Abstract
The outer membrane protein OprM of Pseudomonas aeruginosa is involved in intrinsic and mutational multiple-antibiotic resistance as part of two resistance-nodulation-division efflux systems. The crystal structure of TolC, a homologous protein in Escherichia coli, was recently published (V. Koronakis, A. Sharff, E. Koronakis, B. Luisl, and C. Hughes, Nature 405:914–919, 2000), demonstrating a distinctive architecture comprising outer membrane β-barrel and periplasmic helical-barrel structures, which assemble differently from the common β-barrel-only conformation of porins. Based on their sequence similarity, a similar content of α-helical and β-sheet structure determined by circular dichroism spectroscopy, and our observation that OprM, like TolC, reconstitutes channels in planar bilayer membranes, OprM and TolC were considered to be structurally homologous, and a model of OprM was constructed by threading its sequence to the TolC crystal structure. Residues thought to be important for the TolC structure were conserved in space in this OprM model. Analyses of deletion mutants and previously isolated insertion mutants of OprM in the context of this model allowed us to propose roles for different protein domains. Our data indicate that the helical barrel of the protein is critical for both the function and the integrity of the protein, while a C-terminal domain localized around the equatorial plane of this helical barrel is dispensable. Extracellular loops appear to play a lesser role in substrate specificity for this efflux protein compared to classical porins, and there appears to be a correlation between the change in antimicrobial activity for OprM mutants and the pore size. Our model and channel formation studies support the “iris” mechanism of action for TolC and permit us now to form more focused hypotheses about the functional domains of OprM and its related family of efflux proteins.
Pseudomonas aeruginosa demonstrates high intrinsic resistance to multiple classes of antibiotics, due primarily to a combination of low outer membrane permeability coupled to secondary resistance mechanisms such as an inducible β-lactamase and resistance-nodulation-division (RND) efflux systems (10, 20). The outer membrane protein OprM is involved in two efflux systems that mediate intrinsic antibiotic resistance: the MexA-MexB-OprM system, which is constitutively produced in wild-type strains at a low level and contributes to the intrinsic resistance of the organism to a wide spectrum of structurally unrelated antimicrobial agents and the inducible (by antibiotics) MexX-MexY-OprM system (1), which is involved in intrinsic resistance to aminoglycosides and erythromycin. When these systems are overexpressed, resistance levels become highly increased (1, 15, 18). The pump proteins MexB and MexY are located in the cytoplasmic membrane, whereas the so-called “membrane fusion” proteins MexA and MexX are anchored to the inner membrane but largely located in the periplasm. It was previously assumed that the membrane fusion proteins bridge the periplasm to channel pump substrates toward the outer membrane components, which were assumed to form channels like the porins of many gram-negative bacteria.
The most similar homolog of OprM in Escherichia coli is outer membrane protein TolC, which is also involved in multiple-antimicrobial resistance through an energy-dependent efflux mechanism. It was shown to function with AcrA and AcrB (7) to extrude a wide spectrum of antimicrobial agents. The amino acid sequences of these two proteins are 21% identical and 40% similar, and both proteins were shown to form oligomers (5, 13, 17) and appear to be able to interact with different components from other efflux systems to form functional chimeric complexes (19, 24, 29). Channel activities have also been observed for both proteins (5, 27). Therefore, the two proteins, as well as their numerous homologs in gram-negative bacteria, are likely to share a similar structure. As the outer membrane proteins of the RND efflux systems appeared to form trimers and channels like porins, a structure was predicted for OprM (27) that was similar to that identified for porins, whereby each monomer of the trimer forms a β-barrel. The recent release of the crystal structure for TolC, however, provided a very different vision. It has a distinctive, and previously unknown, structure for any cell membrane protein, a structure comprising three monomers making up one long continuous channel spanning both the outer membrane and the periplasm, where each monomer supplies strands required for channel formation. The outer membrane is traversed by a 12-stranded β-barrel (each monomer supplying 4 strands to this barrel), and this domain sits atop a 12-stranded coiled α-helical channel that spans the periplasm (again, 4 helices are supplied per monomer 14;]. This architecture helps to explain the efflux mechanism of RND systems, in that the cytoplasmic membrane pump can contact this continuous channel, with the “membrane fusion” protein presumably assisting in the interaction of these other two components. This then makes the extrusion of antimicrobial agents to the surroundings more direct and efficient, bypassing of the periplasmic space. Although the specificity of the system has been proposed to reside in the pump component, it was proposed (14) that the helical bundle of TolC that spans the periplasm can open and close like a diaphragm on a camera lens. Since OprM and TolC are similar in sequence and function, we revised here the OprM topology model by threading its sequence to the TolC crystal structure. The creation of defined deletion mutants and data from previously isolated insertion mutants were used to form hypotheses about the functional domains in this model.
MATERIALS AND METHODS
Bacterial strains and plasmids.
For expression experiments and antimicrobial susceptibility assays with P. aeruginosa, the OprM-deficient strain OCR03T (9), a gift from Thilo Köhler (Centre Médical Universitaire, Geneva, Switzerland), was utilized. The oprM gene carried on plasmid pT7-7 (26) (pXZL33/pT7-7::oprM kindly provided by Keith Poole, Queens University, Kingston, Ontario, Canada) was used to create the deletion and insertion mutants, and E. coli strain DH5α was used as the host for transformation of these constructs. The oprM gene and its various mutants were then cloned into plasmid pVLT35 or its derivative pVLT31 (16), kindly provided by Victor de Lorenzo (Centro Nacional de Biotecnología CSIC, Madrid, Spain) for the expression and antimicrobial susceptibility studies.
Media.
Bacterial strains were routinely grown with shaking at 37°C in Luria broth (LB) medium (1% tryptone and 0.5% yeast extract with either 0.5% NaCl for E. coli or 0.05% NaCl for P. aeruginosa strains) or on LB agar with the addition of 2% (wt/vol) Bacto agar. The following antimicrobials were used in selective media: HgCl2 (15 μg/ml), for OprM-deficient P. aeruginosa strain OCR03T; ampicillin (100 μg/ml), spectinomycin (30 μg/ml), and tetracycline (10 μg/ml) for E. coli with constructs made from pT7-7, pVLT35, and pVLT31, respectively; streptomycin (45 μg/ml) and tetracycline (50 μg/ml) for OCR03T with constructs made from pVLT35 or pVLT31, respectively. For expression of oprM and its insertion or deletion mutants, isopropyl-β-d-thiogalactoside (IPTG) was added to mid-log-phase cell cultures to a final concentration of 0.1 mM, and the cells were induced for 2 h before harvest.
DNA methodology.
Restriction endonucleases and T4 DNA ligase purchased from Gibco-BRL and New England Biolabs, Inc., were used in accordance with the protocols supplied by the manufacturers. Plasmid DNA was prepared by the alkaline lysis protocol as described previously (22). Transformations of E. coli and P. aeruginosa with plasmid DNA or ligation products were performed by the CaCl2 and MgCl2 protocols, respectively (22).
Insertion mutagenesis.
Insertion mutagenesis of oprM with malarial epitopes was accomplished as described previously (27).
Deletion mutagenesis.
PCR was used for defined-deletion mutagenesis of oprM. Primers, which were 40 nucleotides long with 20 nucleotides on each side of the 4 or 8 amino acids to be deleted, were designed and then synthesized on an ABI DNA-RNA synthesizer. For direct extension, a deletion-containing primer with a convenient restriction endonuclease site at its 5′ end was used to amplify a part of oprM from pXZL33 (pT7-7::oprM) with a primer annealing to a region of oprM upstream or downstream from the deletion site and also containing a unique restriction endonuclease site. For overlap extension, both the forward and the reverse complementary deletion-containing primers were synthesized. Each of these was used to amplify a part of oprM with a restriction endonuclease site-containing primer further upstream or downstream from the deletion site. The two overlapping products from these two PCRs were purified and subjected to a second PCR containing only the two external primers. All of the final PCR products were purified with a QIAquick kit (Qiagen), digested with the appropriate restriction endonucleases, and ligated to pXZL33 cut with the same enzymes for transformation into E. coli DH5α. After confirmation of the deletions by sequencing, the mutant oprM genes were cloned into pVLT35 and transformed into P. aeruginosa OCR03T.
Outer membrane preparation, SDS-PAGE, and Western immunoblotting.
Outer membranes were isolated by passage through a French press and sucrose density gradient centrifugation or Sarkosyl solubilization, and samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (11). Proteins were stained with Coomassie brilliant blue. Protein concentrations of the isolated outer membrane samples were determined by a modified Lowry assay (23).
For Western immunoblotting, proteins from unstained gels were transferred to Immobilon polyvinylidene difluoride membranes (Millipore, Bedford, Mass.) in cold transfer buffer (25 mM Tris, 0.2 M glycine, 20% [vol/vol] methanol) at 100 V for 1 h. Proteins were then detected as previously described (27), using an OprM-specific murine monoclonal antibody (kindly provided by Naomasa Gotoh, Kyoto Pharmaceutical University, Yamashina, Japan) as the primary antibody and a goat anti-mouse immunoglobulin G alkaline phosphatase-conjugated secondary antibody. The bound antibodies were detected with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium.
DNA sequencing.
Primers annealing to different regions of oprM were synthesized on an ABI DNA synthesizer. Template DNA was prepared with QIAprep spin miniprep kit (Qiagen), and PCR reactions were done according to the protocols provided by Applied Biosystems, Inc. DNA sequencing was performed on an ABI model 373 sequencer using the manufacturer's protocols.
MIC determinations.
MICs were determined by serial twofold dilution in LB medium using the microdilution method described by Amsterdam (2). Results were determined after incubation at 37°C for 24 to 48 h.
Solubilization and purification of protein.
OprM and its mutant forms were isolated from P. aeruginosa OCR03T. Outer membrane proteins isolated from sucrose density gradients were solubilized subsequently in 0.5 and 3% (vol/vol) n-octyl-polyoxyethylene (Octyl-POE; Bachem Bioscience, Inc.) in 10 mM Tris (pH 8). This fraction was dialyzed into buffer A (10 mM Tris, pH 8; 1% [vol/vol] Octyl-POE) and then passed through an anion-exchange MonoQ column (Pharmacia) and eluted with buffer B (buffer A with 1 M NaCl) using fast-protein liquid chromatography. A fraction containing OprM was subjected to SDS-PAGE without heating the sample, and OprM was excised from the gel and eluted in 10 mM Tris (pH 8) with 0.1% SDS at 4°C overnight.
Planar lipid bilayer experiments.
Analysis of the pore-forming ability of proteins was done with the planar lipid bilayer technique as previously described (4). Membranes were composed of 1.0% (wt/vol) diphytanoyl phosphatidylcholine in n-decane. Purified protein was diluted into 0.1% (vol/vol) Triton X-100 prior to addition to various salt solutions bathing the planar bilayer membrane.
CD analysis.
The circular dichroism (CD) spectrum of OprM was obtained from a J810 spectropolarimeter (Jasco, Tokyo, Japan) using a quartz cell with a 1-mm path length. CD spectra were measured at 25°C between 190 and 250 nm at a scanning speed of 10 nm/min in 10 mM Tris buffer (pH 8.0) with 0.1% (wt/vol) SDS. After subtracting the spectrum from background generated from buffer alone, the spectrum for OprM was deconvoluted with the K2D program (3) to determine the percentage of β-pleated sheet and the α-helical structure.
Three-dimensional modeling.
The OprM and TolC protein sequences, without their signal sequences, were aligned using CLUSTALX (http://www.hgmp.mrc.ac.uk/Registered Option/clustalx.html), and then the alignment was manually edited. Using the alignment, the OprM sequence was threaded to the crystal structure of TolC (14) using the Insight II (version 97.2) molecular modeling program Homology (Molecular Simulations, Inc., San Diego, Calif.) by constraining regions that aligned with the α-helical regions or β-strands of TolC and allowing more freedom in the loop regions. The entire structure was then subjected to energy minimization using the Insight II Discover program.
RESULTS
Secondary structure of OprM.
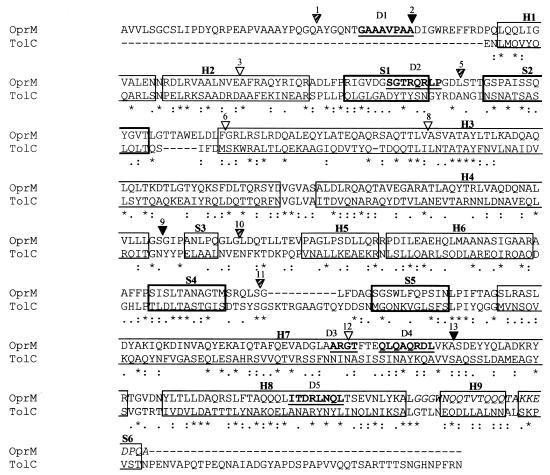
To assist in the modeling of the OprM structure, we first aligned the sequence with that of TolC (Fig. 1). The elements of secondary structure from the TolC crystal structure are indicated as boxes. Most of the similar or identical residues were observed to be within these structural elements. There was a significant sequence gap at the position of a TolC extracellular loop (between S4 and S5), and another lay between β-strand S2 and helical strand H3, while variable extensions were observed at the termini. Interestingly, the 43 residues at the C terminus of TolC were shown to be dispensable for its function (14), as were the last 22 amino acids at the C terminus of OprM (28), and the TolC crystal structure was actually obtained without these 43 amino acids. Overall, these sequences share 40% similarity, which is adequate for modeling purposes.
FIG. 1.
Alignment of the sequences of TolC and OprM. Identical residues (∗) and similar residues (“:” and “.”) are noted, using the similarity defaults of CLUSTALX. Elements of secondary structure from the TolC crystal structure are boxed, H1 to H9 are helices, and S1 to S6 are β-strands where S1, S2, S4, and S5 (thicker borders) are within the β-barrel. The helical barrel is comprised of two long helices (H3 and H7) and two pairs of shorter helices stacking to form pseudocontinuous helices (H2+H4; H6+H8) from each monomer. The equatorial domain is made up of strands S3 and S6 and the three short helices H1, H5, and H9. Sites of malarial epitope insertions into OprM are indicated by solid triangles (fully tolerated), hatched triangles (partially tolerated), and open triangles (nontolerated). Deletions D1 to D5 in OprM are underlined and in boldface. Residues at the C terminus of OprM* that are different from those in wild-type OprM, contained within H9 and S6, are italicized.
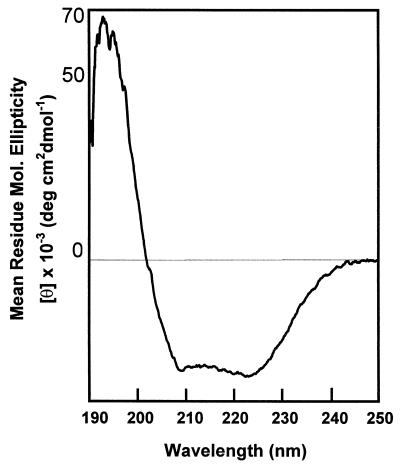
To obtain further evidence that the OprM structure is similar to that of TolC, the purified protein was subjected to CD analysis (Fig. 2). An estimate of secondary structure was obtained by the K2D method (3), yielding estimates of 19% β-sheet and 32% α-helical structure. These data were more comparable to the values of 14% β-sheet and 56% α-helix, determined for the TolC crystal structure, than the high proportions (>50%) of β-sheet observed for several outer membrane proteins, including porins.
FIG. 2.
CD spectral analysis of wild-type OprM in 0.1% SDS.
Three-dimensional model of OprM.
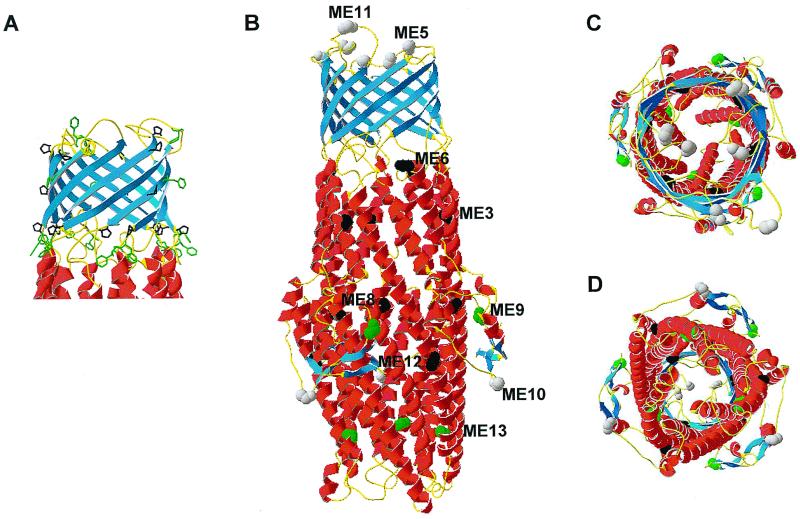
Using the molecular modeling program Homology of the Insight II (version 97.2) program (Molecular Simulations), the OprM amino acid sequence was threaded to the TolC crystal structure based on the alignment shown in Fig. 1. The first 52 residues at the N terminus of OprM were excluded, since there are no corresponding residues in the TolC structure. After the entire structure was subjected to energy minimization using the Insight II Discover program, a model of the architecture of the OprM trimer was generated as shown in Fig. 3. In this model, the OprM channel consists of three monomers, each contributing four β-strands to the β-barrel and four α-helical strands to the helical barrel. Notably, a ring of aromatic residues was evident along the base of the β-barrel at the proposed interface between the lipid bilayer and periplasm (Fig. 3A). This feature was consistent with both the structure of TolC and all outer membrane proteins structures characterized to date, thus supporting the validity of the proposed model. Proline residues also form a ring at the base of the β-barrel (Fig. 3A). As for TolC, these prolines are probably important for disrupting secondary structure for the transition from right-handed β-barrel into left-handed α-helices. Top and bottom views of the structure are shown in Fig. 3C and D. The model is available as a Protein Data Bank file from us, and other images of the model may be seen at www.cmdr.ubc.ca/bobh/oprmmodel.html.
FIG. 3.
Three-dimensional model of OprM, constructed by threading the sequence of OprM on a crystal structure of TolC. (A) β-Barrel domain of the protein, highlighting the proline (black) and phenylalanine (green) residues that predominantly form rings at the base of this barrel. (B) Overview of the molecule, highlighting the amino acids at which the malarial epitopes were inserted (green balls, tolerated; gray, partially tolerated; black, nontolerated). One copy of each insertion in the trimer is labeled for clarity. The insertion sites are highlighted in the same way in the top view (C) and bottom view (D) of the model.
Insertion mutagenesis of oprM.
A number of insertion mutants had been created in oprM previously (27) (Fig. 1, Table 1). The sites of insertions at ME3, ME5, ME6, and ME8 to ME13 are indicated in both the alignment (Fig. 1) and the three-dimensional model (Fig. 3B to D). Malarial epitopes insertion sites at ME1 and ME2 are not shown in the three-dimensional model because they were within the first 52 amino acids excluded from the threading. However, according to this model, malarial epitopes inserted at these two sites would probably be located within an N-terminal extension in the periplasm, and this might explain why they were tolerated. As shown in Fig. 3B to D, all of the insertions that prevented expression of the protein (black, at sites ME3, ME6, ME8, and ME12) were located within the conserved helical structure. In contrast, the insertion at ME13 was fully tolerated. Perhaps insertion of 13 residues at ME13 might be permissive owing to its location close to the more flexible end of helix H7. Another fully tolerated insertion, at ME9, together with a partially tolerated insertion at ME10, were located within the equatorial domain of the α-helical tunnel, suggesting that this domain is more amenable to disruption. Two partially tolerated insertions (at ME5 and ME11, respectively) were each located within the proposed two external loops of the β-barrel in our three-dimensional model. This indicates that these loops are also amenable to some disruption.
TABLE 1.
Antimicrobial susceptibilities of P. aeruginosa OprM-deficient strain OCR03T carrying plasmids expressing wild-type or mutant forms of oprM
| Plasmida (mutant) | Insertion (::) or deletion (Δ) mutation | Protein expressionb | MICc (μg/ml) of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tet | Cml | Nal | Nfx | Mer | Ctx | Cfp | Cfs | Car | |||
| pVLT35 | Vector | − | 0.1 | 0.2 | 1.6 | <0.05 | 0.03 | 0.25 | <0.03 | 0.1 | <0.2 |
| pKW35TM | OprM | +++ | 3.1 | 6.2 | 25 | 0.2 | 0.2 | 2.0 | 0.13 | 0.8 | 25 |
| pKWIN1 (ME1) | ::23 | ++ | 1.6 | 3.1 | 12.5 | 0.4 | 0.1 | 1.0 | 0.06 | 0.2 | 50 |
| pKWIN2 (ME2) | ::37 | +++ | 3.1 | 6.2 | 25 | 0.2 | 0.2 | 2.0 | 0.13 | 0.8 | 25 |
| pKWIN5 (ME5) | ::103 | +++ | 1.6 | 3.1 | 12.5 | 0.1 | 0.1 | 2.0 | 0.13 | 0.2 | 12.5 |
| pKWIN9 (ME9) | ::241 | ++ | 3.1 | 6.2 | 25 | 0.2 | 0.2 | 2.0 | 0.13 | 0.8 | 25 |
| pKWIN10 (ME10) | ::251 | ++ | 0.8 | 1.6 | 6.2 | <0.05 | 0.02 | 0.5 | 0.06 | 0.2 | 6.2 |
| pKWIN11 (ME11) | ::315 | + | 0.4 | 0.4 | 3.1 | <0.05 | 0.02 | 0.25 | <0.03 | 0.1 | 1.6 |
| pKWIN13 (ME13) | ::393 | ++ | 3.1 | 6.2 | 25 | 0.2 | 0.2 | 2.0 | 0.13 | 0.8 | 25 |
| pKWD1 (D1) | ΔG29–A36 | ++ | 1.6 | 6.2 | 25 | 0.2 | 0.2 | 2.0 | 0.13 | 0.8 | 25 |
| pKWD2 (D2) | ΔS93–P100 | ++ | 3.1 | 6.2 | 25 | 0.2 | 0.2 | 2.0 | 0.06 | 0.4 | 25 |
| pKWD3 (D3) | ΔA375–T378 | ++ | 0.3 | 0.8 | 3.1 | 0.1 | 0.05 | 0.25 | 0.1 | 0.4 | |
| pKWD4 (D4) | ΔQ382–L389 | ++ | 0.2 | 0.2 | 3.1 | <0.05 | 0.03 | 0.25 | <0.03 | 0.1 | 0.4 |
| pKWD5 (D5) | ΔI429–L439 | ++ | 0.4 | 0.8 | 6.2 | <0.05 | 0.05 | 0.5 | <0.03 | 0.1 | 1.6 |
| pKW35 (OprM*) | ΔC terminus | +++ | 3.1 | 6.2 | 25 | 0.2 | 0.2 | 2.0 | 0.13 | 0.8 | 25 |
pVLT35 is the control vector, pKW35TM contains wild-type oprM cloned into pVLT35. The pKWIN plasmids carry permissive insertion mutants of oprM with the position of the insertion indicated, e.g., by ::23. The pKWD plasmids carry deletion mutants of oprM, and the positions of the deletions are indicated, e.g., by ΔI429–L439. Position 1 is the N-terminal amino acid of the mature OprM sequence after the putative lipoprotein cleavage site.
Expression levels ranged from undetectable (−) to strong (+++), and results were obtained from SDS-PAGE of outer membranes from P. aeruginosa OCR03T carrying the various plasmids.
All results for the insertion mutants are taken from reference (27) and are reproduced here for comparison. MICs for the other strains were obtained from three repeated experiments. Abbreviations: Tet, tetracycline; Cml, chloramphenicol; Nal, nalidixic acid; Nfx, norfloxacin; Mer, meropenem; Ctx, ceftriaxone; Cfp, cefepime; Cfs, cefsulodin; Car, carbenicillin. Results in boldface indicate a reduction of at least fourfold compared with cells carrying wild-type OprM.
Deletion mutagenesis of oprM.
A series of defined deletion mutants of OprM (Fig. 1) were created by PCR and then transferred into the P. aeruginosa OprM-deficient strain OCR03T for expression studies and antimicrobial susceptibility assays (Table 1). Expression of the OprM deletion mutants was only slightly less than that of wild-type OprM. These deletion mutants exhibited behavior similar to that of the wild-type OprM protein in that a portion of each protein ran as an oligomer on SDS-PAGE and the remainder ran as the monomeric form even without denaturing heat or β-mercaptoethanol treatment (data not shown). OCR03T cells expressing mutants with deletion D1 or D2 showed no significant difference in antimicrobial resistance compared to cells expressing wild-type oprM (Table 1). However, cells expressing the other three oprM mutants with deletions D3, D4, or D5, notably all located in the helical barrel, showed various resistance profiles. Cells expressing the oprM mutant with the deletion D4 had resistance levels similar to those from the cells carrying the control plasmid pVLT35. Cells expressing oprM mutants with deletions D3 and D5 did not increase resistance levels to some antimicrobial agents but partially restored the resistance levels to others, with deletion D3 having a relatively larger impact on the function of OprM. In particular, resistance to the tested β-lactams (meropenem, cefotaxime, cefepime, cefsulodin and, to a somewhat lesser extent, carbenicillin) was eliminated by deletions D3 and D5. Overall, the results with these deletion mutants agreed with those of the insertions. Deletion D3 overlapped insertion site ME12, and the two mutants were poorly tolerated, while deletion D1 overlapped with insertion site ME2 and these two mutants were both fully tolerated.
Channel-forming activities of the mutant proteins.
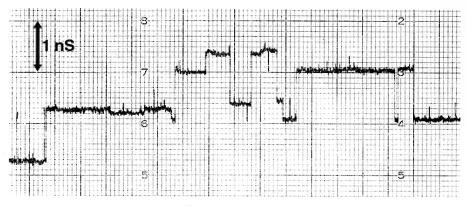
OprM was previously shown to have a single channel conductance of approximately 82 pS, a level very similar to that of TolC (80 pS) at 50 mV of applied voltage and with 1 M KCl (27). However, using several fresh samples of OprM, we observed here a mean single channel conductance in 1 M KCl of 0.85 nS (Fig. 4) at an applied voltage of 50 mV. We believe this discrepancy reflected the age of the sample since older samples gave rise to very noisy current tracings and the apparent channel size varied considerably. We were therefore very careful to use freshly prepared OprM preparations in this study and employed as a lipid the phospholipid, diphytanoyl phosphatidylcholine, rather than oxidized cholesterol, since the latter is very sensitive and can give rise to artifacts.
FIG. 4.
Step increases in single channel conductance after OprM was added to the aqueous phase (1 M KCl, pH 7) bathing the 1% (vol/vol) diphytanoyl phosphatidylcholine membrane dissolved in n-decane. The applied membrane voltage was 50 mV.
OprM molecules tended to form transient channels that were not very stable in the lipid membrane (Fig. 4). Examination of current tracings (Fig. 4) and histograms of the distributions of channel sizes indicated that OprM appeared to form substates in the bilayer membranes, as if the channel partly opened and closed. Studying the single channel conductance of OprM as a function of voltage indicated that OprM did not form voltage-dependent channels.
The single channel conductance of freshly eluted OprM in different salt solutions was measured. It appeared to be a linear function of the KCl concentration between 0.3 and 3.0 M, with mean single channel conductances ± standard errors of 0.20 ± 0.01, 0.85 ± 0.03, and 1.78 ± 0.09 nS in 0.3, 1, and 3 M KCl, respectively, indicating that the channel is water filled. Changing the bathing salt solution to 1 M LiCl (where Li+ is a highly hydrated bulky ion) resulted in a strong decrease in single channel conductance from 0.85 ± 0.03 to 0.23 ± 0.01 nS. In contrast, there was little change in the single channel conductance when the bulky anion acetate replaced the chloride ion (0.76 ± 0.02 nS instead of 0.85 ± 0.03 nS). These data suggested that OprM is cation selective, similar to TolC.
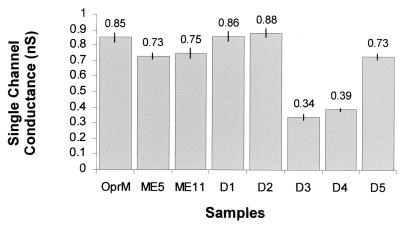
The five deletion mutants, together with two insertion mutants, were also purified from OCR03T for planar lipid bilayer analysis, and the results are shown in Fig. 5. The ME5 insertion mutant in one of the extracellular loops failed to restore resistance to some of the β-lactams but increased the MICs to all of the other antimicrobial agents. The ME11 insertion mutant in the other extracellular loop was expressed relatively poorly, and thus only partly reconstituted resistance levels to all tested antimicrobial agents were observed. The single channel conductances of both of these insertion mutants were similar and were only slightly smaller than that of wild-type OprM. Mutants with the fully tolerated deletion mutants D1 and D2 gave similar single channel conductances to that of wild-type OprM. Deletion mutant D5 had a single channel conductance that was only slightly smaller than wild-type OprM. In contrast, deletion mutants D3 and D4 had small single channel conductances in 1 M KCl that were half to a third of that of OprM.
FIG. 5.
Mean single channel conductances (from 101 to 145 events) of wild-type OprM and its mutant forms after the addition to the aqueous phase (1 M KCl, pH 7) bathing a membrane formed from 1% diphytanoyl phosphatidylcholine dissolved in n-decane. The bars indicate the standard errors.
DISCUSSION
P. aeruginosa OprM and E. coli TolC are outer membrane proteins involved in RND multidrug efflux systems and part of a large, phylogenetically related set of efflux and secretion proteins (21). Their primary sequences share 21% identity and 40% similarity. Functionally, both proteins reconstituted channel-forming activities in planar lipid bilayer experiments (5, 27), and both can form functional chimeric complexes with other linker and pump efflux components for the extrusion of a wide spectrum of antimicrobial agents, with the pump specificity being associated with the latter two components (19, 24, 29). Studies with two-dimensional crystals showed that the structure of TolC was trimeric, similar to that of the nonspecific, channel-forming porins (13). Hence, it was originally proposed that TolC and OprM had a structure similar to that of the porins, and a topology model of OprM (27) was originally predicted based on the classic 16-stranded β-barrel motif observed in the crystal structure of porins (6, 12). However, this was clearly inconsistent with the recently released crystal structure of TolC, which revealed a unique architecture (14), comprising a 12-stranded β-barrel spanning the outer membrane bilayer and a helical barrel spanning the periplasm, in which each member of the trimer contributed strands used to build the resulting barrel structure.
Consistent with the architecture of TolC, our CD spectroscopy data showed that OprM consists of a mixture of α-helical and β-barrel structure. We therefore remodeled OprM by threading its sequence to the published crystal structure of TolC. In this model, only the regions that aligned with TolC were included, and thus the N and C termini that differ in length between the two proteins were excluded. The N terminus of OprM is extended by more than 30 residues relative to TolC, a feature that exists in other members of the RND efflux outer membrane proteins with higher sequence similarity to OprM. Deletion of 8 of these N-terminal residues in mutant D1 or the insertion of 14 residues at ME2 had no effect on the ability of the protein to reconstitute a functional RND efflux system, while the insertion of 19 residues at ME1 was relatively benign, only slightly reducing protein expression and ability to reconstitute a functional efflux system. Thus, we feel that the exclusion of this portion of OprM from the model was justified. According to our proposed OprM model (Fig. 3), the N terminus would be in the periplasm, outside of the more constraining regions which shaped the TolC structure. Similarly, the C terminus of TolC is extended when aligned with OprM. Interestingly, the TolC crystal model was obtained with a mutant deleted for the 43 amino acids at the C terminus which were shown to be dispensable for the function of TolC (14). The C terminus of OprM has also been shown to be nonessential, and deletion and partial replacement of the last 22 residues (Fig. 1), removing a mixed α/β domain that was shown in the TolC structure to form part of an equatorial band around the helical barrel segment, did not affect the function of OprM (27). Therefore, the N and C termini of both TolC and OprM might be considered both structurally and functionally nonessential.
Analysis of our new model of OprM indicated that it had similar specific features to TolC, with the proposed OprM β-strands as amphipathic as those in TolC, with predominantly hydrophobic residues in the β-barrel facing toward the hydrophobic core of the bilayer. Also, a ring of aromatic residues (phenylalanine) at the lipid-water interface between the outer membrane β-barrel and the periplasmic α-helical barrel were clearly evident. Such rings of aromatic residues are found at predominantly at the periplasmic side of all outer membrane β-barrels examined to date (6, 12) and may act to stabilize the barrel. In addition, in our model, we found that proline residues form a notable ring between the β-barrel and the α-helical barrel, an idea consistent with the needed promotion of turns in this region to handle the transition from β-strands into α-helices.
Analyses of our OprM deletion mutants and previously obtained insertion mutants, in the context of this model, provided some insights into the functioning of proposed structures of OprM. Apparently, the external loops of the β-barrel (regions between S1 and S2 and between S4 and S5) and the mixed α/β structure at the equatorial domain of the helical barrel (region between H4 and H6) are flexible, since the insertion of 10 and 18 residues into these external loops (at ME5 and ME11, respectively) were almost fully or were partially tolerated and had only a modest effect on the single channel conductance of the protein. The insertion at ME5 decreased the resistance of the cells to only one of the tested β-lactams, the bulky anionic compound cefsulodin, whereas the ability of ME11 to reconstitute resistance was reduced resulting from its reduced expression. Presumably, the extension of the external loops in ME5 and ME11 might permit them to fold over and partially block the outer membrane channel, which would be consistent with the observed small decrease in channel size. The lack of an apparent significant role of these external loops in the function of OprM contrasts with that for other outer membrane protein porins, which are noted for constricting their β-barrel channels by a long external loop (usually loop 3) that folds into the channel (6, 12). No such external loop is observed in the OprM model or in TolC. However, the OprM surface-exposed loops still apparently contribute to some control of the passage of substrates, according to our functional analysis of the ME5 and ME11 loop mutants.
Two insertions (at ME9 and ME10) were within one region of mixed α/β structure (S3+H5) at the equatorial domain of the helical barrel. The insertion mutant ME9 was fully tolerated, while the insertion mutant ME10 was partially tolerated, with good levels of expression but only partial restoration of resistance to the various antimicrobial agents tested. Similarly, the deletion and/or replacement in the entire C-terminal region contributing to the other proposed mixed α/β structure (H9+S6, Fig. 1) at the equatorial region had no influence on expression or function. It was suggested that the equatorial domain is a possible recognition site for the recruitment of TolC by the inner membrane translocase or that it might be involved in stabilization of the TolC structure (14). Our results appear to indicate that at least part of this domain is unnecessary and that the other part has some structural versatility. The results with the insertion of ME10 could be explained if the portion of the protein (the flexible domain between S3 and H5) is involved in interaction between OprM and the linker protein MexB. The two large periplasmic loops predicted in the MexB topology model may be involved in this interaction (8).
All of the 17- to 19-amino-acid insertions that prevented expression of OprM (ME3 in H2, ME6 and ME8 in H3, and ME12 in H7) were located at sites within the α-helical barrel of the proposed OprM model. Notably, three new OprM deletion mutants that were positioned within the helical barrel were well expressed but either failed to restore resistance (deletion D4 removing 8 amino acids of H7) or partially restored resistance to a few antimicrobial agents (deletion D3 of 4 amino acids straddling insertion site ME12 of H7 and deletion D5 removing 8 amino acids of H8), when compared with cells expressing wild-type OprM. These data indicated that the α-helical barrel core was important for the proper expression and function of OprM. It has been hypothesized that the helical barrel in TolC is coiled in such a way as to form an iris diaphragm that controls substrate passage, by dilating under appropriate conditions to permit the extrusion of substrates (14). These conditions probably include the recruitment of TolC by the AcrA-AcrB linker-pump complex which has been activated by engagement with substrate, and possibly energy from the proton motive force. This interaction is probably cooperative, since chemical cross-linking has demonstrated that the linker protein AcrA of E. coli forms oligomers, probably trimers, as does TolC (30). The single channel conductance measured here for native OprM was 0.85 nS, i.e., larger than that of the E. coli porin OmpF monomer (0.6 nS, i.e., one-third of the trimer conductance (4), and we presume that this represents the open configuration of OprM. In contrast, TolC had a single channel conductance of only 0.08 nS and could be in the closed configuration. Our planar bilayer data also indicated that OprM underwent rapid switching from open to closed states and exhibited several substates (smaller channel sizes), whereas aged OprM stored at 4°C for a month demonstrated channels of 0.08 nS. These data are then consistent with a dynamic channel in which the permeability is controlled by the helical diaphragm. The large channel size of OprM (0.85 nS) is consistent with the established ability of this protein to efflux a wide spectrum of antimicrobial agents, some of which are very bulky. Furthermore, the OprM channel appears to be cation selective and might be more efficient for the efflux of substrates with an overall positive charge.
One completely tolerated deletion D2 removed 8 amino acids, some of which formed the upper half of β-strand S1. The upper portions of β-barrels close to the cell surface are usually more flexible than the rest of the barrel. This is probably why this particular deletion was tolerated. In addition, this deletion fortuitously permitted a continuity of amphipathic sequence, probably pulling part of the following external loop into the β-barrel structure and shortening the length of the loop over the pore.
Results from the planar lipid bilayer experiments of the deletion mutants and the two mutants with insertions at the external loops agreed with results from the antimicrobial susceptibility assays. The mutant proteins containing changes that were well tolerated and did not affect the resistance of the cells showed no or only slight changes in the single channel conductance compared to the wild-type OprM. On the other hand, the mutants that led to decreased resistance of the cells to antimicrobial agents also showed a large decrease in single channel conductance. This indicates that the permeability of the channel is important and that changes in the protein that affect its channel size also affect the proper functioning of the efflux complex.
In general, the three-dimensional model of OprM obtained by threading its sequence to that of TolC appears to be plausible, and trends in the locations of certain residues, such as proline and phenylalanine residues, support the model. In addition, the model is in accordance with the results of Li and Poole (15a). These authors also constructed a series of insertion and deletion mutants and assessed their ability to reconstitute functional efflux pathways in Pseudomonas aeruginosa. The overlaying of these results on an alignment of OprM and TolC was consistent with, and adds further support for, our three-dimensional model of OprM. Similar modeling could also be performed for other related members of the extended OprM-TolC family, 15 of which were recently identified in the genome sequence of P. aeruginosa (25). The TolC structure obtained by Koronakis et al. (14) indicates that these proteins are designed to provide a very efficient mechanism for the direct extrusion of substrates across two membranes and the periplasmic space, and these principles would appear to also be true for OprM. Our insertion and deletion data have provided some insights into the structure-function relationships of OprM. Further mutagenesis studies, guided by the current model, would help to refine the structure and to determine the amino acid residues that are essential for its proper function or for its interaction with the other components and could eventually contribute to the development of inhibitors of these efflux systems.
ACKNOWLEDGMENTS
This work was supported by a grant from the Canadian Cystic Fibrosis Foundation (CCFF) and the Canadian Institutes of Health Research (CIHR) to R.E.W.H. and a grant from the Deutsche Forschungsgemeinschaft (Project B9 of Sonderforschungsbereich 176) and of the Fonds der Chemischen Industrie to R.S.B. K.K.Y.W. was supported by a studentship from CCFF. R.E.W.H. was a recipient of the CIHR Distinguished Scientist Award.
REFERENCES
- 1.Aires J R, Köhler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in laboratory medicine. 3rd ed. Baltimore, Md: The Williams & Wilkins Co.; 1991. pp. 72–78. [Google Scholar]
- 3.Andrade M A, Chacon P, Merelo J J, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 4.Benz R, Jando K, Boos W, Langer P. Formation of large ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim Biophys Acta. 1978;511:305–319. doi: 10.1016/0005-2736(78)90269-9. [DOI] [PubMed] [Google Scholar]
- 5.Benz R, Maier E, Gentschev I. TolC of Escherichia coli functions as an outer membrane channel. Zentbl Bakteriol. 1993;278:187–196. doi: 10.1016/s0934-8840(11)80836-4. [DOI] [PubMed] [Google Scholar]
- 6.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two Escherichia coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 7.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan L, Ehrmann M, Yoneyama H, Nakae T. Membrane topology of the xenobiotic-exporting subunit, MexB, of the MexA,B-OprM extrusion pump in Pseudomonas aeruginosa. J Biol Chem. 1999;274:10517–10522. doi: 10.1074/jbc.274.15.10517. [DOI] [PubMed] [Google Scholar]
- 9.Hamzehpour M M, Pechere J C, Plesiat P, Köhler T. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:2392–2396. doi: 10.1128/aac.39.11.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 11.Hancock R E W, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koebnik R, Locher K P, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 13.Koronakis V, Li J, Koronakis E, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol. 1997;23:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 14.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 15.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Li X-Z, Poole K. Mutational analysis of the OprM outer membrane component of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2001;183:12–27. doi: 10.1128/JB.183.1.12-27.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 17.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:2242–2246. doi: 10.1128/aac.44.9.2242-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole K. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saier M H., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sandermann H, Strominger J L. Purification and properties of C55-isoprenoid alcohol phosphokinase from Staphylococcus aureus. J Biol Chem. 1972;247:5123–5131. [PubMed] [Google Scholar]
- 24.Srikumar R, Li X Z, Poole K. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T, Saier M H, Hancock R E W, Long S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 26.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong K K, Hancock R E W. Insertion mutagenesis and membrane topology model of the Pseudomonas aeruginosa outer membrane protein OprM. J Bacteriol. 2000;182:2402–2410. doi: 10.1128/jb.182.9.2402-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong K K, Poole K, Gotoh N, Hancock R E W. Influence of OprM expression on multiple antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2009–2012. doi: 10.1128/aac.41.9.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoneyama H, Ocaktan A, Gotoh N, Nishino T, Nakae T. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1998;244:898–902. doi: 10.1006/bbrc.1998.8351. [DOI] [PubMed] [Google Scholar]
- 30.Zgurskaya H I, Nikaido H. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J Bacteriol. 2000;182:4264–4267. doi: 10.1128/jb.182.15.4264-4267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]