Abstract
The goal of genomics and systems biologyis to understand how complex systems of factors assemble into pathways and structures that combine to form living organisms. Great advances in understanding biological processes result from determining the function of individual genes, a process that has classically relied on characterizing single mutations. Advances in DNA sequencing has made available the complete set of genetic instructions for an astonishing and growing number of species. To understand the function of this ever-increasing number of genes, a high-throughput method was developed that in a single experiment can measure the function of genes across the genome of an organism. This occurred approximately 10 years ago, when high-throughput DNA sequencing was combined with advances in transposon-mediated mutagenesis in a method termed transposon insertion sequencing (TIS).In the subsequent years, TIS succeeded in addressing fundamental questions regarding the genes of bacteria, many of which have been shown to play central roles in bacterial infections that result in major human diseases. The field of TIS has matured and resulted in studies of hundreds of species that include significant innovations with a number of transposons. Here, we summarize a number of TIS experiments to provide an understanding of the method and explanation of approaches that are instructive when designing a study. Importantly, we emphasize critical aspects of a TIS experiment and highlight the extension and applicability of TIS into nonbacterial species such as yeast.
Keywords: transposon insertion sequencing, Tn-seq, HITS, TraDIS, INSeq, TIS
1. INTRODUCTION
Next-generation sequencing has revolutionized biological research by making whole-genome sequencing relatively cheap and available to the masses. This has resulted in a deluge of available fully sequenced genomes that continues today and has pushed a critically important problem into the limelight: Many of the genetic components that make up a genome have unknown functions. No matter what organism you consider, whether it is human, insect, plant, fungus, or bacterium, we know very little about why most protein-coding genes, and often more than 60% of those in a genome, are there and what they are doing, let alone other genetic components such as noncoding RNAs (ncRNAs). Thus, while the development of next-generation sequencing has been an amazing, liberating achievement, every newly sequenced genome is exacerbating the problem by adding more and more genetic components of unknown function to the pile.
To uncover a gene’s function, a common approach for many decades has been to break a gene (e.g., by mutating it, leading to a loss of function) and see what happens to the organism. For instance, an embryo may stop developing past a certain stage, indicating a gene’s involvement in development, or a bacterium may no longer be able to mend its DNA, pointing to a role in DNA repair. To speed such experiments up, ordered arrays were developed for model organisms such as yeast and Escherichia coli, where every dispensable gene in the genome is knocked out and the strains are stored in 96- or 384-well plates, making them relatively easy to screen by robotics. However, these knockout arrays are time-consuming to produce and have only been created for a limited number of species, including E. coli and the two model yeasts, Saccharomyces cerevisiae and Schizosaccharomyces pombe. About ten years ago, transposon insertion sequencing (TIS) approaches were developed, which enabled the rapid discovery and screening of all genetic components in a bacterial genome. Later, similar approaches were applied in higher organisms such as yeasts, and a promising version of TIS has been developed for human cells.
TIS approaches in hundreds of different strains and species, most of them bacteria, have been used to uncover detailed biology over the years, including many new leads for gene function, ncRNAs, pathways of pathogenesis, antibiotic mechanisms of action, and drug and vaccine targets. The overwhelming number of TIS experiments and the wide range of species studied provide a valuable roadmap for those who wish to adapt this sequencing method to address their research priorities.
The objectives of this review are to describe the powerful capabilities of TIS and provide a guide to apply these methods to any number of biological questions. We describe how TIS is used with bacteria and yeast to identify which genes are essential for viability. We highlight how researchers have varied the conditions of growth to determine which genes of pathogens are necessary for infecting the tissues of plants and animals. We also describe recent innovations of TIS technology, including single-cell methods that can identify genes that are important for growth in communities or for cell phenotypes such as size or shape. In each of these applications, we point out what parameters are key to the success of the experiments and how an approach can be applied to different species and conditions.
2. THE BASIC MECHANICS OF TRANSPOSON INSERTION SEQUENCING
Essential to all TIS studies is an engineered transposon with high integration efficiency and low target specificity. Early studies of molecular genetics identified a number of transposons and characterized their activities, which played a key role in adapting certain transposons for biotechnology. Each transposon comes with its own recombination enzyme, called a transposase, and these have been studied both in vivo and in vitro to define the elements necessary for the excision and integration of the transposon DNAs (see Table 1 for information about the transposons commonly used in TIS).
Table 1.
Transposons
| Transposon | Source | TIS hosts | Sequence specificity |
|---|---|---|---|
| Tn5 | Escherichia coli | Bacteria | 5′-G(CT)(CT)(CT)(AT)(AG)(AG)(AG)C-3′ |
| Tn10 | Shigella flexneri | Bacteria | 5′-GCTNAGC-3′ |
| Himar1 | Horn fly | Bacteria | 5′-TA-3′ |
| Hermes | House fly | Yeasts | 5′- TNNNNA-3′ |
| Ac/Ds | Maize | Yeasts | None reported |
| piggyBac | Cabbage looper moth | Yeasts, Plasmodium | 5′- TTAA-3′ |
The bacterial transposons Tn5 and Tn10 were discovered as a result of their ability to transfer drug resistance, and over 40 years of research have revealed intimate details regarding the transposases and their binding of the terminal inverted repeats at the ends of the transposons (45, 70). The transposon most often used in biotechnology is perhaps Himar1, a member of the Tc1/mariner superfamily reconstructed as a consensus of sequences from the horn fly.Importantly, Himar1 is highly active in bacteria (47).
The TIS method is based on creating high numbers of single transposon insertions in cells that combined make up a dense collection of nearly random disruptions (Figure 1a). After continuous growth, insertions drop out of cultures if they disrupt sequences necessary for viability (Figure 1b). The location and number of insertions are determined before and after growth using high-throughput sequencing (Figure 1c). A key to the success of TIS is that the inserts should be evenly distributed with low sequence bias.Tn5is often used for TISin part because it has relatively low sequence bias at insertion sites. A study of 24,493 insertion events determined a weak consensus sequence of nine bp consisting of 5′-G(CT)(CT)(CT)(AT)(AG)(AG)(AG)C-3′ (81). There are 2 positions where the consensus reaches 70%: the (CT) and the (AG) that flank the central (AT) of the target site. The result is a bias in favor of GC sequences, which allows Tn5 to function well in genomes with high GC content (31). Tn10 has been used in some TIS studies, but it has a stronger sequence bias in its 6-bp consensus, GCTNAGC (6, 36). Himar1 is commonly used in TIS experiments in part because it has relatively low sequence bias, requiring just the TA base pairs at the center of its target sequence to allow for DNA bending (48).
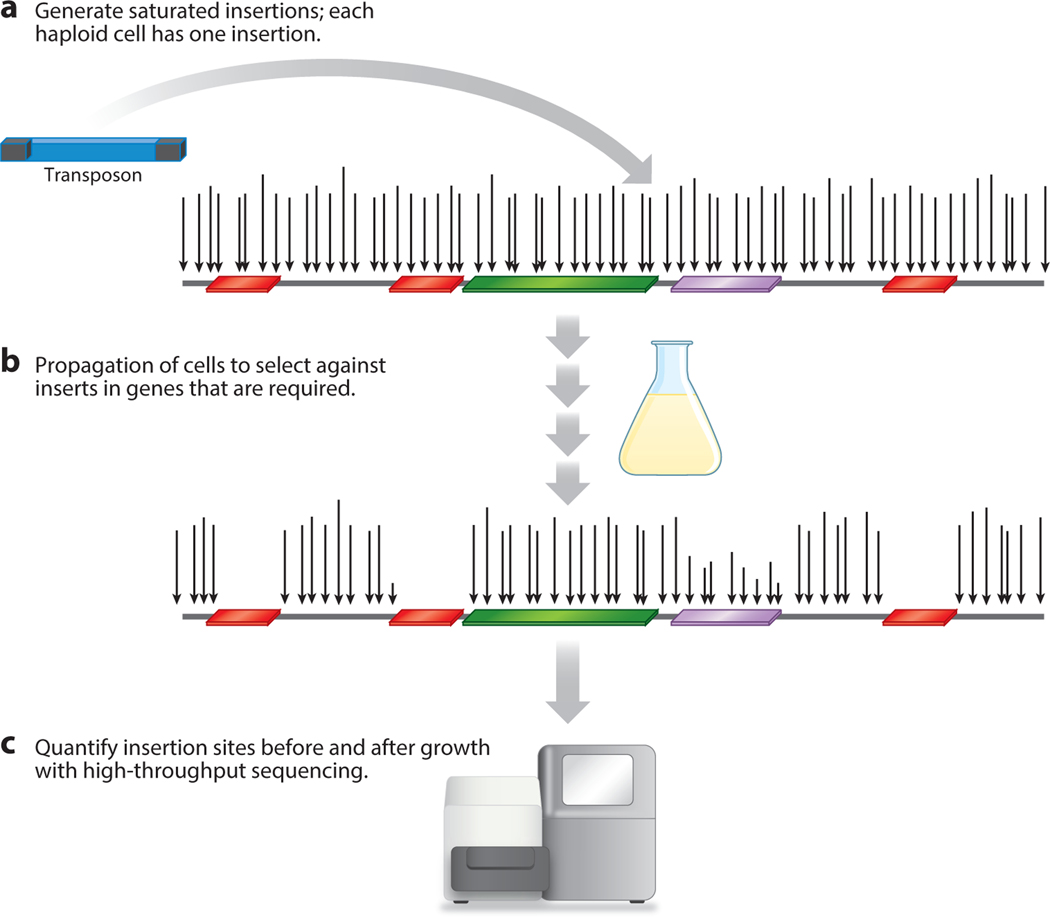
Figure 1.
An overview of transposon insertion sequencing. (a) A saturated and evenly distributed profile of insertions is generated in a population with each cell containing a single insertion. The arrows show the positions of insertion, and their lengths indicate read numbers. Open reading frames are shown as rectangles colored by function (red is essential, green is nonessential, and purple is important). (b) After insertions are produced, the cells are propagated extensively in conditions of interest. Genes important for growth in the defined conditions retain fewer insertions relative to their fitness contribution. (c) The number of reads for each insertion in cultures is quantified with high-throughput sequencing. Sequence reads are measured before and after cells are propagated.
The transposons of bacteria have low integration activity in eukaryotic systems. To conduct TIS in yeast species or the malaria-causing parasite Plasmodium where the genomes are significantly larger than bacteria, transposons isolated from eukaryotes are used. The Hermes transposon, from the housefly, has an insertion target of 8 bp with a strong bias at two positions, NTNNNNAN (34). This recognition sequence is common in the AT-rich genomes of S. pombe and S. cerevisiae, which have been subjected to TIS with Hermes (22, 35, 52, 53). Activator/Dissociation (Ac/Ds) is a highly active transposon from maize that is used in TIS with S. cerevisiae (61) and the human pathogenic yeast Candida albicans (80). One advantage of Ac/Ds in TIS is that, unlike Hermes or piggyBac from the cabbage looper moth, it does not have a significant nucleotide bias (61). A key feature of Ac/Ds with important consequences is that it contains a promoter active in S. cerevisiae that expresses sequences downstream of insertions. This allows disruptions of essential genes to be tolerated when essential domains are expressed downstream of the insertion. In many cases, the location of essential domains within a coding sequence can be visualized as a region of low insertion density (61).While this represents an advance in information provided by TIS, the presence of the promoter allows more insertions to be tolerated in essential genes, which can make it harder to determine which genes are essential. The piggyBac transposon is active in many complex eukaryotes, including mammals (19, 67, 101). Although piggyBac has a four-nucleotide target sequence TTAA, it is able to identify essential genes in Plasmodium falciparum because the genome is >80% AT (105).
For decades, geneticists have been able to generate dense profiles of largely random insertions and select for cells with desired phenotypes in defined conditions. But without a method for mapping high numbers of insertions, it was impossible to know what regions of the genome contributed to growth (1). The development of high-throughput DNA sequencing made it possible to pinpoint the genes and sequences throughout the genome that contribute to growth. This is readily done with maps of approximately 100,000 to 1,000,000 insertion sequences in growing cultures to determine which genes have fewer insertions than expected by statistical measures of the insertion density.
When high-throughput sequencing became available, four groups simultaneously published related methods of TIS with different transposons and sequencing strategies (Figure 2). The TIS methods vary by how the transposon insertions are generated. Transposon-directed insertion site sequencing (TraDIS) relies on Tn5 transposase and transposon DNA that are assembled into transpososomes that are transformed into query strains where integration occurs in vivo (Figure 2a) (50).The high-throughput insertion tracking by deep sequencing (HITS) and Tn-seq methods combine purified Himar1 transposase and miniHimar1 DNA in in vitro reactions with genomic DNA from query strains such as Haemophilus influenzae or Streptococcus pneumoniae (Figure 2b) (26, 91). The resulting insertions are transformed into cells where homologous recombination replaces the wild-type sequence with sequences containing the insertions. It is also possible to create insertions in vivo by expressing the transposase in cells containing a Himar1 donor plasmid, as is done with insertion sequencing (INSeq) (Figure 2c) (30).
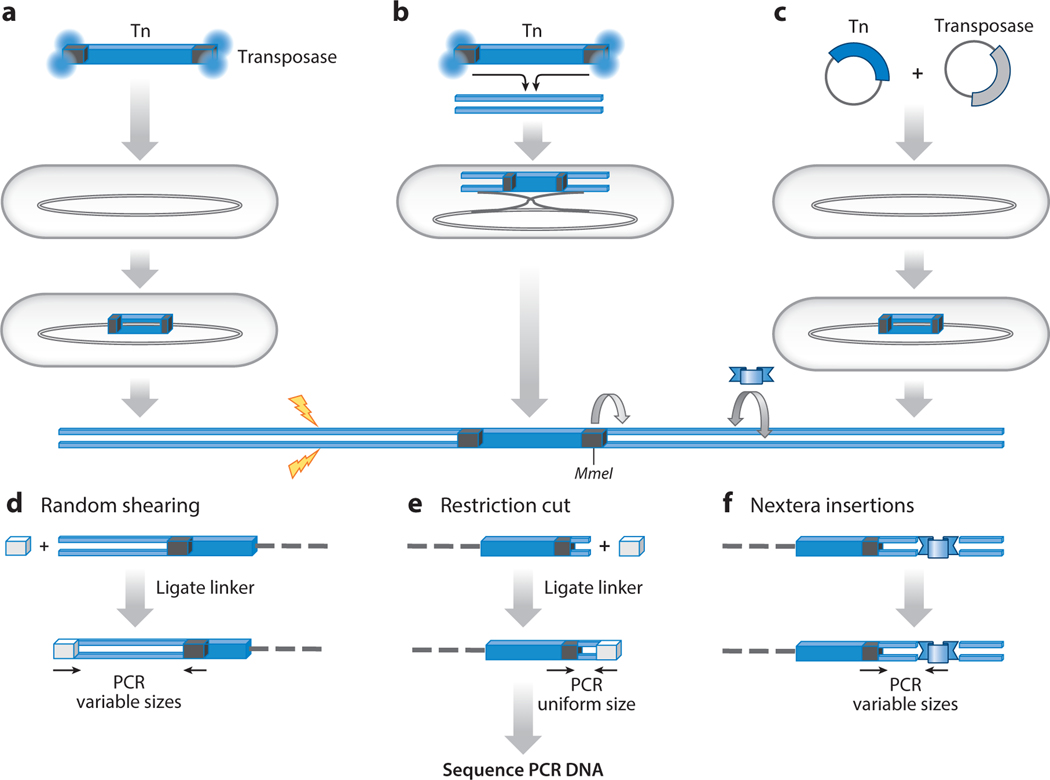
Figure 2.
Three methods for introducing transposon insertions into genomic DNA. (a) Purified transposon-transposase complexes, such as the commercially available EZ-Tn5™, are directly introduced into cells. (b) In vitro integration into genomic DNA is achieved by using purified transposon-transposase complexes. The genomic DNA with insertions is introduced into cells where homologous recombination replaces wild-type DNA with transposon-containing sequences. (c) A transposase is expressed in cells that contain a plasmid copy of a transposon. By introducing plasmids that contain a transposon and express a transposase, insertions are generated in vivo. Once a population of cells contain transposon insertions, there are three methods to sequence their positions. (d) Genomic DNA is sonicated to create random shearing (yellow lightning). The sheared DNA is treated with enzymes to make blunt ends that are ligated to a double-stranded linker. PCR with linker- and transposon-specific oligonucleotides produces variable-sized fragments that are sequenced. (e) Genomic DNA is digested with the type II restriction enzyme MmeI, which binds to a sequence at the end of Himar1 and cuts 16 to 20 bp downstream into the flanking genomic DNA. A linker is ligated to the MmeI cut ends, and primers specific for the linker and transposon PCR amplify uniform-sized fragments that are sequenced. (f) The commercially available Nextera kit (Illumina Inc.), used for whole-genome sequencing, circumvents the need to fragment the DNA by inserting an engineered transposon throughout the genome. Primers specific for the Nextera transposon (blue ribbon) and the TIS transposon are used to PCR amplify variable-sized fragments that are sequenced.
Two TIS approaches, HITS and TraDIS, amplify insertion sites using random shearing of genomic DNA followed by the attachment of linkers (Figure 2d) (26, 50). Two other approaches, INSeq and Tn-seq use Himar1 that was manipulated to contain an MmeI type II restriction enzyme site in the inverted repeats of the transposon (Figure 2e) (30, 91). This is an important innovation because MmeI cuts 16 to 20 bp past the end of the transposon (size varies depending on the version of Himar1). A linker is ligated to the MmeI fragments, and PCR is used to amplify libraries of Himar1 along with 20 bp of the flanking insertion sites. The 16 to 20 bp from the target site identify the position of individual integration events. The advantage of the MmeI method is that the insertion fragments are uniform in size, which reduces PCR bias due to length differences. An advantage of random shearing is that it can be used in conjunction with transposons that lack the MmeI site. Recently, a third approach for fragmenting genomic DNA has been adopted. A commercially available transposon (Nextera) is inserted into the genomic DNA containing the TIS inserts. Primers specific for Nextera and the TIS transposon PCR amplify fragments of varying sizes that are sequenced (Figure 2f).
With different transposons and methods of introducing insertions, it is possible to optimize TIS for a wide range of query species. The primary condition linked to the success of all TIS experiments is that the libraries must achieve a high number of insertions and ideally reach saturation. In particular, the statistical significance as measured by modeling algorithms (described in Section 7) and thereby the predictive value for individual genes can be greatly improved by increasing integration densities. Section 8.1 describes how saturation of dispersed events influences TIS and how the choice of transposon impacts saturation in recipient species. Because saturation is important to the success of TIS, we provide a number of points to optimize library construction in Section 8.2.
To determine the impact of these considerations on an experiment, it is helpful to review the results of published TIS studies. If available for a query species of interest, the examination of integration sites can reveal the level of saturation provided by a specific transposon and number of events. To assist in the review of published studies, we have assembled an extensive reference list of TIS data that have achieved a threshold level of saturation (Supplemental Table 1).
Once the conditions of generating the integration libraries are optimized, there are several types of TIS experiments that can identify the function of genes in a query species. We describe these below with an emphasis on considerations that should be taken into account when planning and performing experiments and computational analyses that follow. If they exist, we describe alternative approaches to answer the same question and the pros and cons of these methods.
3. GENOME-WIDE ESSENTIAL GENE DISCOVERY
TIS has undoubtedly been used most often for the identification of essential genes in a genome (defining essential as agene or genetic region absolutely required for growth or survival in standard lab conditions, for example, rich growth medium). This not only highlights the most critical parts of an organism and carries important information concerning the processes and pathways that make an organism tick, it also forms a gateway to developing a minimal genome. For pathogenic bacteria, essential genes have always been seen as key targets for drug design. However, because transposon disruptions of essential genes are not tolerated, essential genes are defined based on the absence of transposon insertions. Such screens are thus based on the identification of a negative result, which can make them a bit tricky. One obvious issue that comes with such a screen is that the absence of an insertion can also be the result of a gene simply not being hit. When screening for essential genes, it is thus important to create saturated libraries by establishing an even distribution of insertions across the entire genome, with multiple insertions in each gene. Importantly, the denser the distribution, the more accurate the results can become, and they may even lead to the identification of smaller essential genetic components (see Section 8.1).
Over the years, many organisms have been screened with TIS to identify essential genes, including human pathogens like Mycobacterium tuberculosis (32), S. pneumoniae (91, 93), H. influenzae (26), Salmonella typhimurium (50), Staphylococcus aureus (89), Porphyromonas gingivalis (46), Burkholderia cenocepacia (100), Vibrio cholerae (12), and Yersinia pseudotuberculosis (99); the bacterium E.coli K12 (29); commensal gut bacteria like Bifidobacterium breve (75); animal pathogens like Streptococcus equi (13); plant pathogens such as Pseudomonas syringae (60); and bacteria with potential engineering or industrial value such as Methylobacterium extorquens (64). An extensive list of TIS experiments is provided in Supplemental Table 1, which indicates studies that identify essential genes. Highly saturated libraries have been used to go beyond identifying essential genes. For instance, in the cyanobacterium Synechococcus elongatus, a library was created with average insertions for every 11 bp, which, besides essential genes, identified essential intergenic regions, ncRNAs, and regulatory elements (74). In M. tuberculosis, a library of 2.5 × 106 independent insertions, occupying over 84% of the available TA sites in the genome, also resulted in the identification of essential promoters, intergenic regions, and regulatory RNAs (18).
The vast majority of TIS publications describe the function of genes in bacteria. This is in large part because bacterial genomes are well suited, being relatively small, gene dense, and haploid. The genomes of archaeal species are similar in structure to bacteria and have also been studied with TIS to identify essential genes. These species are haploid, are closely related to eukaryotes, and exhibit unusual biology, including growth in extreme temperatures and high salt. TIS studies with Sulfolobus islandicus and Methanococcus maripaludis revealed significant changes in essential gene sets associated with the evolution of Archaea towards eukaryotes (79, 104).
Essential gene screening with TIS thus suffers from the limitation that insertions cannot be directly measured and therefore the function of these genes cannot be studied. A few labs have addressed this challenge by using gain of function insertions with transposons that have outward-facing promoters that can overexpress or repress genes. Measuring changes in insertion frequency when downstream genes are induced by the transposon promoter can identify essential genes because growth depends on the activity of the promoter. Different versions of outward-facing promoters have been used to identify essential genes in Caulobacter crescentus (14) and S. aureus (15, 78). This approach was combined with the traditional TIS method TraDIS to provide the package TraDIS-Xpress (102). The software uses inserts of Tn5 with an inducible PBAD promoter that faces out of a Tn5 combined with detailed transposon inactivation data. As a result, TraDIS-Xpress has successfully identified both essential and nonessential genes and also was applied to reveal genes affecting tolerance to the biocide triclosan. While some studies rely on transposons with inducible outward promoters, others use constitutive promoters with different strengths, which can provide different expression levels in the same culture (15, 78).
The broad success of TIS in determining the function of bacterial genes signaled the possibility that the function of eukaryotic genes might also be revealed with this method, given that some species have a haploid state or can be forced into a haploid state. The genome sizes of model eukaryotes such as yeasts are above 12 Mb, suggesting they could be challenging, given that the typical bacterial genomes studied with TIS range from 2 Mb to 6 Mb. Nevertheless, the identification of highly active eukaryotic transposons and improvements in library production provided the opportunity to use TIS with eukaryotic species. Evolutionarily distant species diverged by 4 × 108 years have been examined, including S.pombe, S.cerevisiae, Saccharomyces uvarum, Pichia pastoris, and C. albicans (35, 61, 76, 80, 108). These studies provide a core set of genes that are essential in eukaryotes over a wide range of evolution (80). The genome of the malaria-causing parasite Plasmodium is 23 Mb and is in reach of TIS to identify the essential genes of its haploid blood stage (105). This research offers the unique opportunity to identify and prioritize therapeutic targets.
4. IDENTIFICATION OF CONDITIONALLY IMPORTANT AND/OR ESSENTIAL GENES IN VITRO
Arguably, the two most obvious extensions of essential gene screening with TIS are changing the screening environment, which enables the identification of conditionally essential genes, and/or exploiting the quantitative nature of TIS, whereby it is possible to identify genetic regions that when disturbed by a transposon insertion may not be strongly detrimental to the organism but has a more subtle negative or positive effect on fitness (see Section 8.3).
Most TIS experiments that screen essential genes or those that are important in a certain condition focus on identifying genetic regions that affect the growth or survival of the organism. These experiments are often performed by growing a transposon library for multiple generations in a specific environment. To determine which transposon insertions exert an effect on fitness, researchers isolate DNA from the transposon library at the start of the experiment (right before the selection regime is initiated) and at the end of the experiment. Sequencing libraries are prepared and sequenced, and the frequency of each transposon insertion is determined at both time points and used to calculate each insertion’s effect on fitness. Genetic regions with insertions that completely disappear from the population are often deemed conditionally essential. But also, more subtle fitness effects can be extracted, as the rate of change in frequency is indicative for the strength of the fitness effect. For instance, transposon insertions that rapidly change in frequency have a larger effect on fitness than those whose change is slower. Depending on how fitness is calculated and what the measurement exactly represents (see Section 8.3), a wide variety of fitness effects can be extracted, including positive, neutral, negative, and essential, and depending on the sensitivity and quantitativeness of the exact TIS approach, even highly sensitive growth rates can be calculated. Finally, these data are used to determine which genes are involved in dealing with the stress experienced in the environment, and this information can be used to obtain leads for new gene function. A very large number of environments have been explored over the years for many different organisms. For instance, nutrient-limiting conditions and sugar and amino acid metabolism have been explored with TIS for a large variety of species to fill in gaps in pathways, leading to the identification of genes with new metabolic roles (88). Other conditions include those that mimic a specific in vivo environment for a bacterial species, such as bile (50), saliva (96), serum (58, 77, 106), and nitric oxide exposure (33). Moreover, ex vivo environments are relatively easy to explore, for instance, to identify Neisseria meningitidis genes and intergenic regions important for host cell colonization (8) or Moraxella catarrhalis genes critical for attachment to epithelial cells (16).
It is important to realize that a TIS experiment always generates a list of genes and/or genetic components that are not entirely straightforward to interpret. On this list will be genes with known and unknown function, of which many on first glance may not seem directly relevant to the exerted stress. This is nicely illustrated when TIS is performed in the presence of antibiotics to identify genes that are involved in intrinsic resistance and thus important in overcoming the stress imposed by the antibiotic. For these types of experiments, transposon libraries are grown for several generations in the presence of a subinhibitory antibiotic concentration that inhibits growth, for instance, by 30–50%.Most antibiotics target specific genes or processes that are often essential for basic growth. Therefore, while a directly targeted essential gene may not always be identifiable with TIS (remember, essential genes do not tolerate transposon insertions), an essential process often depends on a set of genes in which several may not be (completely) essential. For instance, TIS in the presence of cell wall synthesis inhibitors, such as penicillin or vancomycin, highlights the increased importance of nonessential genes that are involved in peptidoglycan synthesis. However, the list of important genes in the presence of cell wall synthesis inhibitors stretches far beyond peptidoglycan synthesis genes and includes many metabolic genes, regulators, transporters, membrane integrity genes, and those involved in energy generation. These results highlight how no single gene or pathway acts just by itself but instead is tightly integrated into a genomic network, interacting with all kinds of other pathways and systems (15, 25, 27, 28, 41, 42, 62, 68, 95). The effects of most stressors, including antibiotics, thus may start at the stressor’s direct target, but because of network interdependencies,the effect of the stress reverberates throughout the organism, and with increasing stress the response becomes more complex, involving more and more genes.
5. IDENTIFICATION OF CONDITIONALLY IMPORTANT AND/OR ESSENTIAL GENES IN VIVO
The studies that introduced INSeq (30) and HITS (26) right away highlighted the application of TIS in vivo. HITS took transposon libraries of H. influenzae to inoculate the mouse lung. This resulted in a list of over 130 genes, dispensable for growth in vitro but critical for lung infection, with diverse functions, including metabolism, oxidative stress, and membrane integrity. INSeq took on a more complex question by creating transposon libraries in the human microbiome species Bacteroides thetaiotaomicron. These libraries were then placed in the mouse gut in the presence and absence of other gut commensals. While this TIS screen identified a very diverse set of genes, including those involved in vitamin B12 synthesis, it also showed how community composition can determine the importance of certain processes. Most subsequent in vivo TIS studies performed with bacterial pathogens have focused on identifying genes that are important for a wide range of infection conditions, including genes important for colonization of the nasopharynx or lung with, for instance, S. pneumoniae (93, 103) or H. influenzae (26), infection of the bloodstream to replicate sepsis by Proteus mirabilis (3) or Acinetobacter baumannii (83), necrotizing fasciitis and myositis in nonhuman primates by group A Streptococcus (109), catheter-associated urinary tract infection from P. mirabilis (4), skin infection with group A Streptococcus (51), intestinal colonization of infant rabbits with V. cholerae (24) or Vibrio parahaemolyticus to simulate diarrheal disease (38), intestinal colonization of infant rabbits and dissemination into the environment (i.e., pond water) with V. cholerae (43), and infection of the spleen by uropathogenic E. coli (84). These data are important because they can give detailed insights into which bacterial mechanisms or pathways are key in establishing an infection or important for the progression of an infection.
Moreover, infections may not always be caused by a single strain or species, and coinfections can make what is important much more complex. For instance, in S. aureus, over 200 genes are essential in a mouse surgical wound infection model. However, when a coinfection is established in the presence of Pseudomonas aeruginosa, 25% of those genes lose their essentiality, indicating that P. aeruginosa somehow changes the environment, rendering a substantial number of processes in S. aureus no longer critical (40). Moreover, the importance of the community on species-specific gene essentiality is further highlighted by a larger study where the oral pathogen Aggregatibacter actinomycetemcomitans was grown in a murine abscess in pairwise coinfections with 25 other oral and nonoral cavity species (54).Interestingly, while some species increased the number of essential genes required for A.actinomycetemcomitans infection,other species decreased the numberof essential genes. These are incredibly intriguing results that highlight how community interactions can affect the manner in which bacteria experience their environment and the enormous complexity hidden in bacterial communities. Moreover, because the importance of some gene products depends on which other organisms are present within the environment, this suggests that drugs or vaccines targeting specific gene products may be less efficient, depending on a possible coinfection.
TIS was also recently used to identify genetic factors that affect the transmission of S. pneumoniae between hosts (73). To accomplish this, a S. pneumoniae TIS library was used to colonize the nasopharynx of ferrets, which were housed in separate cages adjacent to healthy ferrets. The colonized animals were then triggered to sneeze on their neighbors, enabling airborne transmission. Subsequently, bacteria were collected from the possibly newly colonized animals, and transposon mutants were analyzed to determine whether some were overrepresented in the data, indicative of defects in genes that inhibit transmission. Alternatively, underrepresentation of transposon mutants in the data was indicative of those genes and regions that contributed to transmission. This analysis resulted in a detailed fitness landscape of those genes involved in transmission between animals. To ensure whether the identified genes indeed affect transmission, several targeted deletion mutants were created and were used in single-mutant transmission experiments to confirm their phenotype. Nota bene, such validation experiments are critical to assess whether any TIS-generated data are truly of high confidence. Moreover, not only can phenotypically important genes give insight into the mechanisms to which they contribute, in this case transmission, data related to these genes can also be used to design antimicrobial strategies. For instance, putative C3-degrading protease CppA, iron transporter PiaA, and competence regulatory histidine kinase ComD all significantly contribute to transmissibility. Specifically, when any of these genes are knocked out, transmissibility becomes drastically reduced, which suggests that if these genes can be targeted, the transmission of S. pneumoniae between hosts can be reduced. Even more interesting, it turns out that if female mice are vaccinated with the recombinant protein products of these genes, it blocks transmission to their offspring (73).
In vivo experiments can thus be of high value, help us understand how organisms grow and survive in a complex environment, and even direct the design of novel strategies to target these organisms. However, in vivo experiments are in general much more complicated than those performed in vitro and are susceptible to a higher level of experimental variation. This variation is often, at least partially, the result of bottlenecks of the transposon library, which originate from stochastic loss of random transposon insertions due to uncontrolled population reduction during the experiment. Importantly, bottlenecks can and should be taken into account when designing a TIS experiment as well as when analyzing the data (see Section 8.4).
While genes identified as important or essential in a certain environment inform researchers about which genes and pathways the organism uses to overcome the stress, they also simultaneously inform about the composition of the environment or the type of stress the environment applies to the organisms. For instance, TIS in the presence of antibiotics informs researchers about the mechanism of action as well as the secondary consequences of the antibiotic. This is nicely illustrated by fluoroquinolones such as ciprofloxacin that target DNA gyrase and topoisomerase to inhibit DNA replication (28, 93). The secondary effect is that the antibiotic triggers elevated levels of DNA damage because a function of gyrase is to resolve replication knots that occur during DNA replication, which it accomplishes by introducing double-stranded breaks followed by repair. The antibiotic, however, inhibits all functions of gyrase, resulting in elevated levels of DNA damage. This secondary effect of fluoroquinolones is evident in TIS data from the increased importance of DNA repair genes and pathways that are normally mostly inconsequential during growth in rich lab medium.
TIS can also inform researchers about the composition of the in vivo environment. For instance, TIS experiments were performed in healthy mice and mice with sickle-cell disease (SCD) to identify genes that were differentially required in either condition. Children with SCD have an ~400-fold higher probability to acquire S. pneumoniae–induced bacteremia. However, it was unclear what the exact reason behind this is. TIS identified several dozen genes that are required to trigger bacteremia in healthy mice but dispensable in SCD mice.By analyzing the functional roles of these genes, it was possible to identify several environmental factors that are different between hosts, including an increased availability of iron and nucleotides in the SCD host. Most importantly, the gene cppA turned out not to be required in the SCD host, while it normally is critical in establishing infection. CppA is involved in breaking down C3b complement, which tags encapsulated organisms like S.pneumoniae for clearance by the immune system. Indeed, patients with SCD lack C3b complement, thereby increasing the probability of acquiring S.pneumoniae-induced bacteremia (10). Importantly, the TIS data were used to identify targets for vaccine design, and the different requirements between the healthy and immunocompromised hosts were explored for potential differences in vaccine efficacy. Protein-based vaccines could be developed with CppA and with PiaA, a gene product required for iron scavenging. But, while these vaccines had good efficacy in healthy mice, they were not protective in the immunocompromised host, simply because the bacterium could easily escape the pressure against the proteins it no longer needed. This not only highlights the sensitivity and strength of in vivo TIS data but also the importance of conducting experiments across different hosts.
6. INNOVATIVE APPLICATIONS OF TRANSPOSON INSERTION SEQUENCING: CELL PHENOTYPES, SINGLE-CELL ANALYSIS, COMMUNITY-DEPENDENT GENES, INCREASED HIGH-THROUGHPUT APPROACHES, AND EUKARYOTIC SYSTEMS
In the experiments described thus far, TIS was mainly used to measure either growth or survival. In this section, we illustrate examples of how TIS is capable of studying many more processes and phenotypes. For an extensive discussion of advances in this field, we direct you to the recent publication by Cain et al. (7). Forward mutation studies of bacteria identified a number of genes necessary for flagella and flagella-independent motility. Now TIS can be used to measure the contribution of genes to motility using libraries from cells based on their movement across agar plates. A TIS study of biofilm production identified most genes of P. aeruginosa that are known to be important for twitching motility as well as many new factors, such as a sodium-dependent flagellar protein, a flagellar body ring protein, and signal transduction factors (63). To identify genes involved in capsule production, TIS can be used on cells separated by density gradient centrifugation. In a study of Klebsiella pneumoniae, 78 genes linked to capsule production were identified from two clinically significant strains (20).
Fluorescence-activated cell sorting (FACS) provides perhaps a more general means of cell separation that when used with TIS can identify genes that contribute to many different phenotypes. Factors important for efflux activity and potentially multidrug resistance can be identified by sorting cells treated with ethidium bromide, a fluorescent compound that binds DNA (37). Genes that contribute to heterogenous size in cell populations can be identified with TIS by separating cells into pools based on the retention of a fluorescent agent, calcein (69). In this study of Mycobacterium smegmatis, LamA was shown to cause heterogenous cell size by inhibiting growth from nascent cell ends causing asymmetric polar growth. FACS was also used with S. typhimurium to determine which genes are important for the expression of the typhoid toxin subunit pltB, which was replaced with green fluorescent protein (GFP) (23). Bdellovibrio bacteriovorus is capable of killing most gram-negative bacteria by invading their periplasm and consuming nutrients. To identify genes required for the attachment of B. bacteriovorus expressing tdTomato to prey cells expressing GFP (V. cholerae), TIS insertion profiles of cells sorted red were compared to the insertions of bound pairs producing red and green fluorescence. The results of this approach were validated, showing ten mutants were defective in prey attachment, and importantly, the majority of these genes are hypothetical and previously uncharacterized (21).
Each of the examples of TIS discussed above describes processes that occur in populations of cells. Contributions of genes that promote the community of cells or that increase competition cannot be observed. To identify community-specific genes, TIS is performed on single cells individually grown in oil droplets containing growth medium (87). Droplet Tn-seq (dTn-Seq) thereby showed that 1–3% of S. pneumoniae mutants have altered fitness when grown individually, compared to when they are grown in bulk transposon libraries. These genes contribute to a range of activities, including processing of host glycoproteins and community protection against host immune factors such as the serine protease elastase. Moreover, dTn-Seq enables the exploration of phenotypes associated with microcolony formation; FACS sorting of droplets and the repeated encapsulation of droplets creates multiple-layered droplets, which enables the study of interactions (e.g., communication) between bacterial cells or between bacteria and host (immune) cells (87).
The increasing number of TIS systems that link function to networks of genes caused interest in significantly increasing the throughput of the experiments to include hundreds of conditions. The part of TIS that greatly limits the output is the often relatively cumbersome sample preparation for sequencing. To characterize hundreds of fitness tests simultaneously, random barcode Tn-seq (RB-TnSeq) was developed, and it does not rely on shearing genomic DNA and ligating the fragments to linkers (98). With this high-throughput method, random barcodes are cloned as a library into a transposon that is subsequently introduced into cells to create a saturated profile of insertions (Figure 3). Before environment screening can be performed in high-throughput, first each barcode needs to be linked to a specific location in the genome. While this may be slightly complicated, for any subsequent experiments only the barcodes need to be amplified, which is achieved by a single PCR, which is sequenced and quantified. This significantly reduces the hands-on time, and the limited sample preparation can reduce experimental variation. With RB-TnSeq, 130 carbon source combinations were tested with 5 different bacterial species (98). In a separate study, this barcode method was applied to 32 species of bacteria, and by testing large numbers of growth conditions, phenotypes could be associated with 11,779 genes that have no annotated function (65), which can be explored as leads for gene function.
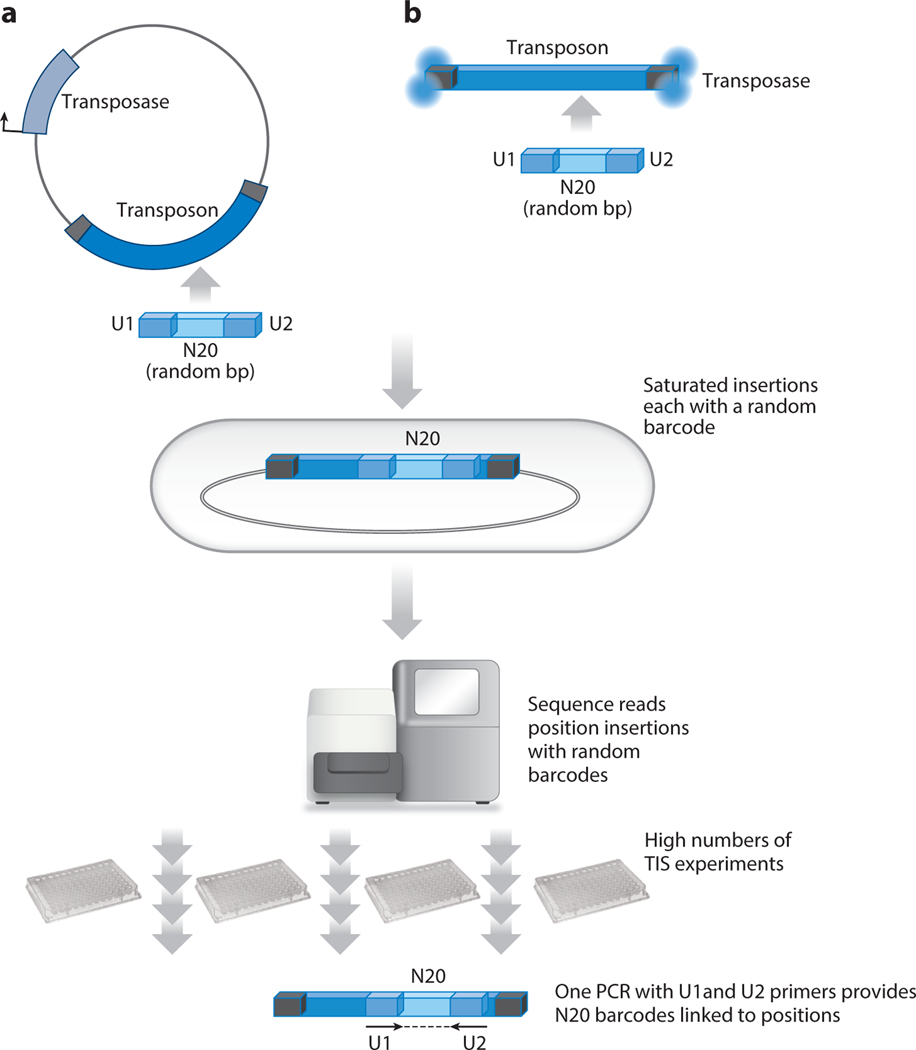
Figure 3.
RB-TnSeq is a powerful method that greatly increases the number of TIS experiments that can be done simultaneously. A large library of transposons with a 20-bp random barcode (N20) is made either in a plasmid (a) or in a transposon fragment assembled into transposomes (b). U1 and U2 are unique sequences that flank the N20 random barcode. Once a saturated collection of insertions is made in cells, one of the three types of sequencing methods (Figure 2) positions each insertion and records the random barcode associated with the position. The culture with insertions can now be used for high numbers of TIS experiments because a single PCR with U1 and U2 primers can be used to measure the barcodes after each growth condition. In turn, the barcodes reveal the position and number of TIS insertions. Abbreviations: RB-TnSeq, random barcode Tn-seq; TIS, transposon insertion sequencing.
Transposons have been identified and TIS methods engineered for the study of eukaryotes, most commonly to determine the essential genes of various yeast species (35, 61, 76, 80, 108). Questions related to human pathogenic fungi can be addressed. C. albicans is a common yeast species of the human microbiome that can cause life-threatening infection in immunocompromised individuals. Methods that produce haploid strains of C. albicans allow TIS to identify approximately 1,200 genes essential for growth under standard laboratory conditions (80). Importantly, 130 of these are also essential in Aspergillus fumigatus, Cryptococcus neoformans, and Histoplasma capsulatum. By identifying a set of essential genes conserved among these four major human pathogens, factors can be identified that lack homologs in humans and can therefore serve as potential targets for the development of antifungal therapies (80).
Prion diseases such as transmissible neurodegenerative disorders result from the propagation of misfolded proteins, a mechanism that is readily studied in S. cerevisiae. Forward genetic screens have identified many factors responsible for the propagation of the prion [URE3], which forms aggregates of a transcriptional repressor and causes dysregulation of nitrogen metabolism. Much less is known about factors that protect cells from the toxic effects of prions. To identify a comprehensive set of genes that compensates for the inhibitory impact of prion propagation, TIS was conducted with a strain of S. cerevisiae that contains [URE3]. Genes that prevent the toxicity of [URE3] can be identified because disruptions are diminished in a strain that carries [URE3] relative to a strain that lacks [URE3] (22). Genes that increase fitness in the presence of the prion include chaperones and several factors involved in the posttranslational attachment to proteins of the short peptides ubiquitin or NEDD8.
The Ac/Ds transposon of maize functions in S.cerevisiae to identify genes essential for growth in standard laboratory medium (61).In addition to its high activity and minimal sequence bias,Ac/Ds is surprising in that it has a promoter that expresses downstream genes. Insertions in nonessential domains of essential genes are tolerated because the promoter expresses the essential domains downstream of the insertion (Figure 4a). These data provide a unique map of the necessary sequences within essential genes.
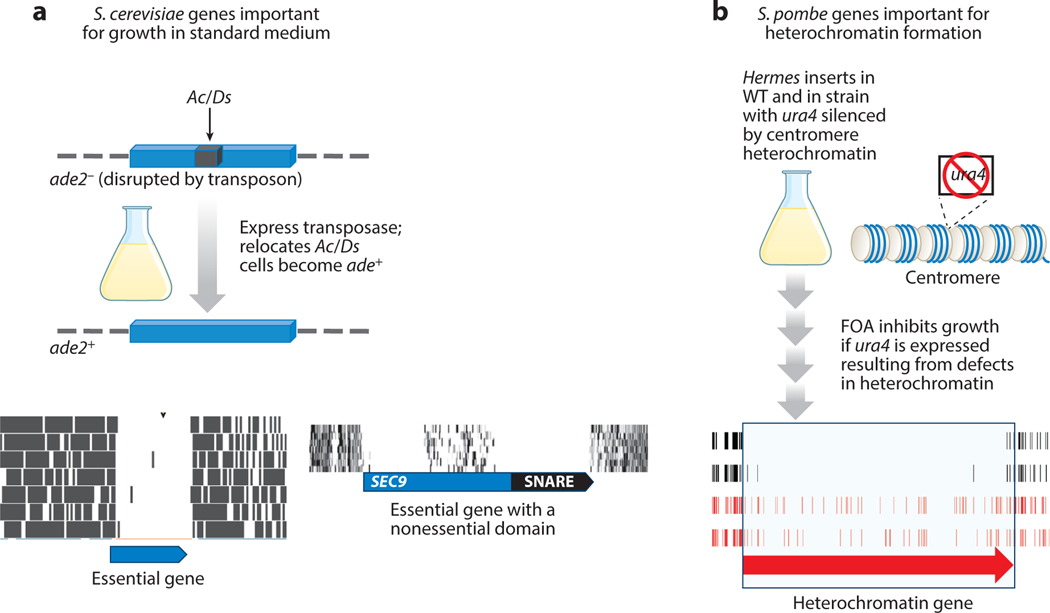
Figure 4.
TIS with yeast identifies unique features of eukaryotic genes. (a) The Ac/Ds transposon in Saccharomyces cerevisiae identifies essential genes and nonessential domains within essential genes. Ac/Ds positioned in ade2 causes cells to be unable to grow in the absence of adenine. Once the Tpase is induced, Ac/Ds is excised and inserts throughout the genome, resulting in the repair of ade2 and ability to grow in the absence of adenine. Insertions in essential genes do not accumulate in cells grown in standard medium. Ectopic transcription from within Ac/Ds can allow the disruption of some essential genes to accumulate if the insertion occurs in a nonessential domain. Such an example occurs in a domain of SEC9. (b) TIS with the Hermes transposon identifies essential and conditionally essential genes in Schizosaccharomyces pombe. Genes important for heterochromatin formation can be identified using FOA, a compound toxic to cells expressing ura4. Mutations in genes important for forming heterochromatin at the centromere allow the expression of ura4 and therefore cannot accumulate in the culture. Abbreviations: Ac/Ds, Activator/Dissociation; FOA, 5-fluoroorotic acid; TIS, transposon insertion sequencing; WT, wild type.
Biological pathways can be identified with S. cerevisiae by combining mutations and testing whether they exhibit interactions, either positive (suppressor) or negative. Typically, a mutation with a phenotype is paired with other mutations in thousands of strains in a deletion set of nonessential genes. This can be a complicated process of mating that typically involves robotics. Alternatively, one can use TIS with S.cerevisiae by making transposon insertions in a pair of strains, one with a mutation of interest and another that lacks the mutation. Genes essential for the growth of S. cerevisiae can be identified in the strain lacking the mutation, and genetic interactions with the query mutation can readily be observed by comparing the insertion profiles of the two strains. Genes that function in the same pathway as the query will typically have fewer insertions in the mutant strain than the wild type. In one TIS experiment, genetic interactions both negative and positive (increased insertions) were detected throughout the genome (61). In addition to identifying genes that function together in pathways, TIS is used to identify genes that are targeted by chemicals or drugs. Chemical interactions with genes were observed with S. cerevisiae by propagating insertion strains in the presence of a biologically active compound. The TORC1 pathway senses nutrients and regulates growth depending on available resources. To identify regulators of TORC1, cultures of insertion cells are grown in various concentrations of rapamycin, a drug that inhibits TORC1 (61). These experiments identified a number of genes known to be important for rapamycin resistance and revealed the factor Pib2 that can provide resistance or sensitivity depending on which domain is expressed. These findings indicate that TIS in S. cerevisiae has the potential to identify pathways of drug resistance and sensitivity.
The saturated profiles of transposon insertions in yeasts provide a significant opportunity to identify genes that contribute to pathways central to the function of eukaryotes. While yeast is a model system key to uncovering fundamental mechanisms of higher organisms, TIS allows the contribution of each gene to be measured in a single experiment. For example, heterochromatin is a highly condensed form of packaged DNA that is key to the function of centromeres, the repression of genes in somatic tissues through development, and the inhibition of transposons. By using a reporter gene silenced in centromere heterochromatin, TIS revealed genes important for the formation of heterochromatin (Figure 4b) (52). Several subunits within the cleavage and polyadenylation factor were implicated as contributing to the formation of heterochromatin. Validation studies of candidates supported their contribution in heterochromatin, including Iss1, a cleavage and polyadenylation factor that promotes heterochromatin formation by bridging the interaction between the RNA-specific binding protein Mmi1 and nuclear exosome factors responsible for recruiting the histone methyltransferase complex (52).
TIS in yeast now allows many critical systems to be probed using the large number of established reporters that are able to measure the output of pathways. Examples of fundamental processes that could be studied include chronological aging, mitochondrial function, stress resistance, and community interaction, to name a few. It is however, early days in applying TIS to yeast, with just a handful of published studies and very little quantitative treatment. A significant opportunity exists to adapt the analysis packages described in Table 2 to measure contributions to fitness or growth rates. These packages provide quantitative rigor that has yet to be incorporated into the study of yeast. In addition, the use of RB-TnSeq with yeast would be a powerful high-throughput framework to investigate a large number of parameters.
Table 2.
A summary of computational tools available for TIS analysis
| Tools/details | ESSENTIALS | Transit | Con-Artist | TraDIS | Tn-seq Explorer | TnseqDiff | MAGenTA | Aerobio |
|---|---|---|---|---|---|---|---|---|
| Raw read processinga | Yes | Through separate tool (TPP) | No, needs separate tools | Yes | No, but enables read mapping with compatible tools | No | Yes | Yes |
| Overall read count Normalizationb | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Genomic location read normalizationc | LOESS correction | No | No | No | No | Yes | No | LOESS correction |
| Data readoutd | Log 2 countsj and ratios | Read counts | Read counts | Log 2 countsj and ratios | Log 2 counts and ratios | Log 2 counts and ratios | Read counts and relative fitness representing the growth ratek | Read counts and relative fitness representing the growth ratek |
| Core model/ approache | Negative binomial distribution | Bayesian or hidden Markov model (HMM) | Mann- Whitney U and HMM | Log-fold changes | Log-fold changes | Construction of confidence distribution function | Fits data to exponential growth model, incorporates population expansion | Fits data to exponential growth model, incorporates population expansion |
| Essential gene/ loci identificationf | Gene | Gene or loci | Gene or loci | Gene | Gene or loci | Gene or defined loci | Gene or loci | Gene or defined loci |
| Conditionally essential gene/loci identificationf | Gene | Gene or loci | Gene or loci | Gene | Gene or loci | Gene | Gene or loci | Gene or defined loci |
| Conditionally important/ quantitative read outs | Semi | Semi | Semi | Semi | Semi | Semi | Yes, fitness is growth rate | Yes, fitness is growth rate |
| Bottleneck calculation & correctiong | No | Nol | Yes | No | No | No | Yes, size calculation and correction | Yes, size calculation and correction |
| Quantitative comparisons across experimentsh | No | Yes, permutation test | Yes, multinomial distribution Simulationm | Yes | No | No | Yes, fitness is relative for each condition and experiment | Yes, fitness is relative for each condition and experiment |
| Operationi | Web-based or CL | GUI | CL | CL | GUI | R-based | CL or Galaxy | CL and server-based |
| Visualization and notes | Several visualization options | Several visualization options | Visualization with Artemisn | Visualization with Artemisn | Several visualization options | - | R-based visualization options | Many visualization options and performs RNA-seq, and whole-genome sequencing analysis. |
| Reference (s) | 110 | 17 | 66 | 5 | 82 | 107 | 59,91 | 2 |
These tools have the integrated capability to perform processes such as barcode clipping, read-quality filtering, and mapping reads to a reference genome.
Are read counts coming from different samples or sequencing runs normalized?
Is there a possibility to account for differences in the number of reads based on the genomic location (e.g., read numbers around the origin of replication in bacterial genomes may sometimes have higher numbers of reads due to increased DNA replication in these locations)?
This is the main type of output provided by this tool.
This is the major approach or model used in the tool that defines data analyses and identifies essential or conditionally essential/important genes or loci.
Each approach identifies essential genes, which are those needed for growth under any condition, and conditionally essential genes, which are those required for growth only in a specific condition. Some use annotation information and are gene centered (gene), some use a sliding window and are annotation independent and can theoretically identify any essential region (e.g., intergenic or even a domain in a gene; loci), with some tools, loci other than genes can be explored, if the loci are specifically defined for instance in the annotation (GenBank) file (defined loci).
Some experiments are affected by bottlenecks, which can be tackled bioinformatically with some tools.
Some tools enable comparisons across experiments and conditions, making it easier to determine whether loci have significant phenotypes in one or more conditions.
The accessibility or user-friendliness of each tool is partially determined by the manner in which they are run: Web-based can be run directly in the browser; CL represents the command line running of scripts in languages such a Perl or Python; GUI is the general user interface and often easy to run; R-based is run in the R-environment; Galaxy is operated in the Galaxy environment; server-based requires extensive expertise to install, while operation is through CL.
These data are generated with the RNA-seq analysis EdgeR package (72).
Fitness in these packages is calculated as the growth rate, as described in detail in References 91, 92, and 94. By incorporating read counts from two time points and the growth expansion or retraction of the population during the experiment into an exponential growth model, the effect of fitness of a single insertion, or a group of insertions in a loci, is represented as the growth rate. Thereby, the measurement becomes independent of time, relative to one (the wt growth rate), and truly quantitative, making cross-experiment, cross-condition, or cross-strain comparisons possible.
The developers of Transit recently explored the zero inflated negative binomial for bottleneck corrections (85). This approach has been used particularly for single-cell RNA-seq analysis, and it could be incorporated into Transit.
Artist developers recently developed CompTIS, a principal component analysis-based approach to analyze multiple data sets (39).
This tool relies on Artemis, a previously developed visualization tool (11).
7. ANALYSIS TOOLS
When the experimental phase, sample preparation, and sequencing have been completed the critical phase of TIS data analysis ensues, for which there are a variety of software packages available (Table 2). Most of these tools can do the same type of analysis but employ (slightly) different approaches to get to a similar result. Choosing what software package to use depends on different factors. It is, of course, important to understand what readout is expected and desired from the analysis. For instance, TIS has from the start mostly been used to identify essential bacterial genes (those genes that are essential for growth under any condition), and all the listed tools have that function. However, not all tools can, for instance, identify genes with a less-dramatic phenotype or make comparisons across experiments or test conditions. Additionally, the experience of the person doing the analysis may be important. While some tools are easier to run than others, they may not be able to perform the desired analysis. In Table 2, the most-used software packages are listed. Below we break the analysis down into several steps and highlight aspects of how the different packages approach them:
In the first step (see raw read processing in Table 2), raw reads that come off of the sequencer often need to be processed before they can be mapped to a reference genome. Several approaches have either developed their own processing tools or incorporated those developed by others, such as the mapping tools bowtie (49) or Burrows-Wheeler alignment (55). There are several important processing steps, including trimming off sequences from each read that are not part of the genomic DNA of the organism and would prevent accurate mapping. Many TIS approaches combine multiple samples into a single sequencing run enabled by adding sample-specific barcodes during library preparation. These samples get split into separate files during raw read processing, and the barcode sequences should be removed from each read as well, because if left untouched, they would prevent mapping to a reference genome. Reads are mapped to a reference genome, the exact location of each insertion is written to a specific file, and the number of reads mapped to each location is recorded.
The number of reads recorded for each insertion is always used downstream to determine whether the insertion has some effect on the phenotype under investigation. The change in number of reads between two time points or two conditions is often used to assess this effect. While it is thus the goal to determine whether the change is caused by the treatment, there are at least two additional reasons these numbers can be affected, and they should be considered in the analysis. First, if more sequence reads are generated for one sample versus another (which is practically always the case) then you may want to normalize the read count. All tools perform such a normalization (see overall read count normalization in Table 2), albeit in different ways. However, for Microbial Assessment by Genome-Wide Tn-seq Analysis (MAGenTA) and Aerobio, this normalization could be excluded when the fitness readout these tools generate is used in downstream analyses. This is because for fitness the frequency of each insertion is calculated over the entire sample, and thus it uses an internal normalization step. A second normalization step (see genomic location read normalization in Table 2), which was first introduced by ESSENTIALS, is to normalize the number of reads for each insertion based on its genomic location by locally weighted scatterplot smoothing (LOESS). This means that a linear regression is performed on a sliding window over the data points in that window, estimating a value that smooths the distribution (the size of the window for a given library can be explored so that it optimizes the normalization). For instance, it turns out that some experimental treatments induce strong DNA replication around the origin of replication of a bacterial genome. This triggers a bias in the number of reads that are sequenced around the origin, which means that an overabundance of reads will map around the origin of replication, represented by a so-called smiley effect when read numbers are plotted on the genome.
All of the packages record the number of reads that are mapped to a certain location (data readout). However, because read counts across different insertions can differ by several orders of magnitude, most packages do additional processing, and most perform a log2 transformation, similar to those performed for RNA expression analyses [e.g., ESSENTIALS and TraDIS use EdgeR to do this (72)]. A log2 ratio of 2 time points (t2/t1; similar to that in RNA expression) is then used to perform statistical analysis on and assess whether the change is significant or not. The packages MAGenTA and Aerobio present read counts and ratios, but their main feature is the ability to calculate the relative fitness effect of each insertion individually and the average fitness effect of each specific loci (e.g., a gene). Fitness in these two packages is determined by fitting the frequency of each insertion in the population before and after treatment to an exponential growth model and incorporating the expansion or retraction the population undergoes during the experiment. One advantage of incorporating this change in the size of the population into the model is that the results thereby become independent of when the second time point is sampled. This is important for growth experiments because the more doublings a population goes through overtime(or the more the population contracts), the larger the differences between time points become, and, for instance, the smaller or larger the ratios. Therefore, unless the timing for different (replicate) experiments is controlled perfectly, they become very difficult to compare. The first paper on Tn-seq (91) recognized this potential caveat and implemented the original Malthusian parameter as a measure of fitness (57, 92), thereby enabling the calculation of the relative growth rate of each insertion or group of insertions, which gives the measurement a real biological meaning (i.e., the growth rate).While for MAGenTA and Aerobio it is irrelevant how long the experiment is performed (i.e., the time between t1 and t2),although measurements do need to be taken during exponential growth (or contraction).Importantly, the sampling of two time points for a growth-based TIS experiment should always be done during exponential growth to avoid the converging of differences that can happen during the stationary phase. This is for the same reason as why quantitative PCR of RNA analyses are done during the exponential amplification of the fluorescent signal, because once it has passed the exponential phase, saturation of the signal leads to the merging of expression differences.
Each package is able to determine whether a gene is essential (see essential gene/loci identification in Table 2 which, in broad strokes, is done by determining whether there is an underrepresentation of insertions in a gene, for instance, compared to surrounding regions, or what would be expected from a mostly random distribution across the genome. The packages ESSENTIALS, Transit, con-Artist, and TnseqDiffuse a variety of different approaches to statistically assess the likelihood that a gene is essential (see core model/approach in Table 2).Of those methods, ESSENTIALS and Transit are arguably the most user-friendly, while Transit has the added advantage of using a sliding window approach, which enables screening the genome for essential regions or domains instead of focusing solely on annotated regions (i.e., genes). Additionally, all packages are able to identify conditionally essential genes, while others are able to employ a sliding window and screen for conditionally essential regions or domains. Thus, the packages that use a sliding window are annotation independent. In general, conditionally essential genes/loci are easier to identify because two time points are used, and in the second time point all insertions in a conditionally essential region should have disappeared. For this, each package uses a (slightly) different approach to assess statistical significance (see core model/approach in Table 2). Importantly, there is no approach that is proven to be clearly superior over any other for any specific transposon system. It is thus left to the researcher to assess what will work best for their system. Sometimes trying out multiple approaches can be helpful; however, only extensive experimental validation can help in evaluating the true confidence of the data.
To some extent, each package is also able to determine more subtle phenotypes, such aswhether a gene/loci is conditionally important. This means that a transposon insertion can have a more subtle phenotype and reduce or increase the growth rate only to a certain extent. For instance, an insertion in a specific region may make a bacterium slightly more sensitive or resistant to an antibiotic. Arguably, after essential genes, these more subtle phenotypesare the most commonly observed and can be incredibly important in obtaining leads for gene function (see Sections 4 and 5). While the first six packages all have the ability to identify semiquantitative phenotypic effects (see conditionally important/quantitative readouts in Table 2), the strength of MAGenTA and Aerobio is the identification of a wide range of quantitative fitness effects due to the calculation of a transposon insertion’s effect on the growth rate, as explained in Analysis Tools number 3. While these packages can calculate very large effects on the growth rate (i.e., a growth rate of 0 means conditionally essential, a rate of 0.5 means that a mutant grows twice as slow as the wild type), depending on the experiment, this can even lead to detailed growth rate estimations within 5% of the wild type [i.e., a doubling time of 30 versus ~32 min (91, 93, 94)].
Only the con-Artist, MAGenTA, and Aerobio packages can calculate and correct for bottleneck effects (see bottleneck calculation and correction in Table 2), which is especially important for in vivo experiments (see also Section 8.4). Moreover, several of the authors of Transit were recently involved in exploring an approach for bottleneck corrections, which could be incorporated in Transit (package 2) in the future (85).
TIS is being used more and more across many different environments. To enable comparisons across experiments and/or conditions or to even enable comparisons between different strains requires approaches that normalize the data (see quantitative comparisons across environments in Table 2), which is done by Transit, con-Artist, TraDIS, MAGenTA, and Aerobio packages.
User-friendliness is also of importance (see operation in Table 2). The packages ESSENTIALS, Transit, and Tn-seq Explorer are among the most user-friendly, due to the manner in which the user interacts with the tool. The other packages are mostly command line–based, which makes them less accessible. Aerobio solely runs on a server environment, and while the operation on command line is mostly straightforward, the installation requires extensive expertise.
Most packages have visualization options (see visualization and notes in Table 2), and some come with the package (ESSENTIALS, Transit, Tn-seq Explorer, and Aerobio),while others use external tools (con-Artist,TraDIS,and MAGenTA).However, most readouts, with some data manipulation, can be easily plotted, for instance, with genome browsers such as the Integrative Genomics Viewer (71) or the University of California, Santa Cruz Genome Browser (44) or combined with other omics data through ShinyOmics (86).
8. KEY CONSIDERATIONS
8.1. Library Saturation
The degree of library saturation is an important factor to ensure a high-quality TIS experiment. Over the years, different computational approaches have been developed that, while taking into account possible transposon site preference, can help determine library saturation (see Table 2). However, there are important aspects and parameters that can and should be considered while designing an experiment.
The sequence bias of the transposon can impact the coverage of insertions across thegenome (see Table 1). For instance, Himar1 almost exclusively inserts in-between TA nucleotides, while Tn5 has only a minor preference for GC sequences. Therefore, if a transposome (i.e., a transposon-transposase complex) has a preference, it is important to determine that the genome has a high prevalence of such sites and that they are evenly distributed.
There can be cold spots in the genome that do not accept insertions, for instance, because sites are heavily occupied by proteins, contain stable secondary structures, or in the case of transposons with a preference, lack preferred insertion sites. In the yeasts S. cerevisiae and S.pombe, nucleosomes reduce integration frequencies so that just 33% of all insertions occur in open reading frames, which nevertheless comprise 60% of the genome (35).While the effects of cold spots cannot always be overcome, in yeast this bias resulting from nucleosomes was dealt with by increasing the number of insertions (35, 61, 80).
The number of insertions in a library should ensure, at least in theory, that all genes in thegenome are hit multiple times. It is therefore important to consider the size of the genome as well as the number of genes and their length. For instance, a library that on average has insertions every 50 nucleotides, and whose insertions are mostly randomly dispersed across the genome, should be strong at indicating whether a gene of ~1,000 nucleotides is essential or not. In such a case you would expect to find on average 25 insertions if the gene is nonessential and basically no insertions if it is essential. However,for a gene of ~200 nucleotides you would only expect ~4 insertions. The absence of these insertions is unlikely to be statistically significant even for genes that are not essential. Increasing library size will thus give a better readout for smaller genes.
Libraries that reach very high saturation enable the identification of smaller and smaller essential genetic components, including promoter regions, ncRNAs, and even essential protein domains.
8.2. Library Construction
A profile of insertions that reach saturation is needed in TIS studies. However, there are many obstacles during the course of the experiment that reduce the number of sites obtained. The following are variables that can significantly increase the density of insertions detected.
Generating the insertions in vivo in the query strain can result in the highest number of insertions because it avoids bottlenecks associated with introducing transpososomes or integrated DNA into query cells. To produce insertions in vivo requires an expression system for the transposase and a plasmid to provide the donor transposon. However, these tools only exist for species that have been developed for genetic studies.
When working with a species that lacks genetic tools, it can be necessary to produce integration events in vitro using purified genomic DNA, transposase, and transposon DNA. The mutagenized DNA is transformed into the query strain, where homologous recombination introduces the insertion. This is a flexible method compatible with many bacteria species, but the size of the libraries is limited by the transformation efficiency. Therefore, every effort should be made to optimize transformation efficiency.
When inducing insertions in vivo, labs often isolate events as independent colonies. In onestudy, colonies were isolated and harvested from 250 plates to pool 1 × 106 colonies that ultimately provided 300,000 independent insertions. Isolating independent colonies can help limit competition but may also cause a substantial process bottleneck. Alternatively, labs have had success in producing large numbers of insertions in liquid culture. The results can far exceed the total numbers of insertions from independent colonies with a fraction of the labor.
The PCR amplification of insertions can also restrict the size of the library. The complexity of the libraries can be greatly reduced when the ligation of linkers is incomplete, especially if a single PCR reaction is used. Controls that measure the efficiency of ligation should be used, and there is benefit in making a library from multiple independent PCR reactions with a minimum number of cycles.
8.3. Phenotypes and Their Readout: Growth, Survival, and Alternative Phenotypes
TIS can be used to screen for a large variety of phenotypes, and the researchers’ imaginations may be the limiting factor. TIS is an especially strong method to identify phenotypes that are genetically encoded and reveal themselves through the manipulation of a single genetic element. This means that highly buffered phenotypes or those encoded by redundant systems are more difficult to untangle using TIS. Although, in those cases, TIS-mediated genetic interaction mapping could be a solution. Below, we highlight some of the most fundamental genotype × phenotype relationships.
Identifying essential genes is fairly straightforward. For this you only need to acquire sequencing data from the starting library population. In an all-around saturated library, essential genes are basically those that lack insertions (but see Section 8.2; Table 2).
To identify conditionally essential genes, sequencing information from the starting library and a second time point are needed, where the library has been grown in, or exposed to, a specific selective environment. Conditionally essential are those genes that when carrying a transposon insertion are unable to grow or survive in a specific environment. If they are unable to grow, then with every cell division, the frequency of such insertions in the library will rapidly decline and they will eventually be diluted out. Those that are unable to survive in an environment may rapidly die and possibly lyse, resulting in an even more rapid loss of these transposon insertions in the population.
Conditionally important genes are those that have a more subtle phenotype compared to essential ones, and this can either have a positive or negative effect on growth or survival. Depending on the accuracy and sensitivity of the chosen TIS approach and subsequent analysis, it is possible to identify insertions along a wide gradient of fitness effects (see Section 7).
Most TIS experiments have focused on determining the effect of transposon insertions on growth or survival in a specific condition. However, due to innovative extensions of TIS, other phenotypes based on different characteristics can be screened for as well, including cell-shape, cell permeability, cell movement, cell wall composition, or cell-cell communication. These alternative phenotypes and their readouts are discussed in Section 6.
8.4. Bottlenecks, How to Deal with Them When You Have Them
A simple TIS experiment, for instance, one where a transposon library is grown in a controlled lab environment, that is carefully executed should in general produce highly reproducible results. However, the more complex an experiment, the higher the probability that stochastic events will have an effect. One of the most important events is when a transposon library is exposed to a bottleneck, resulting in the random loss of transposon insertions, which has especially been shown to occur during in vivo experiments with bacterial infections of animals or plants. Importantly, random loss of transposon insertions can not only significantly increase variation in the data but can lead to the wrongful identification of (conditionally) essential genes. It is thus important to always check the data for the possibility that something random occurred during the experiment that led to a bottleneck. While in many (simple) TIS experiments bottlenecks should not occur and if they do the experiment should probably be disregarded, in some experiments, such as those executed in vivo, bottlenecks are often unavoidable. There are several straightforward ways to determine whether a bottleneck has occurred during an experiment, and if they are deemed acceptable, data can be processed in different ways in an attempt to minimize the bottleneck’s impact on the results (see Section 7; Table 2).
The simplest way to monitor for a bottleneck is by observing the behavior of transposon insertions that occurred in neutral sites in the genome. Neutral sites are those locations that when disturbed will not affect the organism’s fitness, which can include degenerate genes, pseudogenes, or integrated elements with no function. Insertions in such sites should remain stably present during an experiment (from t1 to t2). Thereby, their disappearance from the population is indicative of a bottleneck, while the percentage of disappearance is indicative for the size of the bottleneck.
In general, in a saturated library, the vast majority of insertions will not have an effect onfitness, and the number of insertions that disappear from a library during selection should be negligible with respect to the entire library size. Therefore, as a rule of thumb, the percentage of insertions that do disappear, with respect to the entire library, can also serve as a proxy to estimate the size of the bottleneck.
There are several ways through data analysis to correct for a bottleneck. The first paperto successfully do this (93) determined that a bottleneck occurred using the approaches in Section 8.4, number 1. The size of the bottleneck, as revealed with the method in Section 8.4, number 2, was then used to remove the same percentage of insertions with fitness 0, which are those insertions that disappear from the library, from each genetic region in the genome. For instance, if 50% of neutral site insertions disappear from the library during selection, the bottleneck size is 50%. This means that if a gene has 10 transposon insertions and 5 disappear during selection, those possibly disappeared due to the bottleneck and are removed from the analysis before the overall fitness effect is calculated from the average of the remaining 5 insertions. If 8 insertions had disappeared, 5 would have been disregarded, while 3 insertions with fitness 0 would have been retained to calculate overall fitness. This approach has been used in multiple in vivo experiments and has been successful in reducing the effect of the bottleneck and accurately calculating in vivo growth rates of genetic regions (10, 56, 73, 93).
Recently, other approaches have been developed that use more complex and sophisticated ways to normalize data that have been exposed to a bottleneck (Table 2).These methods include multinomial-based resampling con-Artist –(66), principal component analysis on log fold changes (97), and the use of a zero-inflated negative binomial distribution to test for genes with significant variability in insertion counts across conditions (85), which has similarity to analyses applied to single-cell RNA-seq, which identify transcripts that disappeared for technical reasons (90).
9. CONCLUSION
In ten short years, TIS has made substantial contributions to our understanding of gene function by measuring the role of genes in many different pathways and conditions. The systems-level information not only describes detailed mechanisms of pathogenesis of major medical diseases but also reveals evolutionary strategies of organisms that previously were difficult to study with traditional genetics. The ever-expanding number of species studied with TIS raises the prospect that future innovations will allow TIS to be adopted to study the genes of complex eukaryotes, which may someday include mammals.
The prospect of applying TIS to study multicellular organisms has the potential to make significant advances in our understanding of mechanisms central to human biology, such as development and behavior. But before this can be a reality, two substantial advances must be achieved. The haploid genome of humans is 3,200 Mbp, an expanse approximately 2,700 times larger than yeast (12 Mbp). Achieving TIS saturation of such a large genome is not currently feasible due to limits in the number of insertions that can be generated and cultured. However, one might make real progress in applying TIS in an intermediate genome, such as the 180 Mbp of the Drosophila genome. In addition to solving the challenges of achieving saturation of larger genomes, one must address the problem of ploidy. With diploid cells, transposon insertions are only capable of altering one of the two alleles. True, phenotypes can result if the disruption is dominant or if a 50% reduction of a gene product results in a defect (haploinsufficiency). But to achieve the full potential of TIS, the query genome must be haploid. Here, too, there are opportunities to overcome this problem. A human cell line that is largely haploid was derived from chronic myeloid leukemia cells. Although insertion numbers have not approached saturation, a retroviral vector was able to generate insertions in >98% of expressed genes (9). With continued advances in transposon technology and wise choices in which organisms to study, it is safe to predict that TIS will play an important role in future discoveries related to human biology.
Supplementary Material
ACKNOWLEDGMENTS
H.L.L. acknowledges the support of the Intramural Research Programs of the National Institutes of Health (NIH) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. T.v.O. acknowledges support from the NIH grants U01AI124302, R01AI110724, R01AI148470, and R21AI117247.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Akerley BJ, Lampe DJ. 2002. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 358:100–8 [DOI] [PubMed] [Google Scholar]
- 2.Anthony J, van Opijnen T. 2019. Aerobio. DAG streaming computation server https://github.com/jsa-aerial/aerobio
- 3.Armbruster CE, Forsyth VS, Johnson AO, Smith SN, White AN, et al. 2019. Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection. PLOS Pathog. 15:e1007653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armbruster CE, Forsyth-DeOrnellas V, Johnson AO, Smith SN, Zhao L, et al. 2017. Genome-wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLOS Pathog. 13:e1006434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barquist L, Mayho M, Cummins C, Cain AK, Boinett CJ, et al. 2016. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics 32:1109–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender J, Kleckner N. 1992. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. PNAS 89:7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advancesin transposon-insertion sequencing. Nat. Rev. Genet 21:526–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capel E, Zomer AL, Nussbaumer T, Bole C, Izac B, et al. 2016. Comprehensive identification of meningococcal genes and small noncoding RNAs required for host cell colonization. mBio 7:e01173–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carette JE, Guimaraes CP, Wuethrich I,Blomen VA,Varadarajan M,et al.2011.Global gene disruptionin human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol 29:542–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter R, Wolf J, van Opijnen T, Muller M, Obert C, et al. 2014. Genomic analyses of pneumococci from children with sickle cell disease expose host-specific bacterial adaptations and deficits in current interventions. Cell Host Microbe 15:587–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao MC, Pritchard JR, Zhang YJ, Rubin EJ, Livny J, et al.2013.High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 41:9033–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charbonneau ARL, Forman OP, Cain AK, Newland G, Robinson C, et al. 2017. Defining the ABC ofgene essentiality in streptococci. BMC Genom. 18:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, et al. 2011. The essential genome of a bacterium. Mol. Syst. Biol 7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coe KA, Lee W, Stone MC, Komazin-Meredith G, Meredith TC, et al. 2019. Multi-strain Tn-Seqreveals common daptomycin resistance determinants in Staphylococcus aureus. PLOS Pathog. 15:e1007862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries SP, Eleveld MJ, Hermans PW, Bootsma HJ. 2013. Characterization of the molecular interplay between Moraxella catarrhalis and human respiratory tract epithelial cells. PLOS ONE 8:e72193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT–a software tool forHimar1 TnSeq analysis. PLOS Comput. Biol 11:e1004401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, et al. 2017. Comprehensive essentiality analysis ofthe Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. 2005. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122:473–83 [DOI] [PubMed] [Google Scholar]
- 20.Dorman MJ, Feltwell T, Goulding DA, Parkhill J, Short FL. 2018. The capsule regulatory network ofKlebsiella pneumoniae defined by density-TraDISort. mBio 9:e01863–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan MC, Gillette RK, Maglasang MA, Corn EA, Tai AK, et al. 2019. High-throughput analysis ofgene function in the bacterial predator Bdellovibrio bacteriovorus. mBio 10:e01040–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edskes HK, Mukhamedova M, Edskes BK, Wickner RB. 2018. Hermes transposon mutagenesis shows [URE3] prion pathology prevented by a ubiquitin-targeting protein: evidence for carbon/nitrogen assimilation cross talk and a second function for Ure2p in Saccharomyces cerevisiae. Genetics 209:789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler CC,Galan JE.2018.Decoding a Salmonella Typhi regulatory network that controls typhoid toxin expression within human cells. Cell Host Microbe 23:65–76.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, Waldor MK, Mekalanos JJ. 2013. Tn-seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14:652–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallagher LA, Lee SA, Manoil C. 2017. Importance of core genome functions for an extreme antibiotic resistance trait. mBio 8:e01655–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutantswithin libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. PNAS 106:16422–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisinger E, Mortman NJ, Dai Y, Cokol M, Syal S, et al. 2020. Antibiotic hypersensitivity signatures identify targets for attack in the Acinetobacter baumannii cell envelope. bioRxiv 2020.03.11.987479. 10.1101/2020.03.11.987479 [DOI] [Google Scholar]
- 28.Geisinger E, Vargas-Cuebas G, Mortman NJ, Syal S, Dai Y, et al. 2019. The landscape of phenotypic and transcriptional responses to Ciprofloxacin in Acinetobacter baumannii: Acquired resistance alleles modulate drug-induced SOS response and prophage replication. mBio 10:e01127–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodall ECA, Robinson A, Johnston IG, Jabbari S, Turner KA, et al. 2018. The essential genome of Escherichia coli K-12. mBio 9:e02096–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, et al. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green B, Bouchier C, Fairhead C, Craig NL, Cormack BP. 2012. Insertion site preference of Mu, Tn5,and Tn7 transposons. Mob. DNA 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLOS Pathog. 7:e1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grosser MR, Paluscio E, Thurlow LR, Dillon MM, Cooper VS, et al. 2018. Genetic requirements for Staphylococcus aureus nitric oxide resistance and virulence. PLOS Pathog. 14:e1006907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guimond N, Bideshi DK, Pinkerton AC, Atkinson PW, O’Brochta DA. 2003. Patterns of Hermes transposition in Drosophila melanogaster. Mol. Genet. Genom 268:779–90 [DOI] [PubMed] [Google Scholar]
- 35.Guo Y, Park JM, Cui B, Humes E, Gangadharan S, et al. 2013. Integration profiling of gene function with dense maps of transposon integration. Genetics 195:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halling SM, Kleckner N. 1982. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell 28:155–63 [DOI] [PubMed] [Google Scholar]
- 37.Hassan KA, Cain AK, Huang T, Liu Q, Elbourne LD, et al. 2016. Fluorescence-based flow sortingin parallel with transposon insertion site sequencing identifies multidrug efflux systems in Acinetobacter baumannii. mBio 7:e01200–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel Zur Wiesch P, et al. 2016. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. PNAS 113:6283–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbard TP, D’Gama JD, Billings G, Davis BM, Waldor MK. 2019. Unsupervised learning approachfor comparing multiple transposon insertion sequencing studies. mSphere 4:e00031–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ibberson CB, Stacy A, Fleming D, Dees JL, Rumbaugh K, et al. 2017. Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol 2:17079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jana B, Cain AK, Doerrler WT, Boinett CJ, Fookes MC, et al. 2017. The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci. Rep 7:42483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen PA, Zhu Z, van Opijnen T. 2017. Antibiotics disrupt coordination between transcriptional and phenotypic stress responses in pathogenic bacteria. Cell Rep. 20:1705–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. 2013. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLOS Pathog. 9:e1003800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. 2002. The human genome browser atUCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleckner N, Bender J, Gottesman S 1991.Uses of transposons with emphasison Tn10. Methods Enzymol. 204:139–80 [DOI] [PubMed] [Google Scholar]
- 46.Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. 2012. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genom. 13:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. 1999.Hyperactive transposase mutantsof the Himar1 mariner transposon. PNAS 96:11428–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lampe DJ, Grant TE, Robertson HM. 1998. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149:179–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform 32:11.7.1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, et al. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19:2308–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Breton Y, Belew AT, Freiberg JA, Sundar GS, Islam E, et al. 2017. Genome-wide discovery of novelM1T1 group A streptococcal determinants important for fitness and virulence during soft-tissue infection. PLOS Pathog. 13:e1006584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SY, Hung S, Esnault C, Pathak R, Johnson KR, et al. 2020. Dense transposon integration reveals essential cleavage and polyadenylation factors promote heterochromatin formation. Cell Rep. 30:2686–98.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levitan A, Gale AN, Dallon EK, Kozan DW, Cunningham KW, et al. 2020. Comparing the utilityof in vivo transposon mutagenesis approaches in yeast species to infer gene essentiality. Curr. Genet 10.1007/s00294-020-01096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewin GR, Stacy A, Michie KL, Lamont RJ, Whiteley M. 2019. Large-scale identification of pathogen essential genes during coinfection with sympatric and allopatric microbes. PNAS 116:19685–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mann B, van Opijnen T, Wang J, Obert C, Wang YD, et al. 2012. Control of virulence by small RNAsin Streptococcus pneumoniae. PLOS Pathog. 8:e1002788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maree AF, Keulen W, Boucher CA, De Boer RJ. 2000. Estimating relative fitness in viral competition experiments. J. Virol 74:11067–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy AJ, Stabler RA, Taylor PW. 2018. Genome-wide identification by transposon insertion sequencing of Escherichia coli K1 genes essential for in vitro growth, gastrointestinal colonizing capacity, and survival in serum. J. Bacteriol 200:e00698–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCoy KM, Antonio ML, van Opijnen T. 2017. MAGenTA: a Galaxy implemented tool for completeTn-Seq analysis and data visualization. Bioinformatics 33:2781–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mesarich CH, Rees-George J, Gardner PP, Ghomi FA, Gerth ML, et al. 2017. Transposon insertion libraries for the characterization of mutants from the kiwifruit pathogen Pseudomonas syringae pv. actinidiae. PLOS ONE 12:e0172790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michel AH, Hatakeyama R, Kimmig P, Arter M, Peter M, et al. 2017. Functional mapping of yeast genomes by saturated transposition. eLife 6:e23570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murray JL, Kwon T, Marcotte EM, Whiteley M. 2015. Intrinsic antimicrobial resistance determinants in the superbug Pseudomonas aeruginosa. mBio 6:e01603–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nolan LM, Whitchurch CB, Barquist L, Katrib M, Boinett CJ, et al. 2018. A global genomic approach uncovers novel components for twitching motility-mediated biofilm expansion in Pseudomonas aeruginosa. Microb. Genom 4:e000229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ochsner AM, Christen M, Hemmerle L, Peyraud R, Christen B, Vorholt JA. 2017. Transposon sequencing uncovers an essential regulatory function of phosphoribulokinase for methylotrophy. Curr. Biol 27:2579–88.e6 [DOI] [PubMed] [Google Scholar]
- 65.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, et al. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–9 [DOI] [PubMed] [Google Scholar]
- 66.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, et al. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLOS Genet. 10:e1004782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, et al. 2010. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 330:1104–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rajagopal M, Martin MJ, Santiago M, Lee W, Kos VN, et al.2016.Multidrug intrinsic resistance factors in Staphylococcus aureus identified by profiling fitness within high-diversity transposon libraries. mBio 7:e00950–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rego EH, Audette RE, Rubin EJ.2017.Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546:153–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reznikoff WS. 2008. Transposon Tn5. Annu. Rev. Genet 42:269–86 [DOI] [PubMed] [Google Scholar]
- 71.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, et al. 2011.Integrative genomics viewer. Nat. Biotechnol 29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowe HM, Karlsson E, Echlin H, Chang TC, Wang L, et al. 2019. Bacterial factors required for transmission of Streptococcus pneumoniae in mammalian hosts. Cell Host Microbe 25:884–91.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, et al. 2015. The essential gene setof a photosynthetic organism. PNAS 112:E6634–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruiz L, Bottacini F, Boinett CJ, Cain AK, O’Connell-Motherway M, et al. 2017. The essential genomic landscape of the commensal Bifidobacterium breve UCC2003. Sci. Rep 7:5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanchez MR, Payen C,Cheong F, Hovde BT, Bissonnette S, et al.2019.Transposon insertional mutagenesis in Saccharomyces uvarum reveals trans-acting effects influencing species-dependent essential genes. Genome Res. 29:396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanchez-Larrayoz AF, Elhosseiny NM, Chevrette MG, Fu Y, Giunta P, et al. 2017. Complexity of complement resistance factors expressed by Acinetobacter baumannii needed for survival in human serum. J. Immunol 199:2803–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santiago M, Matano LM, Moussa SH, Gilmore MS, Walker S, Meredith TC. 2015. A new platform for ultra-high density Staphylococcus aureus transposon libraries. BMC Genom. 16:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sarmiento F, Mrazek J, Whitman WB. 2013. Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. PNAS 110:4726–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segal ES, Gritsenko V, Levitan A, Yadav B, Dror N, et al. 2018. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. mBio 9:e02048–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shevchenko Y,Bouffard GG, Butterfield YS, Blakesley RW, Hartley JL, et al.2002.Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 30:2469–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Solaimanpour S, Sarmiento F, Mrazek J. 2015. Tn-seq explorer: a tool for analysis of high-through put sequencing data of transposon mutant libraries. PLOS ONE 10:e0126070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subashchandrabose S, Smith S, DeOrnellas V, Crepin S, Kole M, et al. 2016. Acinetobacter baumannii genes required for bacterial survival during bloodstream infection. mSphere 1:e00013–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HL. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLOS Pathog. 9:e1003788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Subramaniyam S, DeJesus MA, Zaveri A, Smith CM, Baker RE, et al. 2019. Statistical analysis of variability in TnSeq data across conditions using zero-inflated negative binomial regression. BMC Bioinform. 20:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Surujon D, van Opijnen T. 2020. ShinyOmics: collaborative exploration of omics-data. BMC Bioinform. 21:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thibault D, Jensen PA, Wood S, Qabar C, Clark S, et al. 2019. Droplet Tn-Seq combines microfluidics with Tn-Seq for identifying complex single-cell phenotypes. Nat. Commun 10:5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Troy EB, Lin T, Gao L, Lazinski DW, Lundt M, et al. 2016. Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol. Microbiol 101:1003–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Valentino MD, Foulston L, Sadaka A, Kos VN, Villet RA,et al.2014.Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 5:e01729–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van den Berge K, Perraudeau F, Soneson C, Love MI, Risso D, et al. 2018. Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biol. 19:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness andgenetic interaction studies in microorganisms. Nat. Methods 6:767–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Opijnen T, Boerlijst MC, Berkhout B. 2006. Effects of random mutations in the human immunodeficiency virus type 1 transcriptional promoter on viral fitness in different host cell environments. J. Virol 80:6678–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Opijnen T, Camilli A.2012.A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 22:2541–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysisof microorganisms. Nat. Rev. Microbiol 11:435–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Opijnen T, Dedrick S, Bento J. 2016. Strain dependent genetic networks for antibiotic-sensitivityin a bacterial pathogen with a large pan-genome. PLOS Pathog. 12:e1005869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Verhagen LM, de Jonge MI, Burghout P, Schraa K, Spagnuolo L, et al. 2014. Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PLOS ONE 9:e89541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Warr AR, Hubbard TP, Munera D, Blondel CJ, Abel zur Wiesch P, et al. 2019. Transposon-insertion sequencing screens unveil requirements for EHEC growth and intestinal colonization. PLOS Pathog. 15:e1007652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, et al.2015.Rapid quantification of mutant fitnessin diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Willcocks SJ, Stabler RA, Atkins HS, Oyston PF, Wren BW. 2018. High-throughput analysis of Yersinia pseudotuberculosis gene essentiality in optimised in vitro conditions, and implications for the speciation of Yersinia pestis. BMC Microbiol. 18:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wong YC, Abd El Ghany M, Naeem R, Lee KW, Tan YC, et al. 2016. Candidate essential genes in Burkholderia cenocepacia J2315 identified by genome-wide TraDIS. Front. Microbiol 7:1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu S, Ying G, Wu Q, Capecchi MR. 2007. Toward simpler and faster genome-wide mutagenesis inmice. Nat. Genet 39:922–30 [DOI] [PubMed] [Google Scholar]
- 102.Yasir M, Turner AK, Bastkowski S, Baker D, Page AJ, et al. 2020. TraDIS-Xpress: A high-resolution whole-genome assay identifies novel mechanisms of triclosan action and resistance. Genome Res.30:239–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zafar MA, Hammond AJ, Hamaguchi S, Wu W, Kono M, et al. 2019. Identification of pneumococcal factors affecting pneumococcal shedding shows that the dlt locus promotes inflammation and transmission. mBio 10:e01032–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang C, Phillips APR, Wipfler RL, Olsen GJ, Whitaker RJ. 2018. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat. Commun 9:4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang M, Wang C, Otto TD, Oberstaller J, Liao X, et al. 2018. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360:eaap7847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang X, de Maat V, Guzman Prieto AM, Prajsnar TK, Bayjanov JR, et al. 2017. RNA-seq and Tn-seq reveal fitness determinants of vancomycin-resistant Enterococcus faecium during growth in human serum. BMC Genom. 18:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao L, Anderson MT, Wu W, Mobley HLT, Bachman MA. 2017. TnseqDiff: identification of conditionally essential genes in transposon sequencing studies. BMC Bioinform. 18:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhu J, Gong R, Zhu Q, He Q, Xu N, et al. 2018. Genome-wide determination of gene essentiality by transposon insertion sequencing in yeast Pichia pastoris. Sci. Rep 8:10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhu L, Olsen RJ, Beres SB, Eraso JM, Saavedra MO, et al. 2019. Gene fitness landscape of group A streptococcus during necrotizing myositis. J. Clin. Invest 129:887–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zomer A, Burghout P Bootsma HJ, Hermans PW, van Hijum SA. 2012. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLOS ONE 7:e43012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.