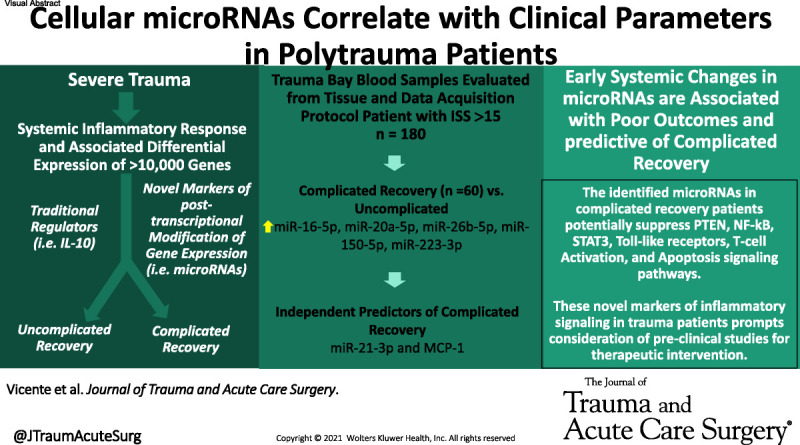
In a large multicenter prospectively collected database, the Surgical Critical Care Initiative's microRNA team characterize novel changes in microRNA expression in response to severe trauma. Systemic levels of microRNAs were associated with recovery complications. #microRNA
KEY WORDS: Trauma, microRNA, complications, inflammation, recovery
INTRODUCTION
The pathophysiology of the inflammatory response after major trauma is complex, and the magnitude correlates with severity of tissue injury and outcomes. Study of infection-mediated immune pathways has demonstrated that cellular microRNAs may modulate the inflammatory response. The authors hypothesize that the expression of microRNAs would correlate to complicated recoveries in polytrauma patients (PtPs).
METHODS
Polytrauma patients enrolled in the prospective observational Tissue and Data Acquisition Protocol with Injury Severity Score of >15 were selected for this study. Polytrauma patients were divided into complicated recoveries and uncomplicated recovery groups. Polytrauma patients' blood samples were obtained at the time of admission (T0). Established biomarkers of systemic inflammation, including cytokines and chemokines, were measured using multiplexed Luminex-based methods, and novel microRNAs were measured in plasma samples using multiplex RNA hybridization.
RESULTS
Polytrauma patients (n = 180) had high Injury Severity Score (26 [20–34]) and complicated recovery rate of 33%. MicroRNAs were lower in PtPs at T0 compared with healthy controls, and bivariate analysis demonstrated that variations of microRNAs correlated with age, race, comorbidities, venous thromboembolism, pulmonary complications, complicated recovery, and mortality. Positive correlations were noted between microRNAs and interleukin 10, vascular endothelial growth factor, Acute Physiology and Chronic Health Evaluation, and Sequential Organ Failure Assessment scores. Multivariable Lasso regression analysis of predictors of complicated recovery based on microRNAs, cytokines, and chemokines revealed that miR-21-3p and monocyte chemoattractant protein-1 were predictive of complicated recovery with an area under the curve of 0.78.
CONCLUSION
Systemic microRNAs were associated with poor outcomes in PtPs, and results are consistent with previously described trends in critically ill patients. These early biomarkers of inflammation might provide predictive utility in early complicated recovery diagnosis and prognosis. Because of their potential to regulate immune responses, microRNAs may provide therapeutic targets for immunomodulation.
LEVEL OF EVIDENCE
Diagnostic Tests/Criteria; Level II.
Trauma remains a leading cause of death worldwide.1 In the United States, injury severity has been increasing, with up to 25% of civilian patients and 54% of combat wounded classified as severely injured.2 Advances in transport times and resuscitation strategies have improved survival rates in several trauma populations.3 Despite this reduction in mortality, severely injured patients have high early and late morbidity including infectious complications (33–52%)2,4,5 and an overall complicated recovery (25%).6
Severe polytraumatic injury can lead to a dysregulated systemic inflammatory response as well as profound immunosuppression, organ dysfunction, and death. This acute, diverse, and excessive nonspecific host response to trauma leads to the massive release of inflammatory mediators that results in suppressed immunity and increased susceptibility to infectious complications, multiple organ failure, and persistent inflammation-immunosuppression catabolism syndrome.7,8 Translational studies have revealed several circulating biomarkers (e.g., interleukin [IL]-6 and monocyte chemoattractant protein [MCP]-1) with well-defined mechanisms in the inflammatory response,9 which correlate with initial tissue damage, ischemia-reperfusion injury, and have also been predictive morbidity and mortality.10–12 Attempts to mitigate the immune response through modulation of these inflammatory mediators have yielded limited clinical benefit and knowledge gaps of the pathophysiology of trauma.3,10 Recent investigations suggest that circulating levels of posttranscriptional modifiers involved in the signaling pathways of these molecules may provide insight into the links between systemic biomarkers and clinical outcomes in inflammatory pathologies.13,14
Microribonucleic acids (miRNAs) are short noncoding RNAs composed of ~21 nucleotides and are involved in posttranscriptional silencing of messenger RNA.13 These microRNAs have been identified in various species, and modeling of miRNA targets suggests that miRNAs are involved in the regulation of more than 60% of human protein coding genes.15 Within blood, these microRNAs travel in extracellular vesicles and are relatively stable.16 Once released from cells in extracellular vesicles, microRNAs can be delivered to recipients' cells where they have the capacity to regulate their gene expression in regional and distant organs/tissues.17
Circulating levels of both viral and cellular microRNAs have been reliably detected in the circulation of patients and have demonstrated prognostic value in infectious, autoimmune, and neoplastic diseases.13,16,18,19 Cellular microRNAs, such as miR-16a-5p and miR-21-3p, target multiple downstream mediators of Toll-like receptors (TLRs) signaling, and variations in circulating levels of these microRNAs are associated with poor outcomes in septic patients.16,20
While there have been no dedicated studies assessing the role of cellular microRNAs in polytrauma patients (PtPs) to date, clinical and preclinical studies have demonstrated differential expression of systemic microRNAs in mild traumatic brain injury (TBI) compared with severe TBI.21 Given the shared signaling pathways between trauma and sepsis3,8,12 and differential expression in TBI patients based on injury severity,21 the authors hypothesized that microRNA profiles may be altered in severely injured PtPs and that variations in systemic levels of these microRNAs will be associated with clinical outcomes.
PATIENTS AND METHODS
Tissue and Data Acquisition Protocol Data Set
The Tissue and Data Acquisition Protocol (TDAP) is a prospectively collected multi-institution data set that contains clinical, proteomic, and genomic molecular data from patients meeting inclusion and exclusion criteria defined by the Surgical Critical Care Initiative, as previously described.14,22 In brief, TDAP included females and males between the ages of 18 and 80 years with an injury or illness requiring surgical, critical, or emergency care.
Patient Selection
Polytrauma patients from the TDAP data set were selected for this subset analysis based on Injury Severity Scores (ISSs) of >15 and available plasma samples from admission to the trauma bay (T0), as previously described.14 To evaluate the prognostic value of early systemic biomarkers for postinjury complications, patients were then further selected for either no recovery complications or severe complications (e.g., ventilator-associated pneumonia [VAP] or organ space infection). Polytrauma patients from the initial selection with only minor complications (i.e., urinary tract infection or surgical site infection) were not included in this analysis.
Polytrauma patients in this subset analysis were categorized into those with a complicated recovery or uncomplicated recovery.14 A complicated recovery was defined as intensive care unit (ICU) admission >14 days, mechanical ventilation >7 days, or mortality within 28 days. The previously described intermediate recovery patients were grouped into uncomplicated recovery in this analysis.
Healthy volunteers (n = 12) between the age of 21 and 40 years without comorbidities and without recent injury served as controls for the biomarker studies. The healthy volunteers included 10 males and 2 females of varied race and ethnicity.
Clinical Data Collection
Variables with prognostic value in trauma were collected from TDAP database and included baseline demographic data (age, sex [determined based on chart review], body mass index, race [determined based on chart review], comorbidities), injury characteristics (mechanism, new ISS,23 Abbreviated Injury Scale [AIS]), physiologic variables (Acute Physiology and Chronic Health Evaluation, and Sequential Organ Failure Assessment), interventions (blood products, surgical interventions), and recovery variables (ICU days, ventilator days, complications, and admission laboratories [platelets (1,000/μL), absolute lymphocyte count (cell/μL), platelet lymphocyte ratio, mortality within 28 days]). Obesity was defined as body mass index of ≥30 kg/m2. Complications were noted throughout hospital admission. Of note, catheter-associated urinary tract infection, urinary tract infection, and superficial surgical site infection were considered less severe complications, and the remainder of the complications was considered severe complications in this analysis.
Blood Sample Collection
Healthy controls underwent collection of blood samples at baseline health. The TDAP study patients underwent blood draws upon arrival to the trauma bay (T0). Whole blood samples were processed as previously described.14
Systemic Biomarkers
Serum and plasma samples were thawed and analyzed for systemic biomarkers (cytokines, chemokines, and microRNAs) as described hereinafter. All laboratory evaluations were conducted by investigators blinded to patient characteristics associated with each sample.
Cytokine and Chemokine Analysis
Cytokine and chemokine expression were evaluated in serum samples using a multiplex assay approach as previously described.5 Serum aliquots were thawed and filtered (0.65 μm; Millipore, Billerica, MS). Human Cytokine Magnetic 35-Plex Panel assays (cat. LHC6005M; ThermoFisher, Frederick, MD) were performed on a Bio-Plex 200 Luminex system with high-throughput fluidics (cat. no. 171000205; Bio-Rad, Hercules, CA). Of the original 35 analytes tested, 11 cytokines and chemokines (IL-1RA, IL-6, IL-8, IL-10, IL-12, hepatocyte growth factor, vascular endothelial growth factor [VEGF], eotaxin, MCP-1, macrophage inflammatory protein-1β, and Regulated upon Activation, Normal T-cell Expressed and Presumably Secreted) were selected for analysis in this study based on at least 65% of the measurements occurring within the linear dynamic range of the target analyte's standard curve.
Multiplex Cellular miRNA Evaluation
Cellular microRNA levels were analyzed by FirePlex miRNA assay (Abcam, Waltham, MA) as previously described.24 For each sample, 20 μL of plasma was mixed with 20 μL of H2O and 40 μL of digestion mix and incubated at 60°C, and then, 25 μL of each sample was then added to a single well of a 96-well plate with 35 μL of Fireplex particles and 25 μL of hybridization buffer and incubated at 37°C shaking. Of note, Fireplex particles were manufactured such that each particle contained capture probes complementary to a single miRNA of interest, and ~100 particles for each miRNA were mixed together in a single well, allowing for multiplexing of each sample. All samples were rinsed, and ligation of samples was performed with 75 μL of 1× labeling buffer and allowed to incubate at 37°C. Samples were rinsed again and then filtered through a vacuum manifold with 65 μL of 95°C RNAse-free water added twice to each well. The filtered Fireplex particles were stored at 4°C, and 30 μL of the eluent was added to a clean polymerase chain reaction (PCR) plate. This eluent was mixed with 20 μL of PCR master mix and underwent 32 cycles of PCR amplification. Suction was then used to remove the previously filtered Fireplex particles, and these particles were suspended in 60 μL of hybridization buffer and then mixed with 20 μL of PCR product. The plate was incubated at 37°C. The wells were then rinsed, and 75 μL of 1× reporting buffer was added to each well. The plate was then incubated at 37°C. The wells were then rinsed, and 175 μL of run buffer was added to each well. At this time, the particles were scanned through a 6HT flow cytometer. microRNAs were recorded in arbitrary units.
MicroRNA targets were selected through literature review of microRNAs involved in various pathways of the early response to inflammatory stimuli and included miR-16-5p, miR-20a-5p, miR-21-3p, miR-23a-3p, miR-26a-5p, miR-26b-5p, miR-92a-3p, miR-103a-3p, miR-150-5p, miR-223-3p, miR-342-3p, and miR-486-5p.13,16,19 The selected microRNAs, associated sequences, and limits of detection are listed in Supplemental Digital Content (Supplementary Table 1, http://links.lww.com/TA/C624). Limit of detection was calculated using background signal in the designated water wells in the multiplex assay. All results were normalized to the geometric mean of all targets detected above a given expression threshold.
STATISTICAL ANALYSIS
The primary outcome of differential expression of microRNAs in polytrauma patients compared with controls and secondary outcomes of differential expression of microRNAs based on outcomes and baseline data were evaluated by standard statistical analysis. Bivariate analyses were conducted with the systemic biomarkers as well as the demographic, hospital course, injury severity, and plasma and serum biomarker data. Categorical variables were evaluated with the χ2 test. Continuous variables were evaluated with Kruskal-Wallis and t tests. Correlations between continuous variables were assessed with Pearson correlation, and ordinal variables with Spearman correlation. Significant correlation was defined as R > 0.3 and p < 0.05.
Independent bivariate analyses were conducted for predictors of complicated recovery (CR) based on the systemic biomarkers (cytokines, chemokines, and microRNAs). Each predictor was evaluated independently for its ability to predict the complicated recovery outcome. Area under the receiver operator characteristic curve analyses were performed to quantify the predictive quality of each biomarker for complicated recovery. Finally, critical biomarker concentrations were determined using threshold analysis for prediction of complicated recovery. The threshold was selected to maximize the product of biomarker predictive sensitivity and specificity.
Multivariable models were fitted to evaluate the predictive value of measured systemic biomarkers at T0 relative to CR, as well as clinical and biomarker values relative to microRNAs at T0. These results were reported in keeping with the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis statement.25 Parsimonious models were preferred for consideration by the clinical investigators; accordingly, we used Lasso regression (LASSO)26 and recursive partitioning via R package rPart (https://CRAN.R-project.org/package=rpart), an R implementation of Classification and Regression Trees.27 Missing values underwent multiple imputations via the random forest algorithm (R package missForest) to reduce bias.28 All models were evaluated with 100-fold cross-validation. All analyses were done in R version 3.6.3.
RESULTS
Polytrauma patients (n = 180) were selected from the TDAP database. From the 620 patients in the TDAP database at the time of analysis, 239 were excluded for ISS of <16, and a subsequent 181 were excluded for complications outside of criteria. Finally, 20 were excluded based on absence of blood sample. These remaining 180 patients were composed of predominantly young (32 [24–49] years) and male patients (77%) with limited comorbidities and high ISS (32 [22–38]) associated with chest or pelvic trauma (Table 1). The complicated recovery (n = 60) group had more female patients (33% vs. 17%) and higher ISS (34 vs. 25) associated with blunt injury (63% vs. 38%). Complicated recovery patients had higher rates of obesity (28% vs. 14%), diabetes (12% vs. 3%), blood transfusion (83% vs. 58%), mortality (10% vs. 0%), and Sequential Organ Failure Assessment >4 (63% vs. 19%). While both the complicated recovery and uncomplicated recovery patients had high abdomen AIS scores, only the complicated recovery patients had elevated chest AIS scores. Complicated recovery patients also had higher Acute Physiology and Chronic Health Evaluation scores on the day of injury (14 vs. 5), as well as longer ICU stay (26 vs. 2 days) and days on mechanical ventilation (18 vs. 0 days). Moreover, as shown in Supplemental Digital Content (Supplementary Table 2, http://links.lww.com/TA/C625), complicated recovery patients had more severe complications largely driven by increased rates of VAP (62% vs. 1%).
TABLE 1.
Demographics, Injury, Management Critical Care, and Recovery Data for Polytrauma Patients With Complicated Recovery and Uncomplicated Recovery
| PtP (n = 180), n (%) or Median (IQR) | Complicated Recovery* (n = 60), n (%) or Median (IQR) | Uncomplicated Recovery (n = 120), n (%) or Median (IQR) | p | |
|---|---|---|---|---|
| Age, y | 32 (24–49) | 37 (25.75–51.25) | 29 (23–45.25) | 0.044 |
| Race (African American) | 115 (63.89%) | 37 (61.67%) | 78 (65%) | 0.78 |
| Sex (female) | 139 (77.22%) | 40 (66.67%) | 99 (82.5%) | 0.03 |
| BMI (≥30 kg/m2) | 34 (18.89%) | 17 (28.33%) | 17 (14.17%) | 0.009 |
| Comorbidity | 116 (64.44%) | 39 (65%) | 77 (64.17%) | >0.099 |
| Diabetes | 10 (5.56%) | 7 (11.67%) | 3 (2.5%) | 0.03 |
| Hypertension | 15 (8.33%) | 8 (13.33%) | 7 (5.83%) | 0.15 |
| ISS | 26 (20–34) | 34 (25–42.25) | 25 (19–29) | <0.01 |
| AIS head and neck (mode) | 0 | 0 | 0 | |
| AIS face (mode) | 0 | 0 | 0 | |
| AIS chest (mode) | 3 | 3 | 0 | |
| AIS extremity (mode) | 0 | 0 | 0 | |
| AIS abdominal (mode) | 4 | 4 | 4 | |
| Injury mechanism (blunt) | 83 (46.11%) | 38 (63.33%) | 45 (37.5%) | <0.01 |
| Crystalloid, mL | 3,000 (2,000–4,500) | 4,000 (2,900–5,000) | 2,500 (1,500–3,900) | <0.01 |
| Blood transfusion (yes) | 120 (66.67%) | 50 (83.33%) | 70 (58.33%) | <0.01 |
| pRBCs, mL | 2,100 (700–4,900) | 5,250 (2,975–7,875) | 1,050 (87.5–2,450) | <0.01 |
| FFP, mL | 380 (0–1,490) | 1,490 (760–2,375) | 0 (0–570) | <0.01 |
| Platelets, mL | 0 (0–230) | 230 (0–460) | 0 (0–0) | |
| Cryoprecipitate, mL | 0 (0–0) | 0 (0–11.5) | 0 (0–0) | |
| WBC (admission, 1,000/μL) | 12.8 (10–18.6) | 11.4 (9.9–17.7) | 13.1 (10.08–18.7) | 0.48 |
| Platelets (admission, 1,000/μL) | 226 (180.5–279) | 233.5 (177–275.75) | 226 (181–284) | 0.68 |
| Absolute lymphocyte count (admission, cells/μL) | 1.73 (1.07–2.94) | 1.61 (1.071–2.95) | 1.75 (1.06–2.74) | 0.31 |
| Platelet lymphocyte ratio | 122.70 (64.66–200.43) | 122.70 (64.92–194.84) | 122.89 (59.82–204.41) | 0.55 |
| Length of ICU stay, d | 5 (1–17.25) | 26 (17.75–42.75) | 2 (0–5) | <0.01 |
| Length of ICU stay (>14 d) | 50 (27.78%) | 50 (83.33%) | 0 (0%) | <0.01 |
| Mechanical ventilation, d | 1 (0–10) | 18.5 (10–28.5) | 0 (0–1.025) | |
| Mechanical ventilation (>7 d) | 52 (28.89%) | 52 (86.67%) | 0 (0%) | <0.01 |
| SOFA >4 | 61 (33.89%) | 38 (63.33%) | 23 (19.17%) | <0.01 |
| APACHE scores | 8 (2–14) | 14 (10–21) | 5 (1–10.25) | <0.01 |
| Mortality | 6 (3.33%) | 6 (10%) | 0 (0%) | <0.01 |
*Complicated recovery (ICU >14 days, ventilator >7 days, mortality within 28 days).
APACHE, Acute Physiology and Chronic Health Evaluation; BMI, body mass index; FFP, fresh frozen plasma; pRBC, packed red blood cell; SOFA, Sequential Organ Failure Assessment; WBC, white blood cell.
MicroRNAs were detected in 99% of PtPs at T0 (median, 158 minutes from injury) as shown in Table 2. Only IL-6 was noted to be elevated in PtPs (60 vs. 54 pg/mL) at this early time point. The microRNAs varied significantly at T0 in PtPs compared with healthy controls (n = 12). miR-16-5p, miR-20a-5p, miR-23a-3p, miR-26a-5p, miR-26b-5p, miR-103a-3p, miR-150-5p, and miR-223-3p were all found to be significantly down regulated in PtP compared with controls, and microRNAs were found significantly lower in complicated recovery group (miR-16-5p, miR-20a-5p, miR-26b-5p, miR-150-5p, and miR-223-3p) and the uncomplicated recovery group (miR-16-5p, miR-20a-5p, miR-23a-3p, miR-26a-5p, miR-103a-3p, miR-150-5p, and miR-223-3p) compared with controls.
TABLE 2.
Plasma Levels of Cellular microRNAs, Cytokines, and Chemokines in Polytrauma Patients Drawn During Admission in Trauma Bay (T0)
| Polytrauma Patients and Controls (n = 192) | Controls (n = 12) | Polytrauma Patient (n = 180) | Complicated Recovery (n = 60) | Uncomplicated Recovery (n = 120) | ||||
|---|---|---|---|---|---|---|---|---|
| Cellular miRNA | Detection Rate,* n (%) | Median (IQR), AU | Median (IQR), AU | p (Controls vs. Polytrauma Patient) | Median (IQR), AU | p (Controls vs. Complicated Recovery) | Median (IQR), AU | p (Controls vs. Uncomplicated Recovery) |
| miR-16-5p | 190 (99) | 8,845.24 (7936.56–9474.97) | 3,231.86 (82.85–158.95) | <0.01 | 3,595.66 (2863.38–4940.65) | <0.01 | 3,036.07 (2322.92–4047.62) | <0.001 |
| miR-20a-5p | 190 (99) | 1,313.49 (7936.56–9474.97) | 463 (351.54–809.06) | <0.01 | 601.03 (356.28–1146.08) | <0.01 | 439.38 (350.20–625.25) | <0.001 |
| miR-21-3p | 190 (99) | 4.16 (0.28–10.55) | 0.72 (0.15–3.56) | 0.41 | 2.59 (0.42–12.14) | 0.79 | 0.5 (0.11–1.48) | 0.17 |
| miR-23a-3p | 190 (99) | 619.88 (556.99–697.78) | 381.13 (206.71–665.69) | 0.02 | 540.72 (254.26–1449.46) | 0.69 | 333.12 (191.49–553.54) | <0.01 |
| miR-26a-5p | 190 (99) | 276.8 (209.38–328.03) | 166.55 (122.93–229.33) | <0.01 | 211.73 (147.67–276.66) | 0.07 | 145.39 (104.52–192.73) | <0.01 |
| miR-26b-5p | 190 (99) | 54.1 (26.0–63.93) | 5.15 (2.96–10.93) | <0.01 | 8.91 (4.8–26.62) | <0.01 | 4.48 (2.76–7.31) | <0.001 |
| miR-92a-3p | 190 (99) | 1,512.67 (1200.75–1705.06) | 1,270.82 (971.64–1861.02) | 0.47 | 1,736.79 (1078.98–2379.69) | 0.38 | 1,206.45 (885.80–1640.25) | 0.13 |
| miR-103a3p | 190 (99) | 397.33 (348.29–482.88) | 224.15 (151.77–345.29) | <0.01 | 317.48 (197.03–487.54) | 0.10 | 191.09 (141.25–269.09) | <0.001 |
| miR-150-5p | 190 (99) | 373.95 (240.54–424.57) | 136.67 (76.80–202.13) | <0.01 | 165.72 (102.67–226.53) | <0.01 | 114.38 (68.57–173.23) | <0.001 |
| miR-223-3p | 190 (99) | 1,643.07 (1513.38–1699.15) | 466.95 (299.95–846.67) | <0.01 | 740.67 (344.95–1360.96) | <0.01 | 415.26 (294.85–587.19) | <0.001 |
| miR-342-3p | 190 (99) | 136.22 (82.85–158.95) | 122.47 (75.87–173.53) | 0.91 | 134.18 (86.36–197.97) | 0.60 | 122.07 (71.44–165.99) | 0.66 |
| Cytokines and Chemokines | Detection Rate,* n (%) | Median (IQR), pg/mL | Median (IQR), pg/mL | p | Median (IQR), pg/mL | p | Median (IQR), pg/mL | p |
| IL-1RA | 136 (71) | 273.2 (273.2–547.5) | 353.1 (227.4–1,405.5) | 0.55 | 871.54 (277.0–3972.22) | <0.05 | 288.88 (222.18–876.28) | 0.85 |
| IL-6 | 132 (69) | 54.3 (26.7–54.2) | 60.4 (54.92–78.1) | <0.01 | 167.26 (58.49–574.95) | <0.01 | 57.82 (51.34–156.25) | <0.05 |
| IL-8 | 136 (71) | 302.7 (87.7–302.7) | 300.0 (238.9–1134.4) | 0.10 | 308.87 (197.71–315.84) | 0.08 | 299.82 (243.83–1086.36) | 0.12 |
| IL-10 | 132 (69) | 19.1 (19.1–19.1) | 23.1 (18.2–110.8) | 0.24 | 59.18 (18.73–174.43) | 0.06 | 19.86 (17.07–72.42) | 0.45 |
| IL-12 | 132 (69) | 55.2 (55.2–68.8) | 55.6 (41.4–60.9) | 0.28 | 56.12 (36.96–69.13) | 0.48 | 55.47 (41.94–59.6) | 0.23 |
| MCP-1 | 136 (71) | 360.1 (360.1–360.1) | 458.1 (356.2–1,462.9) | 0.08 | 1,309.62 (380.86–2773.39) | <0.01 | 411.15 (348.93–840.02) | 0.29 |
| VEGF | 134 (70) | 14.0 (12.0–14.0) | 11.8 (5.3–14.6) | 0.48 | 12.94 (7.54–29.03) | 1.00 | 11.33 (4.88–14.36) | 0.29 |
| MIP-1β | 136 (71) | 66.5 (63.0–66.5) | 73.9 (64.3–109.9) | 0.06 | 78.54 (67.27–150.29) | <0.05 | 73.03 (348.93–840.02) | 0.10 |
| HGF | 134 (70) | 14.0 (12.0–14.0) | 11.8 (5.3–14.6) | 0.48 | 660.81 (425.85–1070.94) | 0.16 | 1,002.06 (527.8–1185.56) | 0.90 |
| RANTES | 134 (70) | 2,339.4 (2,339.4–2,646.4) | 2,324.4 (1,993.1–2,573.1) | 0.27 | 2,335.16 (2086.89–2827.91) | 0.51 | 2,309.75 (1981.02–2469.18) | 0.19 |
| Eotaxin | 132 (69) | 44.3 (44.3–54.8) | 44.9 (39.6–62.7) | 0.62 | 47.61 (38.02–70.57) | 0.87 | 44.49 (39.72–59.89) | 0.41 |
*Detection rate for cellular microRNAs was based on background signal in multiplex assay and for cytokines and chemokines was based on at least 65% of the measurements occurring within the linear dynamic range of the target analyte's standard curve.
Plasma levels also noted for polytrauma patients with complicated recovery and uncomplicated recovery.
AU, arbitrary units; HGF, hepatocyte growth factor; MIP-1β, macrophage inflammatory protein-1β; RANTES, Regulated upon Activation, Normal T-cell Expressed and Presumably Secreted.
Considering the associations between microRNAs and the described baseline and recovery data in Table 1, Figure 1A to C demonstrates significant (p < 0.05) associations including elevated miR-23a-3p and miR-342-3p in younger patients and lower levels of miR-16-5p, miR-26a-5p, and miR-103a-3p in younger patients. miR-342-3p was higher in African American patients, and miR-26a-5p and miR103a-3p were lower in patients with comorbidities. Significant correlations were detected between microRNAs and continuous variables from admission and recovery (data not shown). Sequential Organ Failure Assessment scores positively correlated to miR-26a-5p (R = 0.33, p < 0.01), miR-26b-5p (R = 0.33, p < 0.01), and miR-103a-3p (R = 0.41, p < 0.01). Acute Physiology and Chronic Health Evaluation was positively correlated to miR-21-3p (R = 0.031, p < 0.01) and miR-103a-3p (R = 0.39, p < 0.01).
Figure 1.
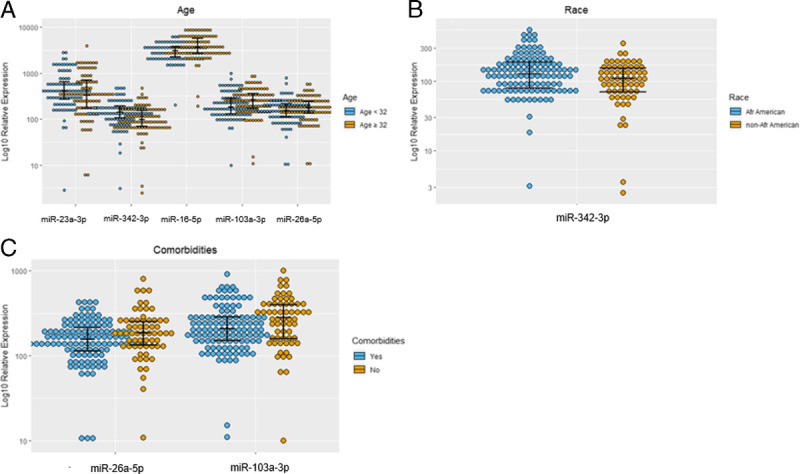
(A–C) Significant (p < 0.05) associations noted between polytrauma patient's plasma levels of cellular miRNAs during admission to the trauma bay and demographic variables.
Table 3 demonstrates the results of multivariable LASSO modeling of predictors of microRNAs at T0 based on systemic inflammatory mediators, available laboratory values, and clinical variables. The largest log odds identified included the predictive value of VEGF for miR-21-3p (log odds, 0.45) and miR-23a-3p (log odds, 0.32), and protective value of Regulated upon Activation, Normal T-cell Expressed and Presumably Secreted for miR-23a-3p (log odds, −0.37).
TABLE 3.
Multivariate LASSO Modeling of Predictors of Polytrauma Patient's Plasma Cellular microRNAs During Admission to the Trauma Bay Based on Systemic Inflammatory Mediators, Available Blood Laboratory Values, and Clinical Variables
| miR-16-5p | miR-20a-5p | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | Log Odds | SE | T | p | Variable | Log Odds | SE | T | p |
| (Intercept) | 3.58 | 0.1 | 36.63 | <0.001 | (Intercept) | 2.95 | 0.1 | 29.98 | <0.001 |
| Age | 0 | 0 | 2.34 | 0.020 | IL-8 | −0.14 | 0.04 | −3.97 | <0.001 |
| Crystalloid | 0 | 0 | 3.31 | 0.001 | VEGF | 0.14 | 0.05 | 2.74 | 0.007 |
| AIS pelvic score | −0.04 | 0.01 | −2.81 | 0.006 | |||||
| Platelets | 0 | 0 | −2.35 | 0.020 | Full data R2 | 0.1 | |||
| XV R2 | 0.13 | ||||||||
| Full data R 2 |
0.17 | ||||||||
| XV R2 | 0.16 | ||||||||
| miR-21-3p | miR-23a-3p | ||||||||
| Variable | Log Odds | SE | T | p | Variable | Log Odds | SE | T | p |
| (Intercept) | −0.7 | 0.12 | −5.71 | <0.001 | (Intercept) | 3.49 | 0.46 | 7.61 | <0.001 |
| VEGF | 0.45 | 0.13 | 3.59 | <0.001 | IL-10 | 0.18 | 0.06 | 3.04 | <0.001 |
| APACHE | 0.03 | 0.01 | 3.5 | <0.001 | VEGF | 0.32 | 0.08 | 3.75 | <0.001 |
| AIS extremity score | 0.04 | 0.02 | 2.18 | 0.031 | |||||
| Full data R2 | 0.17 | IL-8 | −0.13 | 0.05 | −2.58 | 0.011 | |||
| XV R2 | 0.19 | RANTES | −0.37 | 0.14 | −2.61 | 0.010 | |||
| Full data R2 | 0.19 | ||||||||
| XV R2 | 0.15 | ||||||||
| miR-26a-5p | miR-26b-5p | ||||||||
| Variable | Log Odds | SE | T | p | Variable | Log Odds | SE | T | p |
| (Intercept) | 1.98 | 0.06 | 32.06 | <0.001 | (Intercept) | 0.58 | 0.05 | 11.12 | <0.001 |
| SOFA | 0.02 | 0.01 | 2.74 | 0.007 | SOFA | 0.06 | 0.01 | 5.41 | <0.001 |
| IL-10 | 0.09 | 0.04 | 2.33 | 0.021 | |||||
| Full data R2 | 0.14 | ||||||||
| Full data R2 | 0.11 | XV R2 | 0.16 | ||||||
| XV R2 | 0.12 | ||||||||
| miR-92a-3p | miR-103a-3p | ||||||||
| Variable | Log Odds | SE | T | p | Variable | Log Odds | SE | T | p |
| (Intercept) | 3.26 | 0.1 | 33.87 | <0.001 | (Intercept) | 2.16 | 0.04 | 49.23 | <0.001 |
| Platelets | 0 | 0 | −2.96 | 0.004 | SOFA | 0.03 | 0.01 | 4.49 | <0.001 |
| VEGF | 0.09 | 0.04 | 2.22 | 0.028 | Crystalloid | 0 | 0 | 2.01 | 0.046 |
| AIS pelvic score | −0.04 | 0.02 | −2.5 | 0.013 | |||||
| Crystalloid | 0 | 0 | 2.31 | 0.022 | Full data R2 | 0.15 | |||
| XV R2 | 0.16 | ||||||||
| Full data R2 | 0.13 | ||||||||
| XV R2 | 0.1 | ||||||||
APACHE, Acute Physiology and Chronic Health Evaluation; RANTES, Regulated upon Activation, Normal T-cell Expressed and Presumably Secreted; SOFA, Sequential Organ Failure Assessment; XV, cross-validation.
Polytrauma patients with recovery complications had higher levels of microRNAs as seen in Figure 2A to D, and included recoveries complicated by acute respiratory distress syndrome (ARDS) (miR-16-5p, miR-92a-3p, and miR-103a-3p), VAP (miR-21-3p, miR-26a-5p, miR-26b-5p, miR-92a-3p, miR-103a-3p), venous thromboembolic event (VTE) (miR-103a-3p) or PtPs who required blood transfusions (miR-103a-3p, miR-26a-5p, miR-26b-5p, and miR-223-3p). Polytrauma patients with recovery complicated by mortality or PtPs categorized as complicated had higher values of all studied microNRAs except miR-342-3p (Fig. 2E and F).
Figure 2.
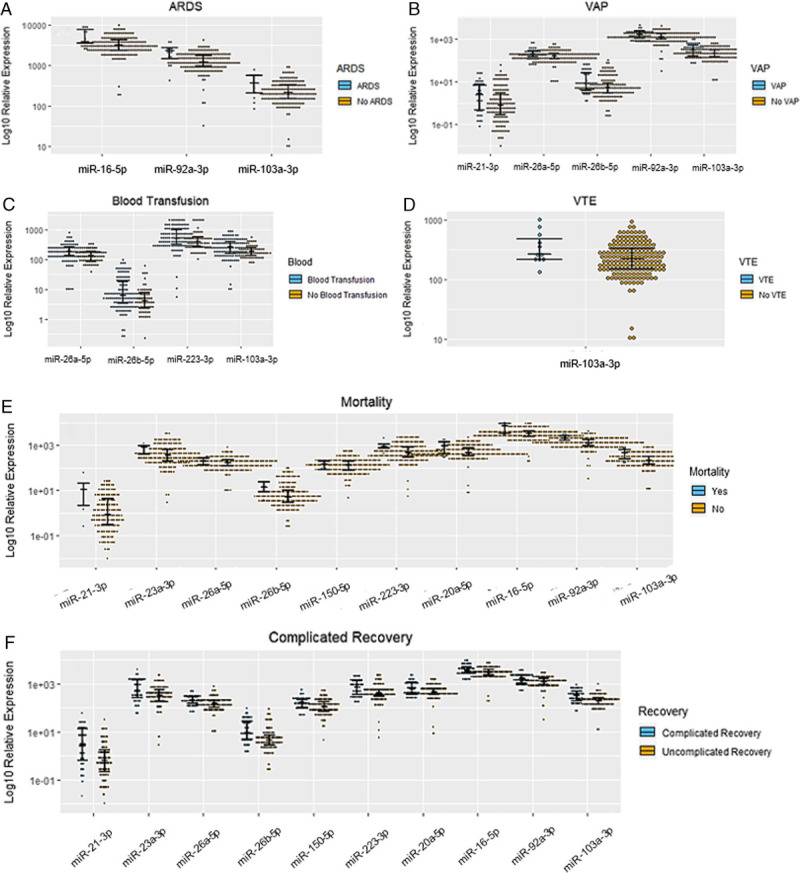
(A–F) Significant (p < 0.05) associations noted between polytrauma patient's plasma levels of cellular miRNAs during admission to the trauma bay and recovery complications.
Independent bivariate analysis of predictors of complicated recovery based on the evaluated systemic mediators of inflammation (microRNAs, cytokines, and chemokines) is listed in Supplemental Digital Content (Supplementary Table 3, http://links.lww.com/TA/C626). The biomarkers with the most pronounced area under the curve to predict complicated recovery include miR-21-3p, miR-26b-5p, and miR-103a-3p as well as MCP-1, VEGF, IL-1Ra, IL-6, and IL-10. The threshold values listed were selected to maximize the product of sensitivity and specificity.
Multivariable LASSO modeling to predict complicated recovery based on the evaluated systemic mediators of inflammation is listed in Table 4 and suggests that miR-21-3p (log odds, 0.96) and MCP-1 (log odds, 1.70) were the most robust predictors. The optimal value of γ was chosen to maximize area under the curve across cross-validations.
TABLE 4.
Multivariate LASSO Modeling to Predict Complicated Recovery Based on the Evaluated Systemic Mediators of Inflammation (Cellular MicroRNAs, Cytokines, and Chemokines)
| Biomarker | Log Odds | SE | Z | p |
|---|---|---|---|---|
| (Intercept) | −5.74 | 1.27 | −4.54 | <0.001 |
| MCP-1 | 1.7 | 0.42 | 4.02 | <0.001 |
| miR-21-3p | 0.96 | 0.24 | 3.97 | <0.001 |
| XV AUC | 0.78 | |||
| XV sensitivity | 0.80 | |||
| XV specificity | 0.72 |
AUC, area under the curve; XV, cross-validation.
DISCUSSION
MicroRNAs are emerging as important systemic biomarkers in inflammatory conditions such as TBI and sepsis.16,19–21 Herein the authors have demonstrated in the first multicenter prospectively collected trauma database that cellular microRNAs, with key mechanisms in regulating the inflammatory response, were reliably detected and differentially expressed in PtPs compared with controls, and early differences in expression levels were predictive of poor outcomes. These novel microRNA findings in PtPs are consistent with previously described early variations in systemic levels of inflammatory mediators, which have demonstrated predictive value of poor outcomes in trauma patients.9 Within this study's subset analysis of PtPs, systemic levels of microRNAs were also differentially expressed based on demographic variables, and higher levels of microRNAs were associated with subsequent diagnosis of ARDS, VAP, and VTE. Limited correlations were noted between microRNAs and other systemic markers of inflammation in admission blood samples, although multivariable models demonstrated that VEGF and IL-10 were predictive of multiple microRNAs and IL-8 was found to be protective of two microRNAs. Among all measured systemic mediators of inflammation, multivariable modeling analysis revealed that miR-21-3p and MCP-1 were the strongest predictors of complicated recovery in polytrauma patients based on blood draws from trauma bay admission.
miR-21-3p is a well conserved microRNA located within the intronic portion of transmembrane protein 49 (TMEM49) gene and associated with an independent promoter region as well as pleiotropic effects.29 miR-21-3p is induced by multiple cytokines, including IL-6 and transforming growth factor β, and promotes immunosuppression and angiogenesis while inhibiting apoptosis by directly targeting programmed cell death 4 and phosphatase and tensin homolog (PTEN).29 Downstream impacts of blocking these pathways have demonstrated suppression of tumor necrosis factor α, IL-12, TLR2 and TLR4 signaling, and T-cell activation as well as increased VEGF, IL-10, and promotion of M2 macrophage phenotypes.17,20,29–31 Interestingly, when released in exosomes, miR-21-3p systemically also functions as a proinflammatory mediator by directly binding TLR8 and inducing release of IL-6 and tumor necrosis factor α in macrophages.17,32 Similar to well established markers of the inflammatory response to trauma (i.e., MCP-1), the described impacted pathways and pleiotropic effects of miR-21-3p would be consistent with an early biomarker to identify trauma patients at risk for a dysregulated immune response and consistent with the findings in our study of miR-21-3p's independent predictive value for complicated recovery in PtPs. Furthermore, miR-21-3p direct targeting of PTEN (a known regulator of hypoxia inducing factor-1 α and VEGF) would be consistent with the independent association with systemic VEGF demonstrated in our multivariable model.9,33
Beyond polytrauma patients with complicated recoveries, miR-21-3p has previously been demonstrated to be elevated in most solid cancers, acute kidney injury patients, septic patients with recovery complications, and TBI patients.17,20,21,29 Similar to the findings in our study, prior studies evaluating miR-21-3p levels in septic patients revealed no significant differences between plasma levels of miR-21-3p in septic patients compared with controls.16 When considering septic patients with recovery complications, however, systemic levels were noted to be elevated in patients with cardiac dysfunction.34 In TBI patients and acute kidney injury patients, systemic miR-21-3p was found to be elevated compared with healthy control patients and elevated in injured tissues.21,30 Taken together with the mechanisms described previously, miR-21-3p is potentially an important biomarker of tissue injury and plays an important role in modulating the inflammatory response with possible deleterious impacts (i.e., prolonged immunosuppression) if dysregulated. Additional studies evaluating miR-21-3p at multiple time points in trauma patients may yield further insight into its role in critically ill trauma patients and potential therapeutic applications.
Although not identified in the multivariable model predicting complicated recovery, plasma levels at T0 of miR-16-5p, miR-20a-5p, miR-23a-3p, miR-26a-5p, miR-26b-5p, miR-92a-3p, miR-103a-3p, miR-150-5p, and miR-223-3p were found, in bivariate analysis, to be elevated in PtPs who died within 30 days of trauma and complicated recovery patients, and select microRNAs were also elevated in patients with recoveries complicated by ARDS, VAP, and VTE. Interestingly, with the exception of miR-92a-3p, all of these microRNAs were lower in complicated recovery patients than healthy controls. Similar to miR-21-3p, these individual microRNAs affect multiple signaling pathways in the inflammatory response and include suppression of PTEN (miR23a-5p, miR-26a-5p, miR-92a-3p), NF-κB (miR-16-5p, miR23a-5p), TLR signaling (miR-16-5p, miR-92a-3p, miR-103a-3p), STAT3 (miR-223-3p), T-cell activation (miR-16-5p, miR-103a-3p, miR-150-5p), and apoptosis (miR-16-5p, miR23a-5p).18,35–40 Patients with uncomplicated recovery demonstrated more significant early suppression of these microRNAs, suggesting that the early inflammatory response to trauma includes downregulation of microRNAs that suppress immune signaling. By contrast, patients with complicated recovery had microRNAs levels closer to healthy controls suggesting limited suppression of these microRNAs, which may, at later time points, be consistent with subsequent immune dysregulation associated with complicated recovery. Prior studies in sepsis have demonstrated similar findings with lower levels of microRNAs (miR-23a-5p, miR-26a-5p, miR-26b-5p, miR-103a-3p, miR-150-5p) in septic patients compared with healthy controls; however, systemic levels of these cellular microRNAs were increased in patients with severe sepsis or sepsis mortality.16,20,37,41,42 Given the concurrent findings between PtPs in our study and sepsis studies, these microRNAs may yield early predictive value for trauma patients at risk for infectious complications and poor outcomes in trauma. Further evaluation of miR-103a-3p as a marker of VTE is merited given that the association between elevated levels of miR-103a-3p and VTE has been previously described in patients with recurrent VTE and potential mechanisms of action have been proposed.43
While some variations in microRNAs at T0 were noted in PtPs with respect to baseline demographic data in this study, there is a paucity of literature available on the associations of variables such as age and race on expression levels of cellular microRNA in response to inflammatory stimuli. In contrast to the findings in our study, which demonstrated elevated miR-342-3p in African American PtPs, Tudor et al.41 demonstrated lower levels of systemic miR-16-5p, miR-21-3p, and miR26b-5p in African American patients with sepsis compared with non–African American patients and no significant difference in miR-342-3p with respect to race in septic patients. These discordant findings could be attributed to the timing of blood draws in trauma studies with defined on-set tissue injury and the less well-defined onset of inflammatory stimuli from sepsis. Studies evaluating subjects at baseline health and with underlying comorbidities have previously demonstrated differential expression of microRNAs based on age and race;44,45 however, given the potential broad implications for microRNAs in the modulation of the early inflammatory response and disparities in trauma outcomes based on demographics, additional studies to consider differences in outcomes based on baseline demographics and microRNAs should be considered in critically ill patients.
There are several limitations to this study. We evaluated a selected group of microRNAs with previously demonstrated variations based on clinical studies in sepsis13,16,19 and TBI patients.21 While prior work has been conducted to evaluate the mechanisms of actions of these microRNAs in in vitro and in vivo models, there is limited literature to interpret the significance of systemic variations of these microRNAs in trauma patients. Furthermore, recent studies have demonstrated that microRNAs are pleuripotent and will have differential impacts on individual cells types based on the clinical context.46 Finally, the complicated recovery and uncomplicated recovery groups considered in this study varied at baseline with respect to age, sex, ISS, and mechanism of injury, as well as incidence of obesity and diabetes. The impact of these differences in baseline data between the groups on systemic microRNAs remains unclear given the overall limited associations noted between baseline variables and microRNAs demonstrated in the multivariate model. Combined clinical and preclinical studies to evaluate whole genome noncoding RNA expression in blood samples from a diverse group of trauma patients and in tissue and blood samples from high fidelity animal trauma models may shed insight into the impact of systemic variations of microRNAs and the utility of microRNAs as biomarkers.12,47 Despite these limitations, the data presented here show improved microRNAs detection rate in trauma patients compared with the previously established systemic cytokines and chemokines, and taken together with the predictive value of microRNAs in this study. MicroRNAs demonstrate significant potential as new biomarkers in evaluating trauma patients at risk for recovery complications (http://links.lww.com/TA/C627).
CONCLUSION
The dysregulated inflammatory response associated with severe injury drastically impacts the recovery of polytrauma patients, and despite an evolving understanding of the underlying mechanisms, the primary therapy continues to be only supportive care for these patients. Herein the authors identified the predictive value of early systemic levels of cellular microRNAs in polytrauma patients with a 99% detection rate of these microRNAs in trauma patients. These microRNAs were found to be associated with multiple demographic and recovery variables as well as other markers of the inflammatory response. Future larger clinical studies in trauma including evaluation of genome wide noncoding miRNAs and a broader biomarker panel at additional time points may yield further insight into the drivers of miRNA transcription. Preclinical experiments to evaluate therapeutic potential of microRNAs in trauma models should be considered in future studies.
Supplementary Material
AUTHORSHIP
D.A.V., S.A.S., S.A., M.J.B., H.R., F.L., M.S., T.G.B., C.J.D., V.K., A.D.K., G.A.C., and E.A.E. conceived and designed the data analysis. S.A., H.H., F.L., V.K., M.S., and G.A.C. collected data. D.A.V., S.A., H.H., F.L., M.J.B., and H.R. performed data analysis. D.A.V., S.A.S., S.A., H.H., F.L., H.R., M.J.B., T.G.B., T.A.D., A.D.K., G.A.C., and E.A.E. wrote the paper,
ACKNOWLEDGMENTS
We thank the hard work and dedication from our various laboratory, data, program management, and regulatory staff in assisting with the various aspects of this work. In particular, we thank Dr. Jaspreet Seth for regulatory support, Sarah Vicente for manuscript preparation, Myra Yusuf for program support, Chandra Almond and Megan Turk for data management support, and Dr. Neal Iwakoshi and Mary-Beth Joshi for bioprocessing and molecular assay work that supported this project.
G.A.C. is the Felix L. Haas Endowed Professor in Basic Science. Work in G.A.C.'s laboratory is supported by the NCI grants 1R01 CA182905-01 and 1R01CA222007-01A1, an NIGMS 1R01GM122775-01 grant, a DoD Idea Award W81XWH2110030, a Team DOD grant in Gastric Cancer, a Chronic Lymphocytic Leukemia Moonshot Flagship project, a CLL Global Research Foundation 2019 grant, a CLL Global Research Foundation 2020 grant, a Mathers Foundation grant, and institutional research grants and development grants associated with the Brain SPORE.
This study was supported, in part, by the Department of Defense's Defense Health Program—Joint Program Committee 6/Combat Casualty Care (USUHS HT9404-13-1-0032 and HU0001-15-2-0001).
DISCLOSURE
G.A.C. is one of the scientific founders of ITHAX Pharmaceuticals.
This work was performed at Uniformed Services University of the Health Sciences, Walter Reed National Military Medical Center, and University of Texas MD Anderson. The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions, or policies of Uniformed Services University, The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., the Department of Defense, and the Departments of the Army, Navy, or Air Force. Mention of trade names, commercial products, or organizations does not imply endorsement by the US Government. This work was prepared by a military (D.A.V.) or civilian (T.A.D., E.A.E) employees of the US Government as part of the individual's official duties and therefore is in the public domain and does not possess copyright protection.
DISCUSSION
MARTIN SCHREIBER, M.D. (Portland, Oregon): Thank you. I just want to point out that the last two papers were both from military sites and highlight the increasing participation of the military in this meeting and increasing collaborations with civilian institutions.
And, in fact, in the last paper from Madigan, the pigs that go to Madigan come from Southern California and they stop in Oregon on the way up there at OHSU and that’s where we get our pigs from, too. And thank you. It saves us a lot of money.
It’s a great collaboration and I appreciate the opportunity to discuss this really interesting and well-done paper.
The authors have performed a substantive analysis of patients enrolled in the tissue and data acquisition protocol, multi-institutional database focusing on patients with an ISS greater than 15 in whom they assessed a number of inflammatory parameters to include cellular microRNAs to test the hypothesis that they are predictive of a complicated recovery.
The authors did confirm an association between cellular microRNAs and poor outcomes. I have the following comments and questions for the authors.
It looks like there are about 1183 patients in the t-DAP database. The authors chose 180 patients or about 6 percent of that database to evaluate for this study. How was this 6 percent of the t-DAP database selected? And could there be bias in that selection?
The complicated recovery group is very different from the uncomplicated recovery group with respect to many demographic characteristics, to include ISS, sex, mechanism of injury, incidence of obesity, diabetes and ISS.
How does this potentially affect the cellular microRNA analysis? And could this also be a bias? Could the authors have matched patients better to take away some of this bias?
The authors refer to the patients in the study as polytrauma patients but only the mode for abdominal AIS is greater than zero in the uncomplicated recovery group.
This also supplies additional potential for bias as the mode for AIS chest and abdomen is higher than zero in the complicated recovery group. Please comment.
Cellular microRNAs were down-regulated in injured patients compared to controls but were up-regulated in complicated recovery patients compared to uncomplicated recovery patients. This seems unusual to me in that if injury down-regulates cellular microRNAs, shouldn’t severe injury down-regulate them more?
In your study, did cellular microRNAs perform better than standard, clinical predictors like ISS and APACHE in predicting outcomes? And in your opinion how realistic is it to believe that they could be used either to predict outcomes in real-life scenarios or to serve as therapeutic targets now or in the future?
Thank you
ROSEMARY A. KOZAR, M.D., Ph.D. (Baltimore, Maryland): Really nice presentation, I’m incredibly jealous of your findings. I’ve been very interested in microRNAs after trauma and my data is definitely not as clean as yours.
I just have a couple of quick questions for you. First, could you how you are reporting changes in microRNAs? Are you reporting the full difference or the CTT value or how are you expressing these?
Second, I was curious where you got your platform from that you chose your different microRNAs. It doesn’t sound like you did a complete array but it was a targeted array.
Really nice work and thank you very much.
DIEGO VICENTE, M.D. (Chicago, Illinois): Thank you so much, everybody. I’m sorry I can’t be there with you all in Atlanta but I really appreciate Dr. Pritts, Dr. Gurney, and the opportunity to present and then Dr. Schreiber and Dr. Kozar for their questions.
I’ll briefly speak to the military-civilian partnerships and collaborations. They’re phenomenally strong and I think this project is one of those settings.
I’ll also note there are a few military folks in the audience there today and I certainly appreciate their support. I really appreciate the AAST entertaining me as a surgical oncologist within your presentations today, and I look forward to continuing this partnership.
To address Dr. Schreiber’s questions, the 180 patients were selected back in 2018 when we had initially conceptualized this idea. And there were about 600 patients in the TDAP database at that point.
Given that there hadn’t been a dedicated study of cellular microRNAs within a trauma population, outside of TBI, we didn’t know if we were going to find a signal at all and so we started looking at which patients are we going to see that are going to demonstrate a signal, if one is available.
With that, we chose specifically patients with ventilator-associated pneumonia or organ space infection or some equivalent severe complication after polytrauma in order to start stratifying our patients and then compared those to patients that had no severe recovery complications.
So cutoff from that initial analysis would have been patients with UTIs or cellulitis after trauma, that would have counted as a complication but not necessarily given the associated inflammatory response which would have been associated with a prolonged ICU stay which was really what we were thinking about in the background.
So from the 600 patients we initially selected out about 240 patients that didn’t meet that complication. Another 180 patients were excluded based on the complication definition and then 20 patients were excluded from not having blood samples available and so that’s how we went from 600 down to 180.
With regard to the question of demographics, this is absolutely an important point and when we were trying to answer the question of well, what do these cellular microRNAs mean in the context of a patient that shows up in the trauma bay and you are able to get an indicator of whether or not they are going to be sick in the ICU for a long time.
We selected the patients initially but then split the groups based on the complicated versus uncomplicated recovery definition.
There were significant differences in the baseline demographics between those two groups. But, interestingly, on the multivariate analysis and then in the standard statistical analysis we didn’t see a lot of associations between those baseline demographics and the resulting cellular microRNAs, even when we were using multivariate modeling.
I think it speaks to the same point as what was found in the Glue Grant Study, which was that we all may look different at baseline on the outside but then when you expose us to severe trauma we do start looking more similar. And so, despite some baseline demographic differences, they would not appear to impact the cellular microRNAs.
With respect to the biases of the chest and abdominal trauma, these studies were conducted at other institutions where the surgical critical care initiative had ongoing protocols and so there was a heavy selection against traumatic brain injury as most of those patients went into another protocol.
I’ve been working on trauma databases for a while and I still don’t have a great way to demonstrate the AIS that reflects the ISS and mode is what our statisticians gave us their best approximation.
Separately, I can comment on the means on each of those but at the end of the day these patients were all severely injured with ISS well above 16.
Regarding to the overall mechanism of cellular microRNAs being lower in the injured patients compared to controls, you would expect to have an ongoing titration effect of the microRNAs being lower in patients that had more severe injury or were going to go on to have complications.
With that, you know, I’ve got to tell you the first time I received this data analysis back from our statisticians I thought that maybe we had flipped something and so I sent it back for another analysis.
And, interestingly, they were absolutely right and no matter which way we ran it the folks that went on to have a complicated recovery had levels of microRNAs that were closer to the healthy controls.
Given that most of these microRNAs regulate early inflammatory pathways, this may speak to an early sign of immune dysregulation where if you are going to have an uncomplicated recovery then maybe your cellular microRNAs are supposed to suppress to a certain level and if they don’t that may be speaking to the underlying mechanism that sets you up for a dysregulated immune response and a prolonged ICU stay.
The therapeutic target applications for microRNAs I think are absolutely huge. In the world of trauma we are developing preclinical models in our lab and we have a lot of potential targets to look at, down-regulating inflammatory response and modulating early responses to injury that may dampen the inflammatory response and maybe give us a chance of mitigating some of the consequences.
I think there are ongoing therapeutic miRNA clinical trials in auto-immune diseases and certainly in the oncology world that are demonstrating some promising results. And so I think that microRNAs are going to make their way into trauma, as well.
To Dr. Kozar’s questions, how we expressed these levels was in arbitrary units. The reason that it’s listed under arbitrary units is how we evaluated them with Abcam in their multiplex assays so it’s not strictly a QPCR assay. I can send you further information from Abcam on how that is conducted.
The reason for the arbitrary units is that we did not follow the typical standard curve analysis but, rather, this was a relative comparison for an exploratory study.
Finally, I think the last question from Dr. Kozar was how did we select these targets. And, basically, I went down the same pathways that I did when I was developing these biomarker panels and I was a resident in Dr. Elster’s lab a few years ago.
I looked at the relevant inflammatory pathways, then most likely microRNAs that would be involved in those pathways, and then I was able to customize a biomarker panel based on these microRNAs with the folks at Abcam.
I think that covers all of the previously-presented questions. Once again, thank you all for this presentation and thank you for the surgical critical care initiative folks for all their support on this project.
Footnotes
Published online: July 7, 2022.
This study was presented at the 80th Annual Meeting of AAST and Clinical Congress of Acute Care Surgery, October 1, 2021, in Atlanta, Georgia.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jtrauma.com).
Contributor Information
Seth A. Schobel, Email: seth.schobel-mchugh.ctr@usuhs.edu.
Simone Anfossi, Email: sanfossi@mdanderson.org.
Hannah Hensman, Email: hannah.hensman@decisionq.com.
Felipe Lisboa, Email: felipe.lisboa.ctr@usuhs.edu.
Henry Robertson, Email: henry.robertson.ctr@usuhs.edu.
Vivek Khatri, Email: vivek.khatari@usuhs.edu.
Matthew J. Bradley, Email: matthew.j.bradley22.mil@mail.mil.
Masayoshi Shimizu, Email: mshimizu@mdanderson.org.
Timothy G. Buchman, Email: tbuchma@emory.edu.
Thomas A. Davis, Email: thomas.davis@usuhs.mil.
Christopher J. Dente, Email: cdente@emory.edu.
Allan D. Kirk, Email: allan.kirk@duke.edu.
George A. Calin, Email: gcalin@mdanderson.org;gecalin2003@yahoo.com.
Eric A. Elster, Email: eric.elster@usuhs.edu.
REFERENCES
- 1.DiMaggio C Ayoung-Chee P Shinseki M Wilson C Marshall G Lee DC, et al. Traumatic injury in the United States: in-patient epidemiology 2000–2011. Injury. 2016;47(7):1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald JR Liang SY Li P Maalouf S Murray CK Weintrob AC, et al. Infectious complications after deployment trauma: following wounded US military personnel into veterans affairs care. Clin Infect Dis. 2018;67(8):1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gentile LF Cuenca AG Efron PA Ang D Bihorac A McKinley BA, et al. Persistent inflammation and immunosuppression: a common syndrome and new horizon for surgical intensive care. J Trauma Acute Care Surg. 2012;72(6):1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belard A Schobel S Bradley M Potter BK Dente C Buchman T, et al. Battlefield to bedside: bringing precision medicine to surgical care. J Am Coll Surg. 2018;226(6):1093–1102. [DOI] [PubMed] [Google Scholar]
- 5.Bradley M Dente C Khatri V Schobel S Lisboa F Shi A, et al. Advanced modeling to predict pneumonia in combat trauma patients. World J Surg. 2020;44(7):2255–2262. [DOI] [PubMed] [Google Scholar]
- 6.Cuenca AG Gentile LF Lopez MC Ungaro R Liu H Xiao W, et al. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Crit Care Med. 2013;41(5):1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stortz JA Murphy TJ Raymond SL Mira JC Ungaro R Dirain ML, et al. Evidence for persistent immune suppression in patients who develop chronic critical illness after sepsis. Shock. 2018;49(3):249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao W Mindrinos MN Seok J Cuschieri J Cuenca AG Gao H, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208(13):2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamparello AJ Namas RA Constantine G McKinley TO Elster E Vodovotz Y, et al. A conceptual time window-based model for the early stratification of trauma patients. J Intern Med. 2019;286(1):2–15. [DOI] [PubMed] [Google Scholar]
- 10.Hotchkiss RS, Opal S. Immunotherapy for sepsis—a new approach against an ancient foe. N Engl J Med. 2010;363(1):87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vicente D Patino M Marcus R Lillmoe H Limani P Newhook T, et al. Impact of epidural analgesia on the systemic biomarker response after hepatic resection. Oncotarget. 2019;10(5):584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicente DA, Bradley MJ, Bograd B, Leonhardt C, Elster EA, Davis TA. The impact of septic stimuli on the systemic inflammatory response and physiologic insult in a preclinical non-human primate model of polytraumatic injury. J Inflamm (Lond). 2018;15:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs — an update. Nat Rev Clin Oncol. 2018;15(9):541–563. [DOI] [PubMed] [Google Scholar]
- 14.Vicente D Schobel SA Anfossi S Hensman H Lisboa F Robertson H, et al. Viral micro-RNAs are detected in the early systemic response to injury and are associated with outcomes in polytrauma patients. Crit Care Med. 2022;50:296–306. [DOI] [PubMed] [Google Scholar]
- 15.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benz F, Roy S, Trautwein C, Roderburg C, Luedde T. Circulating microRNAs as biomarkers for sepsis. Int J Mol Sci. 2016;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber B, Franz N, Marzi I, Henrich D, Leppik L. Extracellular vesicles as mediators and markers of acute organ injury: current concepts. Eur J Trauma Emerg Surg. 2022;48(3):1525–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang M, Li X, Quan X, Li X, Zhou B. MiR-92a family: a novel diagnostic biomarker and potential therapeutic target in human cancers. Front Mol Biosci. 2019;6:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreth S, Hubner M, Hinske LC. MicroRNAs as clinical biomarkers and therapeutic tools in perioperative medicine. Anesth Analg. 2018;126(2):670–681. [DOI] [PubMed] [Google Scholar]
- 20.Giza DE Fuentes-Mattei E Bullock MD Tudor S Goblirsch MJ Fabbri M, et al. Cellular and viral microRNAs in sepsis: mechanisms of action and clinical applications. Cell Death Differ. 2016;23(12):1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atif H, Hicks SD. A review of microRNA biomarkers in traumatic brain injury. J Exp Neurosci. 2019;13:1179069519832286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelbard RB Hensman H Schobel S Khatri V Tracy BM Dente CJ, et al. Random forest modeling can predict infectious complications following trauma laparotomy. J Trauma Acute Care Surg. 2019;87(5):1125–1132. [DOI] [PubMed] [Google Scholar]
- 23.Osler T, Baker SP, Long W. A modification of the injury severity score that both improves accuracy and simplifies scoring. J Trauma. 1997;43(6):922–925; discussion 5–6. [DOI] [PubMed] [Google Scholar]
- 24.Tackett MR, Diwan I. Using FirePlex™ particle technology for multiplex microRNA profiling without RNA purification. Methods Mol Biol. 2017;1654:209–219. [DOI] [PubMed] [Google Scholar]
- 25.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162(1):55–63. [DOI] [PubMed] [Google Scholar]
- 26.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc B Methodol. 1996;58(1):267–288. [Google Scholar]
- 27.Breiman L. Classification and Regression Trees. Fairview, TN: Wadsworth Publishing Group; 1984. [Google Scholar]
- 28.Shah AD, Bartlett JW, Carpenter J, Nicholas O, Hemingway H. Comparison of random forest and parametric imputation models for imputing missing data using MICE: a CALIBER study. Am J Epidemiol. 2014;179(6):764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8(5):706–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison EB Hochfelder CG Lamberty BG Meays BM Morsey BM Kelso ML, et al. Traumatic brain injury increases levels of miR-21 in extracellular vesicles: implications for neuroinflammation. FEBS Open Bio. 2016;6(8):835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sen CK, Roy S. MicroRNA 21 in tissue injury and inflammation: authors' retrospective. Cardiovasc Res. 2012;96(2):230–233. [Google Scholar]
- 32.Fabbri M Paone A Calore F Galli R Gaudio E Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110–E2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong X Chung AC Chen HY Dong Y Meng XM Li R, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56(3):663–674. [DOI] [PubMed] [Google Scholar]
- 34.Wang H Bei Y Shen S Huang P Shi J Zhang J, et al. miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. J Mol Cell Cardiol. 2016;94:43–53. [DOI] [PubMed] [Google Scholar]
- 35.Coronel-Hernandez J Lopez-Urrutia E Contreras-Romero C Delgado-Waldo I Figueroa-Gonzalez G Campos-Parra AD, et al. Cell migration and proliferation are regulated by miR-26a in colorectal cancer via the PTEN-AKT axis. Cancer Cell Int. 2019;19:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gagnon JD Kageyama R Shehata HM Fassett MS Mar DJ Wigton EJ, et al. miR-15/16 restrain memory T cell differentiation, cell cycle, and survival. Cell Rep. 2019;28(8):2169–81.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Liu C, Wang Z, Huang J, Zeng Q. microRNA-23a-5p acts as a potential biomarker for sepsis-induced acute respiratory distress syndrome in early stage. Cell Mol Biol (Noisy-le-Grand). 2016;62(2):31–37. [PubMed] [Google Scholar]
- 38.Pinchi E Frati A Cantatore S D’Errico S Russa R Maiese A, et al. Acute spinal cord injury: a systematic review investigating miRNA families involved. Int J Mol Sci. 2019;20(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahamtan A, Teymoori-Rad M, Nakstad B, Salimi V. Anti-inflammatory microRNAs and their potential for inflammatory diseases treatment. Front Immunol. 2018;9:1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L Boldin MP Yu Y Liu CS Ea CK Ramakrishnan P, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209(9):1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tudor S Giza DE Lin HY Fabris L Yoshiaki K D’Abundo L, et al. Cellular and Kaposi's sarcoma-associated herpes virus microRNAs in sepsis and surgical trauma. Cell Death Dis. 2014;5:e1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M, Zhao L, Sun M. Diagnostic value of miR-103 in patients with sepsis and noninfectious SIRS and its regulatory role in LPS-induced inflammatory response by targeting TLR4. Int J Genomics. 2020;2020:2198308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X Sundquist K Svensson PJ Rastkhani H Palmer K Memon AA, et al. Association of recurrent venous thromboembolism and circulating microRNAs. Clin Epigenetics. 2019;11(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dluzen DF Noren Hooten N Zhang Y Kim Y Glover FE Tajuddin SM, et al. Racial differences in microRNA and gene expression in hypertensive women. Sci Rep. 2016;6:35815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hackl M Brunner S Fortschegger K Schreiner C Micutkova L Muck C, et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9(2):291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai B Wang F Nie X Du H Zhao Y Yin Z, et al. The cell type-specific functions of miR-21 in cardiovascular diseases. Front Genet. 2020;11:563166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valparaiso AP, Vicente DA, Bograd BA, Elster EA, Davis TA. Modeling acute traumatic injury. J Surg Res. 2015;194(1):220–232. [DOI] [PubMed] [Google Scholar]


