. 3.
Schematic diagram of microbial acetate metabolism, electron transport system and electron consumption during denitrification.
Fig. 4.
Effects of CLP concentration on (a) NAR and (b) NIR activities; The variations of (c) MDH, (d) NADH, (e) ATP and (f) ETSA at different phases. Error bars represent standard deviations of triplicate measurements.
3.3.2. Effects of CLP on microbial carbon source metabolism and electron transfer involved in denitrification
Anaerobic denitrification involves two interrelated biochemical reactions: (1) the carbon source (acetate) metabolism to produce NADH and ATP through the tricarboxylic acid (TCA) cycle; (2) the biological reduction of nitrate with the produced NADH (Fig. 3) [46,47]. A previous study showed that MDH was one of the key enzymes in the TCA cycle for catalyzing the critical step to produce NADH for denitrification [19]. Therefore, in the current study, the key enzymes (MDH) in the TCA cycle and the major productions (NADH, ATP) during metabolism were investigated to further reveal the influence mechanism of CLP. As shown in Fig. 4c, compared with the control (2.45 U/g), the MDH decreased to 1.76 U/g as the CLP concentration increased from 0 mg/L to 15 mg/L, then gradually increased to 2.2 U/g at 60 mg/L CLP. The MDH level in phase V was approximately 28% lower than that control. Fig. 4d and e showed that the NADH and ATP in control were 1.88 U/g and 2.57 U/g, respectively, significantly higher than 1.46 U/g and 0.64 U/g in phase V (with 15 mg/L CLP exposure). The change trends of MDH, NADH and ATP followed the same discipline, consistent with denitrification performance and microbial metabolism.
As an essential production of carbon source metabolism, NADH is the primary electron donor for nitrate bio-reduction. Higher NADH production is related to the greater efficiency of denitrification [48]. As reported, more intracellular ATP resulted in greater cell viability under anaerobic conditions [28], which agreed well with the observations in the present study. It was noted that the ATP and NADH in phases VI and VII also gradually increased, which might bring about the recovery of denitrification performance. However, the ATP content remained at a low level after R1 was exposed to CLP. Although it increased slightly in phase VII, it was still 1.67 times lower than control. The decrease in ATP content could be explained by the damage to microbial cells by CLP, such as cell repair and increase in cell synthesis and related proteins expression, which consumed a lot of energy, resulting in relatively low detectable ATP.
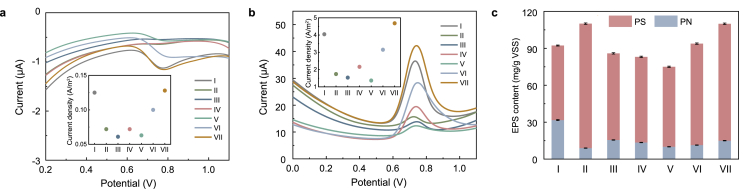
It is well known that the electrons generated during the metabolism of carbon source need to be transferred through a respiratory chain (consisting of quinone pools, cytochrome c, complex I and complex III) to drive the bio-denitrification process (Fig. 3) [21,49]. The previous study has confirmed that ETSA had a great influence on the denitrification process. Increasing electron transfer would enhance denitrification efficiency and reduce the accumulation of intermediates (such as nitrite) [19]. In this study, Fig. 4f showed the ETSA decreased by more than 25% as the CLP concentration increased from 0 mg/L to 15 mg/L. ETSA was kept at a lower concentration level, and then it increased to 2.5 μg/(mg·h) at 60 mg/L CLP. The significant decrease in ETSA might be caused by the inhibition of key enzyme activities in electron transport systems of denitrification by CLP, resulting in low electron transport efficiency and nitrite accumulation. As Chen et al. have documented, a high concentration of CTN accumulation can also affect denitrification performance by inhibiting ETSA [21]. Meanwhile, electrochemical tests (DPV and Tafel) were also performed to reveal the influence of CLP on the electron transfer rate of denitrification. As shown in Fig. 5a and b, the peak current densities of reduction and oxidation at about 0.72 V declined as the CLP concentration increased from 0 mg/L to 15 mg/L, and then gradually enhanced with CLP concentration increasing from 15 mg/L to 60 mg/L. Similarly, the exchange current densities calculated by extrapolation decreased from 3.52 × 10-7 A/cm2 at 0 mg/L CLP to 2.27 × 10-7, 2.20 × 10-7, 2.86 × 10-7 and 1.91 × 10-7 A/cm2 at 0.25, 1, 5 and 15 mg/L CLP, respectively (Fig. S2). Subsequently, the exchange current densities increased gradually when the CLP concentration increased to 35 and 60 mg/L. The changing trend of electron transfer rate was consistent with ETSA, showing that electron transport efficiency was inhibited after denitrifiers were exposed to CLP initially and then gradually recovered.
Fig. 5.
(a, b) DPV curves along a CLP gradient (a) Reduction peak and (b) Oxidation peak, and the insert in (a) and (b) were the peak current densities of reduction and oxidation at 0.72 V, respectively; (c) Changes of EPS at different phase with a gradient concentration of CLP disturbance. Error bars represent standard deviations of triplicate measurements.
3.3.3. Effects of CLP on the EPS secretion in denitrification
The variation of total EPS contents given in terms of PN and PS for this study was shown in Fig. 5c. The initial content of EPS was 92.28 mg/g VSS at 0 mg/L CLP and then increased by 1.2 times as the CLP concentration increased to 0.25 mg/L, indicating that CLP stimulated microorganisms to secret more EPS to resist toxic substances. It is speculated that the thicker EPS layer on the bacteria surface might absorb more CLP, which can delay the spread of poison into the interior of microbes and alleviate the CLP biotoxicity to microbes. Antibiotics (e.g., ofloxacin, sulfadiazine and tetracycline) could also promote the EPS production of activated sludge and biofilm from different bioreactors [41]. From 0.25 to 15 mg/L CLP, the EPS content decreased gradually to 75.00 mg/g VSS at 15 mg/L CLP, indicating that a higher concentration of CLP reduced EPS contents, which was consistent with the overproduction of ROS and the increase of SOD and LDH. Then, it increased to 93.81, 109.97 mg/g VSS at 35 and 60 mg/L CLP, respectively, reflecting the adaptive process of the denitrification system to the pressure of CLP.
The changes in EPS content may be attributed to the following three aspects: Firstly, under the effect of CLP, EPS secreting bacteria were enriched in large quantities, which significantly promoted the production of total EPS; Secondly, carbon source metabolism decreased as the CLP concentration increased, resulting in a decrease in ATP that provided energy for the synthesis and secretion of EPS. Moreover, the biodegradable components of EPS were utilized by bacteria to sustain vital metabolic processes, which further reduced the contents of EPS; Finally, with the restoration of cell metabolic activity and the enrichment of EPS secreting bacteria, sufficient energy and substance conditions were provided for the EPS production. The EPS content was maintained at a high level at 60 mg/L CLP. As noted above, the change of EPS was consistent with the response of microbial metabolic capacity to CLP stress.
3.3.4. Effects of CLP on AHLs release in denitrification
In this study, the AHLs-based QS was further studied in the denitrification process under CLP pressure. As shown in Fig. S3a, the concentrations of C4-HSL, C8-HSL and C14-HSL were detected. They decreased significantly from 22.50, 24.50, 6 μg/g VSS to 1.26, 0.77, 0.22 μg/g VSS respectively as the concentration of CLP increased from 0 to 15 mg/L. It has been reported that AHLs can regulate the expression of key genes in bacteria, thereby enhancing their activities, proliferation, viability and aggregation [50]. Different AHLs have disparate regulatory pathways. For instance, C8-HSL will cause an increase in the production of PN or PS mediated by abundant metabolites (Ala, Val, UDP-Glc, UDP-GlcNAc, etc.) and their metabolic pathways [51], and further enhance microbial flocculation ability and metabolic activity. The results showed that the changing trend of C8-HSL was consistent with PN. Furthermore, C8-HSL could promote heterotrophic metabolism and increase the amount of NADH and ATP [52], and the phenomenon of C8-HSL and C14-HSL was the same with the NADH and ATP. C4-HSL and C14-HSL played a key role in regulating activities of NAR and NIR [35,53], and the change trends of C4-HSL and C14-HSL were also consistent with activities of NAR and NIR. Hence, the reduction of AHLs might negatively affect the synthesis of EPS, carbon source metabolism and nitrite reduction.
In the final stage, the concentration of the AHLs (C4-HSL, C8-HSL, C14-HSL) increased to 9.50, 8.57, 1.43 μg/g VSS, respectively (Fig. S3a). The gradual increase in AHLs could be attributed to three factors: (1) The secretion of large amounts of EPS provided a barrier to reduce the toxic effect of CLP on signal-secreting bacteria; (2) AHLs-secretion bacteria were enriched in large quantities; (3) The enhancement of microbial metabolic activity provided a large amount of energy for the synthesis of AHLs. The concentration of signal molecules will increase with the growth density of the microflora, which will stimulate the expression of the QS system and form a positive feedback loop [35]. QS signal molecules can regulate the metabolism of microorganisms in response to environmental stress, thereby making it easier for microorganisms to adapt to the external environment and multiply to a relatively high abundance.
3.4. Microbial communities in response to CLP
3.4.1. The α-diversity and shift of microbial community
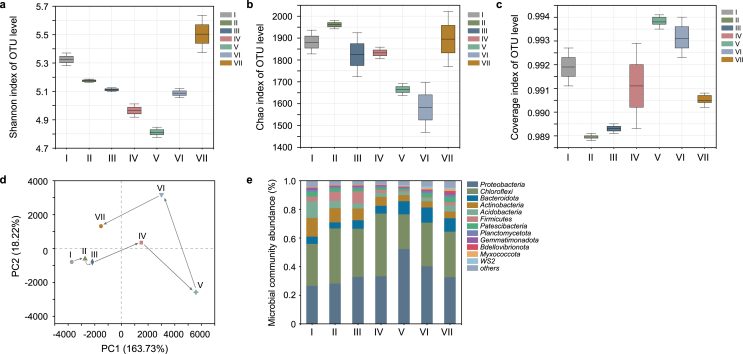
Sequence quantity, OTUs number and alpha diversity index of all samples were listed in Table S1. OTUs of all samples ranged from 1246 to 1533. According to the alpha diversity index, species diversity (Shannon), richness (Chao) and Goods coverages were shown in the boxplot (Fig. 6). Based on these diversity indices, the Shannon index initially declined from phase I to Phase V, and then increased after a further increase in CLP concentration from 15 mg/L to 60 mg/L (Fig. 6a). The variation of Chao values followed a similar pattern to that of diversity (Fig. 6b). The Goods coverages of all samples were more significant than 0.98 (Fig. 6c), suggesting that the obtained sequence libraries covered the microbial diversities. Notably, the CLP concentration had an evident impact on microbial diversity. The biodiversity and richness of the microbial community experienced a U-shaped process. The decrease in species diversity and richness when CLP was initially added indicated that the metabolism of the bacterial community sensitive to CLP toxicity could be inhibited. With the long-term exposure to the gradual increasing CLP, the richness and diversity of phase VII samples were higher than other samples. CLP had no long-term direct toxic effect on the overall microbial metabolic activity and biomass, and diverse communities tended to stabilize and contributed to ecosystem functions. Hura et al. have also reported that CLP has an initial toxic effect on the biomass, which killed some of the microorganisms more susceptible to CLP, while a substantial portion of the biomass survived and actively accommodated CLP without a further reduction in microbial activity [22]. This general decreasing-increasing biodiversity trend was consistent with previous studies on organic pollutants such as dazomet [16] and metam-sodium [54], where microbial populations and diversity reduced after initial exposure of pollutants, but recovered during long acclimatization.
Fig. 6.
Alpha diversity comparisons based on (a) Shannon, (b) Chao and (c) coverage indices; (d) The variation of microbial communities showed by PCA; (e) Fluctuations of major phyla.
To gain better insight into the effect of the CLP on denitrification performance, the microbial community structure was investigated. The phylogenetic classification was carried out at the phylum and genus level. The relative abundances of all samples were illustrated in Figs. 6e and 7. Proteobacteria (26.52% ∼ 56.22%), Chloroflexi (24.25% ∼ 43.75%) were the dominant phyla in all samples, followed by Actinobacteria (4.29% ∼ 13.08%) and Bacteroidetes (4.24% ∼ 10.60%) (Fig. 6e). Proteobacteria was widely reported as major phyla in the denitrification process and included many types of denitrifiers [55]. The samples with CLP added, especially for phase V, showed a high relative abundance of Proteobacteria (52.22%) compared with the phase I sample (26.52%). This indicated that Proteobacteria was resistant to long-term CLP exposure, which was in line with the resistance to other organic pollutants such as sulfamethoxazole and tetracycline hydrochloride [56]. Many other detected phylum, such as Chloroflexi, Bacteroidetes and Firmicutes, are also capable of denitrification. Chloroflexi was another prominent phylum, which has often been observed in heterotrophic nitrogen removal reactors [56]. The relative abundance of Chloroflexi increased from 29.31% at 0 mg/L CLP to 43.75% at 5 mg/L CLP, indicating CLP could stimulate the enrichment of Chloroflexi. Chloroflexi could favor the formation of microbial aggregates by forming a network structure with other bacteria and participate in the degradation of dead bacterial cells [57]. The relative abundance of Proteobacteria and Chloroflexi decreased as CLP concentration increased to 60 mg/L. These results might be attributed to the increased total microbial populations detected, and CLP might not necessarily decrease their absolute abundances, such as the total populations of Proteobacteria and Chloroflexi [58]. The relative abundance of the phylum Bacteroidetes showed a positive change trend with increasing CLP due to the CLP tolerance. It is known that Bacteroidetes is ubiquitous organic nutrient bacteria in various aquatic environments and can degrade organic matters, especially for high molecular weights substances, such as cellulose, proteins and lipids [59]. Li et al. found that Bacteroidetes mainly existed in a hypoxic or anoxic environment and participated in denitrification [60]. Conversely, the abundance of Actinobacteria decreased from 13.08% at 0 mg/L CLP to 4.66% at 60 mg/L CLP, indicating that Actinobacteria was more sensitive to long-term CLP stress.
Fig. 7.
Fluctuations of major genera.
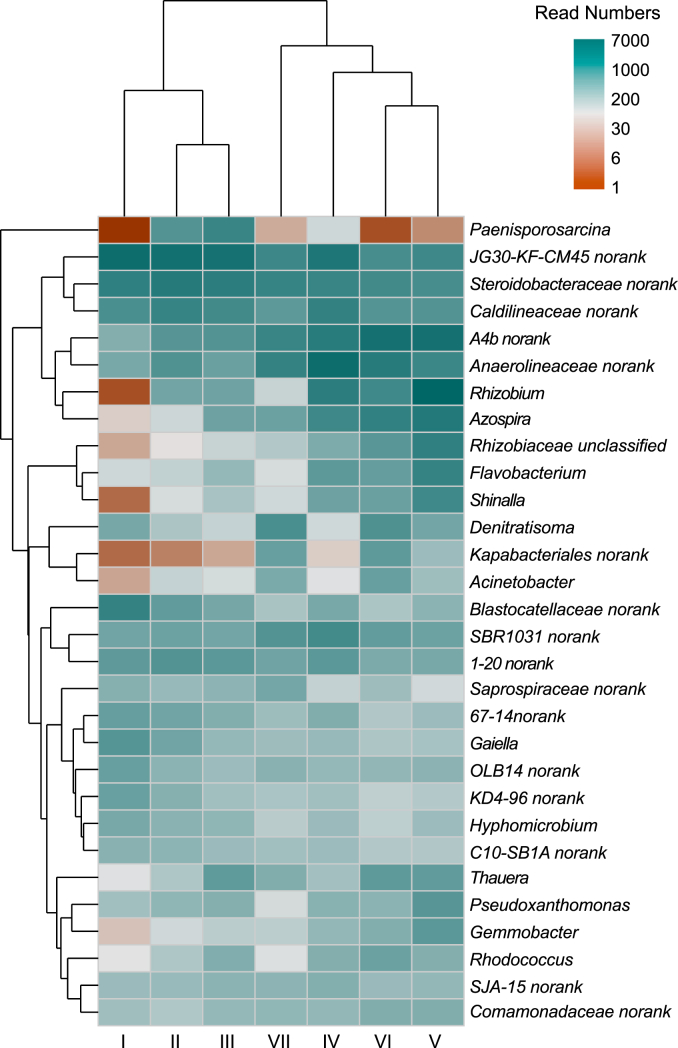
For more details, the species were further classified at the genus level and shown by a clustered heatmap (Fig. 7). The top 30 genera and 7 samples were both clustered based on the Bray-Curtis similarity index. It can be observed that all samples were clustered together separately on the columns. The results of PCA analysis (Fig. 6d) also directly proved this affinity relationship. Overall, microbial communities in CLP initially diverged from the phase I sample, and then tended to recover, while the greatest genetic distance between the control and treatment groups was at 15 mg/L CLP. The observed changes in the bacterial community may have resulted from the microbial community's resistance (insensitivity to disturbance) and resilience (recovery rate after disturbance) to CLP exposure [61]. It was consistent with the results of previous studies on different extrinsic stresses [62]. Furthermore, the response of different species of microorganisms to chemicals was different, and the competition and change in microbial community exist, resulting in the disturbed microbial community changing [63].
3.4.2. Functional species shift
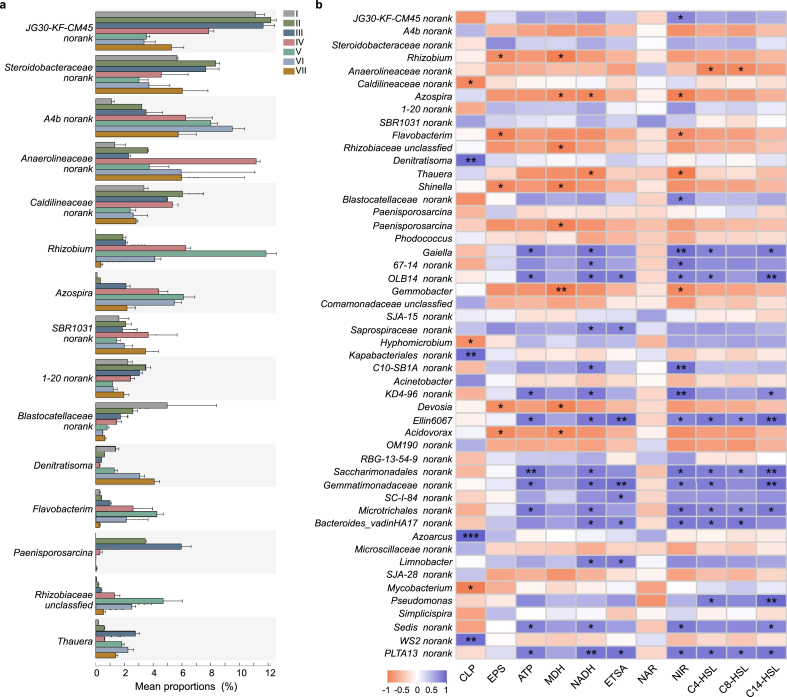
To examine the response mechanism of the bacterial community to elevated CLP, the relative abundance variations of denitrification-, carbon metabolism-, EPS- and AHLs-associated microorganisms were evaluated. They were essential functional bacteria and determined the denitrification performance. As shown in Fig. 8a and b, the predominant and potential denitrifying genera were JG30-KF-CM45 (class Chloroflexia), Steroidobacteraceae and Denitratisoma at phase I, which were positive-correlated with NIR, MDH, ATP and ETSA. As the concentration of CLP increased from 0.25 mg/L to 15 mg/L, the abundance of the above genus decreased mainly. This significant decrease was consistent with the results of the widespread accumulation of large amounts of nitrite. Instead, genera A4b (class Anaerolineae), Anaerolineaceae, Rhizobium and Azospira showed increased abundance. These bacteria might have strong competitiveness to adapt to a specific concentration of CLP and have potential pesticide resistance to the exposure dose gradient of CLP. For example, Azospira is a kind of heterotrophic denitrifier and grew in the coexistence of refractory organics and nitrate [64]. The species Anaerolineaceae can degrade refractory pollutants (e.g., aromatic hydrocarbons, phenols and quinoline compounds) [65]. They might play a vital role in the restoration of denitrification performance.
Fig. 8.
(a) The average relative abundance of dominant genera along a CLP gradient; (b) Pearson correlation between microbiota abundances and environment conditions or physiological and biochemical indexes.
The inhibition of NIR activity resulted in a decline in the metabolic activity of partial denitrifying genera (Denitratisoma, JG30-KF-CM45 and Steroidobacteraceae), with appearing of several new genera. The relative abundance of Thauera (Figs. 7 and 8a) increased from 0.18% at phase I to 2.24% at phase VI. Some studies showed that Thauera could adapt to the long-term antibiotic pressure and secrete EPS [66]. The increase of Thauera was consistent with the increase of EPS and microbial community succession, and this might be essential for protecting the microbial cells against acute damage. Shinella and Acinetobacter were reported to be AHLs secreting bacteria. The relative abundance of Acinetobacter and Shinella (Fig. S4) increased from 0.03%, 0.01% at 0 mg/L CLP to 2.49%, 1.76% at 35 mg/L CLP, respectively. It could increase AHLs and microbial community succession, promoting EPS synthesis, carbon source metabolism and denitrification process.
All the results indicated that the composition, abundance and structure of bacterial communities were altered by CLP stress. The relative abundance of denitrification- and carbon metabolism-associated microorganisms which were sensitive to CLP decreased and then recovered with the enrich of EPS-, and AHLs-associated microorganisms. These microorganisms protected microorganisms from toxic substances and regulated interactions among inter- and intra-species. Denitrification is primarily dominated by biological effects. The changes in microbial diversity, taxonomic composition and microbial community structure altered denitrification system functions. The dynamic change and succession of the microbial community under CLP stress could also be explained by analogous r/k-strategy [67]. The community was identified as the r-strategists because of initial adaption and competition during the early part of CLP exposure. Then, more thriving species increased and outcompeted weak ones, tending to be equitable with higher richness and biodiversity. Thus, the community transferred from r-to k-strategy with higher resistance capability and less competition, leading to a microbial and metabolic activity recovery.
3.5. The multiple responses of denitrifying microorganisms to CLP exposure
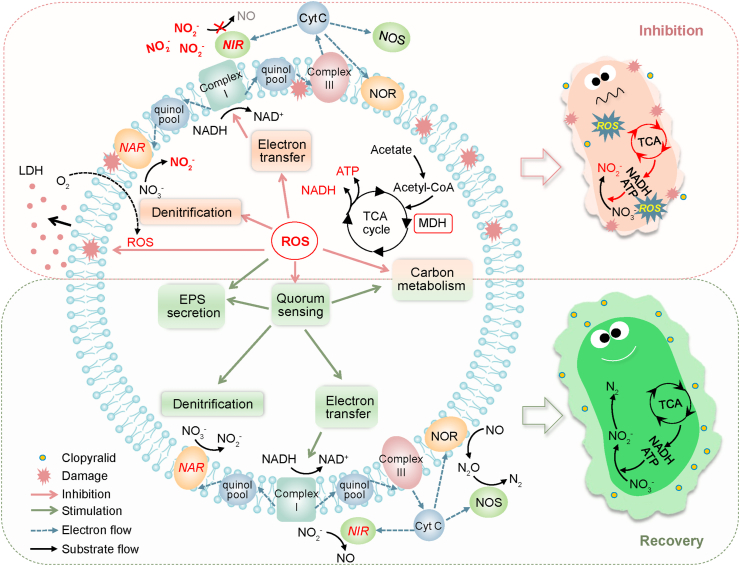
Summarizing the inhibition and recovery effects of CLP on denitrification and the microbial community evolution results, the biological response mechanisms of long-term exposure to CLP were proposed in Fig. 9.
Fig. 9.
A proposed model for the responses of denitrifying microorganisms to CLP gradient exposure.
Firstly, the negative effect of CLP on membrane damage via excessive production of ROS inhibited the NIR activity and caused the accumulation of nitrite. Meanwhile, the destruction of the microenvironment inevitably resulted in a decline in the metabolic activity of microorganisms. Enzymes related to carbon metabolism (MDH) and denitrification electron transfer (ETSA) were significantly inhibited by exposure to low concentrations of CLP. Furthermore, the positive effect of CLP on EPS and microbial community succession alleviated the accumulation of nitrite in denitrification. To better understand the relationships between the denitrification performance and the related indicators studied above (such as ROS, NIR activity, ETSA, NADH, ATP and EPS), the correlation analysis was established (Table 2). The LDH release and SOD activity were positively correlated with ROS production (0.91, 0.78, respectively), confirming the damage of CLP exposure to microbial membranes. As mentioned above, the main targets of ROS were in the cytoplasm, and ROS can directly react with enzymes located in the periplasm, leading to cytoplasmic enzyme dysfunction [44]. NIR is freely distributed in the periplasm of denitrifying bacteria, and NAR is stably embedded in the membrane [19], suggesting that the activity of NIR is more susceptible to ROS than NAR. In addition to the attack from ROS, the deterioration of NIR is also limited, which might be due to the lower competitive capability of NIR than NAR for electrons [68]. It is well known that denitrifiers need to degrade extracellular carbon sources to provide electrons to completely reduce nitrate into nitrogen [30]. The microbial metabolic activity and electron transfer efficiency declined due to cell membrane damage, which can be confirmed by the negative correlation between ROS and MDH (-0.39), NADH (-0.49), ATP (-0.61), ETSA (-0.62), as shown in Table 2. The decrease in electron generation and transfer capabilities further aggravated the accumulation of nitrite.
Table 2.
Pearson correlations of physiological indices of denitrifiers. Asterisks indicate the level of significance as follows: ∗p < 0.05, ∗∗p < 0.01.
| ROS | LDH | SOD | MDH | NADH | ATP | ETSA | NIR | EPS | |
|---|---|---|---|---|---|---|---|---|---|
| ROS | 1 | ||||||||
| LDH | 0.91∗∗ | 1 | |||||||
| SOD | 0.78∗ | 0.824∗ | 1 | ||||||
| MDH | 0.39 | 0.69 | 0.53 | 1 | |||||
| NADH | 0.39 | 0.71 | 0.51 | 0.83∗ | 1 | ||||
| ATP | 0.49 | 0.77∗ | 0.62 | 0.92∗∗ | 0.91∗∗ | 1 | |||
| ETSA | 0.61 | 0.87∗ | 0.78∗ | 0.89∗∗ | 0.92∗∗ | 0.94∗∗ | 1 | ||
| NIR | 0.29 | 0.63 | 0.36 | 0.86∗ | 0.98∗∗ | 0.92∗∗ | 0.86∗ | 1 | |
| EPS | 0.36 | 0.58 | 0.27 | 0.55 | 0.57 | 0.43 | 0.55 | 0.55 | 1 |
After more than 80 days of acclimation, a recovery trend appeared due to the improved NIR and metabolic activities of microorganisms due to the increased EPS. As shown in Table 2, NIR activity and MDH/NADH/ATP/ETSA were positively correlated with the total EPS (0.55, 0.55, 0.57, 0.43, 0.55). The increase in EPS concentration could protect microbes from CLP and weaken the membrane damage and ROS generation to maintain the higher metabolic activity [69]. Meanwhile, EPS showed a positive correlation with C4-HSL, C8-HSL and C14-HSL in the recovery stage of denitrification (0.86, 0.45, 0.90) (Fig. S3b, S3c and S3d). The increase of AHLs stimulated the expression of the QS system, which stimulated the metabolism of microorganisms, including carbon consumption, denitrification and EPS synthesis. This feedback loop made it easier for denitrifying microorganisms to adapt to the stress of CLP. Overall, our study deeply reveals the biological responses to denitrification under CLP stress, which is of great significance to the treatment of herbicide-containing wastewater from WWTPs.
4. Conclusions
The study explores the long-term perturbation of CLP on bio-denitrification and nitrite accumulation at microbial metabolism and the community level. The meaningful relationships between key indicators and denitrification performance, membrane damage, metabolism activity and microbial community were investigated. The study showed that CLP reduced the key enzyme activity (NIR) and inhibited metabolic activity via causing oxidative stress and membrane damage, resulting in nitrite accumulation. The decrease of MDH involved in carbon metabolism decreased the generation of NADH and ATP. Thus, ETSA and electrons for denitrifying enzymes were inhibited, and nitrite was accumulated. Furthermore, the increased secretion of EPS protected denitrifying microorganisms from CLP by preventing oxidative stress and membrane damage and improved NIR activity and metabolic activities. Meanwhile, the relative abundance of denitrification- and metabolism-associated microorganisms sensitive to CLP initially decreased and then recovered with the enrichment of EPS- and AHLs-associated microorganisms protecting microorganisms from toxic substances and regulating interactions among inter- and intra-species.
Conflicts of interest
There are no conflicts of interest to declare.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 52000134); China Postdoctoral Science Foundation (No. 2020M680894); Tianjin Science and Technology Program (No. 21YDTPJC00700) and Natural Science Foundation of Tianjin Science and Technology Correspondent Project (No. 19JCQNJC07800); Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (No. TSBICIP-CXRC-007).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2021.100144.
Contributor Information
Ya-Nan Hou, Email: houyn2013@163.com.
Jianbo Guo, Email: jianbguo@163.com.
Appendix B. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ferreira M.B., Souza F.L., Munoz-Morales M., Saez C., Canizares P., Martınez-Huitle C.A., Rodrigo M.A. Clopyralid degradation by AOPs enhanced with zero valent iron. J. Hazard Mater. 2020;392 doi: 10.1016/j.jhazmat.2020.122282. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L.J., Qian L., Ding L.Y., Wang L., Wong M.H., Tao H.C. Ecological and toxicological assessments of anthropogenic contaminants based on environmental metabolomics. Environ. Sci. Ecotechnol. 2021;5 doi: 10.1016/j.ese.2021.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M.C., Cernava T. Overhauling the assessment of agrochemical-driven interferences with microbial communities for improved global ecosystem integrity. Environ. Sci. Ecotechnol. 2020;4 doi: 10.1016/j.ese.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang A.J., Shi K., Ning D.L., Cheng H.Y., Wang H.C., Liu W.Z., Gao S.H., Li Z.L., Han J.L., Liang B., Zhou J.Z. Electrical selection for planktonic sludge microbial community function and assembly. Water Res. 2021;206 doi: 10.1016/j.watres.2021.117744. [DOI] [PubMed] [Google Scholar]
- 5.Contreras M.B.C., Fourcade F., Assadi A., Amrane A., Fernandez-Morales F.J. Electro Fenton removal of clopyralid in soil washing effluents. Chemosphere. 2019;237 doi: 10.1016/j.chemosphere.2019.124447. [DOI] [PubMed] [Google Scholar]
- 6.Hao T., Wang Y.Y., Tang X., Li L.S., Feng F., Mahmood Q., Wu D., Tang C.J. Quantitative determination of cavitation formation and sludge flotation in Anammox granules by using a new diffusion-reaction integrated mathematical model. Water Res. 2020;174 doi: 10.1016/j.watres.2020.115632. [DOI] [PubMed] [Google Scholar]
- 7.Yang X.R., Ding X., Zhou L., Fan H.H., Wang X.B., Ferronato C., Chovelon J.M., Xiu G.L. New insights into clopyralid degradation by sulfate radical: pyridine ring cleavage pathways. Water Res. 2020;171 doi: 10.1016/j.watres.2019.115378. [DOI] [PubMed] [Google Scholar]
- 8.Carboneras M.B., Cañizares P., Rodrigo M.A., Villaseñor J., Fernandez-Morales F.J. Improving biodegradability of soil washing effluents using anodic oxidation. Bioresour Technol. 2018;252:1–6. doi: 10.1016/j.biortech.2017.12.060. [DOI] [PubMed] [Google Scholar]
- 9.Raschitor A., Llanos J., Rodrigo M.A., Cañizares P. Is it worth using the coupled electrodialysis/electro-oxidation system for the removal of pesticides? Process modelling and role of the pollutant. Chemosphere. 2020;246:1257. doi: 10.1016/j.chemosphere.2019.125781. [DOI] [PubMed] [Google Scholar]
- 10.Park H., May A., Portilla L., Dietrich H., Münch F., Rejek T., Sarcletti M., Banspach L., Zahn D., Halik M. Magnetite nanoparticles as efficient materials for removal of glyphosate from water. Nat. Sustain. 2020;3:129–135. [Google Scholar]
- 11.Johnson B.W., Poulton S.W., Goldblatt C. Marine oxygen production and open water supported an active nitrogen cycle during the Marinoan Snowball Earth. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris E., Diaz-Pines E., Stoll E., Schloter M., Schulz S., Duffner C., Li K., Moore K.L., Ingrisch J., Reinthaler D., Zechmeister-Boltenstern S., Glatzel S., Brüggemann N., Bahn M. Denitrifying pathways dominate nitrous oxide emissions from managed grassland during drought and rewetting. Science. 2021;7(6) doi: 10.1126/sciadv.abb7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seeley M.E., Song B., Passie R., Hale R.C. Microplastics affect sedimentary microbial communities and nitrogen cycling. Nat. Commun. 2020;11(1):2372. doi: 10.1038/s41467-020-16235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y.X., M A., Fan F., Chai X.L., Wang S., Wang Y.Y., Tang C.J. Development and kinetics of a granule-based ANAMMOX UASB reactor under short HRTs. J. Cent. S. Univ. 2020;27:1197–1210. [Google Scholar]
- 15.Su X.X., Chen Y., Wang Y.Y., Yang X.Y., He Q. Impacts of chlorothalonil on denitrification and N2O emission in riparian sediments: microbial metabolism mechanism. Water Res. 2019;148:188–197. doi: 10.1016/j.watres.2018.10.052. [DOI] [PubMed] [Google Scholar]
- 16.Fang W.S., Yan D.D., Wang X.L., Huang B., Wang X.N., Liu J., Liu X.M., Li Y., Ouyang C., Wang Q.X., Cao A. Responses of nitrogen-cycling microorganisms to dazomet fumigation. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du P.Q., Wu X.H., Xu J., Dong F.S., Liu X.G., Zhang Y., Zheng Y.Q. Clomazone influence soil microbial community and soil nitrogen cycling. Sci. Total Environ. 2018;644:475–485. doi: 10.1016/j.scitotenv.2018.06.214. [DOI] [PubMed] [Google Scholar]
- 18.Wu L.L., Zhu G.S., Zhang X.X., Si Y.B. Silver nanoparticles inhibit denitrification by altering the viability and metabolic activity of pseudomonas stutzeri-sciencedirect. Sci. Total Environ. 2020;706 doi: 10.1016/j.scitotenv.2019.135711. [DOI] [PubMed] [Google Scholar]
- 19.Liu S.Q., Wang C., Hou J., Wang P.F., Miao L.Z. Effects of Ag NPs on denitrification in suspended sediments via inhibiting microbial electron behaviors. Water Res. 2020;171 doi: 10.1016/j.watres.2019.115436. [DOI] [PubMed] [Google Scholar]
- 20.Zheng X.Y., Lu D., Chen W., Gao Y.J., Zhou G., Zhang Y., Zhou X., Jin M.Q. Response of aerobic granular sludge to the long-term presence of CuO NPs in A/O/A SBRs: nitrogen and phosphorus removal, enzymatic activity and the microbial community. Environ. Sci. Technol. 2017 doi: 10.1021/acs.est.7b02768. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y., Su X.X., Wang Y.Y., Zhao S.Y., He Q. Short-term responses of denitrification to chlorothalonil in riparian sediments: process, mechanism and implication. Chem. Eng. J. 2019;358:1390–1398. [Google Scholar]
- 22.Hura A.K. University of Canterbury; Christchurch, New Zealand): 2019. Biological Treatment of Industrial Strength Clopyralid in Wastewaters: Biodegradation & Toxicity. Doctoral thesis. [Google Scholar]
- 23.Zhu J.L., Ding G.L., Liu Y., Wang B.T., Zhang W.B., Guo M.C., Geng Q.Q., Cao Y.S. Ionic liquid forms of clopyralid with increased efficacy against weeds and reduced leaching from soils. Chem. Eng. J. 2015;279:472–477. [Google Scholar]
- 24.Solís R.R., Rivas F.J., Gimeno O., Pérez-Bote J.L. Photocatalytic ozonation of clopyralid, picloram and triclopyr. Kinetics, toxicity and influence of operational parameters. J. Chem. Technol. Biotechnol. 2016;91:51–58. [Google Scholar]
- 25.Guo H.X., Chen Z., Guo J.B., Lu C.C., Song Y.Y., Han Y., Li H.B., Hou Y.N. Enhanced denitrification performance and biocatalysis mechanisms of polyoxometalates as environmentally-friendly inorganic redox mediators. Bioresour. Technol. 2019;291 doi: 10.1016/j.biortech.2019.121816. [DOI] [PubMed] [Google Scholar]
- 26.Guo Q., Shi Z.J., Xu J.L., Yang C.C., Huang M., Shi M.L., Jin R.C. Inhibition of the partial nitritation by roxithromycin and Cu(II) Bioresour. Technol. 2016;214:253–258. doi: 10.1016/j.biortech.2016.04.116. [DOI] [PubMed] [Google Scholar]
- 27.Xiao Y., Zhang E.H., Zhang J.D., Dai Y.F., Yang Z.H., Christensen H.E.M., Ulstrup J., Zhao F. Extracellular polymeric substances are transient media for microbial extracellular electron transfer. Sci. Adv. 2017;3(7) doi: 10.1126/sciadv.1700623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y., Guo J.B., Song Y.Y., Chen Z., Lu C.C., Han Y., Li H.B., Hou Y.N., Zhao R. Acceleration mechanism of bioavailable Fe(III) on Te(IV) bioreduction of Shewanella oneidensis MR-1: promotion of electron generation, electron transfer and energy level. J. Hazard Mater. 2021;403 doi: 10.1016/j.jhazmat.2020.123728. [DOI] [PubMed] [Google Scholar]
- 29.Koju R., Miao S.Y., Luo J., Wang D.L., Joshi D.R., Bai Y.H., Liu R.P., Liu H.J., Qu J.H. Effects of 1-hydroxyethane-(1,1-bisphosphonic acid) on heterotrophic denitrification performance: impact of denitrifying microbial communities variation. Chem. Eng. J. 2020 [Google Scholar]
- 30.Lu J., Zhang S., Gao S.H., Wang P., Bond P.L., Guo J.H. New insights of the bacterial response to exposure of differently sized silver nanomaterials. Water Res. 2020;169 doi: 10.1016/j.watres.2019.115205. 115205.1-115205.11. [DOI] [PubMed] [Google Scholar]
- 31.Zhao R., Zhang H.M., Zou X., Yang F.L. Effects of inhibiting acylated homoserine lactones (AHLs) on anammox activity and stability of granules. Curr. Microbiol. 2016;73(1):108–114. doi: 10.1007/s00284-016-1031-y. [DOI] [PubMed] [Google Scholar]
- 32.Wang T., Guo J.B., Song Y.Y., Lian J., Li H.B., Lu C.C., Han Y., Hou Y.N. Efficient nitrogen removal in separate coupled-system of anammox and sulfur autotrophic denitrification with a nitrification side-branch under substrate fluctuation. Sci. Total Environ. 2019;696 doi: 10.1016/j.scitotenv.2019.133929. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X.J., Chen T., Zhang J., Zhang H., Zheng S.H., Chen Z., Ma Y.P. Performance of the nitrogen removal, bioactivity and microbial community responded to elevated norfloxacin antibiotic in an anammox biofilm system. Chemosphere. 2018;210:1185–1192. doi: 10.1016/j.chemosphere.2018.07.100. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Y.F., Li G.F., Ma W.J., Xue Y., Liu Q., Zhang Z.Z., Jin R.C. Resistance of anammox granular sludge to copper nanoparticles and oxytetracycline and restoration of performance. Bioresour. Technol. 2020;307 doi: 10.1016/j.biortech.2020.123264. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Q., Xie X.Y., Feng F.L., Huang S.J., Sun Y.X. Impact of acyl-homoserine lactones on the response of nitrogen cycling in sediment to florfenicol stress. Sci. Total Environ. 2021;785 doi: 10.1016/j.scitotenv.2021.147294. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q., Wu J., Yu Y.Y., He Y.J., Huang Y., Fan N.S., Huang B.C., Jin R.C. Microbial and genetic responses of anammox process to the successive exposure of different antibiotics. Chem. Eng. J. 2021;420 [Google Scholar]
- 37.Padma R., Yalavarthy P.D. Screening of diclofenac for antibacterial activity against pathogenic microorganisms. Int. J. Adv. Pharm. Biol. Chem. 2015;4(3):554558. [Google Scholar]
- 38.Han X.M., Wang Z.W., Wang X.Y., Zheng X., Ma J.X., Wu Z.C. Microbial responses to membrane cleaning using sodium hypochlorite in membrane bioreactors: cell integrity, key enzymes and intracellular reactive oxygen species. Water Res. 2016;88:293–300. doi: 10.1016/j.watres.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 39.Kim S.Y., Park C., Jang H.J., Kim B., Bae H.W., Chung I.Y., Kim E.S., Cho Y.H. Antibacterial strategies inspired by the oxidative stress and response networks. J. Microbiol. 2019;57(3):203–212. doi: 10.1007/s12275-019-8711-9. [DOI] [PubMed] [Google Scholar]
- 40.Esposti M.D., McLennan H. Mitochondria and cells produce reactive oxygen species in virtual anaerobiosis: relevance to ceramide-induced apoptosis, Fed. Eur. Biochem. Soc. 1998;430:338–342. doi: 10.1016/s0014-5793(98)00688-7. [DOI] [PubMed] [Google Scholar]
- 41.Yu N.L., Zhao C.K., Ma B.R., Li S.S., She Z.L., Guo L., Zhang Q., Zhao Y.G., Jin C.J., Gao M.C. Impact of ampicillin on the nitrogen removal, microbial community and enzymatic activity of activated sludge. Bioresour. Technol. 2018;272:337–345. doi: 10.1016/j.biortech.2018.10.048. [DOI] [PubMed] [Google Scholar]
- 42.Ray P.D., Huang B.W., Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan R., Chen Y.G., Zheng X., Su Y.L., Li M. Effect of CO2 on microbial denitrification via inhibiting electron transport and consumption. Environ. Sci. Technol. 2016;50:9915–9922. doi: 10.1021/acs.est.5b05850. [DOI] [PubMed] [Google Scholar]
- 44.Fu H.H., Yuan J., Gao H.C. Microbial oxidative stress response: novel insights from environmental facultative anaerobic bacteria. Arch. Biochem. Biophys. 2015;584:28–35. doi: 10.1016/j.abb.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Han X.M., Wang Z.W., Chen M., Zhang X.R., Tang C.Y.Y., Wu Z.C. Acute responses of microorganisms from membrane bioreactors in the presence of NaOCl: protective mechanisms of extracellular polymeric substances. Environ. Sci. Technol. 2017;51(6):3233–3241. doi: 10.1021/acs.est.6b05475. [DOI] [PubMed] [Google Scholar]
- 46.Wang J., Liu X.L., Jiang X.B., Zhang L.B., Hou C., Su G.Y., Wang L.J., Mu Y., Shen J.Y. Facilitated bio-mineralization of N, N-dimethylformamide in anoxic denitrification system: long-term performance and biological mechanism. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116306. [DOI] [PubMed] [Google Scholar]
- 47.Wan R., Chen Y.G., Zheng X., Su Y.L., Huang H.N. Effect of CO2 on NADH production of denitrifying microbes via inhibiting carbon source transport and its metabolism. Sci. Total Environ. 2018;627:896–904. doi: 10.1016/j.scitotenv.2018.01.315. [DOI] [PubMed] [Google Scholar]
- 48.Wan R., Wang L., Chen Y.G., Zheng X., Chew J.W., Huang H.N. Tetrabromobisphenol A (TBBPA) inhibits denitrification via regulating carbon metabolism to decrease electron donation and bacterial population. Water Res. 2019;162:190–199. doi: 10.1016/j.watres.2019.06.046. [DOI] [PubMed] [Google Scholar]
- 49.Li M., Su Y.L., Chen Y.G., Wan R., Zheng X., Liu K. The effects of fulvic acid on microbial denitrification: promotion of NADH generation, electron transfer, and consumption. Appl. Microbiol. Biotechnol. 2016;100:5607–5618. doi: 10.1007/s00253-016-7383-1. 12. [DOI] [PubMed] [Google Scholar]
- 50.Prescott R.D., Decho A.W. Flexibility and adaptability of quorum sensing in nature. Trends Microbiol. 2020;28(6) doi: 10.1016/j.tim.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raju M.N., Binbin S., Yuan S.S., Zhou Z.B., Villamar-Torres R., Meng F.G. Roles of quorum sensing in biological wastewater treatment: a critical review. Chemosphere. 2019;221:616–629. doi: 10.1016/j.chemosphere.2019.01.064. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z.M., Cao R.J., Jin L.N., Zhu W.T., Ji Y.T., Xu X.Y., Zhu L. The regulation of N-acyl-homoserine lactones (AHLs)-based quorum sensing on EPS secretion via ATP synthetic for the stability of aerobic granular sludge. Sci. Total Environ. 2019;673:83–91. doi: 10.1016/j.scitotenv.2019.04.052. [DOI] [PubMed] [Google Scholar]
- 53.Toyofuku M., Nomura N., Fujii T., Takaya N., Maseda H., Sawada I., Nakajima T., Uchiyama H. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2007;189(13):4969–4972. doi: 10.1128/JB.00289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J., Huang B., Wang Q.X., Li Y., Fang W.S., Han D.W., Yan D.D., Guo M.X., Cao A.C. Effects of fumigation with metam-sodium on soil microbial biomass, respiration, nitrogen transformation, bacterial community diversity and genes encoding key enzymes involved in nitrogen cycling. Sci. Total Environ. 2017;598:1027–1036. doi: 10.1016/j.scitotenv.2017.02.058. [DOI] [PubMed] [Google Scholar]
- 55.Ji J.T., Peng Y.Z., Wang B., Mai W.K., Li X.Y., Zhang Q., Wang S.Y. Effects of salinity build-up on the performance and microbial community of partial-denitrification granular sludge with high nitrite accumulation. Bioresour. Technol. 2018;209:53–60. doi: 10.1016/j.chemosphere.2018.05.193. [DOI] [PubMed] [Google Scholar]
- 56.Zhu Y.J., Wang Y.Y., Jiang X.X., Zhou S., Wu M., Pan M.L., Chen H. Microbial community compositional analysis for membrane bioreactor treating antibiotics containing wastewater. Chem. Eng. J. 2017;325 [Google Scholar]
- 57.Wang Y.Y., Chen J., Zhou S., Wang X.D., Chen Y., Lin X.M., Yan Y., Ma X., Wu M., Han H.C. 16S rRNA gene high-throughput sequencing reveals shift in nitrogen conversion related microorganisms in a CANON system in response to salt stress. Chem. Eng. J. 2017;2:96. [Google Scholar]
- 58.Zhang Y.T., Wei W., Huang Q.S., Wang C., Wang Y., Ni B.J. Insights into the microbial response of anaerobic granular sludge during long-term exposure to polyethylene terephthalate microplastics. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115898. [DOI] [PubMed] [Google Scholar]
- 59.Si Z.H., Song X.S., Wang Y.Y., Cao X., Zhao Y.F., Wang B.D., Chen Y., Arefe A. Intensified heterotrophic denitrification in constructed wetlands using four solid carbon sources: denitrification efficiency and bacterial community structure. Bioresour. Technol. 2018;267:416–425. doi: 10.1016/j.biortech.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 60.Li L., Dong Y.H., Qian G.S., Hu X.M., Ye L.L. Performance and microbial community analysis of bio-electrocoagulation on simultaneous nitrification and denitrification in submerged membrane bioreactor at limited dissolved oxygen. Bioresour. Technol. 2018;258:168–176. doi: 10.1016/j.biortech.2018.02.121. [DOI] [PubMed] [Google Scholar]
- 61.Pishgar R., Dominic J.A., Sheng Z.J., Tay J.H. Denitrification performance and microbial versatility in response to different selection pressures. Bioresour. Technol. 2019;281:72–83. doi: 10.1016/j.biortech.2019.02.061. [DOI] [PubMed] [Google Scholar]
- 62.Pishgar R., Dominic J.A., Sheng Z.J., Tay J.H. Denitrification performance and microbial versatility in response to different selection pressures. Bioresour. Technol. 2019;281:72–83. doi: 10.1016/j.biortech.2019.02.061. [DOI] [PubMed] [Google Scholar]
- 63.Du P.Q., Wu X.H., Xu J., Dong F.S., Liu X.G., Zheng Y.Q. Effects of trifluralin on the soil microbial community and functional groups involved in nitrogen cycling. J. Hazard Mater. 2018;353:204–213. doi: 10.1016/j.jhazmat.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 64.Wu C.Y., Zhou L.M., Zhou C., Zhou Y., Xia S.Q., Rittmann B.E. Co-removal of 2,4-dichlorophenol and nitrate using a palladized biofilm: denitrification-promoted microbial mineralization following catalytic dechlorination. J. Hazard Mater. 2022;422 doi: 10.1016/j.jhazmat.2021.126916. [DOI] [PubMed] [Google Scholar]
- 65.Wu B.T., Wang J., Hu Z.H., Yuan S.J., Wang W. Anaerobic biotransformation and potential impact of quinoline in an anaerobic methanogenic reactor treating synthetic coal gasification wastewater and response of microbial community. J. Hazard Mater. 2019;384 doi: 10.1016/j.jhazmat.2019.121404. [DOI] [PubMed] [Google Scholar]
- 66.Pishgar R., Dominic J.A., Sheng Z.J., Tay J.H. Denitrification performance and microbial versatility in response to different selection pressures. Bioresour. Technol. 2019;281:72–83. doi: 10.1016/j.biortech.2019.02.061. [DOI] [PubMed] [Google Scholar]
- 67.Miao Y., Johnson N.W., Gedalanga P.B., Adamson D., Newell C., Mahendra S. Response and recovery of microbial communities subjected to oxidative and biological treatments of 1,4-dioxane and cocontaminants. Water Res. 2019;149:74–85. doi: 10.1016/j.watres.2018.10.070. [DOI] [PubMed] [Google Scholar]
- 68.Du R., Peng Y.Z., Cao S.B., Li B.K., Wang S.Y., Niu M. Mechanisms and microbial structure of partial denitrification with high nitrite accumulation. Appl. Microbiol. Biotechnol. 2016;100:2011–2021. doi: 10.1007/s00253-015-7052-9. [DOI] [PubMed] [Google Scholar]
- 69.Shi S.N., Liu J.X., Xu J., Zeng Q.Z., Hou Y., Jiang B. Effects of biochar on the phenol treatment performance and microbial communities shift in sequencing batch reactors. Water Res. 2019;161:1–10. doi: 10.1016/j.watres.2019.05.097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.