Abstract
In the enteric bacteria Escherichia coli and Salmonella enterica, sulfate is reduced to sulfide and assimilated into the amino acid cysteine; in turn, cysteine provides the sulfur atom for other sulfur-bearing molecules in the cell, including methionine. These organisms cannot use methionine as a sole source of sulfur. Here we report that this constraint is not shared by many other enteric bacteria, which can use either cysteine or methionine as the sole source of sulfur. The enteric bacterium Klebsiella aerogenes appears to use at least two pathways to allow the reduced sulfur of methionine to be recycled into cysteine. In addition, the ability to recycle methionine on solid media, where cys mutants cannot use methionine as a sulfur source, appears to be different from that in liquid media, where they can. One pathway likely uses a cystathionine intermediate to convert homocysteine to cysteine and is induced under conditions of sulfur starvation, which is likely sensed by low levels of the sulfate reduction intermediate adenosine-5′-phosphosulfate. The CysB regulatory proteins appear to control activation of this pathway. A second pathway may use a methanesulfonate intermediate to convert methionine-derived methanethiol to sulfite. While the transsulfurylation pathway may be directed to recovery of methionine, the methanethiol pathway likely represents a general salvage mechanism for recovery of alkane sulfide and alkane sulfonates. Therefore, the relatively distinct biosyntheses of cysteine and methionine in E. coli and Salmonella appear to be more intertwined in Klebsiella.
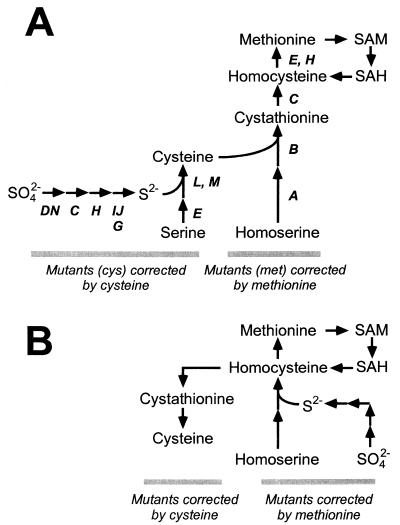
The enteric bacteria Escherichia coli and Salmonella enterica serve as model systems for the physiology and regulation of numerous metabolic processes in bacteria, including sulfur assimilation. Sulfur is a constituent of several indispensable biomolecules, including cysteine, methionine, thiamine, biotin, lipoic acid, and coenzyme A; in these contexts, sulfur is used in its fully reduced state (S2−). In many environments, sulfur is found primarily in the oxidized state of sulfate (SO42−) and must be reduced to sulfide before assimilation into organic material. While the reduction of sulfate to sulfide follows a common pathway in all organisms studied to date, the assimilation of sulfide itself into an organic molecule may take one of two routes. In most bacteria, including enteric bacteria, sulfide is incorporated into activated forms of serine to form cysteine (18); cysteine then serves as the sulfur group donor, either directly or indirectly, for the synthesis of all other sulfur-bearing molecules in the cell (Fig. 1A). Alternatively, yeast and some bacteria assimilate sulfide primarily into activated homoserine to form homocysteine, an intermediate in methionine biosynthesis (30); homocysteine is used for biosynthesis of cysteine, methionine, and other sulfur-bearing molecules (Fig. 1B). Some organisms, such as Pseudomonas putida, can assimilate sulfide into either compound (31).
FIG. 1.
Pathways for sulfur assimilation. (A) Sulfur assimilation in enteric bacteria E. coli and S. enterica. Genes whose products contribute to each step are noted. (B) Sulfur assimilation in yeast Saccharomyces cerevisiae. SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine.
In E. coli and S. enterica, auxotrophs corrected by the addition of sulfur-bearing amino acids fall neatly into two classes (Fig. 1A). Mutants defective in sulfate reduction or sulfide assimilation are corrected by the addition of cysteine to the growth medium; by definition the associated mutations affect cys genes and their defects are not corrected by the addition of methionine (18). Mutants defective in the synthesis of methionine (affecting met genes) are corrected by methionine but not by cysteine (9). One unusual class of mutants with leaky point mutations (termed cym) which are corrected by the addition of either cysteine or methionine has been reported in S. enterica (14, 15, 29); these mutations mapped to various cysteine biosynthetic genes. Their leaky behavior sustained very slow growth rates when the cell was spared the need to use cysteine to synthesize methionine (which comprises 40% of the reduced sulfur requirement for the cell), which then provided just enough cysteine to satisfy other requirements.
The metabolism of E. coli and S. enterica are often used as paradigms in unraveling the physiology of diverse and unrelated organisms, wherein detection of enzyme homologues in these species implies their implementation in specific metabolic pathways. The validity of these inferences relies heavily on a satisfactory understanding of these processes within E. coli. The enteric bacterium Klebsiella aerogenes is related to E. coli and S. enterica and therefore serves as an outside reference taxon for understanding the evolution of the metabolic capabilities in these organisms (2). As reported below, Klebsiella expresses at least two pathways that allow the sulfur atom of methionine to be recycled back into cysteine. Although present in many enteric bacteria, this capability appears to have been lost from the E. coli/Salmonella lineage. Hence, the cysteine and methionine biosynthetic pathways of Klebsiella are far more intertwined than suspected, being only two portions of a complex series of metabolic cycles. Initial genetic and biochemical characterization of these pathways is presented, and their implication in the evolution of enteric bacteria is discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains used in these studies (Table 1) were constructed from K. aerogenes W70 derivative KC2668 (hsdR suc+ hutC515(Con) dadA lac Δbla-2), kindly provided by R. Bender. Plasmid pTAS1 bears the lamB region from E. coli and renders Klebsiella sensitive to bacteriophage λ. Plasmid pTAS1 was constructed from pTROY11 (6) by cleavage with HindIII and BamHI, treatment with T4 DNA polymerase to create blunt ends, and ligation with T4 DNA ligase according to the manufacturer's instructions; this treatment removed a portion of the tetracycline resistance gene and eliminated the appearance of spontaneous tetracycline-resistant colonies. Plasmid pMMK1 (17) is pTAS1 with the gene encoding the altered-target specificity (ATS) Tn10 transposase from plasmid pNK2881 (16) inserted into the EcoRI site.
TABLE 1.
Bacteria strains and plasmids
| Strain | Species | Relevant genotype | Source or reference |
|---|---|---|---|
| LD561 | K. aerogenes | hsdR suc+ hutC515(Con) dadA lac Δbla-2 | KC2668 from R. Bender |
| LD806 | K. aerogenes | pTAS1 | This study |
| LD821 | K. aerogenes | PMMK1 | 17 |
| LD807 | K. aerogenes | metE4021::Tn10dKn | This study |
| LD822 | K. aerogenes | metA4004::Tn10dTc | This study |
| LD823 | K. aerogenes | zgb-4002::Tn10dCm | This study |
| LD824 | K. aerogenes | cysQ4007::Tn10dTc | This study |
| LD825 | K. aerogenes | cysT4071::Tn10dCm (cymX) | This study |
| LD826 | K. aerogenes | cysDN4070::Tn10dTc (cymY) | This study |
| LD827 | K. aerogenes | cysIJ4009::Tn10dTc | This study |
| LD828 | K. aerogenes | cysC4022::Tn10dCm | This study |
| LD829 | K. aerogenes | cysH4005::Tn10dTc | This study |
| LD830 | K. aerogenes | cysB4020::Tn10dTc | This study |
| LD831 | K. aerogenes | zgb-4001::Tn10dCm | This study |
| LD832 | K. aerogenes | rha-4001::Tn10dKn | This study |
| LD833 | K. aerogenes | cob-4009 arg-4007::Tn10dTc | This study |
| LD834 | K. aerogenes | metB4031::Tn10dCm | This study |
| LD835 | K. aerogenes | srl-4001::Tn10dKn | This study |
| LD836 | K. aerogenes | fuc-4001::Tn10dKn | This study |
| LD837 | K. aerogenes | trp-4002::Tn10dKn | This study |
| LD838 | K. aerogenes | cysJ4034::Tn10dCm | This study |
| LD839 | K. aerogenes | cysD4072::Tn10dCm | This study |
| LD840 | K. aerogenes | cysQ4046::Tn10dCm | This study |
| LD841 | K. aerogenes | cysD4072::Tn10dCm cysIJ4009::Tn10dTc | This study |
| LD842 | K. aerogenes | metA4004::Tn10dTc mtc-4007::Tn10LK | This study |
| LD843 | K. aerogenes | cysD4072::Tn10dCm mtc-4008::Tn10LK | This study |
| LD849 | K. aerogenes | zig-4004::Tn10dTc | This study |
| LD850 | K. aerogenes | metC4029::Tn10dCm | This study |
| LD856 | K. aerogenes | metB4031::Tn10dCm metC4010::Tn10dTc | This study |
| LD507 | K. pneumoniae | M5aL from V. Stewart | |
| LD482 | E. coli K-12, W3110 | Laboratory collection | |
| ECOR1 | E. coli | 28 | |
| ECOR16 | E. coli | 28 | |
| ECOR47 | E. coli | 28 | |
| LD633 | S. enterica serovar Typhimurium LT2 | Laboratory collection | |
| SARB3 | S. enterica serovar Branderburg | 3 | |
| SARB9 | S. enterica serovar Derby | 3 | |
| SARB19 | S. enterica serovar Enteritidis | 3 | |
| LD118 | Enterobacter aerogenes | ATCCa | |
| LD126 | Escherichia vulneris | ATCC | |
| LD130 | E. fergusonii | ATCC | |
| LD137 | Serratia marcescens | ATCC |
ATCC, American Type Culture Collection.
Media and antibiotics.
The rich medium used was Luria-Bertani (LB) medium; minimal defined medium was E (32). Sulfur-free E medium (NSE) was created by substituting MgCl2 for MgSO4; glucose was used as a carbon source. Solid medium was made by the addition of Bacto agar (Difco) to 1.2%, agarose (Gibco-BRL) to 1.3%, Gelrite (Schweizer Hall) to 1.3%, or Phytagel (Sigma) to 1.3%. P1 buffer contained 5 mM CaCl2 and 10 mM MgSO4. Ampicillin was used at 200 μg/ml; kanamycin was used at 20 μg/ml; tetracycline was used at 10 μg/ml for selection of transductants and at 20 μg/ml otherwise; chloramphenicol was used at 50 μg/ml for selection of Tn10dCm transposition mutants and at 20 μg/ml otherwise. For growth curves, 5 ml of NSE–0.2% glucose was inoculated with 50 μl of a fresh, overnight culture that had been washed and resuspended in an equal volume of NSE.
Genetic methods.
Transposition-defective derivatives of Tn10 Tn10dKn (kanamycin resistance), Tn10dTc (tetracycline resistance), and Tn10dCm (chloramphenicol resistance) were delivered into pTAS1-bearing strains of K. aerogenes using bacteriophage λ delivery vectors λNK1316, λNK1323, and λNK1324 as described by Kolko et al. (17). Transduction was mediated by bacteriophage P1 vir as described by Kolko et al. (17).
Enzyme assays.
Cells were prepared by growth to mid-log phase, concentration by centrifugation, and resuspension in 1/10 volume of 100 mM KxPO4, pH 8.0. Cells were lysed by sonication in a 30-ml Corex tube using eight 10-s pulses on ice. Debris was removed by centrifugation, and the protein-bearing supernatant was desalted on a PD-10 gel filtration column (Pharmacia) and eluted in 10 mM KxPO4, pH 8.0. Enzyme extracts were used immediately in assays for cystathionine-γ-lyase (26) or methionine-γ-lyase (8) as described; assays were performed in triplicate. Assays for cystathionine-γ-lyase activity used 50 mM homoserine, 8.4 mM cystathionine (all four stereoisomers), and 2.1 mM ll-cystathionine (all are final concentrations) as substrates. In all assays, α-ketobutyrate was detected as a product. Protein concentration was determined by a Bradford assay (4).
Cloning and sequencing.
Chromosomal DNA was prepared from Tn10dCm-bearing cells and partially digested with the Sau3AI restriction endonuclease. DNA was size fractionated on agarose gels; high-molecular-weight fragments were purified using the JetSorb kit (Promega) and ligated into pNEB193 prepared by BamHI digestion and phosphatase treatment. Ligations were introduced into XL2-Gold-Kn competent cells (Stratagene) according to the manufacturer's instructions, and transformants were selected on ampicillin-containing media. Chloramphenicol-resistant colonies were isolated by replica printing. Positive clones were identified by DNA sequencing using an ABI 310 sequencer and primers directing replication out of Tn10dCm. Homology to E. coli genes was inferred from BLAST analysis (1).
RESULTS
Klebsiella cym mutants are corrected by cysteine or by methionine.
To isolate Klebsiella mutants defective for biosynthesis of sulfur-bearing amino acids, insertion mutagenesis was performed with transposition-defective derivatives of Tn10 (Tn10dTc, Tn10dCm, and Tn10dKn). Mutants were isolated on LB plates bearing the appropriate antibiotic and screened for auxotrophs by replica printing to minimal media and to minimal media containing both cysteine and methionine; a total of 130 independent mutants were isolated in four different mutagenic screens. Mutants were sorted into nutritional groups by determining which of the two amino acids suppressed their growth defect. As noted above, auxotrophs of E. coli and S. enterica corrected by the addition of sulfur-bearing amino acids fall into two classes: those corrected by the addition of cysteine and those corrected by the addition of methionine. In contrast, auxotrophs of K. aerogenes corrected by sulfur-bearing amino acids defined three distinct classes (Table 2). In addition to the expected cys and met gene classes, up to 40% of insertion mutations (corrected by the combination of cysteine and methionine in the original screen) were corrected by the addition of either amino acid to the growth medium (Table 2). We termed this class of mutations cym (corrected by cysteine or methionine).
TABLE 2.
Classes of sulfur amino acid auxotrophs in K. aerogenes on solid medium
| Phenotype | Growtha on indicated sulfur source
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Without SeO4−2
|
With 150 μM SeO42−
|
|||||||
| SO42− | Cysteine | Methionine | Cysteine + methionine | SO42− | Cysteine | Methionine | Cysteine + methionine | |
| Wild type | + | + | + | + | − | + | − | + |
| Cys | − | + | − | + | − | + | − | + |
| Met | − | − | + | + | − | − | − | + |
| Cym | − | + | + | + | − | + | + | + |
| Cys Cymb | − | + | + | + | − | + | + | + |
Phenotypes were evident after 18 h of growth; −, incubation for up to 5 days did not reveal any further growth.
The strains with this phenotype were created by P1 transduction.
Klebsiella cym mutations confer gain-of-function phenotypes.
Klebsiella cym mutations are not leaky cys mutations, like those described for S. enterica (14, 15, 29). Rather, cym mutations confer two gain-of-function phenotypes that are inconsistent with leaky cys mutations. First, cys cym double mutants are corrected by methionine (Table 2). Therefore, the cym mutation is not a leaky cys mutation since it allows tight cys mutants to utilize methionine as a sulfur source; the cym mutation uncovered a metabolic activity that was masked in the cys mutant on this growth medium, i.e., growth on methionine as the sole sulfur source. Second, cym mutations confer selenate resistance (Table 2). Selenate (SeO42−) is an analogue of sulfate that is toxic when metabolized into selenide (subsequent assimilation into selenocysteine results in tRNASer mischarged with selenocysteine). In E. coli, wild-type cells are sensitive to selenate unless they are growing in the presence of excess cysteine, which prevents CysB activation (10, 11) of genes for sulfate and selenate transport (23), reduction, and assimilation. In wild-type Klebsiella, the presence of methionine does not mitigate the toxic effects of selenate (Table 2). In contrast, Klebsiella cym mutants grow on methionine as the sole sulfur source even in the presence of selenate (Table 2). As discussed below, selenate resistance likely results from the lack of selenate transport or activation in cym mutants.
Genetic mapping and identification of cys, cym, and met loci.
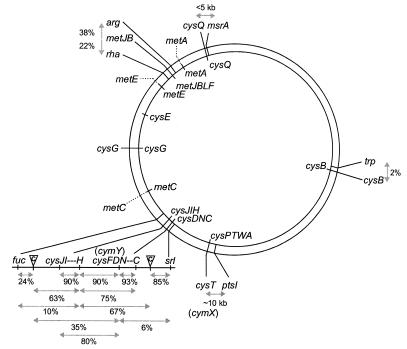
To identify the cym loci and to infer their mode of action, mutations conferring cys, cym, and met nutritional phenotypes were sorted into 11 groups by transductional analysis and nutritional profiling (Table 3, Fig. 2). The likely identities of these groups were assigned by linkage to loci which flank the corresponding genes in E. coli and S. enterica and/or by distinctive growth and nutritional phenotypes.
TABLE 3.
Phenotypes of classes of sulfurbearing amino acid auxotrophs of K. aerogenes on solid medium
| Classb | No. of mutations isolated | Growthc on specified sulfur sourcea
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aerobic
|
Anaerobic
|
|||||||||||||
| SO42− | SO32− | S2− | Cys | Met | SO42− + Cyst | SO42− + HCys | SO42− + Met | SO42− + B12 | SO42 | Cys | Met | SO42− + Met | ||
| Wild type | n/ad | ++ | ++ | ++ | ++ | − | ++ | ++ | ++ | ++ | ++ | ++ | − | ++ |
| cysQ | 9 | − | ++ | ++ | ++ | − | − | − | − | − | + | ++ | − | ++ |
| cysB | 1 | − | − | − | + | − | − | − | − | − | − | ++ | − | − |
| cysIJ | 20 | − | − | ++ | ++ | − | − | − | − | − | − | ++ | − | − |
| cysH | 10 | − | + | + | + | − | − | − | − | − | − | ++ | − | − |
| cysC | 5 | − | ++ | ++ | ++ | − | − | − | − | − | − | ++ | − | − |
| cymY (cysDN) | 20 | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − | − | ++ | + | ++ |
| cymX (cysPTWA) | 22 | +/= | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +/= | − | ++ | ++ | ++ |
| metA | 23 | − | − | − | − | − | ++ | ++ | ++ | − | − | − | − | ++ |
| metB | 3 | − | − | − | − | − | ++ | ++ | ++ | − | − | − | − | ++ |
| metC | 9 | − | − | − | − | − | − | ++ | ++ | − | − | − | − | ++ |
| metE | 6 | − | − | − | − | − | − | − | ++ | ++ | − | − | − | ++ |
Cys, cysteine; Met, methionine; Cyst, cystathionine; HCys, homocysteine.
Class is defined by nutritional profiling, linkage analysis, and DNA sequence information (Fig. 2).
−, no growth; +/=, very poor growth; +, moderate growth; ++, strong growth. Phenotypes were evident after 18 (aerobic) to 36 h (anaerobic) of growth; incubation for up to 5 days did not reveal any further growth.
n/a, not applicable.
FIG. 2.
Linkage groups for cys, cym, and met genes in K. aerogenes. Corresponding genes on the E. coli chromosome are noted on the inner circle. Gene identifications were made by linkage analysis and/or DNA sequence. Dotted lines denote linkage groups that have not yet been cloned from Klebsiella. Linkages were calculated on the basis of cotransduction frequencies using bacteriophage P1 vir. Tn10dCm insertions in strains LD823 and LD831 are noted between cysC and srl and between cysJ and fuc, respectively.
The cysIJ locus was identified since mutants defective at this site failed to reduce sulfite (Table 3) and since mutations are linked to the fuc locus (Fig. 2); this assignment has been verified by the DNA sequence at the insertion site of the Tn10dCm in strain LD828 (codon 2 of the cysJ gene). The cysH locus was identified by three-factor cross analysis (Fig. 2)—mutations are 90 to 95% linked to cysIJ, distal to the fuc operon—and by very poor growth on sulfite in the presence of sulfate due to accumulation of the toxic intermediate 3′-phosphoadenosine-5′-phosphosulfate (PAPS). Three-factor cross analysis between cys, srl, and fuc loci identified cymY mutations as likely affecting the cysDN genes, encoding adenosine-5′-phosphosulfate (APS) synthase (Fig. 2). This assignment has been verified by the DNA sequence of the insertion site for the Tn10dCm in strain LD839 (codon 95 of the cysD gene). The cysC locus was identified by linkage analysis as being downstream of the cysDN (cymY) genes; DNA sequences from the Tn10dCm-bearing clones isolated from LD839 verify the presence of a cysC homologue downstream of the cysN gene. Mutations at the cymX locus affect the cysPTWA operon encoding the sulfate transport apparatus; this identification was made by linkage to the ptsI gene and by the DNA sequence of the insertion site of the Tn10dCm in strain LD825 (codon 106 of the cysT gene). The cysQ locus was identified by linkage to the msrA gene, encoding methionine sulfoxide reductase, and by the DNA sequence at the site of insertion of the Tn10dCm in strain LD840 (codon 110 in the cysQ gene). Moreover, Klebsiella cysQ mutants have no auxotrophic phenotype when grown under anaerobic conditions, as seen for mutations affecting the E. coli cysQ gene (27). The cysB locus was identified by its failure to use any inorganic sulfur source on solid media and by its linkage to the trp locus (Fig. 2). Assignment of the cysG linkage group was complicated by the presence of the functionally redundant cysF gene; the characterization of these loci is discussed elsewhere (17).
Four linkage groups of met mutations were uncovered. The metB locus, which includes the metJBLF genes in E. coli, was identified by linkages to the rha and arg loci; the phenotype produced by insertions in this group are consistent with defects in the metB gene. This assignment was confirmed by the sequence flanking the site of insertion of the Tn10dCm in strain LD834 (codon 188 of the metB gene), which also identified a homologue of the metJ regulatory gene. Mutations defining the metA linkage group are linked to each other but not to the rha or arg loci; like metB mutants, these strains are corrected by cystathionine (Table 2). Mutants defining the metC linkage group are not corrected by cystathionine but are corrected by homocysteine. Mutations in the metE gene, encoding the cobalamin-independent methionine synthase, were identified as those conferring methionine auxotrophy that was not corrected by homocysteine but that was correctable by coenzyme B12; B12 is a required cofactor for the alternative methionine synthase MetH. Since Klebsiella synthesizes B12 de novo (22), metE mutations were isolated in a cob mutant, which fails to synthesize B12.
Although 130 mutants were isolated, not all genes involved in cysteine and methionine biosynthesis have been identified in this screen. We did not isolate mutants lacking acetylserine thiolyase activity, which would only be corrected by cysteine; this enzyme may be encoded by redundant genes, since in E. coli and Salmonella both the cysM or cysK genes encode acetylserine thiolyases. Similarly, we did not isolate mutations in the cysG gene in this screen due to the presence of the functionally redundant cysF gene (17). In addition, we did not isolate mutations in the cysE gene, encoding serine transacetylase; this gene may be duplicated, may provide an additional function essential to Klebsiella, thereby preventing the isolation of cysE null mutants, or we may have been unlucky. Considering that the cysB locus is defined by a single mutation (poor growth of cysB mutants precluded facile identification), it is likely that saturation mutagenesis was not achieved.
Klebsiella uses methionine as a sole sulfur source in liquid medium.
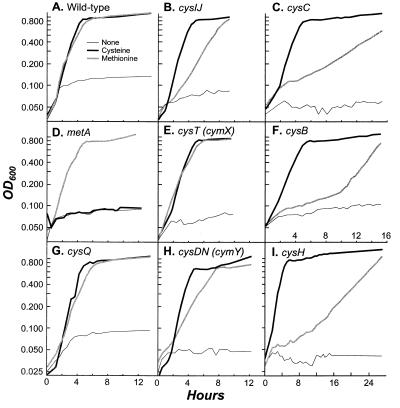
The behavior of cym mutants on solid media suggests that methionine can be employed as a sole sulfur source by K. aerogenes. Since agar media contain trace amounts of sulfate and usable alkane sulfates, this hypothesis was tested by growing wild-type cells and cys, met, and cym mutants on a variety of sulfur sources in sulfur-free liquid media (Fig. 3). Growth of wild-type K. aerogenes when cysteine was provided as the sole source of sulfur was similar to that with methionine as the sole source; no significant growth was detected on unsupplemented NSE media, attesting to the low sulfur source content of this medium (Fig. 3A). In addition, cym mutants grew well on methionine as a sole sulfur source in liquid media (Fig. 3D and E). Surprisingly, cys mutants, corrected only by cysteine on solid medium, were readily corrected by methionine in liquid medium (Fig. 3C and F to I), although both cysH and cysB mutants showed a substantial lag before utilizing methionine that is not seen when cysteine is used as a sulfur source.
FIG. 3.
Growth of K. aerogenes strains on sulfur sources. (A) Wild-type strain LD561; (B) LD827 (cysIJ4009::Tn10dTc); (C) LD828 (cysC4022::Tn10dCm); (D) LD822 (met-4004::Tn10dTc); (E) LD825 (cysT4071::Tn10dCm [cymX]); (F) LD830 (cysB4020::Tn10dTc); (G) LD824 (cysQ4007::Tn10dTc); (H) LD826 (cysDN4070::Tn10dTc [cymY]); (I) LD829 (cysH4005::Tn10dTc).
These growth curves demonstrate two salient points. First, the phenotype of cym mutants is consistent with the utilization of methionine as a sole sulfur source, rather than the activation of a cryptic pathway for use of an alternative sulfur source on solid medium (derived from agar). Second, cys mutants are unable to degrade methionine on solid medium but are not inhibited in liquid medium. This difference is not attributable to a decreased oxygen tension during growth in liquid media, since cys mutants are not corrected by methionine on solid medium under anaerobic conditions.
Methionine utilization does not require assimilation of sulfite or sulfide.
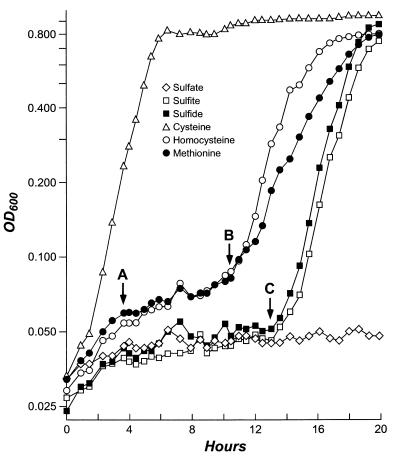
It is possible that methionine is degraded and that free sulfide, or an oxidized form of sulfur, is released. Alternatively, methionine may be converted to cysteine via a cystathionine intermediate, similar to the transsulfurylation pathway of yeasts and some bacteria (Fig. 1). To test the hypothesis that methionine utilization proceeds through a sulfite intermediate, a cymY (i.e., cysDN) cysIJ double mutant was created (LD841) by P1 transduction. This mutant fails to use sulfite as a sulfur source but does use methionine, implying that sulfite is not a requisite intermediate in methionine utilization. Since mutants lacking cysteine synthase activity were not isolated (likely because Klebsiella bears two cysteine synthases homologous to the E. coli CysM and CysK enzymes), we examined the growth of a cysB mutant in liquid media (Fig. 4). Lacking the requisite positive activator, cysB mutants cannot assimilate any form of inorganic sulfur on solid media and are cysteine auxotrophs (Table 3). However, they do assimilate alternative sulfur sources in liquid media after a substantial lag period; the growth rate after the lag phase is comparable to the growth rate of wild-type cells (serial dilution and plating experiments verify that this apparent lag is not attributable to contamination of the culture or reversion of the cysB mutation).
FIG. 4.
Growth curves of LD830 (cysB::Tn10dTc) in NSE-glucose medium. Compounds serving as sole sources of sulfur are noted. The doubling time of cells grown on cysteine is 76.7 min. Between points A and B, the doubling times of cells grown on homocysteine and methionine are 583 and 775 min, respectively. Between points A and C, the doubling times of cells grown on sulfate, sulfite, and sulfide are 7,180, 1,530, and 1,410 min, respectively. OD600, optical density at 600 nm.
Although this effect is highly repeatable, it is not clear what factors mediate the lag or the recovery. Close inspection of Fig. 3 and 4 shows that the cysB mutant grows very slowly on methionine (and its precursor homocysteine) during its initial lag (∼600-min doubling time; period A to B in Fig. 4), during which time no growth is seen on sulfate, sulfite, or sulfide (∼2,000- to 7,000-min doubling time; period A to C). The cells regain the ability to utilize methionine and homocysteine about 3 h before they are able to assimilate sulfide (points B and C, respectively). These data suggest that methionine utilization does not proceed via a requisite sulfide intermediate; the admittedly small lag in the cysB growth curve leading to this conclusion is reproducible but does not rule out the possibility that methionine recycling proceeds via a sulfide intermediate. In addition, the more-effective growth on homocysteine than on methionine (Fig. 4), even in the face of poorer transport of homocysteine into the cell, suggests that homocysteine is converted directly to cysteine and that a rate-limiting step in the utilization of methionine via the homocysteine intermediate may be present. These data support the hypothesis that methionine utilization can occur via a direct transsulfurylation pathway, mediating the transfer of the sulfhydryl group from homocysteine to serine to form cysteine.
Klebsiella expresses a cystathionine-γ-lyase.
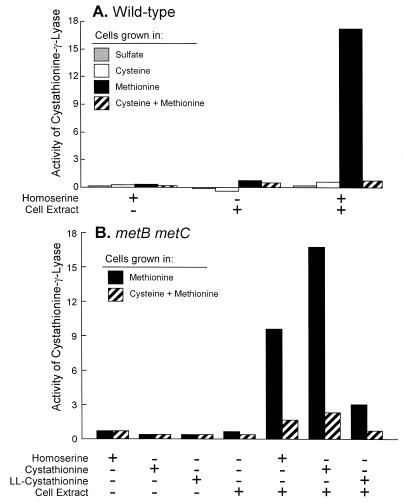
The conversion of homocysteine to cysteine by transsulfurylation proceeds in fungi via an l-allo-cystathionine intermediate, followed by its subsequent cleavage to yield cysteine, α-ketobutyrate, and ammonia (Fig. 1B). To test for cystathionine-γ-lyase activity in Klebsiella, enzyme extracts were prepared from wild-type K. aerogenes grown on sulfate, cysteine, methionine, or cysteine plus methionine as the sulfur source (Fig. 5A). Cystathionine-γ-lyase activity was detected in Klebsiella, but it was present only in cells grown with methionine as the sole sulfur source; sulfate or cysteine in the growth medium, regardless of the presence of methionine, eliminated this activity. Cystathionine-γ-lyase activity was detected using either homoserine or cystathionine as the substrate. When assaying for cystathionine utilization, we employed a metB metC double mutant, eliminating any contribution of the associated methionine-biosynthetic enzymes, which could act on cystathionine as a substrate. Although the mixture of all four stereoisomers of cystathionine served as a substrate, the ll-cystathionine stereoisomer alone appears to be unsuitable (Fig. 5B). We attribute the residual activity seen on ll-cystathionine to contaminants in the ll-cystathionine preparation, which is reported to be 90% pure; alternatively, cystathionine-γ-lyase could act on more than one stereoisomer of cystathionine. Therefore, we conclude that an inducible cystathionine-γ-lyase activity allows K. aerogenes to utilize methionine as a sole sulfur source in the absence of cysteine or sulfate. We should note that although α-ketobutyrate was effectively detected as a product of cystathionine cleavage, cysteine was not reproducibly detected. This result may reflect difficulties with the assay or may be due to intact cysteine not being released as a product. Although cystathionine-γ-lyase has been reported to use the l-allo-cystathionine isomer in other organisms, from these data we can only conclude that ll-cystathionine is not the substrate. Assays of E. coli and S. enterica protein extracts failed to detect cystathionine-γ-lyase activity, even when cells were grown (as well as they could be, see below) on methionine as the sole potential source of sulfur.
FIG. 5.
Assays for cystathionine-γ-lyase in K. aerogenes LD561 grown in different sulfur sources. Specific activities were calculated as increases in absorbance at 320 nm per minute per milligram of protein. Activities were calculated for substrate only, extract only, and complete reaction mixtures (for calculation of substrate-only activities, the protein concentration of the corresponding extract was used). Values are mean activities for four assays. (A) Strain LD561. (B) LD856.
Evidence for a second pathway for methionine recycling.
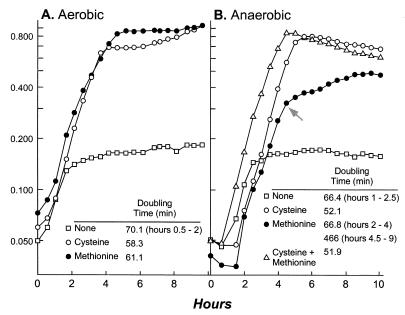
Although the data presented above provide compelling evidence for a pathway converting homocysteine to cysteine in K. aerogenes, these data do not preclude the presence of an additional pathway for methionine recycling. Since methionine serves as a poor sulfur source under anaerobic conditions (Fig. 6), we reasoned that a second pathway, proceeding via methanethiol (CH3SH) and methanesulfonate (CH3SO3) intermediates, could contribute to aerobic methionine recycling. Production of methanethiol via 3-methylthiopropionate uses the oxygen-dependent enzyme aci-reductone oxidase (CO forming) (33, 34). Methanethiol would be oxidized to methanesulfonate, and the sulfur would be recovered as sulfite, through the action of an oxygen-dependent alkane sulfonatase (5, 13), which has been described for Klebsiella (7).
FIG. 6.
Growth of wild-type K. aerogenes (LD561) on different sulfur sources under aerobic and anaerobic conditions. The residual growth observed in media without added sulfur can be attributed to the use of internal sulfur stores and the scavenging of residual sulfate. The addition of methionine extends the period of stored- and scavenged-sulfur use under anaerobic growth conditions. After this point (arrow), methionine utilization results in slower growth than that under aerobic conditions.
To test for the presence of a second methionine-recycling pathway, we conducted a search for mutants that fail to degrade methionine as a sulfur source. A cymY::Tn10dCm strain (LD839), which grows on methionine as a sole sulfur source on solid media, was mutagenized with Tn10dTc and Tn10dLK as described above; mutants defective in methionine recycling were isolated as strains that grew on minimal medium when supplemented with cysteine but not with methionine. In this preliminary screen, 10 mutants were isolated and termed mtc (defective for conversion of methionine to cysteine); transductional analysis sorted these mutants into at least four different linkage groups. The mtc mutations did not affect methionine transport into the cell, as methionine corrected the auxotrophy of mtc met double mutants (e.g., LD842). However, all of the mtc mutants that were isolated were leaky and merely grew poorly on methionine as a sole source of sulfur. These data suggest that two pathways contribute to methionine recycling and that any one mutation cannot eliminate both routes; on their own, mtc mutants had no phenotype.
Klebsiella does not express a methionine-γ-lyase.
Methanethiol can be produced from methionine in two ways. First, Klebsiella is known to use an aci-reductone oxidase to produce 3-methylthiopropionate from an intermediate generated during the recycling of methylthioadenosine via methylthioribose (33, 34); methanethiol is formed upon 3-methylthiopropionate degradation. Alternatively, a methionine-γ-lyase may produce methanethiol and α-ketobutyrate directly. We assayed for methionine-γ-lyase activity and found no significant activity in cells grown with methionine as the sole sulfur source, or under any other growth condition. We conclude that either our strains of K. aerogenes do not encode a methionine-γ-lyase or we have failed to induce its production. Since Klebsiella is known to produce 3-methylthiopropionate from methylthioadenosine, we favor this route as the likely source of methanethiol in Klebsiella.
The ability to recycle methionine as the sulfur source was lost from the E. coli/Salmonella lineage.
To determine if strains of E. coli or S. enterica could recycle methionine into cysteine, we tested both standard laboratory strains of each species (K-12 and LT2, respectively), as well as a number of isolates from representative collections of natural isolates (the ECOR [28] and SARB [3] collections, respectively), for their ability to use methionine as the sole source of sulfur; representative data are shown in Fig. 7. No strain of E. coli or S. enterica, either a laboratory strain or a natural isolate, could degrade methionine as the sole source of sulfur; the initial increase in cell density on methionine-grown cells can be attributed to the utilization of cytoplasmic stores of sulfur, which are rapidly exhausted. Long-term growth experiments show that strains of E. coli and Salmonella never reach a higher cell density, so it is unlikely that a cryptic pathway has remained uninduced, as in Klebsiella cysB mutants (Fig. 4). In addition, no cystathionine-γ-lyase activity was detected in enzyme extracts of E. coli or Salmonella cells.
FIG. 7.
Growth of enteric bacteria on sulfur sources. (A) Klebsiella aerogenes LD561; (B) Klebsiella pneumoniae M5aL; (C) Escherichia vulneris LD126; (D) Enterobacter cloacae LD118; (E) Serratia marcescens LD137; (F) E. fergusonii LD130; (G) E. coli ECOR-1; (H) E. coli ECOR-16; (I) E. coli ECOR-47; (J) S. enterica SARB-3; (K) S. enterica SARB-9; (L) S. enterica SARB-19. The residual growth observed in media without added sulfur can be attributed to the use of internal sulfur stores and scavenging of residual sulfate. The addition of methionine extends the period of stored- and scavenged-sulfur use for strains of E. coli and S. enterica. OD600, optical density at 600 nm.
To determine the distribution of methionine utilization among enteric bacteria, strains of enteric bacteria were grown in liquid minimal medium with either sulfate, cysteine, or methionine as the sole sulfur source. E. coli and S. enterica appear to be atypical in not being able to degrade methionine as the sole source of sulfur, as virtually all other species of enteric bacteria have the capability of growing on methionine as the sole sulfur source in liquid media (representative data are shown in Fig. 7). Since E. coli, Escherichia. fergusonii, and S. enterica are sister species (21), the absence of methionine recycling in these taxa likely reflects its absence from their common ancestor (Fig. 8). Similarly, this capability of Klebsiella and other enteric bacteria can be attributed most parsimoniously to a pathway ancestral to the enteric bacteria which was lost in the ancestor of E. coli and Salmonella.
FIG. 8.
Distribution of methionine recycling among enteric bacteria. Open bars, organisms that fail to use methionine as a sulfur source; solid bars, organisms that use methionine as the sole sulfur in liquid media. Growth curves for representative strains are shown in Fig. 7. The dendrogram is based on sequences of the gapA, ompA, and trpR loci, as presented in reference 21. Despite their nomenclature, other strains of Escherichia are not necessarily closely related to E. coli (21) and were classified as Escherichia based on their ability to degrade lactose.
DISCUSSION
Assimilation of sulfur in Klebsiella is similar to the E. coli model.
Nutritional analysis of Klebsiella mutants shows that sulfur is assimilated into cysteine, as in E. coli and S. enterica (green pathway in Fig. 9), and not into homocysteine, as in Saccharomyces (Fig. 1). That is, all mutants failing to reduce sulfate are corrected by sulfide and cysteine and no mutants corrected only by methionine are not corrected by sulfide. These data, as well as the preliminary correspondence between the E. coli and Klebsiella genes for sulfur assimilation, justify our use of the E. coli models for sulfate reduction, cysteine biosynthesis, and methionine biosynthesis. Yet, in E. coli and S. enterica, methionine cannot serve as the sole sulfur source, and these organisms appear to contain a single transsulfurylation pathway, allowing conversion of cysteine to homocysteine (and then to methionine; blue pathway in Fig. 9) but not of homocysteine to cysteine. Therefore, a comprehensive model for sulfur-bearing amino acid metabolism in Klebsiella requires additional metabolic pathways not found in E. coli or Salmonella.
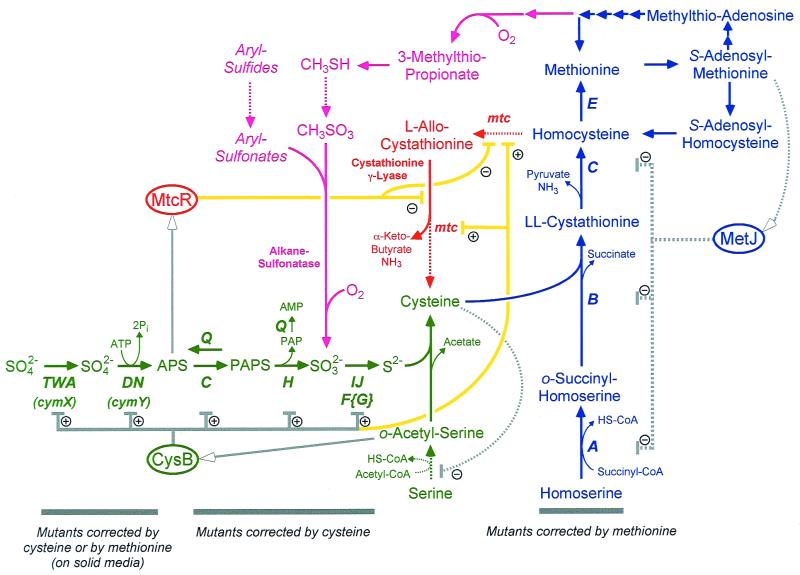
FIG. 9.
Model for methionine recycling pathways in K. aerogenes. The formal cysteine-biosynthetic pathway is shown in green, with genes identified in Klebsiella noted. Similarly, the methionine-biosynthetic pathway is shown in blue. The inferred pathway of homocysteine recycling via the transsulfurylation pathway is shown in red, and a possible pathway for methionine recycling via methanethiol is shown in magenta; both these pathways await rigorous biochemical confirmation. Regulatory interactions inferred from studies of E. coli and S. enterica are shown in gray, and regulatory interactions inferred from this work are shown in yellow. Ellipses, regulatory proteins. Arrows with open heads direct compounds serving as effectors to their cognate proteins. Dotted lines, proteins, steps, and interactions without experimental evidence for Klebsiella. SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; CoA, coenzyme A.
Evidence for the transsulfurylation pathway of methionine utilization in Klebsiella.
K. aerogenes, like yeasts (30) and some Archaea (35), appears to have both transsulfurylation pathways, allowing either cysteine or methionine to serve as the sole sulfur source (red pathway in Fig. 9). Evidence for this pathway comes from four sources. (i) Klebsiella can use methionine as the sole source of sulfur, although a cym mutation is required to allow its use on solid medium. (ii) A cysIJ cym double mutant still uses methionine as the sole source of sulfur, meaning sulfite is not a requisite intermediate. (iii) A cysB mutant utilizes methionine and homocysteine as sulfur sources during a period when sulfide, sulfite, and thiosulfate cannot be used. These data suggest that sulfide assimilation is not required for methionine utilization but are not conclusive. (iv) Cystathionine-γ-lyase activity has been detected in Klebsiella cells. This activity is induced by methionine and is repressed by cysteine.
Evidence for the methanethiol pathway.
The transsulfurylation pathway is not sufficient to explain all phenotypes associated with Klebsiella's growth on methionine as the sole source of sulfur. We propose that a second pathway is employed, entailing the utilization of 3-methylthiopropionate. This compound is produced by the enzyme aci-reductone oxidase, which has been described for Klebsiella (33, 34) and which acts on an intermediate in the recycling of methylthioadenosine, a compound produced during spermidine synthesis. The utilization of 3-methylthiopropionate would proceed via methanethiol (CH3SH) and methanesulfonate (CH3SO3) intermediates; methanesulfonate utilization has been reported for Klebsiella (7). Three pieces of evidence suggest, albeit only indirectly, that a second pathway allows for methionine utilization in Klebsiella. (i) Methionine utilization is partially impaired under anaerobic growth conditions (Fig. 6). While the transsulfurylation pathway has no requirement for molecular oxygen, the methanethiol pathway requires molecular oxygen both in the production of 3-methylthiopropionate and in the utilization of alkane sulfonates (magenta pathway in Fig. 9). (ii) Klebsiella mtc mutants, which have defects in methionine utilization, are uniformly leaky, suggesting that any one mutant cannot eliminate methionine recycling activity entirely. (ii) Klebsiella cysB mutants grown on methionine show two distinct growth phases (Fig. 4), consistent with immediate deployment of an ineffective pathway for methionine recycling and the late deployment of a more effective means. These data are clearly speculative, and confirmation that two pathways operate for methionine recycling in Klebsiella and the nature of these pathways await the further characterization of mtc mutants.
Regulation of methionine utilization involves the CysB protein.
CysB acts as a positive activator to allow transcription of genes involved in sulfate assimilation in enteric bacteria (11, 12). This is accomplished by sensing high levels of o-acetylserine (via the isomer N-acetylserine), the form of activated serine into which sulfide is assimilated (green pathway in Fig. 9). Two lines of evidence suggest that CysB is involved, either directly or indirectly, in activating the transsulfurylation pathway of methionine utilization. First, activity of cystathionine-γ-lyase is repressed by the presence of cysteine in the media. Cysteine inhibits the activity of the CysE serine transacetylase (19, 20), thereby decreasing the production of N-acetylserine and preventing transcription activation by CysB. Second, cysB mutants fail to use methionine as a sulfur source in liquid media until after a very long lag (Fig. 4), whereas wild-type cells use it immediately (compare Fig. 3A with 3I and 4). Since defects in sulfur reduction and assimilation (also affected by a cysB mutation) are not required by the transsulfurylation pathway (although they are required for the methanethiol pathway), these data suggest that CysB activity is required for the transsulfurylation pathway and that the repression of cystathionine-γ-lyase activity is the result of CysB-mediated transcriptional control (yellow interactions in Fig. 9). This regulation is not unexpected if sulfate or cysteine is preferred as a sulfur source over methionine. Activation of methionine recycling would require both the absence of sulfate (lowering APS levels; see below) and the absence of cysteine.
APS as a signal molecule and the origin of the cym phenotype.
Although a requirement for CysB activation may prevent methionine utilization when cysteine is abundant, CysB does not respond directly to the concentration of sulfate in the environment. Naturally, CysB activates transcription of the sulfur assimilation genes when cysteine concentration is low, independent of the concentration of sulfate. Therefore, sulfate concentrations must be sensed if methionine, or any other alternative sulfur source, is to be used by Klebsiella when sulfate concentrations are low and sulfate cannot be assimilated efficiently.
Klebsiella likely senses sulfate levels via the activated form, APS. APS is formed by the CysDN ATP sulfurylase after sulfate transport; this compound serves not only to capture sulfate and prevent its exit from the cell but also to reduce the midpoint potential for sulfate reduction. We hypothesize that the levels of APS are sensed by a novel regulatory protein, termed MtcR (Fig. 9). If APS levels are high, then sulfate is present in sufficient concentrations and the methionine recycling pathway is repressed. However, if APS levels are low, this pathway would be derepressed and methionine utilization could occur. Since the CysB protein would be required, expression of this pathway would require low levels of cysteine as well. Since PAPS is toxic at a high concentration and since its concentration in the cell is regulated by the preemptive phosphatase CysQ, the APS concentration is more likely to be an accurate reporter of available sulfate pools.
This hypothesis is supported by the nature of cym mutations, which affect sulfate transport (cymX lesions map to the cysPTWA operon, encoding the sulfate transport system) and the formation of APS (cymY mutations affect the CysDN ATP sulfurylase). In these cells, APS concentrations would be low, thereby allowing the proposed regulatory protein to activate the methionine recycling pathways. These functions also explain selenate resistance in cym mutants, since selenate is transported into the cell by the CysPTWA system (23) and is activated by CysDN. However, cysC, cysH, and cysIJ mutants would have higher concentrations of APS, which would continue to effect repression of methionine assimilation pathways and lead to strict cysteine auxotrophy (at least on solid medium; see below).
Failure to use methionine on solid media.
Although both wild-type Klebsiella and cys mutants use methionine as a sulfur source in liquid media quite effectively (Fig. 3), they cannot use methionine as a sulfur source on solid medium. We don't believe that agar possesses a compound that inhibits this activity since (i) this inhibition is also seen on agarose-, Phytagel-, and Gelrite-based media and (ii) agar does not inhibit methionine utilization when the bacteria are grown embedded within an agar matrix (data not shown). Moreover, the high oxygen tension experienced on plates does not prevent methionine utilization, as plates incubated anaerobically show the same effect. In fact, oxygen assists in methionine utilization (both in liquid for all cells and on plates for cym mutants), likely by allowing production of 3-methylthiopropionate and subsequent employment of the methanethiol pathway (magenta pathway in Fig. 9).
At least one factor preventing methionine utilization on solid media appears to be the accumulation of sulfate-derived APS, a compound which is absent in both cymX and cymY mutants (green pathway in Fig. 9). Sulfate is present in low concentrations as a contaminant even in “sulfate-free” solid media, which may prevent methionine utilization of some cys mutants. However, this cannot be the sole regulatory factor, as methionine is used quite handily by cys mutants in liquid media, even in the presence of 1 mM sulfate. Therefore, an additional regulatory input, possibly detecting growth on a surface, likely mediates methionine utilization depending on the nature of the environment.
Why two pathways?
While both the transsulfurylation pathway and the methanethiol pathway serve to recycle methionine into cysteine under laboratory conditions, they likely serve different purposes in natural environments. The repression of cystathionine-γ-lyase by CysB implies that sulfate is a preferred sulfur source and that other compounds are used only under sulfate-limiting conditions. A similar behavior is seen in the regulation of alkane sulfatase activity in Klebsiella (24, 25). Methionine can be recovered from proteins, and we envision that it is recycled primarily through the transsulfurylation pathway, which is specific for the transfer of sulfhydryl groups from homocysteine to serine to form cysteine.
In contrast, the methanethiol pathway appears to be more general in scope, passing through methanethiol and methanesulfonate intermediates. Methanesulfonate is likely degraded by an alkane sulfonatase, which has been described for Klebsiella (7); many compounds may serve as substrates for sulfite production by this enzyme. While we can only speculate on the conversion of methanethiol to methanesulfonate, it is possible that other alkane sulfides may serve as sulfur sources as well. In this way, the methanethiol pathway may represent a broadly acting sulfur-scavenging system which is induced when cysteine concentration is low.
ACKNOWLEDGMENTS
We thank Robert Bender and Valley Stewart for strains, Amy Wieczenski for technical assistance in the isolation of metE mutations, and David Wolfe for assistance with isolation of mutations in the rha and trp genes.
This work was supported by grants from the Alfred P. Sloan Foundation and the David and Lucille Packard Foundation.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bender R A. Variation on a theme by Escherichia. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 4–9. [Google Scholar]
- 3.Boyd E F, Wang F-S, Baltran P, Plock S A, Nelson K, Selander R K. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J Gen Microbiol. 1993;139:1125–1132. doi: 10.1099/00221287-139-6-1125. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Cook A M, Laue H, Junker F. Microbial desulfonation. FEMS Microbiol Rev. 1998;22:399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 6.de Vries G E, Raymond C K, Ludwig R A. Extension of bacteriophage λ host range: selection, cloning, and characterization of a constitutive λ receptor gene. Proc Natl Acad Sci USA. 1984;81:6080–6084. doi: 10.1073/pnas.81.19.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley M W, Frost J W. Biocatalytic desulfurization of arylsulfonates. Bioorg Med Chem. 1994;2:681–690. doi: 10.1016/0968-0896(94)85018-6. [DOI] [PubMed] [Google Scholar]
- 8.Esaki N, Soda K. Methionine-γ-lyase from Pseudomonas putida and Aeromonas. Methods Enzymol. 1987;143:459–465. doi: 10.1016/0076-6879(87)43081-4. [DOI] [PubMed] [Google Scholar]
- 9.Greene R C. Biosynthesis of methionine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 542–560. [Google Scholar]
- 10.Jones-Mortimer M C. Positive control of sulfate reduction in Escherichia coli: isolation, characterization and mapping of cysteineless mutants of E. coli K12. Biochem J. 1968;110:589–595. doi: 10.1042/bj1100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones-Mortimer M C. Positive control of sulfate reduction in Escherichia coli: the nature of the pleiotropic cysteineless mutants of E. coli K12. Biochem J. 1968;110:597–602. doi: 10.1042/bj1100597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones-Mortimer M C, Wheldrake J R, Pasternak C A. The control of sulfate reduction in Escherichia coli by o-acetyl-serine. Biochem J. 1968;107:51–53. doi: 10.1042/bj1070051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kertesz M A. Riding the sulfur cycle: metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol Rev. 2000;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 14.Kingsman A J, Smith D A. The nature of genetic instability in auxotrophs of Salmonella typhimurium requiring cysteine or methionine and resistant to inhibition by 1,2,4-triazole. Genetics. 1978;89:439–451. doi: 10.1093/genetics/89.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingsman A J, Smith D A, Hulanicka M D. Genetic instability in auxotrophs of Salmonella typhimurium requiring cysteine or methionine and resistant to inhibition by 1,2,4-triazole. Genetics. 1978;89:419–437. doi: 10.1093/genetics/89.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 17.Kolko M M, Kapetanovich L A, Lawrence J G. Alternative pathways for siroheme synthesis in Klebsiella aerogenes. J Bacteriol. 2001;183:328–335. doi: 10.1128/JB.183.1.328-335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kredich N. Biosynthesis of cysteine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 514–527. [Google Scholar]
- 19.Kredich N M, Becker M A, Tomkins G M. Purification and characterization of cysteine synthase, a bifunctional protein complex, from Salmonella typhimurium. J Biol Chem. 1969;244:2428–2439. [PubMed] [Google Scholar]
- 20.Kredich N M, Tomkins G M. The enzymatic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966;241:4955–4965. [PubMed] [Google Scholar]
- 21.Lawrence J G, Ochman H, Hartl D L. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991;137:1911–1921. doi: 10.1099/00221287-137-8-1911. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence J G, Roth J R. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics. 1996;142:11–24. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindblow-Kull C, Kull F J, Shrift A. Single transporter for sulfate, selenate, and selenite in Escherichia coli K-12. J Bacteriol. 1985;163:1267–1269. doi: 10.1128/jb.163.3.1267-1269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murooka Y, Adachi T, Okamura H, Harada T. Genetic control of arylsulfatase synthesis in Klebsiella aerogenes. J Bacteriol. 1977;130:74–81. doi: 10.1128/jb.130.1.74-81.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murooka Y, Ishibashi K, Yasumoto M, Sasaki M, Sugino H, Azakami H, Yamashita M. A sulfur- and tyramine-regulated Klebsiella aerogenes operon containing the arylsulfatase (atsA) gene and the atsB gene. J Bacteriol. 1990;172:2131–2140. doi: 10.1128/jb.172.4.2131-2140.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagasawa T, Kanzaki H, Yamada H. Cystathionine-γ-lyase from Streptomyces phaeochromogenes. Methods Enzymol. 1987;143:486–493. doi: 10.1016/0076-6879(87)43087-5. [DOI] [PubMed] [Google Scholar]
- 27.Neuwald A F, Krishnan B R, Brikun I, Kulakauskas S, Suziedelis K, Tomcsanyi T, Leyh T S, Berg D E. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J Bacteriol. 1992;174:415–425. doi: 10.1128/jb.174.2.415-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qureshi M A, Smith D A, Kingsman A J. Mutants of Salmonella typhimurium responding to cysteine or methionine: their nature and possible role in the regulation of cysteine biosynthesis. J Gen Microbiol. 1975;89:353–370. doi: 10.1099/00221287-89-2-353. [DOI] [PubMed] [Google Scholar]
- 30.Thomas D, Surdin-Kerjan Y. Metabolism of sulphur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermeij P, Kertesz M A. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J Bacteriol. 1999;181:5833–5837. doi: 10.1128/jb.181.18.5833-5837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 33.Wray J W, Abeles R H. A bacterial enzyme that catalyzes formation of carbon monoxide. J Biol Chem. 1993;268:21466–21469. [PubMed] [Google Scholar]
- 34.Wray J W, Abeles R H. The methionine salvage pathway in Klebsiella pneumoniae and rat liver. J Biol Chem. 1995;270:3147–3153. doi: 10.1074/jbc.270.7.3147. [DOI] [PubMed] [Google Scholar]
- 35.Zhou D, White R H. Transsulfuration in archaebacteria. J Bacteriol. 1991;173:3250–3251. doi: 10.1128/jb.173.10.3250-3251.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]