Muscle junctophilins 1 and 2 and neuronal junctophilins 3 and 4 differ in sequence and in interactions with RYR1. Junctophilin 2 has been shown to support voltage-induced calcium release. Perni and Beam show that junctophilins 1 and 3 also support such a release, but that junctophilin 4 does not.
Abstract
In skeletal muscle, depolarization of the plasma membrane (PM) causes conformational changes of the calcium channel CaV1.1 that then activate RYR1 to release calcium from the SR. Being independent of extracellular calcium entry, this process is termed voltage-induced calcium release. In skeletal muscle, junctophilins (JPHs) 1 and 2 form the SR–PM junctions at which voltage-induced calcium release occurs. Previous work demonstrated that JPH2 is able to recapitulate voltage-induced calcium release when expressed in HEK293 cells together with CaV1.1, β1a, Stac3, and RYR1. However, it is unknown whether JPH1 and the more distantly related neuronal JPH3 and JPH4 might also function in this manner, a question of interest because different JPH isoforms diverge in their interactions with RYR1. Here, we show that, like JPH2, JPH1 and JPH3, coexpressed with CaV1.1, β1a, Stac3, and RYR1 in HEK293 cells, cause colocalization of CaV1.1 and RYR1 at ER–PM junctions. Furthermore, potassium depolarization elicited cytoplasmic calcium transients in cells in which WT CaV1.1 was replaced with the calcium impermeant mutant CaV1.1(N617D), indicating that JPH1, JPH2, and JPH3 can all support voltage-induced calcium release, despite sequence divergence and differences in interaction with RYR1. Conversely, JPH4-induced ER–PM junctions contain CaV1.1 but not RYR1, and cells expressing JPH4 are unable to produce depolarization-induced calcium transients. Thus, JPHs seem to act primarily to form ER–PM junctions and to recruit the necessary signaling proteins to these junctions but appear not to be directly involved in the functional interactions between these proteins.
Introduction
The junctophilin (JPH) proteins were first discovered and characterized by Takeshima and collaborators ∼20 yrs ago (Takeshima et al., 2000; Nishi et al., 2003). JPHs are embedded in the ER membrane by their C-terminal transmembrane (TM) domain and interact with the plasma membrane (PM) through their eight N-terminal membrane occupation and recognition nexus (MORN) motifs (Takeshima et al., 2000). As a result, the JPHs tether the ER and PMs in close proximity (∼12 nm) to one another. The resulting ER–PM junctions allow voltage- and ligand-gated Ca2+ channels, and Ca2+-activated K+ channels, in the PM to interact functionally with ER Ca2+-releasing channels, mostly RYRs, and mediate a variety of Ca2+-signaling events (Takeshima et al., 2015).
JPH has four isoforms, two of which (JPH3 and JPH4) are widely expressed in the brain (Nishi et al., 2003). The other two isoforms, JPH1 and JPH2, are both expressed in skeletal muscle, whereas only JPH2 is expressed in cardiac muscle (Takeshima et al., 2000). The junctions induced by JPH1 and JPH2 largely occur between the SR and PM invaginations, termed t-tubules. Depolarization of the PM (t-tubules) is translated into calcium release from the SR, which results in contraction, a process known overall as excitation–contraction coupling. Although sharing many features, excitation–contraction coupling calcium release differs in cardiac and skeletal muscle. In cardiac muscle, calcium entry through CaV1.2 voltage-gated calcium channels in the t-tubules triggers a larger calcium release from the type 2 RYR in the SR (Eisner et al., 2017). In skeletal muscle, the voltage-gated calcium channel CaV1.1 in the t-tubules activates calcium release via the type 1 RYR (RYR1) without the requirement for calcium entry (Armstrong et al., 1972; Dayal et al., 2017), a process that has been termed voltage-induced calcium release.
In skeletal muscle, JPH1 and JPH2 provide the SR–PM junctions at which voltage-induced calcium release occurs, with the stability required to withstand the mechanical stress imposed by the stretching and contraction of the muscle fiber, even in extreme conditions (Rome et al., 1996; Close et al., 2014). In addition to forming and stabilizing the junctions, JPH1 and JPH2 may also play a more direct role in the coupling between CaV1.1 and RYR1. In particular, JPH1 and JPH2 both coimmunoprecipitate with CaV1.1 (Golini et al., 2011), an interaction that appears to involve a 20-residue sequence in the C terminus of CaV1.1 (Nakada et al., 2018). Although JPH1 and JPH2 interact similarly with CaV1.1, they are dissimilar with respect to RYR1, in that RYR1 coimmunoprecipitates with JPH1 but not with JPH2 (Phimister et al., 2007). Thus, the question arises as to whether JPH1 and JPH2 are equivalent in their ability to support voltage-induced calcium release. The ability of JPH2 to support such release seems clear. First, the expression of JPH2 in mouse skeletal muscle substantially precedes the expression of JPH1, and knockout mice lacking JPH1 retain their ability to contract their muscles, although less efficiently than WT mice (Ito et al., 2001). Second, voltage-induced calcium release can be reconstituted in tsA201 cells by the expression of JPH2, CaV1.1, β1a, Stac3, and RYR1 (Perni et al., 2017). Whether JPH1 can also support voltage-induced calcium release remains an open question because the knockout of JPH2 is embryonically lethal due to cardiac defects (Takeshima et al., 2000). To surmount this difficulty, we have determined whether JPH1 could functionally substitute for JPH2 in the tsA201 system. We also tested neuronal JPH3 and JPH4; the former has been shown to interact strongly with RYR1 via a motif that is absent in both JPH1 and JPH2, while the latter showed no detectable interaction with RYR1 (Perni and Beam, 2021).
Materials and methods
Expression plasmids
JPHs
cDNAs encoding human JPH1, JPH2, JPH3, and JPH4 were obtained from GenScript (clones OHu03384, OHu07425, OHu27485, and OHu31404, respectively). All the constructs contain the eight-residue sequence DYKDDDDK (FLAG tag) either at the N terminus (JPH1 and JPH2) or at the C terminus (JPH3 and JPH4) based on sequencing by GenScript and confirmed in our laboratory. mCherry-JPH1 and mCherry-JPH2 were made from the intermediate constructs YFP-JPH1 and YFP-JPH2, which were made by cutting YFP-RYR1 (Polster et al., 2018) with HindIII and XbaI and inserting the fragment produced from the GenScript clones for either JPH1 or JPH2 cut with the same two enzymes (thus replacing the RYR1 coding sequence). mCherry-JPH1 and mCherry-JPH2 were then obtained by excising the PciI-BsrGI fragment of YFP-JPH1 or YFP-JPH2, which includes the cytomegalovirus enhancer-promoter and YFP, and replacing it with the PciI-BsrGI fragment from mCherry-C1 (ref. #PT3975-5; Takara; provided by Dr. M. Tamkun, Colorado State University, Fort Collins, CO). The construction of mCherry-JPH3 and mCherry-JPH4 was described previously (Perni and Beam, 2021).
Calcium channels and accessory subunits
The expression plasmids for CFP- and YFP-CaV1.1 (Papadopoulos et al., 2004), unlabeled β1a and Stac3 (Polster et al., 2015), and YFP-RYR1 (Polster et al., 2018) were previously described.
Plasmids with three or four separated coding sequences
The three-sequence plasmid contained CFP-β1a, CaV1.1 (with N617D mutation; Schredelseker et al., 2010), and Stac3-YFP, with linking amino acids of (β1a to CaV1.1): MSTSGSGATNFSLLKQAGDVEENPGPGSVDGTETSQVAPAGGGFD and (CaV1.1 to Stac3): STVPKLASGSGATNFSLLKQAGDVEENPGPGSAT. The four-sequence plasmid contained CFP-β1a, CaV1.1(N617D), JPH3, and Stac3-YFP, with linking amino acids of (β1a to CaV1.1): MSTSGSGATNFSLLKQAGDVEENPGPGSVDGTETSQVAPAGGGFD; (CaV1.1 to JPH3): STVPKLASGSGATNFSLLKQAGDVEENPGPGSAT; and (JPH3 to Stac3): DYKDDDDKVPRARDPPDLELRVASGSGATNFSLLKQAGDVEENPGPGSEF. Underlining indicates the self-cleaving P2A sequence.
Construction of the three-sequence plasmid
The sequence encoding the second P2A peptide in the pcDNA3.0-TRPV4-p2A-ferritin-p2A-mCherry plasmid (Addgene Plasmid #74309; provided by Dr. John Bankston, University of Colorado, Anschutz Medical Campus) was amplified with the primers forward 5′-GTGGAGGGAGCTCCCGAGTGGCTAGC-3′ and reverse 5′-GCCCTTGAATTCGGATCCGGGACCGG-3′ to introduce a SacI and an EcoRI site at the 5′ and 3′ ends of the P2A sequence, respectively. The amplicon was then digested with SacI and EcoRI and ligated at the 5′ end of the Stac3 sequence in the Stac3-YFP plasmid, cut with the same enzymes, to obtain the P2A-Stac3-YFP.
The P2A-Stac3-YFP construct was then amplified with the primers forward 5′-GACTCAGATCTAGAGCCCCGAGTGGC-3′ and reverse 5′-CAGGTTCAGGGGGAGGTGTGGGAGG-3′ to add an XbaI site at the 5′ end of the P2A sequence to pair with an already-existing XbaI site at the 3′ end of the YFP sequence. The entire P2A-Stac3-YFP sequence was excised with XbaI and ligated into a plasmid encoding CFP-β1a-CFP-CaV1.1 (kindly provided by Dr. Symeon Papadopoulos, University of Cologne, Cologne, Germany) also cut with XbaI, to yield CFP-β1a-CFP-CaV1.1-P2A-Stac3-YFP. To replace the WT CaV1.1 with CaV1.1(N617D), one of the two SalI restriction sites was removed from the previously described YFP-CaV1.1(N617D) plasmid (see supplemental methods in Perni et al., 2017) with the primers forward 5′-GCCTTGACCAGTCCACGGTACCGCGGGC-3′ and reverse 5′-GCCCGCGGTACCGTGGACTGGTCAAGGC-3′, which introduced a silent mutation (TCG>TCC, underlined) that ablates the SalI restriction site at the 3′ end of CaV1.1(N617D) sequence, leaving the construct with a unique SalI site between the YFP and CaV1.1(N617D) sequences. This construct was digested with SalI and KpnI, and the CaV1.1(N617D) sequence was then used to replace the entire CFP-CaV1.1 sequence in the CFP-β1a-CFP-CaV1.1-P2A-Stac3-YFP plasmid, cut with the same enzymes. The resulting plasmid encoded CFP-β1a-CaV1.1(N617D)-P2A-Stac3-YFP. Finally, to introduce the P2A sequence between β1a and CaV1.1(N617D), the second P2A sequence in the pcDNA3.0-TRPV4-p2A-ferritin-p2A-mCherry plasmid was amplified with the primers forward 5′-GCTCCTCCCGAGTCGACTAGCGGCAGCGG-3′ and reverse 5′-CCTCGCCCTTGTCGACGGATCCGGGACCGGGG-3′ designed to add an SalI restriction site at each end of the P2A sequence. The amplicon was then digested with SalI and inserted at the 3′ end of the β1a sequence in the CFP-β1a-CaV1.1(N617D)-P2A-Stac3-YFP plasmid, also digested with SalI, to obtain the final construct CFP-β1a-P2A-CaV1.1(N617D)-P2A-Stac3-YFP.
Construction of the four-sequence plasmid
The P2A from the P2A-Stac3-YFP plasmid, created in the making of the three-sequence plasmid (see above), was amplified with the primers forward 5′-GACTCAGATCTAGAGCCCCGAGTGGC-3′ and reverse 5′-TTTCTGTCATAAGCTTGGATCCGGGACCGGGG-3′ designed to introduce an NheI and a HindIII site at the 5′ and 3′ ends of the P2A sequence, respectively. The amplified DNA was then inserted 5′ to the sequence encoding JPH2 using NheI and HindIII to obtain P2A-JPH2. The P2A-JPH3 construct was made by replacing JPH2 with JPH3 in the P2A-JPH2 plasmid by digestion with BamHI and AvrII. Finally, the P2A-JPH3 sequence was amplified with the primers forward 5′-CTATAGGGGTACCCAAGCTGGCTAGCGG-3′ and reverse 5′-GCAGAATGGTACCCAGGTCAGGAGGTGAACAAAGAGG-3′ designed to introduce a KpnI site at each end of the sequence. The KpnI enzyme was then used to introduce P2A-JPH3 between CaV1.1(N617D) and Stac3-YFP in the three-sequence plasmid (described above) to create the final four-sequence plasmid CFP-β1a-P2A-CaV1.1(N617D)-P2A-JPH3-P2A-Stac3-YFP.
Cell culture and cDNA transfection
tsA201 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (Mediatech) supplemented with 10% (vol/vol) FBS and 2 mM glutamine in a humidified incubator with 5% (vol/vol) CO2. The generation of tsA201 and spiking HEK293 cells stably expressing RYR1 were described previously (Perni et al., 2017; Perni and Beam, 2021). The spiking HEK293 cells, which stably express KIR2.1 and NaV1.3 (Park et al., 2013), were provided by Dr. Adam Cohen (Harvard University, Boston, MA). The stable cell lines were cultured in the same medium as described for tsA201 cells with the addition of 300 μg/ml of Hygromycin, for both lines, and 2 μg/ml Puromycin and 500 μg/ml Geneticin (G418) for the spiking HEK293 cells stably transfected with RYR1.
Cells at ∼70% confluence were transiently transfected by exposure for 3.5 h to the jetPRIME reagent (Polyplus-transfection Inc.) containing either 1.5 μg (multicoding sequence plasmids), 1 μg (individual CaV1.1 and RYR1 constructs), or 0.5 μg (individual β1a, Stac3, and JPH constructs) of cDNA per 35-mm plastic culture dish (Falcon). After the 3.5 h of transfection, the cells were detached from the dish using trypsin-EDTA (Mediatech) and replated at ∼2.5 × 104/cm2 in glass-bottom microwell dishes (35-mm dish, 14-mm microwell diameter; MatTek) previously coated with collagen type III (Sigma-Aldrich) or ECL (Millipore) for confocal imaging or at ∼2 × 103/cm2 on plastic 35-mm dishes for perforated patch clamp. Experiments were performed the following day.
Imaging
Cells were superfused with physiological saline (in mM: 146 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, and 10 HEPES, pH 7.4, with NaOH) and imaged at room temperature (∼22°C) using a Zeiss 710 confocal microscope. Images were obtained as single optical sections (∼0.9-μm thick) with a 63×/1.4 NA oil immersion objective. Fluorescence excitation (Ex) and emission (Em) were as follows: Ex, 440 nm and Em, 454−508 nm for CFP; Ex, 514 nm and Em, 519−577 nm for YFP; Ex, 543 nm and Em, 592−696 nm for mCherry; and Ex, 488 nm and Em 503–549 nm for Fluo3.
KCl stimulation
Spiking HEK293 cells stably transfected with RYR1 and transiently transfected with the three-sequence plasmid, either alone or together with plasmids for either mCherry-JPH1, mCherry-JPH2, mCherry-JPH3, or mCherry-JPH4, were loaded with 5 μM Fluo3-AM in serum-free cell medium for 20 min at 37°C and then washed and superfused with physiological saline (composition reported above). Cells showing CFP fluorescence (from CFP-β1a), as well as mCherry fluorescence in cells also transfected with mCherry-JPHs, were chosen for KCl stimulation. Transients were evoked in individual cells by focal application of physiological saline via a patch clamp glass pipette containing 80 mM KCl (K+ replacing Na+) for 0.5 s using a Picospritzer. For KCl stimulation in nominal Ca2+-free solution, CaCl2 in the regular and high-KCl physiological saline was replaced by MgCl2. Fluo3 transients were acquired at a frequency of 100 frames/s (10 ms/frame). The baseline Fluo3 fluorescence (F0) was taken as the average fluorescence in the 500-ms interval immediately preceding the beginning of the KCl application.
Perforated patch clamp
tsA201 cells stably expressing RYR1, transfected with the four-sequence plasmid, were loaded with 5 μM Fluo3-AM in cell medium for 20 min at 37°C and then washed and superfused with a bath solution containing (in mM) 137 NaCl, 5.6 KCl, 2.6 CaCl2, 1.2 MgCl2, 10 glucose, and 10 HEPES, pH 7.4, with NaOH. Cells to be patch clamped were chosen based on the expression of CFP. Borosilicate pipettes with a resistance of 2–3.5 MΩ were front filled by immersing the tip for ∼8–9 s in a pipette solution containing (in mM) 137 Cs-aspartate, 10 NaCl, 1 CaCl2, 1 MgCl2, and 10 HEPES, pH 7.2, with CsOH. The pipette was then backfilled with the same solution that additionally contained a 1:100 dilution of a 20 mg/ml stock of amphotericin B and 0.5% pluronic acid in DMSO. After forming a gigaseal (>1 GΩ), the negative pressure on the pipette was released, and the test steps were applied, from a holding potential of −60 mV, once the access resistance decreased to ≤20 MΩ. A Nikon Diaphot epifluorescence microscope equipped with a fluorometer apparatus (Biomedical Instrumentation Group, University of Pennsylvania, Philadelphia, PA) with fluorescein optics was used to measure changes in Fluo3 fluorescence (ΔF) from baseline in response to 50-ms depolarizations to +30 mV and +90 mV from a holding potential of −60 mV. The baseline was taken as the average fluorescence in the 5-ms interval immediately preceding the test depolarization.
Statistical analysis
One-way ANOVA with multiple comparisons and post hoc Tukey’s test was used to evaluate significant differences among the datasets reported in Fig. 3 B. Paired t test was used to evaluate significant differences between the two sets of data reported in Fig. 4 B. All statistical analyses were performed using GraphPad Prism software.
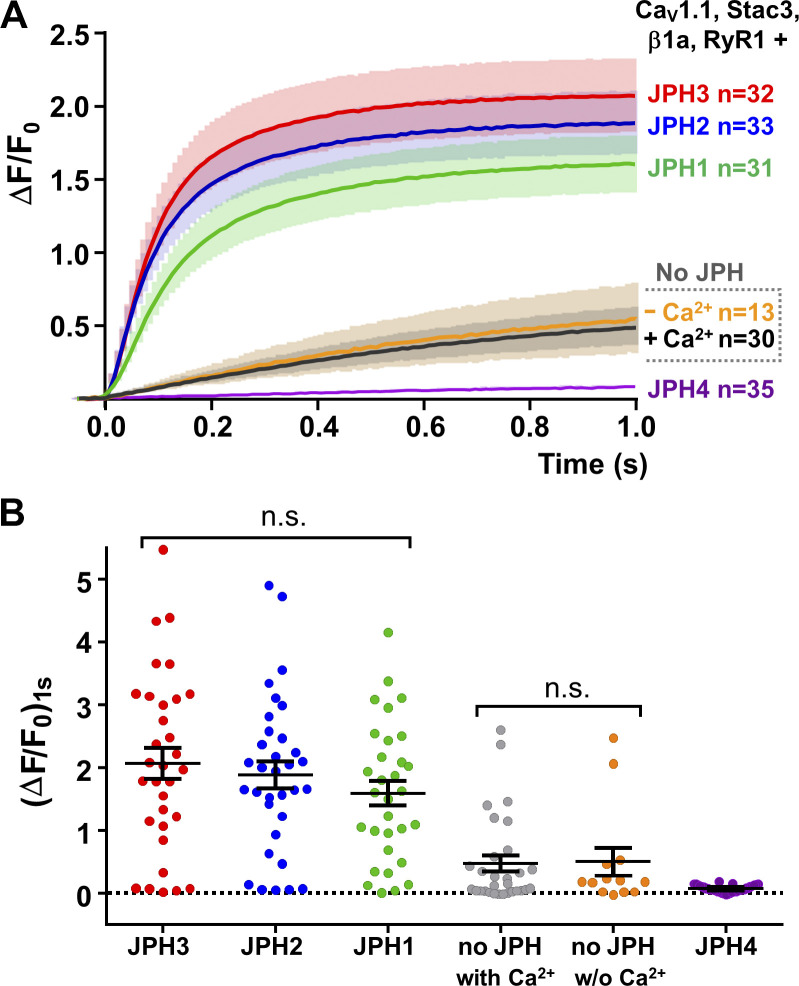
Figure 3.
JPH1, JPH2, and JPH3 can support voltage-induced calcium release. HEK293 cells that stably expressed NaV1.3 and KIR2.1 were stably transfected with RYR1 (Perni and Beam, 2021) and transiently transfected with a three-sequence plasmid containing the coding sequences for CFP-β1a, CaV1.1(N617D), and Stac3-YFP and no JPHs or individual cDNAs for either mCherry-JPH1, mCherry-JPH2, mCherry-JPH3, or mCherry-JPH4. Individual cells were depolarized by focal application of 80 mM KCl for 0.5 s. (A) Comparison of average calcium transients (solid lines) ± SEM (semitransparent envelopes). The Ca2+ transients for cells transfected with JPH1 (green), JPH2 (blue), or JPH3 (red) were large and rapidly activating and not a result of calcium entry via CaV1.1 because the N617D mutation eliminates its calcium permeability. In cells not transfected with any JPH, the transients were small and slowly activating and were similar when Ca2+ was present or absent in the extracellular medium (black and orange, respectively); thus, these transients also appear to represent voltage-induced calcium release, which could be explained if the cells endogenously express JPH3 at low levels. The Ca2+ transients were very small, or absent, in cells transfected with JPH4 (purple), which can be explained if JPH4 recruited CaV1.1 to junctions lacking RYR1. (B) Calcium transient amplitudes recorded 1 s after onset of KCl application (ΔF/F0)1s. Each point represents a single cell, and mean ± SEM is indicated by the longer and shorter horizontal lines, respectively. Values of (ΔF/F0)1s were not significantly different for JPH1, JPH2, and JPH3 (P ≥ 0.374) or for no JPH ± Ca2+ (P > 0.99). Values of (ΔF/F0)1s for JPH1, JPH2, and JPH3 were significantly (P < 0.0001) larger than those for either JPH4 or for no JPH ± Ca2+. Statistical significance was calculated using one-way ANOVA with Tukey’s post hoc test. Data were recorded from one transfection in the case of “no JPH 0 Ca2+” and from two separate transfections for all the other construct combinations. n.s., not significant; w/o, without.
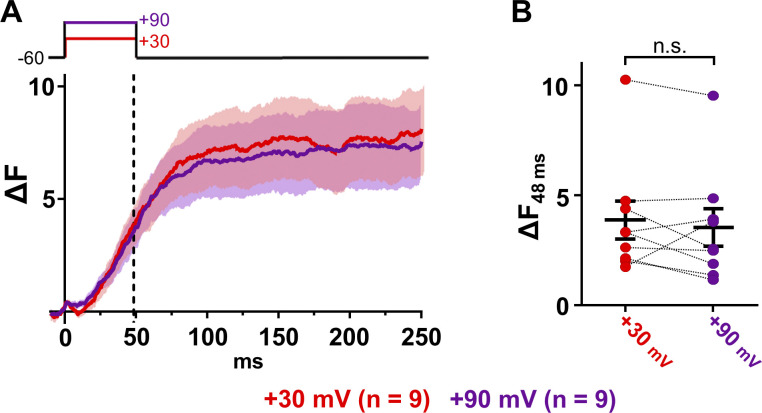
Figure 4.
Perforated patch clamp recording confirms voltage-induced calcium release for JPH3. (A) Mean transients (solid lines) ± SEM (semitransparent envelopes) recorded from tsA201 cells stably expressing RYR1 and transiently transfected with a four-sequence plasmid containing the coding sequences for CFP-β1a, CaV1.1(N617D), JPH3, and Stac3-YFP. Transients were elicited by depolarizing individual cells from the holding potential (−60 mV) to +30 mV (red) and to +90 mV (purple) for 50 ms. (B) ΔF measured 48 ms after the onset of the depolarization (vertical dashed line in A) for each analyzed cell. Each pair of points, connected by the dashed line, represents a single cell depolarized to +30 mV (red) and to +90 mV (purple). Mean ± SEM is indicated by the longer and shorter horizontal lines, respectively. The mean transient amplitude at 48 ms did not differ significantly between +30 mV and +90 mV (paired t test P = 0.415). n.s., not significant.
Results
JPH1, JPH2, and JPH3 recruit CaV1.1 into ER–PM junctions
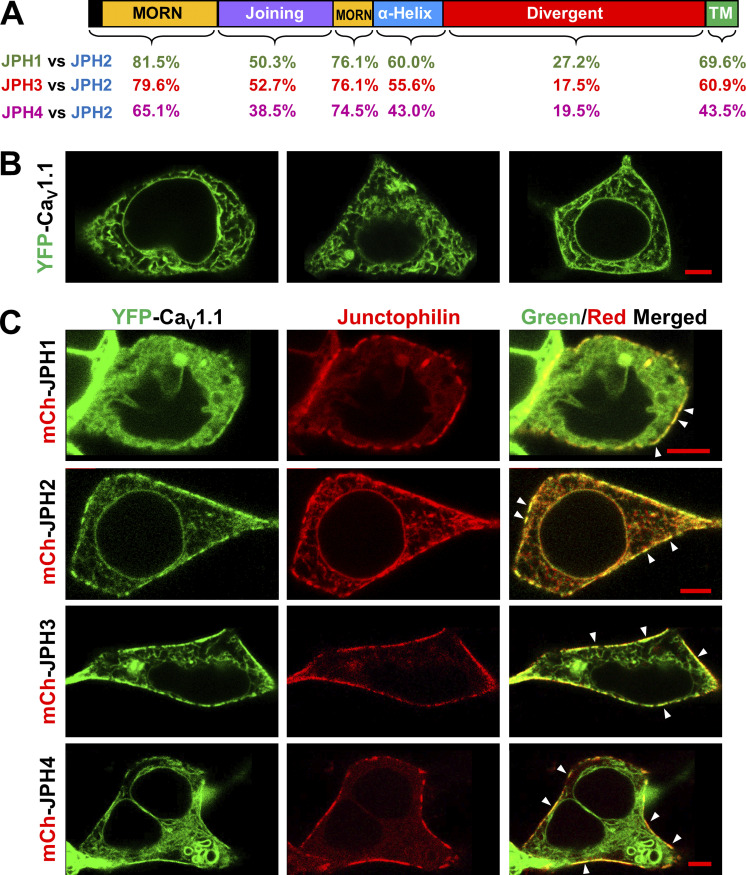
Fig. 1 illustrates schematically the domain structure shared by all the JPHs (Takeshima et al., 2000) and compares the extent of sequence identity between JPH2 and either JPH1, JPH3, or JPH4. For JPH1 and JPH3, the sequence identity with JPH2 is high (>75%) within the regions containing the MORN motifs that are important for association with the PM. The sequence identity is also substantial (>60%) for the short segment (TM) that traverses the ER membrane and moderate (>50%) for the α-helical and joining domains. JPH4, being the most phylogenetically distant isoform (Landstrom et al., 2014), shows the least conservation with JPH2, having 65.1% and 74.5% identity in the first and second MORN repeats, respectively, and only 38.5%, 43%, and 43.5% identity for the joining domain, α-helical domain, and TM domain, respectively. As indicated by its designation, the divergent domains of JPH1, JPH3, and JPH4 differ substantially from that of JPH2.
Figure 1.
Sequence conservation of JPH1, JPH3, and JPH4 with JPH2 and comparison of the ability of the four JPH isoforms to recruit CaV1.1 into ER–PM junctions. (A) Schematic representation of the domain architecture of JPHs, including the MORN domains, responsible for the association with the PM and the C-terminal TM domain that embeds these proteins in the ER membrane. The numbers beneath the schematic indicate the percentage of identical residues for each domain between JPH1 and JPH2 (green), JPH3 and JPH2 (red), and JPH4 and JPH2 (purple). A detailed sequence alignment is illustrated in Fig. 5. (B) Mid-level confocal optical sections of tsA201 cells transfected with cDNAs for YFP-CaV1.1 (green), β1a, and Stac3. A substantial amount of the YFP fluorescence appears to be ER associated in the cell interior, and for the cells illustrated in the left and center panels, there is no clear association of YFP with the cell surface. In other cells (right panel), there was a fairly continuous band of YFP-CaV1.1 closely associated with the cell surface. (C) Mid-level optical sections of tsA201 cells transfected with cDNAs for YFP-CaV1.1 (green) and mCherry (mCh)-tagged JPH1, JPH2, JPH3, and JPH4 (red). cDNAs for β1a and Stac3 were also transfected. Colocalization, visible as yellow areas in the rightmost merged images, is primarily found in discrete spots (some indicated by arrowheads) at the cell’s periphery, presumptive ER–PM junctions. The total number of cells analyzed for each condition was no JPH, 14 from one transfection; JPH1, 16 from one transfection; JPH2, 36 from two transfections; JPH3, 37 from two transfections; and JPH4, 17 from one transfection. Scale bars, 5 μm.
Previously, we showed that JPH2 was able to induce ER–PM junctions in tsA201 cells and to recruit the L-type Ca2+ channel CaV1.1 into such junctions (Perni et al., 2017). To test qualitatively whether JPH1, JPH3, and JPH4 could also recruit CaV1.1, we transfected tsA201 cells with YFP-tagged CaV1.1, its auxiliary subunit β1a, and the adapter protein Stac3 either without or with mCherry-tagged JPHs. Fig. 1 B shows three examples of tsA201 cells expressing YFP-CaV1.1, β1a, and Stac3 in the absence of exogenously expressed JPH. In ∼80% of the analyzed cells, CaV1.1 showed no clear-cut association with the cell surface (leftmost and center images), likely because these cells were not expressing β1a and/or Stac3, without which little or no channel traffics to the PM (Polster et al., 2015). In the remaining ∼20% cells, which were likely expressing β1a and Stac3 at higher levels, CaV1.1 appeared to be present in a nearly continuous stripe outlining the cell periphery (Fig. 1 B, rightmost image).
In cells expressing any of the four isoforms of JPH (Fig. 1 C, indicated in red), CaV1.1 (indicated in green) appeared to be present in discontinuous segments at the cell periphery, and these overlapped with similarly arrayed segments of the JPH (yellow regions in the merged images). Because they are segmented, the regions of overlap at the cell periphery (arrowheads in rightmost panels) likely correspond to ER–PM junctions (Perni et al., 2017; Perni and Beam, 2021). The ability of JPH1 to recruit CaV1.1 is consistent with previous work on muscle cells (Nakada et al., 2018).
JPH1, JPH2, and JPH3, but not JPH4, cause junctional accumulation of RYR1 in tsA201 cells expressing CaV1.1
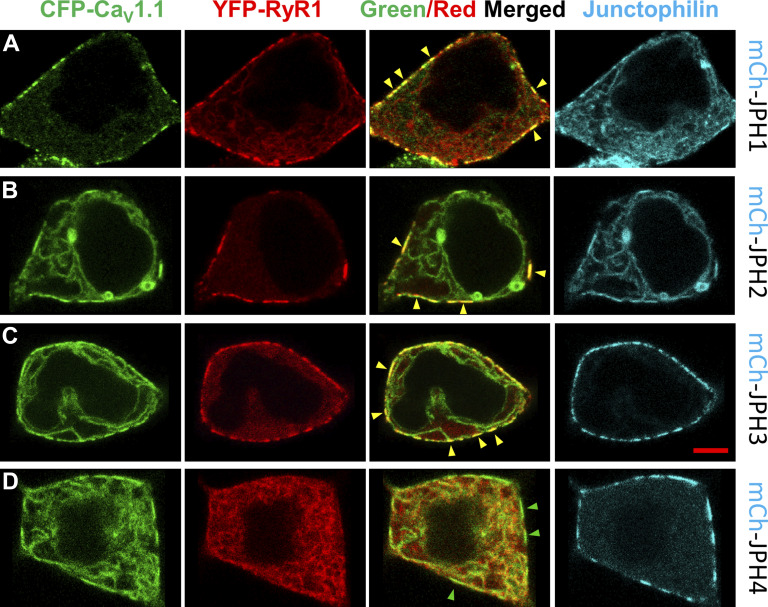
Before determining whether JPH1, JPH3, and JPH4 could support voltage-induced calcium release, we tested their ability to cause RYR1 to colocalize with CaV1.1 at ER–PM junctions, an essential prerequisite for the reconstitution of this type of release. Thus, we transfected tsA201 cells with cDNAs for CFP-tagged CaV1.1 (plus β1a and Stac3), YFP-tagged RYR1, and mCherry-tagged JPHs. Representative images are shown in Fig. 2. For JPH1 (Fig. 2 A), JPH2 (Fig. 2 B), and JPH3 (Fig. 2 C), RYR1 (shown in red) colocalized with CaV1.1 (shown in green) at presumptive ER–PM junctions as indicated by a segmented pattern at the cell periphery that was similar to that of the JPHs (shown in cyan). Although JPH4 caused junctional accumulation of CaV1.1, RYR1 was not present in these regions (Fig. 2 D), which is reminiscent of previous results showing that JPH4 does not recruit RYR1 to ER–PM junctions containing CaV2.1 or CaV2.2 (Perni and Beam, 2021).
Figure 2.
RYR1 colocalizes with CaV1.1 at ER–PM junctions induced by JPH1, JPH2, and JPH3 but not by JPH4. (A–D) Mid-level confocal optical sections of tsA201 cells transfected with cDNAs encoding for CFP-CaV1.1 (represented in green), YFP-RYR1 (represented in red), and mCherry (mCh)-tagged JPH1 (A), JPH2 (B), JPH3 (C), or JPH4 (D) where the JPHs are represented in cyan. cDNAs for β1a and Stac3 were also transfected. For JPH1, JPH2, and JPH3, the fluorescence signals for CFP-CaV1.1 and YFP-RYR1 were colocalized in discrete spots at the cell periphery, visible as yellow in the red/green overlay, some of which are indicated by yellow arrowheads. These occur in regions also containing JPH. In cells expressing JPH4, there was no colocalization between CaV1.1 and RYR1, despite a visible accumulation of CaV1.1 in discrete spots at the cell periphery, some indicated by green arrowheads, where JPH4 is also present. The total number of cells analyzed for each condition was JPH1, 17 from one transfection; JPH2, 18 from one transfection; JPH3, 17 from one transfection; and JPH4 38 from two transfections. Scale bar, 5 μm.
Voltage-induced calcium release is supported by JPH1, JPH2, and JPH3
To test for voltage-induced calcium release, we used spiking HEK293 cells, which stably express NaV1.3 and KIR2.1 (Park et al., 2013), and stably transfected them with RYR1 (Perni and Beam, 2021). These cells were then transiently transfected with, or without, JPHs and with a three-sequence plasmid containing the coding sequences for CFP-β1a, Stac3-YFP, and a mutant CaV1.1 construct, CaV1.1(N617D), and subsequently loaded with Fluo3-AM. The presence of KIR2.1 meant that these cells could be depolarized by a brief (0.5-s) focal application of elevated potassium, and the use of the nonconductive CaV1.1(N617D) mutant (Schredelseker et al., 2010) ensured that any changes in cytoplasmic Ca2+ were not a consequence of extracellular Ca2+ influx through the L-type channel. A significant contribution of any small Ca2+ permeability of NaV1.3 also seems unlikely because these channels inactivate almost completely in ∼10 ms (Cummins et al., 2001).
Fig. 3 A illustrates the mean (solid line) ± SEM (semitransparent envelope) Ca2+ transients elicited by depolarization of spiking HEK293 cells stably expressing RYR1 and transiently transfected with CFP-β1a, Stac3-YFP, CaV1.1(N617D), and either no JPH or JPH1, JPH2, JPH3, or JPH4. The cells additionally transfected with JPH1 (green), JPH2 (blue), or JPH3 (red) produced robust Ca2+ transients, but depolarization under identical conditions of cells not transfected with a JPH also elicited transients that were smaller and slower (Fig. 3 A, black and orange traces). Thus, it appears that either voltage-induced calcium release does not require the expression of an exogenous JPH in these cells or depolarization activated the entry of extracellular calcium. The former seems plausible because the transcript for JPH3 has been shown by PCR to be present in tsA201 cells (Sahu et al., 2019), which are HEK293 cells stably expressing an SV40 temperature-sensitive T antigen. However, we tested directly for the possibility of calcium entry by using a physiological saline that was nominally Ca2+ free (equimolar replacement of Ca2+ by Mg2+). The transient in these cells—RYR1-stable spiking HEK293s transiently transfected with CFP-β1a, Stac3-YFP, CaV1.1(N617D)—differed little in the presence and absence of extracellular Ca2+ (Fig. 3 A, black and orange traces, respectively). Thus, these transients seem likely to be a consequence of voltage-induced calcium release, and their small size and slow activation might be a consequence of a low number of ER–PM junctions induced by endogenously expressed JPH3. In this regard, it is noteworthy that expression of exogenous JPH4 caused the size of the depolarization-evoked Ca2+ transients (Fig. 3 A, purple trace) to be much smaller than those in the absence of a transfected JPH. If it is correct that junctions containing endogenous JPH3, together with CaV1.1 and RYR1, underlie the transients in the cells not transfected with exogenous JPH, then one could speculate that the expressed JPH4 recruits CaV1.1 to junctions not containing RYR1 (Fig. 2 D), which would largely abolish voltage-induced calcium release.
Fig. 3 B illustrates the amplitudes of the calcium transients measured in individual cells at 1 s after KCl application ((ΔF/F0)1s), with the values of mean ± SEM superimposed. Except for JPH4, there was considerable variability for each of the construct combinations to which variable expression of the channel proteins undoubtedly contributed. In the case of JPH4, the transients were uniformly small, with the largest value of (ΔF/F0)1s from 35 cells being 0.19. There were no significant differences between JPH1, JPH2, and JPH3 (P ≥ 0.374). Likewise, for no transfected JPH, there was no significant difference between the transients with, or without, extracellular calcium (P > 0.999). The amplitudes for JPH1, JPH2, and JPH3 were all significantly (P < 0.0001) larger than those for either no JPH (as a group) or JPH4.
Because JPH3 is not expressed in skeletal muscle, it was surprising that it appeared able to support voltage-induced calcium release. Thus, we further investigated JPH3 using perforated patch clamping applied to tsA201 cells that stably expressed RYR1 (Perni et al., 2017) and were transiently transfected with a four-sequence plasmid encoding CFP-β1a, CaV1.1(N617D), JPH3, and Stac3-YFP. Fig. 4 A illustrates the mean (solid lines) ± SEM (semitransparent areas) transients elicited in transfected cells loaded with Fluo3-AM and depolarized to +30 mV (red) and +90 mV (purple). The mean transient at +90 mV, where there is little electrochemical gradient for entry of extracellular calcium, was almost identical to that at +30 mV, consistent with voltage-induced calcium release. This behavior was also evident in individual cells, as shown in Fig. 4 B, which plots the amplitudes 48 ms after the voltage steps to +30 mV and +90 mV. The mean amplitudes at +30 mV and +90 mV did not significantly differ from one other (paired t test P = 0.415). Note that the transients in JPH3-expressing cells recorded with perforated patch clamp are not directly comparable to those with focal application of KCl because they were recorded with different acquisition setups and depolarizing conditions (see Materials and methods).
Discussion
Here, we show that JPH1 and JPH3, but not JPH4, can replace JPH2 in supporting voltage-induced calcium release when expressed together with CaV1.1, β1a, Stac3, and RYR1 in HEK293-derived cells. Using colocalization of fluorescently tagged proteins, we first established that JPH1, JPH3, and JPH4 could recruit CaV1.1 to ER–PM junctions (Fig. 1 B). As a possible basis for this recruitment, it has been shown that a peptide corresponding to JPH1 residues 232–369 (Fig. 5, underlined in red) coimmunoprecipitates with CaV1.1 (Golini et al., 2011). The corresponding regions of JPH2, JPH3, and to a lesser extent JPH4 contain scattered stretches of residues that are highly conserved with JPH1. Thus, it may be that one or more of these conserved stretches account for the ability of JPH2, JPH3, and JPH4 to recruit CaV1.1. Interestingly, JPH3 and JPH4 appear to have a broad ability to recruit high-voltage activated calcium channels other than CaV1.1, including CaV1.2, CaV1.3 (Sahu et al., 2019), CaV2.1, and CaV2.2 (Perni and Beam, 2021).
Figure 5.
Sequence alignment of human JPH2, JPH1, JPH3, and JPH4. The different domains are color-coded in orange (MORNs), purple (joining domain), blue (α-helical domain), red (divergent domain), and green (TM domain). Residues that are identical or conserved among the four isoforms are indicated by asterisks or colons, respectively. The red underlining indicates the region of JPH1 that contains a binding site for CaV1.1 (Golini et al., 2011). The green underlining indicates the cysteine residues of JPH1 whose reactivity depends on the gating state of RYR1 (Phimister et al., 2007). Note that Cys 626 in the human JPH1 sequence reported here corresponds to Cys 627 of the rabbit JPH1 sequence described by Phimister et al. (2007). The residues highlighted in yellow near the C terminus of JPH3 indicates the region that appears to support a direct interaction of JPH3 with the cytoplasmic domain of RYR1.
RYR1 colocalized with CaV1.1 at junctions induced by JPH1, JPH2, and JPH3, but not by JPH4 (Fig. 2). The inability of JPH4 to recruit RYR1 was already described in earlier work from our laboratory (Perni and Beam, 2021) in which it was also shown that the recruitment of RYR1 by JPH3 likely involves a sequence of 28 residues located toward the C-terminal end of the divergent domain (Fig. 5, highlighted in yellow) because this region appears to bind strongly to the cytoplasmic domain of RYR1. Although lacking this 28-residue segment, JPH1 coimmunoprecipitates with RYR1 (Phimister et al., 2007). Furthermore, JPH1 contains three cysteines (Fig. 5, underlined in green) whose reactivity depends on the gating state of RYR1 (Phimister et al., 2007), which is further suggestive of a close association of these two proteins. JPH2 does not coimmunoprecipitate with RYR1 (Phimister et al., 2007). Nonetheless, in contrast with JPH4-induced ER–PM junctions that show no accumulation of RYR1, JPH2 junctions show the simultaneous presence of both CaV1.1 and RYR1, suggesting that JPH2 still plays a crucial role in the recruitment of RYR1 into the junctions. It is possible that JPH2 interacts weakly with RYR1 and depends on additional interactions between RYR1 and other junctionally recruited proteins, specifically CaV1.1, β1a, and Stac3, to cause RYR1 to localize to junctions. Thus, the available evidence indicates that the mechanisms underlying recruitment of RYR1 to ER–PM junctions differ among JPH1, JPH2, and JPH3.
To test for voltage-induced calcium release like that occurring in skeletal muscle, we used either spiking HEK293 cells (to compare all four JPHs) or tsA201 cells (to further characterize JPH3). Because the HEK293 cells had been stably transfected with KIR2.1, they could be depolarized by the focal application of a high-potassium solution. This focal application to spiking HEK293 cells transfected with RYR1, CaV1.1(N617D), which is calcium impermeable, β1a, Stac3, and either JPH1, JPH2, or JPH3, elicited large calcium transients (Fig. 3). By contrast, calcium transients were barely detectable for JPH4, which indicates that JPH4 cannot support voltage-induced calcium release and that the PM of spiking HEK293 cells contains few calcium entry pathways activated by depolarization. The near absence of Ca2+ transients for JPH4 also provides support for the idea that calcium entry was not important for triggering the calcium transients for JPH1, JPH2, and JPH3 and that these were a consequence of voltage-induced calcium release. Using perforated patch clamp recordings applied to transfected tsA201 cells, we directly confirmed that calcium entry was not important for JPH3 (Fig. 4) and in our previous work for JPH2 (Perni et al., 2017). Although not tested directly, it also seems unlikely that calcium entry was important for JPH1, unless the expression of JPH1 caused the upregulation of a population of calcium-permeable channels activated by depolarization.
Somewhat surprisingly, depolarization elicited calcium transients in spiking HEK293 cells transfected with RYR1, CaV1.1(N617D), β1a, and Stac3 but not with an exogenous JPH (Fig. 3). These transients appeared to be a consequence of voltage-induced calcium release because they were little affected by removal of extracellular calcium, suggesting that the cells contained ER–PM junctions at which CaV1.1 controlled the activation of RYR1. Several lines of evidence indicate that small numbers of ER–PM junctions likely form in these cells, even when they are not transfected with an exogenous JPH. First, tsA201 cells express the transcript for JPH3; second, transfected CaV1.3, KCa3.1, and RYR1 colocalize in these cells at ER–PM junctions based on electrophysiology, ultraresolution microscopy, and FRET (Sahu et al., 2019); and third, electron microscopy revealed the presence of two or more ER–PM junctions in ∼8% of untransfected cells, although these junctions were both smaller and scarcer than in cells transfected with JPH2 (Perni et al., 2017). Since the 70–80-nm-thick sections examined with electron microscopy would capture only a fraction of the junctions in a cell, it seems likely that a significant percentage of nontransfected cells would contain at least a few junctions. After transfection with the other necessary components (RYR1, CaV1.1(N617D), β1a, and Stac3), voltage-induced calcium release at these relatively scarce junctions could trigger additional release from RYR1 not at junctions, which could explain why the transients activated so slowly compared with when either JPH1, JPH2, or JPH3 was also transfected (Fig. 3 A). The small size and relative scarcity of the endogenous ER–PM junctions would make them difficult to detect with standard-resolution confocal microscopy. Furthermore, the scarcity of endogenous ER–PM junctions indicates that endogenous JPH3 is present only at low levels compared with that of the transfected, exogenous JPHs. As a result, in cells transfected with exogenous JPHs, it seems likely that minimal voltage-induced calcium release would have occurred at junctions containing endogenous JPH3 because mass action would have caused the vast majority of CaV1.1 to be associated with the exogenous JPH.
Obviously, there are many differences between calcium movements in skeletal muscle cells and those in transfected HEK293 cells. In skeletal muscle, for example, the time course of the myoplasmic calcium transient is strongly affected by the binding of the released calcium to troponin C and parvalbumin and by transport proteins that are expressed at high levels. Additionally, the release process itself is subject to regulation by the large complement of proteins localized in triad junctions. Thus, important differences in the behavior of JPH1 and JPH2 may be detectable only when these proteins are in their native environments. For this analysis, comparison of calcium release in specific muscles after inducible knockouts of JPH1 or JPH2 seems likely to be highly informative.
In conclusion, our data indicate that all four JPHs may behave similarly with respect to CaV1.1 but clearly differ in their interactions with RYR1. JPH1, JPH2, and JPH3 recruit both CaV1.1 and RYR1 to ER–PM junctions in transfected tsA201 cells and can similarly support voltage-induced calcium release. JPH4 is unable to recruit RYR1 together with CaV1.1 and cannot support voltage-induced calcium release. These data suggest that the primary roles of the JPHs are to cause junctions to form between the PM and ER/SR and to recruit the channels that are important for signaling at these junctions. Any JPH able to fulfill both of these roles with respect to CaV1.1 and RYR1 can support voltage-induced calcium release.
Acknowledgments
Eduardo Ríos served as editor.
This work was supported by National Institutes of Health grant AR070298 to K. Beam.
The authors declare no competing financial interests.
Author contributions: K. Beam and S. Perni were responsible for conceptualization and manuscript writing. S. Perni provided data curation, formal analysis, and investigation. K. Beam was responsible for funding acquisition.
Footnotes
This work is part of a special issue on excitation–contraction coupling.
References
- Armstrong, C.M., Bezanilla F.M., and Horowicz P.. 1972. Twitches in the presence of ethylene glycol bis(β-aminoethyl ether)-N,N′-tetracetic acid. Biochim. Biophys. Acta. 267:605–608. 10.1016/0005-2728(72)90194-6 [DOI] [PubMed] [Google Scholar]
- Close, M., Perni S., Franzini-Armstrong C., and Cundall D.. 2014. Highly extensible skeletal muscle in snakes. J. Exp. Biol. 217:2445–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins, T.R., Aglieco F., Renganathan M., Herzog R.I., Dib-Hajj S.D., and Waxman S.G.. 2001. Nav1.3 sodium channels: rapid repriming and slow closed-state inactivation display quantitative differences after expression in a mammalian cell line and in spinal sensory neurons. J. Neurosci. 21:5952–5961. 10.1523/JNEUROSCI.21-16-05952.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayal, A., Schrötter K., Pan Y., Föhr K., Melzer W., and Grabner M.. 2017. The Ca2+ influx through the mammalian skeletal muscle dihydropyridine receptor is irrelevant for muscle performance. Nat. Commun. 8:475. 10.1038/s41467-017-00629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner, D.A., Caldwell J.L., Kistamás K., and Trafford A.W.. 2017. Calcium and excitation-contraction coupling in the heart. Circ. Res. 121:181–195. 10.1161/CIRCRESAHA.117.310230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golini, L., Chouabe C., Berthier C., Cusimano V., Fornaro M., Bonvallet R., Formoso L., Giacomello E., Jacquemond V., and Sorrentino V.. 2011. Junctophilin 1 and 2 proteins interact with the L-type Ca2+ channel dihydropyridine receptors (DHPRs) in skeletal muscle. J. Biol. Chem. 286:43717–43725. 10.1074/jbc.M111.292755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., Komazaki S., Sasamoto K., Yoshida M., Nishi M., Kitamura K., and Takeshima H.. 2001. Deficiency of triad junction and contraction in mutant skeletal muscle lacking junctophilin type 1. J. Cell Biol. 154:1059–1068. 10.1083/jcb.200105040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstrom, A.P., Beavers D.L., and Wehrens X.H.. 2014. The junctophilin family of proteins: from bench to bedside. Trends Mol. Med. 20:353–362. 10.1016/j.molmed.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, T., Kashihara T., Komatsu M., Kojima K., Takeshita T., and Yamada M.. 2018. Physical interaction of junctophilin and the CaV1.1 C terminus is crucial for skeletal muscle contraction. Proc. Natl. Acad. Sci. USA. 115:4507–4512. 10.1073/pnas.1716649115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, M., Sakagami H., Komazaki S., Kondo H., and Takeshima H.. 2003. Coexpression of junctophilin type 3 and type 4 in brain. Brain Res. Mol. Brain Res. 118:102–110. 10.1016/S0169-328X(03)00341-3 [DOI] [PubMed] [Google Scholar]
- Papadopoulos, S., Leuranguer V., Bannister R.A., and Beam K.G.. 2004. Mapping sites of potential proximity between the dihydropyridine receptor and RyR1 in muscle using a cyan fluorescent protein-yellow fluorescent protein tandem as a fluorescence resonance energy transfer probe. J. Biol. Chem. 279:44046–44056. 10.1074/jbc.M405317200 [DOI] [PubMed] [Google Scholar]
- Park, J., Werley C.A., Venkatachalam V., Kralj J.M., Dib-Hajj S.D., Waxman S.G., and Cohen A.E.. 2013. Screening fluorescent voltage indicators with spontaneously spiking HEK cells. PLoS One. 8:e85221. 10.1371/journal.pone.0085221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni, S., and Beam K.. 2021. Neuronal junctophilins recruit specific CaV and RyR isoforms to ER-PM junctions and functionally alter CaV2.1 and CaV2.2. eLife. 10:e64249. 10.7554/eLife.64249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni, S., Lavorato M., and Beam K.G.. 2017. De novo reconstitution reveals the proteins required for skeletal muscle voltage-induced Ca2+ release. Proc. Natl. Acad. Sci. USA. 114:13822–13827. 10.1073/pnas.1716461115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phimister, A.J., Lango J., Lee E.H., Ernst-Russell M.A., Takeshima H., Ma J., Allen P.D., and Pessah I.N.. 2007. Conformation-dependent stability of junctophilin 1 (JP1) and ryanodine receptor type 1 (RyR1) channel complex is mediated by their hyper-reactive thiols. J. Biol. Chem. 282:8667–8677. 10.1074/jbc.M609936200 [DOI] [PubMed] [Google Scholar]
- Polster, A., Perni S., Bichraoui H., and Beam K.G.. 2015. Stac adaptor proteins regulate trafficking and function of muscle and neuronal L-type Ca2+ channels. Proc. Natl. Acad. Sci. USA. 112:602–606. 10.1073/pnas.1423113112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polster, A., Perni S., Filipova D., Moua O., Ohrtman J.D., Bichraoui H., Beam K.G., and Papadopoulos S.. 2018. Junctional trafficking and restoration of retrograde signaling by the cytoplasmic RyR1 domain. J. Gen. Physiol. 150:293–306. 10.1085/jgp.201711879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome, L.C., Syme D.A., Hollingworth S., Lindstedt S.L., and Baylor S.M.. 1996. The whistle and the rattle: the design of sound producing muscles. Proc. Natl. Acad. Sci. USA. 93:8095–8100. 10.1073/pnas.93.15.8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu, G., Wazen R.M., Colarusso P., Chen S.R.W., Zamponi G.W., and Turner R.W.. 2019. Junctophilin proteins tether a Cav1-RyR2-KCa3.1 tripartite complex to regulate neuronal excitability. Cell Rep. 28:2427–2442.e6. 10.1016/j.celrep.2019.07.075 [DOI] [PubMed] [Google Scholar]
- Schredelseker, J., Shrivastav M., Dayal A., and Grabner M.. 2010. Non-Ca2+-conducting Ca2+ channels in fish skeletal muscle excitation-contraction coupling. Proc. Natl. Acad. Sci. USA. 107:5658–5663. 10.1073/pnas.0912153107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima, H., Komazaki S., Nishi M., Iino M., and Kangawa K.. 2000. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell. 6:11–22. [DOI] [PubMed] [Google Scholar]
- Takeshima, H., Hoshijima M., and Song L.S.. 2015. Ca2+ microdomains organized by junctophilins. Cell Calcium. 58:349–356. 10.1016/j.ceca.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]