Abstract
Lung fibrosis can be observed in systemic sclerosis and in idiopathic pulmonary fibrosis, two disorders where lung involvement carries a poor prognosis. Although much has been learned about the pathogenesis of these conditions, interventions capable of reversing or, at the very least, halting disease progression are not available. Recent studies point to the potential role of micro messenger RNAs (microRNAs) in cancer and tissue fibrogenesis. MicroRNAs are short non-coding RNA sequences (20–23 nucleotides) that are endogenous, evolutionarily conserved and encoded in the genome. By acting on several genes, microRNAs control protein expression. Considering the above, we engaged in a systematic review of the literature in search of overlapping observations implicating microRNAs in the pathogenesis of both idiopathic pulmonary fibrosis (IPF) and systemic sclerosis (SSc). Our objective was to uncover top microRNA candidates for further investigation based on their mechanisms of action and their potential for serving as targets for intervention against lung fibrosis. Our review points to microRNAs of the -29 family, -21-5p and -92a-3p, -26a-5p and let-7d-5p as having distinct and counter-balancing actions related to lung fibrosis. Based on this, we speculate that readjusting the disrupted balance between these microRNAs in lung fibrosis related to SSc and IPF may have therapeutic potential.
Short abstract
miR-21-5p and the miR-29 family group cluster in systemic sclerosis and idiopathic pulmonary fibrosis http://ow.ly/D6B030bg2vn
Introduction
From the discovery of the first microRNA (miRNA) in 1993 [1], these short non-coding RNA sequences (20–23 nucleotides), which are endogenous, evolutionarily conserved and encoded in the genome, have been explored for a potential role as therapeutic targets or disease biomarkers in a wide range of conditions. These conditions include cancer [2] and fibrotic disorders [3]. Most miRNA genes derive from independent transcription units, but a minority are located in the intronic region [4].
Briefly, miRNA biogenesis follows a stepwise process. First, the precursor transcript (pri-miRNA), made by RNA polymerase II, is converted to precursor miRNA (pre-miRNA) by Drosha (an RNaseIII-like enzyme) complex. Second, the pre-miRNA is transported into the cytoplasm from the nucleus by the Exportin5 complex. Third, an RNA duplex structure (miRNA/miRNA) is generated by Dicer, an enzyme with RNaseIII-like activity. Subsequently, miRNA binds to the RNA-induced silencing complex (RISC), which recognises and binds messenger RNA in a sequence-specific mode, finally resulting in degradation or translation termination of the corresponding mRNAs [5, 6].
miRNAs biogenesis is strictly regulated since they exert a wide range of regulatory function, modulating different crucial biological pathways of specific cell types and tissues. Indeed, one miRNA can influence the activity of several genes, and one gene possessing multiple partially complementary binding sites can be regulated by several miRNAs [7].
Young et al. [8] suggest that miRNAs are more biologically complex than just endogenous regulators of mRNA stability, and may also provide inflammatory signals as cytokines do, further extending the role of miRNAs to that of direct regulators of immune processes involved in the pathogenesis of fibrotic disorders. They use the term “mirokine” to designate a group of miRNAs shown to have such cytokine-like function.
In addition, an elegant review by Vettori et al. [9] categorised the evidence available on the role of miRNAs across a wide range of fibrotic conditions, including idiopathic pulmonary fibrosis (IPF) and systemic sclerosis (SSc). This review highlighted the importance of the miR-29 group in the regulation of extracellular matrix (ECM) synthesis and, in particular, its antifibrotic effects. Interestingly, the miR-29 group is one of the most studied miRNAs in SSc and IPF. We were interested in extending the same wide-ranging review technique to focus on a comparison of the roles of miRNAs in lung fibrosis related to two particular fibrotic disorders: SSc and IPF.
The increased proliferation and activation of fibroblasts and their altered and exuberant deposition of extracellular matrices, leading to effacement of the original lung architecture, is a feature shared by both conditions, although it is organ specific for IPF and generalised for SSc.
Considering the similarities observed in the histology of lung fibrosis and in disease progression in these two entities, we speculate that certain miRNAs play important roles in both conditions. Furthermore, we believe that the identification of miRNAs aberrantly expressed in both conditions may unveil new targets for intervention in the management of lung fibrosis in both SSc and IPF. We therefore systematically reviewed the literature in search of overlapping observations implicating miRNAs in the pathogenesis of both IPF and SSc. Our objective was to uncover top miRNA candidates for further investigation based on their mechanisms of action and their potential for serving as targets for intervention against lung fibrosis. Our review points to miR-29a-3p, miR-29b-3p, miR-29c-3p, miR-21-5p, miR-92a-3p, miR-26a-5p and let-7d-5p as having distinct and counter-balancing actions related to lung fibrosis in these disorders.
Methods
The protocol for this review has been registered within PROSPERO and can be accessed under registration no. CRD42016030028.
Data sources and eligibility criteria
A comprehensive collection of reviews and other relevant publications on microRNAs related to IPF and SSc was compiled according to PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines. This pooled-data, hypothesis-generating exercise is a systematic review falling short of a formal meta-analysis. We performed a systematic search in the electronic databases MEDLINE, EMBASE, ScienceDirect and the Cochrane Library for articles published in English. The search covered the interval between January 2010 and October 2016, and used the following MeSH entry terms: “microRNAs”, “systemic sclerosis” and “idiopathic pulmonary fibrosis”. The compilation included human studies.
Inclusion and exclusion criteria
As noted, the search strategy included the following four terms: “microRNAs” and “idiopathic pulmonary fibrosis”; “microRNAs” and “systemic sclerosis”. The resulting papers were then collected, and “overlapping” miRNAs appearing in both IPF and SSc literature were analysed. Once overlapping papers were identified, they were categorised based on significance, study specimen, miRNA detection method, study design, subjects studied and target mRNA, when available. Only publications in English were considered. No other fibrotic disorders were included in the data analysis. Data related to other fibrotic disorders are summarised in the 2012 systematic review by Vettori et al. [9], which employs similar methods.
Outcomes of interest and data collection
The outcome of interest is represented by all the miRNAs that overlapped, i.e. changes in expression of which are reported in both SSc and IPF literature. Brief paragraphs or précis were prepared summarising the most consistent finding reported for relevant miRNAs. This format allowed us to discuss and compare current reported data and speculate on a role in fibrogenesis for each miRNA analysed.
The miRNA nomenclature used in the current review was obtained from www.mirbase.org. A capitalised “miR-” refers to the mature form of the miRNA, while the uncapitalised “mir-” refers to the miRNA precursor. miRNAs with nearly identical sequences except for a few nucleotides are expressed with an additional lower case letter. The use of a three-letter prefix for the species of origin is avoided because the review will focus only on human data.
A precursor mir has two mature products originating from opposite arms of the same precursors, which are denoted with a -3p or -5p suffix. Although an asterisk after the number of a mature miR product has previously been used to indicate that it was found in the cell at a lower concentration than the other mature miR, we decided to follow the www.mirbase.org notation to refer to mature products as miR-number-3p or -5p according to the available information on each relevant miRNA [10].
Results
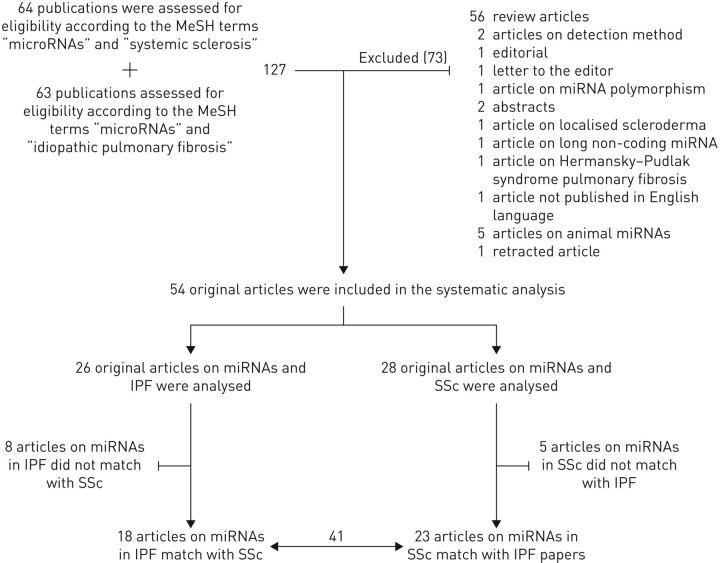
Our search findings are presented in figure 1. The flow diagram shows the numbers of studies screened, assessed for eligibility and included in the review, with reasons for exclusions at each stage. The search linking miRNAs and SSc yielded 64 studies, and the search linking miRNAs and IPF revealed 63 studies. Review articles were then excluded, as were retracted articles, those not published in English, or those with abstracts only. As depicted in figure 1, a total of 54 original articles resulted from this systematic analysis. Of these, 13 articles focused on miRNAs not reported in both diseases and, therefore, were not overlapping. In the end, 41 articles examined miRNAs that were studied in both IPF and SSc. 18 of these primarily concerned SSc and 23 concentrated on IPF. These 41 articles represent the focus of this review.
FIGURE 1.
Flow diagram of the systematic research method for detecting matching microRNAs (miRNAs) in systemic sclerosis (SSc) and idiopathic pulmonary fibrosis (IPF). The upper part of the flow diagram shows the number of papers reviewed and the detailed list of excluded manuscripts. The lower part shows the number of included papers and the remaining part of those after excluding those related to miRNAs not overlapping for SSc and IPF.
Subsequent analysis revealed that there were seven main overlapping miRNAs (miR-29a-3p, 29b-3p, 29c-3p, miR-21-5p, miR-92a-3p, miR-26a-5p and let-7d-5p) consistently found in the peer-reviewed publications and reported in at least 10 publications. We also identified other less well studied, but still overlapping miRNAs altered in both diseases. Those miRNAs that have been analysed in at least six publications are described in the section “Minor overlapping miRNAs”.
An overview of the functions of the most relevant miRNAs is reported in table 1, and the complete list of overlapping miRNAs is presented in table 2, where overlapping papers for SSc and IPF are described according to the main overlapping miRNAs, specimen tested, study design, detection method for tissue or circulatory miRNA, subjects studied and the reference number reported in the manuscript.
TABLE 1.
Overview of putative functions of the most relevant microRNAs (miRNAs) overlapping in systemic sclerosis and idiopathic pulmonary fibrosis
| Putative functions | |
| miR-29a-3p | Collagen gene expression – apoptosis |
| miR-29b-3p | Collagen gene expression |
| miR-29c-3p | ECM-related genes expression |
| miR-21-5p | EMT – collagen expression – SMAD7 |
| miR-92a-3p | MMP-1 expression |
| miR-26a-5p | EMT – collagen expression |
| let-7d-5p | EMT |
The table shows candidate functions for each major overlapping miRNA in systemic sclerosis and idiopathic pulmonary fibrosis, and provides an overview of the specific functions in fibrogenesis regulated by a specific miRNA. ECM: extracellular matrix; SMAD7: small mother against decapentaplegic protein 7; MMP-1: matrix metalloproteinase 1; EMT: epithelial–mesenchymal transition.
TABLE 2.
Overlapping microRNAs (miRNAs) in systemic sclerosis (SSc) and idiopathic pulmonary fibrosis (IPF)
| miRNAs | Specimen | Method | Target mRNA | Design | Subjects n | [Ref.] |
| 29a-3p | SSc and normal dermal fibroblasts | miR-29a-3p mimic/antimir | Bcl-2 Bax | Case–control | dcSSc=10, controls=10 | [18] |
| 29a-3p, 29b-3p, 29c-3p, pre-mir-29a, pre-mir-29b | Dermal fibroblasts | qPCR, pre-mir29a/b mimic | N/A | Cross-sectional | Controls=9 | [17] |
| 29a-3p, 29b-3p, 29c-3p, pre-mir-29a, pre-mir-29b, pre-mir-29c | SSc and normal skin biopsy; dermal fibroblasts | qPCR, pre-miR29a/b/c mimic/antimir | COL1A1, COL3A1 | Case–control | SSc=12, controls=11 | [15] |
| 29c-3p, pre-mir-29c | IPF and normal lung biopsy; primary mesenchymal cells and controls; pulmonary fibroblasts | qPCR, miR-29c-3p overexpression/knockdown lentiviral transfection | N/A | Case–control | IPF=11, controls=10 | [22] |
| 29c-3p | IPF and normal lung biopsy; pulmonary fibroblasts; decellularised primary human lung; ECM | qPCR, miR-29c-3p lentiviral transfection | COL6A2, LAMA2, COL4A2, COL1A2, LAMC1, COL4A1, COL1A1, COL5A1, NID1, MFAP2 | Case–control | IPF=5, controls=5 | [23] |
| 21-5p | SSc and normal skin biopsy; dermal fibroblasts | qPCR, in situ hybridisation, miR-21-5p mimic/antimir | FN1, ACTA2, COL1A1, COL1A2 | Case–control | dcSSc=4, lcSSc=2, controls=4 | [28] |
| 9-5p, 10a-5p, 15a-5p, 21-5p, 31-5p, 125a-5p, 125b-5p, 137-3p, 181a-5p, 203a-3p, 218-5p, 381-3p, 410, let-7a-5p, let-7b-5p, let-7c | Serum | Array, initial qPCR for miR-146a-5p, 155-5p, let-7a-5p, 181a-5p, 454-5p, let-7b-5p, 28-3p and miR-885-5p; confirmatory qPCR for miR-21-5p, 199a-5p, 200c-3p, 31-5p, let-7a-5p and let-7d-5p | N/A | Case–control | Rapidly progressive=12, slowly progressive IPF=12, controls for profiling=12; rapidly progressive IPF=20, slowly progressive IPF =24, controls=20 for qPCR confirmation | [24] |
| 21-5p, 155-5p | Serum | qPCR | N/A | Case–control | IPF=65, controls=65 | [30] |
| 21-5p | IPF and normal lung biopsy; MRC-5 and IMR-90, HEK-293 cells | Northern blotting, in situ hybridisation, miR-21-5p mimic/antimir | N/A | Case–control | IPF=8, controls=8 | [31] |
| 21-5p | IPF and normal lung biopsy; human alveolar type 2 cells | qPCR, in situ hybridisation | N/A | Case–control | IPF=3, controls=3 | [32] |
| 21-5p, 29a-3p, 92a-3p, 101-3p, 106a-5p, 142-3p, 155-5p, 181b-5p-5p, 184-3p, 223-3p, 342-3p, 409-3p, let-7c | Serum | qPCR | N/A | Cross-sectional | dcSSc=32, lcSSc=63 | [21] |
| 17-5p, 20a-5p, 29b-3p, 92a-3p, 106a-5p, 142-3p, 145-5p, 146b-5p, 181b-5p-5p, 221-3p, 223-3p, 342-3p | Serum | qPCR | N/A | Case–control | dcSSc=41, lcSSc=79, SLE=29, controls=40 | [20] |
| 26a-5p, 30a-5p, 30b-5p, 30d-5p, 92a-3p, 101-3p, 125a-5p, 126-3p, 184-3p, 203a-3p, 218-5p, 222-3p, 375-3p, let-7d-5p, let-7g-5p | IPF and normal lung biopsy; pulmonary fibroblasts | qPCR, miR-30a-5p, 30d-5p, 92a-3p mimic/antimir | WISP1 | Case–control | IPF=8, controls=7 | [37] |
| 92a-3p | SSc and normal skin biopsy; dermal fibroblasts; serum | qPCR, miR-92a-3p mimic | N/A | Case–control | dcSSc=23, lcSSc=38, SSD=12, DM=7, SLE=7, controls=18 | [34] |
| 17∼92 cluster (17-5p, 18a-5p, 19a-3p, 19b-3p, 20a-5p, 92a-3p) | IPF and COPD lung biopsy; normal and IPF pulmonary fibroblasts | qPCR, in situ hybridisation, miR-17∼92 mimic/antimir | N/A | Case–control | >80% FVC IPF+COPD=7, 50–80% FVC IPF+COPD=8, <50% FVC IPF+COPD=9, controls=10 | [38] |
| 26a-5p, 27b-3p, 30a-5p, 30d-5p, 92a-3p, 125a-5p, 125b-5p, 140-5p, 145-5p, 197-3p, 214-3p, 377-3p, 381-3p, 486-5p, let-7g-5p | Skin biopsy | Array, qPCR let-7g-5p, 206-3p, 125b-5p | N/A (identified by computational prediction algorithms) | Case–control | SSc=3, controls=3 | [36] |
| 21-5p, 29b-3p, 31-5p, 145-5p, 146a-5p, 503-5p | SSc and normal skin biopsy; dermal fibroblasts | Array, qPCR for miR-21, 31, 503, 146, 145, 29b | SMAD7, SMAD3 COL1A1 | Case–control | dcSSc=5, lcSSc=2, controls=7 | [16] |
| 30a-3p | SSc and normal skin; rheumatoid arthritis and normal synovial biopsy; dermal fibroblasts, fibroblasts-like synoviocytes | qPCR, miR-30a-3p mimic/antimir | BAFF | Case–control | FLS: RA=5, controls=5; DF: SSc=4, controls=3 |
[50] |
| 9-5p, 10a-5p, 15a-5p, 17-5p, 18a-5p, 20a-5p, 21-5p, 22-3p, 26a-5p, 92a-3p, 93-5p, 96-5p, 100-5p, 101-3p, 103a-3p, 106b-5p, 125a-5p, 126-3p, 130a-3p, 132-3p, 133b-3p, 134, 137-3p, 141-3p, 142-3p, 142-5p, 146a-5p, 155-5p, 181a-5p, 182-5p, 205-5p, 210, 214-3p, 222-3p, 223-3p, 301a-3p, 302c-3p, 345-5p, 370, 375-3p, 424-5p, 520g, let-7a-5p, let-7b, let-7c, let-7d-5p, let-7g-5p | Serum; dermal fibroblasts; skin tissues | Array, qPCR for let-7a-5p, in situ hybridisation, let-7a mimic/antimir | N/A | Case–control | Serum: dcSSc=20, lcSSc=19, LSc=39, SLE=8, DM=8, controls=17; skin tissues: SSc=7, LSc=7, keloid=5, controls=7 |
[43] |
| 19a-5p, 26a-5p, 30b-5p, 106b-5p, 181a-5p, 181b-5p, 203a-3p, 205-5p, 302c-3p, 409-3p, 410 let-7a-5p, let-7b-5p, let-7c, let-7d-5p | SSc and normal skin biopsy; dermal fibroblasts; serum | qPCR, miR-30b-5p mimic | PDGFR-β, COL1A2, ACTA2 | Case–control | dcSSc=18, lcSSc=32, controls=24 | [45] |
| 10a-5p, 15a-5p, 17-5p, 18a-5p, 20a-5p, 21-5p, 93-5p, 96-5p, 101-3p, 103a-3p, 106b-5p, 130a-3p, 132-3p, 133b-3p, 134, 137-3p, 141-3p, 142-5p, 142-3p, 155-5p, 182-5p, 210, 301a-3p, 370, 424-5p, 520g, let-7a-5p, let-7b-5p, let-7c, let-7d-5p, let-7g-5p | SSc and normal skin biopsy; dermal fibroblasts | Array, qPCR for miR-150, in situ hybridisation, miR-150 mimic | N/A | Case–control | Serum: dcSSc=20, lcSSc=20, DM=5, SLE=5, controls=20; skin tissues: dcSSc=5, controls=5 |
[46] |
| 1-3p, 9-5p, 10a-5p, 15a-5p, 17-5p, 18a-5p, 20a-5p, 21-5p, 22-3p, 26a-5p, 92a-3p, 93-5p, 96-5p, 100-5p, 101-3p, 103a-3p, 106b-5p, 125a-5p, 126-3p, 128-3p, 130a-3p, 132-3p, 133b-3p, 134, 137-3p, 142-5p, 142-3p, 155-5p, 181a-5p, 182-5p, 205-5p, 210, 214-3p, 218-5p, 222-3p, 223-3p, 301a-3p, 302c-3p, 370, 375-3p, 424-5p, 520g, let-7a-5p, let-7b-5p, let-7c, let-7d-5p, let-7g-5p | SSc and normal skin biopsy; dermal fibroblasts | Array, qPCR for miR-196a, miR-196a mimic/antimir | N/A | Case–control | Serum: dcSSc=20, lcSSc=20, controls=25; skin tissues: SSc=5, TGF-β-stimulated=5, controls=5 |
[49] |
| 1-3p | SSc and normal dermal fibroblasts | Array | N/A | Case–control | Skin tissues: dcSSc=6, controls=8 | [54] |
| 142-3p | Serum | qPCR | N/A | Case–control | dcSSc=23, lcSSc=38, SSD=12, controls=20, SLE=8, DM=8 | [51] |
| 9-5p | IPF and normal lung biopsy; human fetal lung fibroblasts; human mesothelial cells | Array, qPCR for miR-9-5p, in situ hybridisation, pre-miR-9-5p mimic/antimir | TGFBR2, NOX4, ACTA2, COL1A1, FN1 | Case–control | IPF=7, controls=3 | [55] |
| 31-5p, 424-5p | TGF-β1 stimulated and normal human lung epithelial cells (A549) | Array, miR-424-5p, 1224-5p and 23b-3p overexpression/knockdown lentiviral transfection | N/A | Case–control | Cells | [57] |
| 96-5p | IPF and normal lung biopsy; pulmonary fibroblasts | qPCR, in situ hybridisation, miR-96-5p mimic/antimir | FoxO3a, p27, p21, BIM | Case–control | IPF=8, controls=8 | [53] |
| 210 | IPF and normal lung biopsy; lung mesenchymal cells |
qPCR, in situ hybridisation, miR-210 overexpression/knockdown lentiviral transfection | N/A | Case–control | IPF=7, controls=6 | [48] |
| 26a-5p | Human lung epithelial cells (A549) | Array (downloaded from Gene Expression Omnibus database), miR-26a-5p mimic/antimir | Human genes annotated in the biological process of “EMT” were downloaded from the Gene Ontology database (GO:0001837) | Case–control | Cells | [41] |
| 26a-5p, 30a-5p, 30b-5p, 133b-3p | IPF and normal lung biopsy; human fetal lung fibroblasts | Array, miR-26a-5p mimic | COL1A1, COL3A1 | Case–control | IPF=8, controls=10 cells | [40] |
| let-7d-5p | Human fetal lung fibroblasts, human lung fibroblasts, human fetal foreskin fibroblasts |
let-7d-5p overexpression lentiviral transfection | PAI-1, HMGA2, ACTA2, CDH2, ID1, ID2, SLUG, FSP-1, FN1, KRT19 | Case–control | Cells | [47] |
| 140-5p | Human type II alveolar epithelial cells (A549) | qPCR, miR-140-5p mimic/antimir | Case–control | IPF=5, controls=5 cells | [56] | |
| 1-3p, 10a-5p, 17-5p, 27b-3p, 30a-3p, 30a-5p, 30b-3p, 30d-5p, 126-3p, 133b-3p, 181a-5p, 184-3p-3p, 203a-3p, 210, 222-3p, 370, 375-3p, 377-3p, 381-3p, 409-3p, 410, 520g, let-7d-5p | IPF and normal lung biopsy; IPF, TGF-β-stimulated and normal human lung fibroblasts | Array, miR-154-5p mimic/antimir | MEG3, DKK2, DIXDC1, PPP2CA, FZD 4/5/6, LRP, KREMEN1, β-CATENIN, WISP1 | Case–control | IPF=13, controls=12; IPF=32, controls=28 for qPCR confirmation | [44] |
| 15a-5p, 17-5p, 18a-5p, 19a-5p, 20a-5p, 21-5p, 22-3p, 27b-3p, 29a-3p, 29b-3p, 29c-3p, 30a-5p, 30b-5p, 92a-3p, 93-5p, 96-5p, 100-5p, 101-3p, 103a-3p, 106b, 125a-5p, 126-3p, 128, 130a, 140-5p, 141, 142-3p, 142-5p, 155, 181b-5p, 210, 222-3p, 223-3p, 302c-3p, 424-5p, let-7c, let-7d-5p | IPF and normal lung biopsy, IPF and normal lung fibroblasts | Array | N/A | Case–control | Rapidly progressive IPF=9, slowly progressive IPF=6, controls=10 | [29] |
| 26a-5p, 30a-3p, 30b-5p, 30d-5p, 92a-3p, 125a-5p, 126-3p, 132-3p, 134, 155-5p, 182-5p, 184-3p, 197-3p, 203a-3p, 205-5p, 214-3p, 409-3p, let-7d-5p | IPF and normal lung tissues, A549 cells | Array, qPCR for 26a-5p, 30a-3p, 30b-5p, 30d-5p, 92a-3p, let-7d-5p mimic/inhibitor | HMGA2, CDH2, VIM, ACTA2 | Case–control | IPF=10, controls=10 cells | [39] |
| 130a-3p, 142-5p | BAL fluid samples; IPF and normal macrophages | qPCR | N/A | Case–control | IPF=9, control=7 | [52] |
| 29a-3p | SSc and normal skin biopsy; dermal fibroblasts | qPCR, mir-29a mimic | COL1A1, TIMP1, TAB1 | Case–control | SSc=4–6, controls=4–6 | [19] |
| 130b-3p | SSc and normal skin biopsy; dermal fibroblasts | qPCR, miR-130b-3p mimic/antimir | COL1A1, COL1A2, FN1, ASMA | Case-control | SSc=14, controls=14 | [58] |
| 130b-3p | IPF and normal lung biopsy; human primary type II alveolar epithelial and lung fibroblasts | qPCR, miR-130b-3p mimic/antimir | IGF1 | Case–control | IPF=4, controls=3 | [59] |
| 26a-5p, let-7d-5p | Human alveolar basal epithelial cells | qPCR, miR-26a-5p mimic/antimir | LIN28B | Case–control | IPF=106, controls=50 (microarray data set of GSE32538) | [42] |
| 29b-3p | IPF lung biopsy | qPCR, miR-29b-3p mimic | COL1A1, COL3A1 | Case–control | IPF=16 | [25] |
The table describes all the manuscripts that reported overlapping miRNAs between SSc and IPF. The manuscripts have been categorised according to the significance, specimen analysed, miRNAs detection method employed and, where described, the mRNA target. Moreover, the design of the study and the number of subjects involved in each study, together with the reference, are reported. Bcl-2: B-cell lymphoma 2; BAX: bcl-2-like protein 4; dcSSc: diffuse cutaneous SSc; COL1A1, COL1A2, etc.: collagen, type I, alpha 1, collagen, type I, alpha 2, etc.; ECM: extracellular matrix; LAMA2: laminin, alpha 2; LAMC1: laminin, gamma 1; NID1: nidogen 1; MFAP2: microfibrillar-associated protein 2; FN1: fibronectin 1; ACTA: alpha smooth muscle actin; lcSSc: limited cutaneous SSc; MRC-5: Medical Research Council cell strain 5; HEK-293: human embryonic kidney cells 293; SLE: systemic lupus erythematosus; WISP1: WNT1-inducible-signalling pathway protein 1; SSD: scleroderma spectrum disorders; DM: dermatomyositis; COPD: chronic obstructive pulmonary disease; N/A: not available; FVC: forced vital capacity; SMAD: small mother against decapentaplegic protein; BAFF: B-cell activating factor; FLS: fibroblast-like synoviocytes; RA: rheumatoid arthritis; DF: dermal fibroblasts; LSc: localised scleroderma; PDGFR-β: platelet-derived growth factor receptor β; TGF-β: transforming growth factor-β; NOX4: NADPH oxidase 4; FoxO3a: Forkhead box O3; BIM: Bcl-2-like protein 11; EMT: epithelial–mesenchymal transition; PAI-1: plasminogen activator inhibitor-1; HMGA2: high-mobility group AT-hook 2; CDH2: cadherin-2; ID1–ID2: DNA-binding protein inhibitor 1–2; SLUG: protein snail homolog 2; FSP-1: fibroblast-specific protein 1; KRT19: keratin 19; MEG3: maternally expressed 3; DKK2: Dickkopf WNT signaling pathway inhibitor 2; DIXDC1: DIX domain containing 1; PP2CA: protein phosphatase 2CA; FZD4-5-6: frizzled-4 frizzled-5 frizzled-6; LRP: lipoprotein receptor-related protein; KREMEN: Kringle-Containing Protein Marking the Eye and the Nose; VIM: vimentin; BAL: bronchoalveolar lavage; TIMP: tissue inhibitor of metalloproteinase 1; TAB1: TGF-β activated kinase 1/MAP3K7 binding protein 1; ASMA: a smooth muscle actin; IGF: insulin-like growth factor.
miR-29 family in IPF and SSc fibrogenesis
The miR-29 family in humans includes miR-29a, miR-29b-1, miR-29b-2 and miR-29c. miR-29b-1 and miR-29b-2 have identical mature sequences, which are together called miR-29b. The gene encoding the precursors of miR-29b-1 and miR-29a is located on chromosome 7q32.3 in humans, and the gene encoding miR-29b-2 and miR-29c is on chromosome 1q32.2 [11]. The miR-29 family has been implicated in a wide range of experimental fibrotic disorders [12–14]. This miRNA is one of the most consistently found among IPF and SSc studies, and its downregulation is associated with increased fibrogenesis, suggesting an antifibrotic role for this miRNA.
Maurer et al. [15] highlighted that the expression of miR-29a-3p in the skin of subjects with SSc is reduced compared to that in healthy controls. Importantly, they found that its overexpression in SSc fibroblasts is associated with decreased levels of mRNA and protein for type I and type III collagen, supporting a role for miR-29a-3p in the regulation of collagen expression [15]. In contrast, the downregulation of miR-29b-3p correlated with the upregulation of type I collagen in SSc fibroblasts compared to controls [16]. The relationship between type I collagen and miR-29 was explored in another study. These experiments demonstrated that the overexpression of pre-mir-29a and pre-mir-29b in normal dermal fibroblasts abrogates the synergistic stimulation of collagen gene expression induced by the natural ligand of TLR4, lipopolysaccharide (LPS), in the presence of transforming growth factor (TGF)-β1 [17]. Apart from the connection with collagen type I, there is evidence of apoptotic-inducing properties of miR-29a-3p in fibrotic fibroblasts. The transfection of a miR-29a-3p mimic was sufficient to decrease anti-apoptotic Bcl-2 mRNA expression and increase the Bax:Bcl-2 ratio, leading to increased fibroblast susceptibility to apoptosis, both in SSc and TGF-β-stimulated fibroblasts [18]. Another provocative study delineated a novel role of miR-29a-3p, demonstrating that enhancing the expression of miR-29a-3p reverses the profibrotic phenotype of SSc fibroblasts by reducing the secretion of the tissue inhibitor of metalloproteinase 1 (TIMP-1) via TGF-β activated kinase (MAP3K7) binding protein 1 (TAB1) repression and collagen type I expression. These changes increased functional matrix metalloproteinase 1 (MMP-1) production and promoted collagen degradation [19].
Despite the above data derived from tissue compartments, measurements of circulating serum levels of miR-29 in SSc patients remain scarce [20], apart from the evidence that miR-29a-3p serum levels were higher in anticentromere antibody (ACA) positive subjects compared to anti-U1 ribonucleoprotein (RNP) positive subjects among the limited cutaneous group of SSc patients [21].
Data on circulating serum levels of miR-29 are also lacking in IPF-related papers, and most of the evidence implicating it comes from ex vivo studies. Of note, among the miR-29 family members, miR-29c-3p appears to have a prominent role in IPF. In fact, Khalil et al. [22] demonstrated that IPF fibroblasts, seeded on polymerised type I collagen, express low miR-29c-3p levels and high type I collagen. Furthermore, they show that the suppression of miR-29c-3p involves low protein phosphatase 2A function and subsequent decreased nuclear translocation of histone deacetylase 4. In another study, miR-29c-3p overexpression abrogated pathological ECM gene expression, such as collagen and laminin gene expression, in IPF fibroblasts [23]. miR-29 family members are downregulated in IPF lung samples from patients with both rapidly and slowly progressing disease, with no significant differences found between the two clinical conditions [24]. Also, miR-29b-3p was able to reduce collagen type 1 and type 3 mRNA synthesis when administered in primary pulmonary fibroblasts of IPF patients [25].
miR-21-5p in IPF and SSc fibrogenesis
miR-21-5p has been mapped at chromosome 17q23.2, where it overlaps with the gene encoding vacuole membrane protein 1 (VMP1) [26]. A previous report suggests that miR-21-5p facilitates epithelial–mesenchymal transition (EMT), but the fundamental molecular mechanisms involved remain unclear [27]. EMT represents one of the main processes underlying the dissemination of a fibrotic injury, and increased expression of this miRNA could facilitate the transition process and also collagen deposition by interfering with the inhibitory effect of small mother against decapentaplegic protein 7 (SMAD7) on the SMAD2/3 fibrotic pathway.
Studies focusing on miR-21 have been performed both in SSc and in IPF assessing circulating and tissue concentrations. The overexpression of miR-21-5p in SSc fibroblasts decreases the levels of SMAD7, whereas knockdown of miR-21-5p increases its expression, further regulating fibronectin (FN), α1 type I collagen (COL1A1), α2 type I collagen COL1A2 and α smooth muscle actin (α-SMA) mRNA expression. Although a study shows that miR-21-5p expression in SSc is decreased compared to controls [28], there is also one study reporting the opposite results [15].
IPF patients with rapidly progressive disease have higher circulating concentrations of miR-21-5p than both IPF patients with slowly progressive disease and healthy controls [24–29].
The clinical relevance of miR-21-5p in IPF patients was further supported by the work of Li et al. [30], who described an association with both worsening forced vital capacity and radiological features. Further studies confirmed that miR-21-5p expression is upregulated in the diseased lungs of IPF patients [31]. In particular, miR-21-5p functions have been explored in IPF alveolar type 2 cells. This study supports the role of miR-21-5p in promoting EMT in lung epithelial cells from IPF patients [32].
miR-92a-3p in IPF and SSc fibrogenesis
The miR-17-92 cluster (oncomiR-1) is located on chromosome 13 (13q31.3). The primary transcript is processed into different mature miRNAs: miR-17-5p, miR-18a-5p, miR-19a-3p, miR-19b-3p, miR-20a-5p and miR-92a-3p [33].
The miR-92a family regulates fundamental processes in the development of mammalian organs including heart, blood vessel formation, lungs and the immune system. Among these, miR-92a-3p acquires specific relevance lacking for other miRNAs, because decreased circulating levels have been associated with interstitial lung disease (ILD) in SSc patients [21]. However, this association was not confirmed in another study. Indeed, contradictory associations have been described for this miRNA in SSc, depending upon whether the measurements are from serum or from tissue.
Evidence suggests that miR-92a downregulates MMP-1 expression, which is an enzyme that degrades the ECM [21, 24, 34–36]. Increased levels can therefore lead to reduced collagen degradation. Also, in IPF studies of lung biopsies, there are conflicting reports regarding the expression of this miRNA in diseased lung [30, 37, 38]. Although very controversial, results on this miRNA remain the most promising because it was closely associated with pulmonary involvement in SSc. Further studies are needed to confirm the potential role as a biomarker of lung involvement in SSc and IPF.
miR-26a-5p in IPF and SSc fibrogenesis
miR-26a-5p expression is downregulated in lung specimens from IPF patients [37, 39] and in bleomycin-treated murine lungs [31] compared to controls. miR-26a-5p overexpression partly reduces the exaggerated deposition of collagen, as evidenced in experiments where human fetal lung fibroblasts were exposed to antagomirs (also known as anti-miRs), used to silence endogenous miR-26a-5p, and to chemically synthesised and optimised nucleic acids designed to mimic miR-26a-5p [40]. In a subsequent study, the forced overexpression of miR-26a-5p in human alveolar type 2 cells reduced EMT [41]. In addition, a recent work suggests that miR-26a-5p overexpression can suppress EMT in alveolar epithelial cells. miR-26a-5p can target the LIN28B gene, which induces EMT by inhibiting let-7d expression. By targeting and blocking LIN28B expression, miR-26a-5p disrupts the Lin28B/let-7d circuit. This enhances the biogenesis of let-7d, which consequently inhibits EMT and pulmonary fibrosis [42]. Similar results, with the exception of one original article [37], have been observed in SSc skin tissues, with decreased expression of miR-26a-5p [36, 43]. Similar to what happens with miR-21a-5p, overexpression of miR-26a-5p could reduce EMT in alveolar epithelial cells, further extending the potential action of this miRNA at a different cellular level.
let-7d-5p in IPF and SSc fibrogenesis
The let-7 miRNA is a founding member of the miRNA family, one of the first to be discovered. The data collected on let-7d-5p demonstrate that its expression is downregulated both in the skin of SSc and in the lungs of IPF patients [35, 37, 44–46]. In IPF, both with rapid and with slowly progressive disease, its expression is downregulated compared to controls [30]. In human fetal lung and foreskin and human adult lung fibroblasts, the transfection of cells with let-7d-5p reduces the expression of mesenchymal markers [47]. The translational potential of let-7d administration justifies a search for more evidence to confirm its efficacy as an antifibrotic agent, given that both in SSc and IPF, reduced levels have been observed.
Minor overlapping miRNAs
Several other overlapping miRNAs are of potential interest, but we found limited published evidence supporting their roles. Among the let-7 family, in addition to let-7d-5p evidence, let-7a-5p serum levels are lower in SSc patients and they inversely correlate with skin severity [43], while in rapidly progressive IPF patients, serum levels of let-7a-5p are significantly lower than in controls [24]. In addition, confirming the prominent role of this miRNA family in fibrotic disorders, let-7b-5p, let-7c and let-7g-5p have been found to be downregulated in both SSc and in IPF tissues [24, 37, 44–46].
For example, miR-210, miR-17-5p, miR-155-5p and miR-101-3p show contradictory results [21, 29, 30, 37, 42, 46, 48, 49]. Of note, opposite results have been described for miR-125a-5p and 133b-3p: both are downregulated in SSc tissues [24, 37] and upregulated in IPF lungs [40, 42].
miR-30 family members have been studied in SSc. Serum levels of miR-30b-5p are significantly depressed in SSc compared to controls and in diffuse cutaneous SSc (dcSSc) compared to limited cutaneous SSc (lcSSc). miR-30b-5p mimic induces a significant decrease in platelet-derived growth factor (PDGF) receptor β, COL1A1 and α-SMA mRNA expression in dermal fibroblasts. The transfection of miR-30a-3p mimic in Poly(I:C)- and interferon (IFN)-γ-activated rheumatoid arthritis fibroblast-like synoviocytes and in SSc human dermal fibroblasts reduced B-cell activating factor (BAFF) synthesis and release, further extending the reach of this miRNA family group into lymphocytes biology [50].
In a study comparing patients affected by SSc to disease controls with systemic lupus erythematosus, dermatomyositis and also to subjects with a scleroderma spectrum disorders, miR-142-3p serum levels were higher in the SSc group. No differences were observed in SSc disease subtypes or clinical parameters [51].
In IPF, the blockade of miR-142-5p in macrophages modulates the switch of the phenotype of macrophage to M2 and subsequent profibrotic activation [53]. However, other papers related to IPF and SSc have reported opposite findings for miR-142-3p and 142-5p [20, 50, 52, 53]. Other papers analysed a wide panel of miRNAs, performing specific confirmatory analysis on miRNAs not relevant to the aim of the present review comparing IPF and SSc [35, 53–59].
Table 3 recaps, by miRNA and tissue, the detailed literature review, which is summarised in the right-hand side of figure 2 using an abstract fibroblast.
TABLE 3.
Major overlapping miRNAs in systemic sclerosis (SSc) and idiopathic pulmonary fibrosis (IPF)
| SSc | IPF | |||||||
| Fibroblasts | Serum expression | Skin expression | Fibroblasts | Serum expression | Lung expression | |||
| Expression | Effect | Expression | Effect | |||||
| 29a-3p | ↓ | Mimic induces apoptosis, abrogates collagen expression, increases collagen degradation | ↑ in RNP+ compared to ACA+ | ↓ | ↓ | |||
| 29b-3p | ↓ | Mimic abrogates collagen expression | ↑ | ↓ | ↓ | |||
| 29c-3p | ↓ | Mimic abrogates ECM expression | ↓ | ↓ | Overexpression abrogates collagen expression | ↓ | ||
| 21-5p | ↔ | ↑ in RNP+ compared to ACA+ | ↑ | Knockdown abrogates collagen expression and EMT | ↑ in rapid progressive disease - associated with worse FVC | ↑ | ||
| 92a-3p | ↔ | Overexpression induces collagen degradation | ↔ in SSc and in SSc-ILD | ↔ | ↔ | |||
| 26a-5p | ↓ | ↔ | Overexpression abrogates collagen expression and EMT | ↓ | ||||
| let-7d-5p | ↓ | ↓ | ↓ | Overexpression reduces EMT | ↓ in slowly and rapid progressive disease | ↓ | ||
The table compares the expression levels and the effects of major overlapping miRNAs in SSc skin and fibroblasts with those observed in IPF lung and fibroblasts. Circulating serum levels for SSc and IPF patients are also reported. RNP+: SSc patients with anti-U1 ribonucleoprotein antibodies; ACA+: SSc patients with anticentromere antibodies; ECM: extracellular matrix; EMT: epithelial–mesenchymal transition; FVC: forced vital capacity; SSc-ILD: systemic sclerosis-associated interstitial lung disease. ↑: high levels; ↓: low levels; ↔: controversial results.
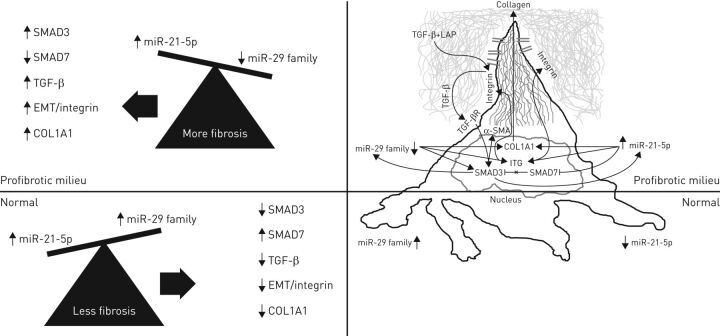
FIGURE 2.
Fibroblast responses to changes in miR-21-5p and miR-29 family levels. miR-21-5p and miR-29 family members can act synergistically to potentiate fibroblast functions in fibrotic conditions. The figure shows complex but established relationships supported by the literature review. Above the horizontal line is the activation of a profibrotic fibroblast in both idiopathic pulmonary fibrosis (IPF) and systemic sclerosis (SSc). Below the line represents the normal state. Highlighted on the right surrounding the abstract fibroblast is the mechanism by which both miRNAs can regulate transforming growth factor (TGF)-β release from its latency-associated peptide (LAP) by stimulating integrin expression and collagen synthesis. The right-hand fibroblast image also shows the effect of miRNAs 21-5p and the 29 family on α-SMA expression. Small mother against decapentaplegic protein (SMAD)3 has a pivotal role in the maintenance of the loop of this finely regulated system: SMAD3 stimulates the secretion of miR-21-5p, which blocks the inhibitory effect of SMAD7 on SMAD3; in addition, SMAD3 reduces miR-29 family levels, which reinforces SMAD3 activation.
Discussion
Summary of results
A growing body of evidence supports a prominent role for miRNAs in regulating fibroblast functions, including ECM deposition in a wide range of fibrotic diseases [3, 57, 60]. Seven miRNAs appearing to have important roles in both SSc and IPF, based on studies at both the tissue and circulatory level, were identified in this systematic literature review. The putative functions of the most relevant overlapping miRNAs in SSc and IPF are summarised in table 1.
Among these, members of the miR-29 family have been widely investigated as one of the main antifibrotic mediators in fibrotic disorders, as they can directly regulate collagen synthesis [15]. Importantly, the number of proven regulatory pathways influenced by these miRNAs is increasing. These pathways include those of TGF-β, TIMP, BCL-2, TNF-α [18, 20] and PP2A/HDAC4.
miR-21-5p appears to have functions opposite to those of the miR-29 family. Also in contrast to the miR-29 group, miR-21-5p is highly expressed in both IPF lung fibroblasts and SSc skin fibroblasts [28, 30]. The expression of miR-21-5p is also high in alveolar type 2 cells and is positively associated with rapidly progressive disease in IPF patients. These observations suggest a wider distribution of miR-21-5p, thereby supporting potential roles for miR-21-5p in diverse cell functions and in differentiation. Consistent with this interpretation, miR-21-5p elevation has been associated with increased expression of mesenchymal markers of differentiation in skin fibroblasts harvested from subjects with SSc [28].
We hypothesise that the balance between miRNAs 21 and 29 determines or influences lung fibrogenesis (figure 2). The figure emphasises that these two overlapping miRNAs exert most of the effects through the pivotal cell in orchestrating fibrotic damage, the fibroblast. In this cell, miRNAs can influence cell cycle, through proliferation and apoptosis, as well as cell function, including collagen deposition and degradation and also myofibroblast transformation.
More ambiguous results were obtained for miR-92a-3p, but it too has been found to be decreased in serum in association with ILD in SSc [21]. However, no studies on circulating levels of this miRNA are yet available in IPF, while lung biopsy studies on IPF tend to describe downregulated expression [37]. Although the robustness of evidence concerning let-7d-5p and miR-26a-5p is lower compared to miR-21-5p, the miR-29 family and miR-92a-3p, decreased levels of both of the former miRNAs have been associated with increased mesenchymal markers expression, suggesting a prominent role in EMT in both SSc and IPF. Several other miRNAs have been investigated in both diseases at least once, but the small number of publications and lack of consistency in results in one disease or the other leaves us unable to assess the overlap. The complexity of miRNA systems and the number of individual potential mediators involved may require a computational algorithmic approach.
Implications with regards to regulation of profibrotic pathways and SMADs
Together, these observations suggest that miR-21-5p and the miR-29 family are pluripotent miRNAs participating in a variety of pathological mechanisms [61–63]. In the two fibrotic conditions examined here, one can envision these miRNAs acting synergistically to potentiate fibroblast activation and transformation and, thereby, fibrogenesis. Both miRNAs can directly influence collagen synthesis and α-SMA expression, which is a typical feature of myofibroblasts. Moreover, each of these two miRNAs can independently induce integrin expression, favouring the release of TGF-β from its latency peptide (LAP). In this case, both miRNAs are acting on the same physiological targets, miR-29 family decrease and miR-21-5p increase both activating integrin genes transcription. This action allows TGF-β to bind to its receptor, activating SMAD3, which further stimulates collagen synthesis and α-SMA expression. Interestingly, SMAD2 and SMAD3 are main regulators of the induction of both miRNAs.
Indeed, lung fibroblasts lacking the SMAD3 gene are protected from the loss of miR-29 and fibrosis in vivo and in vitro in response to TGF-β [64]. Similarly, it has been demonstrated that TGF-β is an inducer of miR-21 expression. NIH-3T3 fibroblasts transfected with pre-mir-21 responded to TGF-β1 stimulation by overexpressing miR-21-5p and the overexpression of SMAD2/3 resulted in higher production of mature miR-21-5p [65]. By way of contrast, in kidney fibrosis, SMAD2 and SMAD3 differentially regulated the post-transcriptional modifications of miR-21-5p, SMAD2 being a negative regulating factor for miR-21-5p [66]. Similar results have been described for miR-21-5p in liver fibrosis [67].
At the cellular level, high levels miR-21-5p block the inhibitory effect of SMAD7 on SMAD3, while the miR-29 family directly stimulates SMAD3 activity. It appears, then, that these two miRNAs, are potential candidates for targeted therapies in diseases characterised by an abnormal collagen deposition and myofibroblast transdifferentiation The ratio between them could contribute to a candidate biomarker index for fibrosis or end organ damage in fibrotic disease. Interpretation of these relationships may be confounded by the importance of absolute levels, as well as ratios. For example, higher levels of both miRNA 21-5p and the 29 group may amplify TGF-β, while their ratio remains the same.
Recent experiments further support the role of SMAD3 in regulating the processing and maturation of miRNAs. Briefly, in miRNA biogenesis, a precursor transcript (pri-miRNA), made by RNA polymerase II, is converted to the precursor miRNA (pre-miRNA) by Drosha (RNaseIII-like enzyme). Drosha acts as part of a larger complex, to determine the precise location of miRNAs processing, orchestrating specific miRNA maturation. Several proteins interact with this miRNA processing complex, including the DEAD-box RNA helicase protein p68, which is also a Smad3 interacting protein [68, 69].
Limitations
The computational biology approach to these data and the field of miRNAs in fibrotic disease is a large enough undertaking to initially justify a survey approach (such as in this review). The systematic method used in the current review has some limitations, however. Many of these limitations are inherent to the heterogeneity of the primary data (table 2).
The first major limitation is related to the heterogeneity of the compartments in which miRNAs can be measured. The SSc studies reported here are from skin biopsies and fibroblasts, but none from the lung, because no studies to date have analysed miRNAs from that tissue compartment in SSc. On the other hand, the majority of IPF studies are from lung and lung fibroblasts, with no studies from the skin. Although it has been documented that each tissue can have cell-specific functions and responses to given miRNAs, a survey or overview may help in building a general picture of miRNAs in fibrotic conditions.
Secondly, the method of comparing overlapping miRNAs between diseases is highly biased by the number of publications found for each miRNA in SSc and IPF. This method allows a wide collection without exclusion of papers according to restrictive exclusion criteria. It therefore allows us to collect up-to-date evidence for both SSc and IPF. However, such a broad summary might lead to mistaking momentum in the literature for biological importance. Are miR-21-5p and the 29 family the most important miRNAs in fibrotic lung disease, or merely the most studied thus far? Due to the rapidly increasing complexity of the area of miRNA measurement in fibrosis, it may be of value to periodically compile a survey of information on various miRNAs in these emblematic fibrotic lung disesases, IPF and scleroderma-ILD.
Lastly, the fact that SSc and IPF share only the usual interstitial pneumonia (UIP) pattern, commonly associated with a worse prognosis among ILDs, and not the non-specific interstitial pneumonia (NSIP) pattern, must be included as a limitation of the review. In fact, in SSc, UIP accounts for only 25–30% of cases, while in IPF it is theoretically 100%. Despite some degree of overlap in their clinical features and pathogenesis, several divergent features reflect differences in the pathways regulating fibrogenesis, repair mechanisms and altered cellular response. These differences may carry implications for diagnosis, evaluation and management. Yet, the relevance of identifying overlapping regulators of fibrosis comparing SSc and IPF remains.
Perspectives
The detection of specific circulating miRNAs that correlate with fibrosis in SSc and IPF could address the unmet need for reliable and accurate biomarkers of fibrogenesis. Larger and more detailed studies will be required to better evaluate the potential role of these miRNAs as diagnostic or prognostic biomarkers and to tailor targeted therapies. There are several various levels at which a single miRNA can act in the fibrotic milieu. These levels encompass the induction of fibroblast proliferation, transition into myofibroblasts, collagen synthesis and degradation, mechanotransduction signalling and adhesion. The hypothesis that modulation of the miRNA 21/29 ratio can lead to significant changes in profibrotic gene activation needs experimental testing in order to unravel how cells integrate the complex repertoire of fine-tuned TGF-β signalling pathways with miRNA expression patterns in fibrotic diseases. The emerging role of miRNAs in gene expression at the post-transcriptional level in pathological conditions offers a fascinating insight and may offer novel therapeutic targets. However, some entirely new roles have been attributed to these small noncoding miRNAs, such as the ability to act directly as inflammatory molecules, suggesting that their complex activity can have direct immune function apart from their indirect influence via specific gene activation, perhaps meriting the “mirokine” designation that some have assigned [8]. Indeed, one of the most immediate challenges is to assess whether there are patterns of miRNAs that closely associate with pathological conditions and/or with common end-organ damage, e.g. cancer or fibrosis.
It is possible to hypothesise that different patterns or shared clusters of miRNA aberrations might influence different organs in either an organ-specific manner or a disease-specific manner. The large number of miRNAs, any differences between the 3′ and 5′ miRNAs, the number of genes possibly influenced by a single miRNA, and the possibility of “U” or “J” shaped dose–response curves all potentially multiply complexity and suggest a computational biology approach might be applicable.
Disclosures
W.N. Roberts ERR-0125-2016_Roberts (1.2MB, pdf)
J. Roman ERR-0125-2016_Roman (1.2MB, pdf)
Footnotes
Conflict of interest: Disclosures can be found alongside this article at err.ersjournals.com
Provenance: Submitted article, peer reviewed.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, Dumitru CD, Shimizu M, et al. . Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002; 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Reilly S. MicroRNAs in fibrosis: opportunities and challenges. Arthritis Res Ther 2016; 18: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 2005; 24: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009; 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 8.Young NA, Wu LC, Amici S, et al. . Estrogen-regulated STAT1 activation promotes TLR8 overexpression and facilitates mirokine signaling via exosomes containing MIR-21 endogenous ligand: a novel innate inflammatory pathway in systemic lupus erythematosus. Ann Rheum Dis 2015; 74: 906–907. [Google Scholar]
- 9.Vettori S, Gay S, Distler O. Role of microRNAs in fibrosis. Open Rheumatol J 2012; 6: 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Issler O, Chen A. Determining the role of microRNAs in psychiatric disorders. Nat Rev Neurosci 2015; 16: 201–212. [DOI] [PubMed] [Google Scholar]
- 11.Kriegel AJ, Liu Y, Fang Y, et al. . The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genom 2012; 44: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rooij E, Sutherland LB, Thatcher JE, et al. . Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 2008; 105: 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin W, Chung AC, Huang XR, et al. . TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol 2011; 22: 1462–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roderburg C, Urban GW, Bettermann K, et al. . MicroRNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011; 53: 209–218. [DOI] [PubMed] [Google Scholar]
- 15.Maurer B, Stanczyk J, Jüngel A, et al. . MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 2010; 62: 1733–1743. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Li Y, Qu S, et al. . MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol 2012; 32: 514–522. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharyya S, Kelley K, Melichian DS, et al. . Toll-like receptor 4 signaling augments transforming growth factor-β responses: a novel mechanism for maintaining and amplifying fibrosis in scleroderma. Am J Pathol 2013; 182: 192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jafarinejad-Farsangi S, Farazmand A, Mahmoudi M, et al. . MicroRNA-29a induces apoptosis via increasing the Bax: Bcl-2 ratio in dermal fibroblasts of patients with systemic sclerosis. Autoimmunity 2015; 48: 369–378. [DOI] [PubMed] [Google Scholar]
- 19.Ciechomska M, O'Reilly S, Suwara M, et al. . MiR-29a reduces TIMP-1 production by dermal fibroblasts via targeting TGF-β activated kinase 1 binding protein 1, implications for systemic sclerosis. PLoS ONE 2014; 9: e115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steen SO, Iversen LV, Carlsen AL, et al. . The circulating cell-free microRNA profile in systemic sclerosis is distinct from both healthy controls and systemic lupus erythematosus. J Rheumatol 2015; 42: 214–221. [DOI] [PubMed] [Google Scholar]
- 21.Wuttge DM, Carlsen AL, Teku G, et al. . Specific autoantibody profiles and disease subgroups correlate with circulating microRNA in systemic sclerosis. Rheumatology (Oxford) 2015; 54: 2100–2107. [DOI] [PubMed] [Google Scholar]
- 22.Khalil W, Xia H, Bodempudi V, et al. . Pathologic regulation of collagen I by an aberrant protein phosphatase 2A/histone deacetylase C4/microRNA-29 signal axis in idiopathic pulmonary fibrosis fibroblasts. Am J Respir Cell Mol Biol 2015; 53: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker MW, Rossi D, Peterson M, et al. . Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest 2014; 124: 1622–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang G, Yang L, Wang W, et al. . Discovery and validation of extracellular/circulating microRNAs during idiopathic pulmonary fibrosis disease progression. Gene 2015; 562: 138–144. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery RL, Yu G, Latimer PA, et al. . MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med 2014; 6: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekar D, Hairul Islam VI, Thirugnanasambantham K, et al. . Relevance of miR-21 in HIV and non-HIV-related lymphomas. Tumour Biol 2014; 35: 8387–8393. [DOI] [PubMed] [Google Scholar]
- 27.Han M, Liu M, Wang Y, et al. . Re-expression of miR-21 contributes to migration and invasion by inducing epithelial–mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Mol Cell Biochem 2012; 363: 427–436. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, Luo H, Li Y, et al. . MicroRNA-21 in scleroderma fibrosis and its function in TGF-β-regulated fibrosis-related genes expression. J Clin Immunol 2013; 33: 1100–1109. [DOI] [PubMed] [Google Scholar]
- 29.Oak SR, Murray L, Herath A, et al. . A micro RNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PLoS ONE 2011; 6: e21253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, Zhao GQ, Chen TF, et al. . Serum miR-21 and miR-155 expression in idiopathic pulmonary fibrosis. J Asthma 2013; 50: 960–964. [DOI] [PubMed] [Google Scholar]
- 31.Liu G, Friggeri A, Yang Y, et al. . miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 2010; 207: 1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada M, Kubo H, Ota C, et al. . The increase of microRNA-21 during lung fibrosis and its contribution to epithelial–mesenchymal transition in pulmonary epithelial cells. Respir Res 2013; 14: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuziwara CS, Kimura ET. Insights into regulation of the miR-17-92 cluster of miRNAs in cancer. Front Med (Lausanne) 2015; 2: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sing T, Jinnin M, Yamane K, et al. . microRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford) 2012; 51: 1550–1556. [DOI] [PubMed] [Google Scholar]
- 35.Etoh M, Jinnin M, Makino K, et al. . microRNA-7 down-regulation mediates excessive collagen expression in localized scleroderma. Arch Dermatol Res 2013; 305: 9–15. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Yang R, Fan X, et al. . MicroRNA array analysis of microRNAs related to systemic scleroderma. Rheumatol Int 2012; 32: 307–313. [DOI] [PubMed] [Google Scholar]
- 37.Berschneider B, Ellwanger DC, Baarsma HA, et al. . miR-92a regulates TGF-β1-induced WISP1 expression in pulmonary fibrosis. Int J Biochem Cell Biol 2014; 53: 432–441. [DOI] [PubMed] [Google Scholar]
- 38.Dakhlallah D, Batte K, Wang Y, et al. . Epigenetic regulation of miR-17∼92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 2013; 187: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandit KV, Corcoran D, Yousef H, et al. . Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2010; 182: 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang H, Xu C, Pan Z, et al. . The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Mol Ther 2014; 22: 1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang H, Gu Y, Li T, et al. . Integrated analyses identify the involvement of microRNA-26a in epithelial-mesenchymal transition during idiopathic pulmonary fibrosis. Cell Death Dis 2014; 5: e1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang H, Liu S, Chen Y, et al. . miR-26a suppresses EMT by disrupting the Lin28B/let-7d axis: potential cross-talks among miRNAs in IPF. J Mol Med 2016; 94: 655–665. [DOI] [PubMed] [Google Scholar]
- 43.Makino K, Jinnin M, Hirano A, et al. . The downregulation of microRNA let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. J Immunol 2013; 190: 3905–3915. [DOI] [PubMed] [Google Scholar]
- 44.Milosevic J, Pandit K, Magister M, et al. . Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol 2012; 47: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka S, Suto A, Ikeda K, et al. . Alteration of circulating miRNAs in SSc: miR-30b regulates the expression of PDGF receptor β. Rheumatology (Oxford) 2013; 52: 1963–1972. [DOI] [PubMed] [Google Scholar]
- 46.Honda N, Jinnin M, Kira-Etoh T, et al. . miR-150 down-regulation contributes to the constitutive type I collagen overexpression in scleroderma dermal fibroblasts via the induction of integrin β3. Am J Pathol 2013; 182: 206–216. [DOI] [PubMed] [Google Scholar]
- 47.Huleihel L, Ben-Yehudah A, Milosevic J, et al. . Let-7d microRNA affects mesenchymal phenotypic properties of lung fibroblasts. Am J Physiol Lung Cell Mol Physiol 2014; 306: L534–L542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bodempudi V, Hergert P, Smith K, et al. . miR-210 promotes IPF fibroblast proliferation in response to hypoxia. Am J Physiol Lung Cell Mol Physiol 2014; 307: L283–L294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honda N, Jinnin M, Kajihara I, et al. . TGF-β-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol 2012; 188: 3323–3331. [DOI] [PubMed] [Google Scholar]
- 50.Alsaleh G, François A, Philippe L, et al. . MiR-30a-3p negatively regulates BAFF synthesis in systemic sclerosis and rheumatoid arthritis fibroblasts. PLoS ONE 2014; 9: e111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makino K, Jinnin M, Kajihara I, et al. . Circulating miR-142-3p levels in patients with systemic sclerosis. Clin Exp Dermatol 2012; 37: 34–39. [DOI] [PubMed] [Google Scholar]
- 52.Su S, Zhao Q, He C, et al. . miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat Commun 2015; 6: 8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nho RS, Im J, Ho YY, et al. . MicroRNA-96 inhibits FoxO3a function in IPF fibroblasts on type I collagen matrix. Am J Physiol Lung Cell Mol Physiol 2014; 307: L632–L642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kajihara I, Jinnin M, Yamane K, et al. . Increased accumulation of extracellular thrombospondin-2 due to low degradation activity stimulates type I collagen expression in scleroderma fibroblasts. Am J Pathol 2012; 180: 703–714. [DOI] [PubMed] [Google Scholar]
- 55.Fierro-Fernández M, Busnadiego Ó, Sandoval P, et al. . miR-9-5p suppresses pro-fibrogenic transformation of fibroblasts and prevents organ fibrosis by targeting NOX4 and TGFBR2. EMBO Rep 2015; 16: 1358–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Song X, Li Y, et al. . Low-dose paclitaxel ameliorates pulmonary fibrosis by suppressing TGF-β1/Smad3 pathway via miR-140 upregulation. PLoS ONE 2013; 8: e70725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao X, Huang C, Zhao C, et al. . Regulation of myofibroblast differentiation by miR-424 during epithelial-to-mesenchymal transition. Arch Biochem Biophys 2015; 566: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo H, Zhu H, Zhou B, et al. . MicroRNA-130b regulates scleroderma fibrosis by targeting peroxisome proliferator-activated receptor γ. Mod Rheumatol 2015; 25: 595–602. [DOI] [PubMed] [Google Scholar]
- 59.Li S, Geng J, Xu X, et al. . miR-130b-3p modulates epithelial-mesenchymal crosstalk in lung fibrosis by targeting IGF-1. PLoS ONE 2016; 11: e0150418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang X, Tsitsiou E, Herrick SE, et al. . MicroRNAs and the regulation of fibrosis. FEBS J 2010; 277: 2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zanotti S, Gibertini S, Curcio M, et al. . Opposing roles of miR-21 and miR-29 in the progression of fibrosis in Duchenne muscular dystrophy. Biochim Biophys Acta 2015; 1852: 1451–1464. [DOI] [PubMed] [Google Scholar]
- 62.Wang D, Fan Z, Liu F, et al. . Hsa-miR-21 and Hsa-miR-29 in tissue as potential diagnostic and prognostic biomarkers for gastric cancer. Cell Physiol Biochem 2015; 37: 1454–1462. [DOI] [PubMed] [Google Scholar]
- 63.Wang G, Kwan BC, Lai FM, et al. . Urinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosis. Am J Nephrol 2012; 36: 412–418. [DOI] [PubMed] [Google Scholar]
- 64.Xiao J, Meng XM, Huang XR, et al. . miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther 2012; 20: 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.García R, Nistal JF, Merino D, et al. . p-SMAD2/3 and DICER promote pre-miR-21 processing during pressure overload-associated myocardial remodeling. Biochim Biophys Acta 2015; 1852: 1520–1530. [DOI] [PubMed] [Google Scholar]
- 66.Zhong X, Chung AC, Chen HY, et al. . Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol 2011; 22: 1668–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noetel A, Kwiecinski M, Elfimova N, et al. . microRNA are central players in anti- and profibrotic gene regulation during liver fibrosis. Front Physiol 2012; 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett 2012; 586: 1906–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Warner DR, Bhattacherjee V, Yin X, et al. . Functional interaction between Smad, CREB binding protein, and p68 RNA helicase. Biochem Biophys Res Commun 2004; 324: 70–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
W.N. Roberts ERR-0125-2016_Roberts (1.2MB, pdf)
J. Roman ERR-0125-2016_Roman (1.2MB, pdf)