Abstract
Facilitated transport is necessitated for large size, charged, and/or hydrophilic drugs to move across the membrane. The drug transporters in the solute carrier (SLC) superfamily, mainly including organic anion-transporting polypeptides, organic anion transporters, organic cation transporters, organic cation/carnitine transporters, peptide transporters, and multidrug and toxin extrusion proteins, are critical facilitators of drug transport and distribution in human body. The expression of these SLC drug transporters is found in tissues throughout the body, with high abundance in the epithelial cells of major organs for drug disposition such as intestine, liver, and kidney. These SLC drug transporters are clinically important in drug absorption, metabolism, distribution, and excretion. The mechanisms underlying their regulation have been revealing in recent years. Epigenetic and nuclear receptor–mediated transcriptional regulation of SLC drug transporters has particularly attracted much attention. This review focuses on the transcriptional regulation of major SLC drug transporter genes. Revealing the mechanisms underlying the transcription of these critical drug transporters will help us understand pharmacokinetics and pharmacodynamics, ultimately improving drug therapeutic effectiveness while minimizing drug toxicity.
SIGNIFICANCE STATEMENT
It has become increasingly recognized that solute carrier drug transporters play a crucial and sometimes determinative role in drug disposition and response, which is reflected in decision making during not only clinical drug therapy but also drug development. Understanding the mechanisms accounting for the transcription of these transporters is critical to interpret their abundance in various tissues under different conditions, which is necessary to clarify the pharmacological response, adverse effects, and drug-drug interactions for clinically used drugs.
Introduction
Drug transporter is a general term for proteins that undertake the function of transporting drugs across the cell membrane (International Transporter Consortium et al., 2010). These proteins usually belong to two super families: solute carrier (SLC) and ATP-binding cassette (ABC) proteins. Although there are exceptions, most SLC transporters are influx transporters, which mostly mediate the uptake of their substrates into cells, and ABC transporters are usually efflux transporters, which transport substrates out of cells by hydrolyzing ATP (Hediger et al., 2004). SLCs are widely distributed in various tissues and organs in human body. They usually use the ion concentration gradient across the cell membrane as the driving force to transport their substrates (Anderson and Thwaites, 2010). Most SLC transporter proteins contain 7 to 14 transmembrane helices and have an intracellular and/or extracellular flexible domain, which can assist transmembrane helices to form transmembrane channels (Anderson and Thwaites, 2010). The SLC proteins include over 400 members belonging to 66 families (Perland and Fredriksson, 2017).
In recent years, many SLC members have been characterized as drug transporters because their substrates include structurally diverse clinically used drugs. The genes encoding these SLC drug transporters mainly include SLC21/SLCO gene family (encoding for organic anion-transporting polypeptides, OATPs), SLC22A gene subfamily (for organic anion transporters, OATs; organic cation transporters, OCTs; and organic cation/carnitine transporters, OCTNs), SLC15A gene subfamily (for peptide transporters, PEPTs), and SLC47A gene subfamily (for multidrug and toxin extrusion proteins, MATEs) (Liu, 2019) (Fig. 1). It is notable that although MATEs belong to the SLC superfamily, they may function as efflux transporters in polarized epithelial cells such as hepatocytes and renal proximal tubular cells (Lončar et al., 2016). Due to their critical role in drug absorption, distribution, and elimination, alteration in the activities of these SLC drug transporters may result in changes in pharmacokinetics and consequently in drug response, potentially leading to reduced therapeutic effectiveness, developed drug resistance, increased susceptibility to drug-induced tissue injuries, and endogenous toxin-mediated diseases (Niemi, 2010; Huang et al., 2020; Yang and Shu, 2021). The activity of a drug transporter is subject to regulation by various factors such as genetic polymorphisms, comedication, environmental toxins, food, and disease conditions.
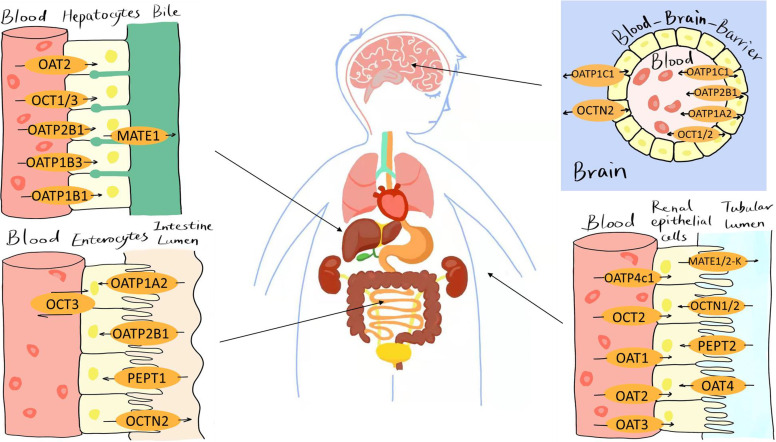
Fig. 1.
Location of major SLC drug transporters in human liver, intestine, blood-brain barrier (BBB), and kidney.
The molecular biology, substrates, inhibitors, and role in drug disposition and response of these SLC drug transporters have been comprehensively reviewed elsewhere (Roth et al., 2012; César-Razquin et al., 2015; Lin et al., 2015; Bhutia et al., 2016; Murray and Zhou, 2017; Rives et al., 2017; Nigam, 2018; Schlessinger et al., 2018; Liu, 2019; Ayka and Şehirli, 2020; Bednarczyk and Sanghvi, 2020; Huang et al., 2020; Koepsell, 2020; Pácha et al., 2021; Pizzagalli et al., 2021; Yang and Shu, 2021). These major SLC drug transporters are also briefly summarized in Table 1. In contrast to their well recognized clinical importance, the regulatory mechanisms accounting for the activities of SLC drug transporters remain less characterized in general. However, there have been important progresses in recent years on regulation of individual SLC drug transporters via the mechanisms at different levels, including transcription, post-transcription, translation, and post-translation. Herein we choose to review the transcriptional regulation of those major SLC drug transporters. Currently, the evidence supporting various transcriptional regulation of an individual SLC drug transporter is predominantly obtained from cellular studies and animal models. Although further clinical studies with a sufficient sample size are needed to validate those preclinical findings, certain transcriptomic analyses with patient tissue samples, pharmacogenetic studies, and characterization of probe drug disposition and effects in human subjects have begun to reveal clinical relevance of such transcriptional regulation. Below we first provide a brief overview of the general mechanisms involved in transcriptional regulation of SLC drug transporters, which is followed by our review on the research progress of transcriptional regulation on those important SLC drug transporters and then our prospective on future research in this area.
TABLE 1.
Major solute carrier (SLC) drug transporters
Only those SLC drug transporters reviewed in the text are presented.
| SLC Subfamily | Species | Transporter Gene Name (Protein Name) |
Organ with Highest Expressiona | Common Substrateb |
|---|---|---|---|---|
| OATP | Human | SLCO1A2 or SLCO1A3 (OATP-A or OATP1A2) | Brain | Bromsulfthalein (BSP), methotrexate, enalapril, levofloxacin |
| SLCO1B1 (OATP1B1) | Liver | Cobimetinib, lovastatin, clotrimazole, valsartan, cyclosporine, rifampicin | ||
| SLCO1B3 (OATP1B3) | Liver | Rifampicin, olmesartan, methotrexate, digoxin, pioglitazone | ||
| SLCO1B3-SLCO1B7 or LST-3TM12 (OATP1B3-1B7) | Liver, breast | Taurocholic acid, lithocholic acid, ezetimibe | ||
| SLCO2A1 (OATP2A1) | Lung | Alprostadil, latanoprost, iloprost | ||
| SLCO2B1 (OATP2B1) | Liver | Azilsartan, balsalazide, gavestinel, valsartan | ||
| SLCO3A1 (OATP3A1) | Brain | Benzylpenicillin, safinamide, iloprost | ||
| SLCO4A1 (OATP4A1) | Lung | Liothyronine, levothyroxine, benzylpenicillin | ||
| SLCO4C1 (OATP4C1) | Kidney | Liothyronine, digoxin, saxagliptin, ouabain | ||
| Rodent | Slco1a1 (Oatp1a1 or Oatp1) | Liver, kidney | Estradiol 17beta-d-glucuronide | |
| Slco1a4 (Oatp1a4 or Oatp2) | Brain, liver, kidney | Sulforhodamine-101 (SR-101) | ||
| Slco1a5 (Oatp1a5) | Kidney | Fexofenadine, celiprolol | ||
| Slco1a6 (Oatp1a6) | Kidney | Taurocholic acid (TCA) | ||
| Slco1b2 (Oatp1b2) | Liver | Pitavastatin | ||
| Slc16a2 (Oatp1c1) | Liver, kidney | Sulforhodamine-101 (SR-101), thyroxine (T4) | ||
| Slco2a1 (Oatp2a1) | Lung, liver | Phenolsulfonphthalein | ||
| Slco2b1 (Oatp2b1) | Ubiquitous | Fexofenadine rosuvastatin | ||
| Slco3a1 (Oatp3a1) | Brain | Sodium fluorescein | ||
| Slco4c1 (Oatp4c1) | Kidney, lung | Digoxin | ||
| OAT | Human | SLC22A6 (OAT1) | Kidney | Cimetidine, methotrexate, indomethacin, probenecid, oxytetracycline, hydrochlorothiazide |
| SLC22A7 (OAT2) | Liver | Oxytetracycline, cimetidine, methotrexate, salicylic acid | ||
| SLC22A8 (OAT3) | Kidney | Taurocholic acid, benzylpenicillin, cimetidine, tetracycline | ||
| SLC22A11 (OAT4) | Kidney | Oxytetracycline, aminohippuric acid, methotrexate, relebactam | ||
| SLC22A10 (OAT5) | Liver | None reported | ||
| SLC22A9 (OAT7) | Liver | Estrone sulfate, dehydroepiandrosterone, butyrate | ||
| SLC22A12 (URAT1) | Kidney | Urate | ||
| Rodent | Slc22a6 (Oat1) | Kidney | Same as SLC22A6 | |
| Slc22a7 (Oat2) | Liver, kidney | Same as SLC22A7 | ||
| Slc22a8 (Oat3) | Kidney | Same as SLC22A8 | ||
| Slc22a12 (Urat1) | Kidney | Urate | ||
| OCT | Human | SLC22A1 (OCT1) | Liver | Metformin, levofloxacin, cimetidine, codeine |
| SLC22A2 (OCT2) | Kidney | Metformin, cimetidine, cisplatin, quinidine | ||
| SLC22A3 (OCT3) | Skeletal muscle | Metformin, cimetidine, oxaliplatin | ||
| Rodent | Slc22a1 (Oct1) | Liver, kidney, intestine | Same as SLC22A1 | |
| Slc22a2 (Oct2) | Kidney | Same as SLC22A2 | ||
| OCTN | Human | SLC22A5 (OCTN2) | Skeletal muscle, kidney, heart, placenta | Amphetamine, nicotine, formoterol, colistin, oxaliplatin |
| Rodent | Slc22a5 (Octn2) | Kidney, liver, testis | Same as SLC22A5 | |
| LOC303140 (Octn3) | Testes | Acetylcarnitine | ||
| PEPT | Human | SLC15A1 (PEPT1) | Intestine | Cephalexin, enalapril, benzylpenicillin |
| Rodent | Slc15a1 (Pept1) | Intestine | Same as SLC15A1 | |
| MATE | Human | SLC47A1 (MATE1) | Adrenal gland | Metformin, levofloxacin, cimetidine, cisplatin |
| SLC47A2 (MATE2 or MATE2-K) | Kidney | Metformin, cimetidine, cisplatin, quinidine | ||
| Rodent | Slc47a1 (Mate1) | Kidney, liver | Same as SLC47A1 | |
| Slc47a2 (Mate2) | Testis | Same as SLC47A2 |
aThe information is extracted from the Human Protein Atlas (https://www.proteinatlas.org/) and UniProt (https://www.uniprot.org/).
bOnly selected compounds are presented; please refer to the literatures for a more comprehensive list of the substrates (Roth et al., 2012; César-Razquin et al., 2015; Lin et al., 2015; Bhutia et al., 2016; Murray and Zhou, 2017; Rives et al., 2017; Nigam, 2018; Schlessinger et al., 2018; Liu, 2019; Ayka and Şehirli, 2020; Bednarczyk and Sanghvi, 2020; Huang et al., 2020; Koepsell, 2020; Pácha et al., 2021; Pizzagalli et al., 2021; Yang and Shu, 2021).
Overview of the Mechanisms Involved in Transcriptional Regulation of SLC Drug Transporters
Transcriptional regulation refers to alteration in the level of gene expression by changing the rate of transcription. It is a vital process in regulation of gene function in eukaryotes that plays an important role in the accuracy and diversity of genetic information transmission. Transcriptional regulation in eukaryotes includes various processes that are highly related to each other such as DNA methylation, histone modification, chromatin remodeling, and control of transcription via different regulatory factors (Miller and Grant, 2013).
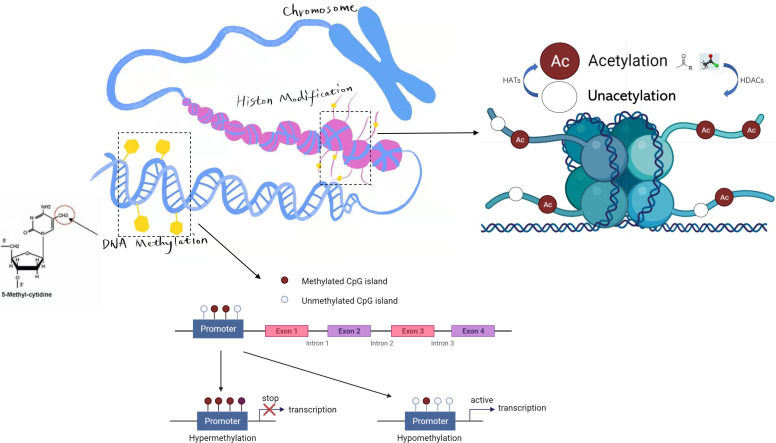
DNA methylation is a form of chemical modification for DNA that alters gene expression without altering the DNA sequence (Fig. 2). The process results in the addition of methyl (CH3) groups to the DNA strands, often to the fifth carbon atom of a cytosine ring (Jin et al., 2011). DNA methylation can be found in all vertebrates. The methylation of specific cytosines can be determined by using bisulfite sequencing. In eukaryotes, DNA methylation predominantly occurs at the dinucleotide cytosine-phosphate-guanine (CpG). The methylation of CpG dinucleotides in the promoter region may suppress the transcription of the corresponding gene (Sperling, 2007). Sometimes, it may fully silence gene expression, which in turn can render functional deficiency of the corresponding gene.
Fig. 2.
Epigenetic regulation of gene expression by DNA methylation and histone acetylation. As shown, the degree of methylated CpG islands in a promoter is usually negatively correlated with the level of gene expression. Histone acetylation is also an important epigenetic mechanism for gene expression regulation. The histone acetylases (HATs) may catalyze the transfer of acetyl groups to the lysine residues on the histone tail and activate gene transcription, whereas histone deacetylases (HDACs) can deacetylate histone and inhibit gene transcription instead.
The chromosomes of eukaryotes are mainly composed of DNA and proteins that include histones and nonhistones. Posttranslational modification of the DNA-binding histones has been characterized as a common mechanism in regulation of gene expression (Fig. 2). It refers to chemical modification of histones such as methylation, acetylation, phosphorylation, adenylation, ubiquitination, and ADP ribosylation under the action of related enzymes such as histone acetylase (HAT), deacetylase (HDAC), methyltransferase, and demethylase (Wang et al., 2016; Zhang et al., 2021). Histone modification could regulate gene expression by affecting the binding between histones and DNA double strands and altering the conformation of nucleosomes, which may result in chromatin remodeling, or by affecting the binding between transcription factors and the promoter of a gene (Moore et al., 2013).
Chromatin remodeling refers to the molecular mechanism by which the packaging state of chromatin, histones in nucleosomes, and the corresponding DNA molecules are changed during the replication and recombination (Lorch and Kornberg, 2017). ATP-dependent chromatin remodeling factors play a critical role in this process. They regulate chromatin conformation by altering the assembly, disassembly, and rearrangement of nucleosomes on chromatin, thereby improving the accessibility of transcription factors and other transcription-related factors to local chromatin DNA (Hota and Bruneau, 2016).
Transcription factors are a set of proteins that bind to specific regions of DNA molecules that control transcription, such as the sequences of promoters and enhancers, promoting or suppressing the transcription of genetic information from DNA into RNA (Lambert et al., 2018). The function of transcription factors can be accomplished alone or by forming complexes with other proteins or molecules. The transcriptional factors are encoded by a variety of genes. About 1600 genes have been predicted in the human genome to encode transcription factors. The nuclear receptors (NRs) are a group of transcription factors in cells. Members of the nuclear receptor superfamily play important roles in cell growth, development, differentiation, and metabolism and are particularly known to positively or negatively regulate the transcription of SLC drug transporter genes (Geier et al., 2007; Stieger and Geier, 2011; Svoboda et al., 2011; Stieger and Hagenbuch, 2014).
The transcriptional regulation of a transporter gene can be complicated. Multiple mechanisms described above could be involved in regulation of an individual transporter gene. For example, a single NR may be involved in the regulation of multiple hepatic transporter genes, whereas a transporter gene can be regulated by multiple NRs in the liver (Staudinger et al., 2013; Amacher, 2016). Those major regulatory mechanisms and/or the related regulatory molecules for transcription of major SLC drug transporter genes are briefly summarized in Table 2.
TABLE 2.
Regulatory mechanisms and protein factors involved in transcription of solute carrier (SLC) drug transporter genes
Only those regulatory mechanisms/protein factors reviewed in the text are presented. The uppercase letters represent human proteins, whereas the others are rodent proteins.
| Regulatory Mechanism/ Protein Factor | Transporter | Representative Agonist/Activator | Representative Antagonist/ Inhibitor |
|---|---|---|---|
| DNA methylation | OATP1B1, OATP1B3, OATP3A1, OATP4A1, OATP3A1, OATP4C1, OATP2A1, Oatp1b2, Oatp1a1, Oatp1a6, Oatp1c1, Oatp1a4, OAT1, OAT3, Oat1, Oat3, OCT1, OCT2, OCT3, OCTN2, PEPT1, MATE1 | 5-Aza-2'-deoxy cytidine (5azadC), decitabine, metformin | |
| Histone modification | OATP2A1, OATP2A1, OATP1B1, OATP1B3, Oatp1b2, Oatp1a1, Oatp1a6, Oatp1c1, Oatp1a4, OAT2, OCT2, PEPT1, MATE2 | Histone deacetylase inhibitor: trichostatin, vorinostat (SAHA) | |
| PXR | Oatp2, OATP1B1, OATP1B3, OATP2B1, OATP2A1, OATP-A, OATP1A2, Oat1, Oat3, OCT1 | PCN, rifampicin, phenobarbital, doxorubicin, spironolactone | Methotrexate |
| AhR | Oatp2b1, Oatp3a1, OATP4C1, OATP1B1, OATP2B1, Oatp1a1, Oatp2, OAT1, OAT2 | TCDD, polychlorinated biphenyl 126, beta-naphthoflavone, diesel exhaust particles (DEPs), indoxyl sulfate, 3-methylcholanthrene | |
| CAR | OATP1A2, OATP1B3, OATP2B1, OATP1B3, Oatp1a1, Oatp1a4, Oatp2, OAT2, Oat1, Oat3 | Diallyl sulfide, phenobarbital, 6-(4-chlorophenyl)-imidazo [2,1-b] thiazole-5-, carbaldehyde-O-(3,4- dichlorobenzyl)oxime (CITCO) |
Methotrexate |
| FXR | OATP1B1, OATP1B3, OAT2, PEPT1, Pept1, MATE1, MATE2-K | Vitamin A, bile acids, CDCA, GW4064, fexaramine, berberine | DY268, Z-guggulsterone, glycine-β-muricholic acid, LPS |
| LXR | OATP1B1, Oatp1b2, OAT1, OAT2 | Oxysterols, berberine, TO-901317, GW3965, 22(R)-hydroxy-cholesterol, 22(S)-hydroxy-cholesterol, 9-cis retinoic acid (CRA), 25-hydroxy-cholesterol (HC) | |
| HIF-1α | OATP1B3, OATP1B3, OATP2B1 | ||
| HNF | OATP1B1, OATP1B3, Oatp1b2, Oatp1, Oatp2, Oatp1a1, Oatp1a4, Oatp1a5, Oatp1b2, Oatp2b1, Oatp1a1, Oatp1a6, Oatp2b1, Oatp3a1, Oatp4c1, Oatp2a1, OAT1, OAT2, OAT3, Oat1, Oat3, OCT1, Mate1 | LPS, UDCA | |
| Nrf2 | Oatp1a1, Oatp2b1, Oatp1b2, Oatp1a6, OAT2, Oat2, PEPT1, MATE1, MATE2-K | Sulforaphane | Retinoic acid, oltipraz |
| RXR | OATP1B1, OATP2B1, OCT1, OCTN2 | atRA, CRA | |
| RAR | OATP1B1, OATP2B1, OCT1 | atRA, CRA | |
| VDR | OATP1A2 | 1,25-Dihydroxyvitamin D3 (D3) | |
| PPARα | Oatp1a1, Oatp1a4, Oatp1b2, Oatp2a1, Oatp2b1, OCT1, OCT2, Oct2, OCTN2, Octn2, OCTN3, PEPT1, PepT1, Mate1 | Perfluorooctanoic acid, perfluorodecanoic acid, clofibrate, ciprofibrate, diethylhexylphthalate, fenofibrate | WY-14,643 |
| PPARγ | OCTN2 | Thiazolidinediones (troglitazone and rosiglitazone), luteolin | Bisphenol A diglycidyl ether (BADGE), GW9662 |
| Androgen receptor (AR) | Oct2 | Testosterone | |
| Estrogen receptor (ER) | OAT1, OCTN2 | Estrogen |
Transcriptional Regulation of OATPs
Studies have shown that the expression of OATPs is subject to the regulation via DNA methylation and histone modification (Imai et al., 2013a). A recent analysis on DNA methylation of SLCO genes showed that the CpG dinucleotides around the transcription start site (TSS) of SLCO1B3 have multiple methylation patterns (Ichihara et al., 2010). These methylation patterns have differential impact on the expression of SLCO1B3 in various human cancer cell lines (Imai et al., 2013b). Furthermore, it has been reported that the promoter region of SLCO3A1 was hypermethylated whereas that of SLCO4A1 was hypomethylated in colorectal cancer tissues and cell lines, which resulted in a low mRNA expression of SLCO3A1 and high expression of SLCO4A1, respectively (Rawłuszko-Wieczorek et al., 2015). Additionally, in prostate tumor tissues, the promoter region of SLCO4C1 was found to be significantly hypermethylated, which was negatively correlated with the expression of SLCO4C1 and the prognosis of patients after radical prostatectomy (Li et al., 2019). Interestingly, the use of demethylating agents or histone deacetylase inhibitor could partially restore the expression of some SLCOs such as SLCO2A1 in colorectal neoplasia (Holla et al., 2008) and in human head and neck squamous cell carcinoma (Zolk et al., 2013) and SLCO1B3 in hepatic carcinoma (Ichihara et al., 2010). A genome-wide DNA methylation profiling has revealed that the expression of Slco1b2 in mouse liver was regulated by DNA methylation in the promoter and by histone acetylation (Imai et al., 2009). In subsequent analyses, the epigenetic profiles of DNA methylation and histone acetylation around the TSS of mouse and human SLCOs (Slco1a1, Slco1a6, Slc16a2, Slco1a4, SLCO1B1, and SLCO1B3) were found to be associated with tissue-specific expression of these transporters (Imai et al., 2013a). The DNA methylation may be a determinant of the specific tissue distribution of OATPs.
The role of transcription factors in regulation of OATP expression has been widely characterized. The NRs involved in the transcription of SLCO genes include: 1) the typical xenobiotic responsive NRs such as pregnane X receptor (PXR), aryl hydrocarbon receptor (AhR), and constitutive androgen receptor (CAR) and 2) other NRs of both physiologic and pharmacological importance such as farnesoid X receptor (FXR), liver X receptor (LXR), hypoxia-inducible factor 1 alpha (HIF-1α), hepatocyte nuclear factors (HNFs), the nuclear factor erythroid 2-related factor 2 (Nrf2), retinoid X receptor (RXR), retinoic acid receptor (RAR), vitamin D receptor (VDR), and peroxisome proliferator-activated receptor alpha (PPARα) (Guo et al., 2002a; Cheng et al., 2005; Jigorel et al., 2006; Meyer zu Schwabedissen and Kim, 2009; Klaassen and Aleksunes, 2010; Aleksunes and Klaassen, 2012; Eloranta et al., 2012; Wang et al., 2012b).
PXR is a ligand-dependent transcription factor with a broad spectrum of ligands, including many endogenous and exogenous compounds (Masuyama et al., 2005). After binding with ligand, PXR experiences conformation change and binds to the promoter region of target genes to regulate gene expression (Wang et al., 2014). In animals, it was reported early that treatment with PXR ligand pregnenolone 16α-carbonitrile (PCN) could induce the protein level of rat Oatp2 by more than 3-fold (Guo et al., 2002b). In addition, the mRNA expression of rodent Slco1a4 and the hepatic uptake of OATP substrates could be upregulated by several PXR ligands (phenobarbital, PCN, and spironolactone) (Hagenbuch et al., 2001; Cheng et al., 2005). In human subjects, carbamazepine (CBZ), which is a PXR agonist, was reported to increase the mRNA expression of OATP-A (SLCO1A2), OATP-B (SLCO2B1), OATP-C (SLCO1B1), and OATP-8 (OATP1B3) in the livers of CBZ-treated patients (Oscarson et al., 2006). Atorvastatin, a possible PXR agonist, has been shown to increase the mRNA expression of SLCO2B1 in patient livers as well (Björkhem-Bergman et al., 2013). The in vivo findings are supported by abundant in vitro evidence. For example, treatment with the PXR ligand rifampicin was found to upregulate the mRNA expression of SLCO1B1, SLCO1B3, and SLCO2B1 in human primary hepatocytes (Rodrigues et al., 2020). PXR also plays a significant role in regulating the mRNA expression of OATPs in certain cancer cells, including SLCO2A1 in head and neck squamous cell carcinoma cells (Zolk et al., 2013), OATP-A (SLC21A3) in breast carcinoma (Miki et al., 2006), and SLCO1A2 in breast cancer cells (Meyer zu Schwabedissen et al., 2008). Notably, as a possible effect of OATP induction on drug disposition, rifampicin administration could alter the pharmacokinetics of statins that are OATP substrates (Rodrigues et al., 2020). However, we should be careful while interpreting the drug-drug interactions (DDIs) involved those PXR ligands that may have multiple pharmacological mechanisms or targets in different species. For example, there are controversial reports in which rifampicin had no direct effect on the gene expression of SLCO in hepatocytes in vitro and in monkey livers (Meyer Zu Schwabedissen et al., 2010; Benson et al., 2016; Niu et al., 2019). The molecular mechanism of PXR signaling and the effects by PXR ligands in regulation of SLCO transcription in vivo thus remains to be fully characterized.
AhR and CAR are also ligand-dependent transcription factors that regulate the transcription of downstream target genes and particularly those xenobiotic response genes including SLCO genes (Kawamoto et al., 1999; Murray et al., 2014; Kersten and Stienstra, 2017). The effects of AhR and CAR activation on specific SLCO gene expression may be opposite. Studies have shown that AhR ligands (2,3,7,8-tetrachlorodibenzo-p-dioxin, TCDD; polychlorinated biphenyl 126; beta-naphthoflavone; and 3-methylcholanthrene) could increase the mRNA expression of certain OATPs such as Slco2b1 and Slco3a1 in mouse liver (Cheng et al., 2005). Treatment with statins, which have not been characterized as direct AhR ligands, were reported to induce the mRNA expression of SLCO4C1 through AhR activation in human kidney proximal cells (Toyohara et al., 2009; Suzuki et al., 2011). Furthermore, treatment with the natural product shikonin has been reported to effectively upregulate the transcription of SLCO1B1 and SLCO2B1 through activation of AhR in hepatocytes as well (Huang et al., 2018). On the other hand, it has been reported that TCDD could repress the mRNA expression of SLCO2B1 (Le Vee et al., 2015), Slco1a1 (Aleksunes and Klaassen, 2012), and Slco1a4 (Guo et al., 2002a). Whereas CAR activation has been reported to cause decreased expression of SLCO1B3, SLCO2B1 (Jigorel et al., 2006), and Slco1a1 (Cheng et al., 2005; Aleksunes and Klaassen, 2012), it results in an enhanced activity of SLCO1A2 gene promoter (Meyer zu Schwabedissen et al., 2008) and an increased expression of Slco1a4 (Aleksunes and Klaassen, 2012). In addition, the CAR ligand diallyl sulfide could increase the mRNA expression of mouse Slco1a4 (Guo et al., 2002a) but decrease that of SLCO1B3 in human liver slices (Jigorel et al., 2006).
FXR acts as an intrahepatic “bile acid receptor,” regulating bile acid synthesis, detoxification, and transport and maintaining the homeostasis balance between bile acid and liposome (Glastras et al., 2015). FXR can also regulate the transcription of SLCO indirectly or directly (Ohtsuka et al., 2006; Godoy et al., 2013). On the one hand, the accumulation of bile acids can activate FXR, which in turn promotes the transcription of the FXR-responsive gene encoding small heterodimer partner 1 (SHP) protein. The SHP could repress the expression of HNF4α, eventually reducing HNF4-mediated expression of SLCO1B1 and SLCO1B3 (Kullak-Ublick et al., 2004). On the other hand, the activated FXR could directly bind to the promoter of SLCO genes and regulate its transcription. This is exemplified by several lines of evidence. The genetic polymorphism of FXR promoter has been associated with a decreased expression of SLCO1B1 (Marzolini et al., 2007). Treatment with the bile acid chenodeoxycholic acid (CDCA), an FXR ligand, could increase the mRNA expression of SLCO1B3 in human hepatocellular carcinoma cells (Jung et al., 2002). The synthetic FXR agonists GW4064 and fexaramine could significantly increase the transcription activity of SLCO1B1 gene (Meyer Zu Schwabedissen et al., 2010). In addition, berberine could induce the nuclear translocation of FXR and LXRα, which in turn increased the expression of SLCO1B1 (Liu et al., 2020). Both agonists and antagonists of FXR could also regulate the transcription of other SLCO such as SLCO1A2 and SLCO1B3-1B7 (LST-3TM12) (Yang et al., 2014; Malagnino et al., 2019). SLCO1B3-1B7 is a splice variant of SLCO1B3 and SLCO1B7. For OATP1B3-1B7 protein, the N-terminal part of its mRNA derives from SLCO1B3, whereas the remaining part of the transcript originates from the neighboring SLCO1B7 gene locus. Notably, the regulation of SLCO transcription by FXR signaling is often involved with other NRs.
LXR is also activated by ligands, which could consequently initiate and regulate the transcriptional expression of its target genes that are important in endogenous metabolism (Wang et al., 2015). It has been reported that treatment with the LXRα agonists TO-901317 and GW3965 could upregulate the mRNA expression of SLCO1B1 in Huh-7 cells (Meyer Zu Schwabedissen et al., 2010). LXRα collaborates with FXR to coregulate SLCO1B1 transcription. There is an interaction among LXRα, FXR, and the specific DNA-binding module of the 5′ noncoding region of the SLCO1B1 gene.
HIFs are transcription factors that respond to oxygen content in cellular environment and are activated in the presence of reduced oxygen levels or hypoxia (Ziello et al., 2007). Several lines of evidence have indicated a role of HIF-1α in SLCO transcriptional regulation. Studies have shown that HIF-1α could bind to the promoter sequences of certain cancer-specific OATP1B3 variants and regulate their mRNA expression (Han et al., 2013). Consistently, HIF-1α was found to regulate the transcription of SLCO1B3 via two HIF response elements in intron 1 of SLCO1B3 gene (Ramachandran et al., 2013). In addition, the mRNA expression of SLCO1B3 and SLCO2B1 has been found to be induced by HIF-1α stabilizers but reduced by HIF-1α knockdown (Shi et al., 2014).
HNFs are a group of phylogenetically unrelated transcription factors that regulate the transcription of a wide range of genes (Lau et al., 2018). Functional HNF1α response elements have been identified in the proximal promoter of SLCO1B1 (Jung et al., 2001; Maher et al., 2006). The regulation of SLCO transcription by HNFs seems to be related to disease conditions. Lipopolysaccharide (LPS) could downregulate the mRNA expression of Slco1b2, which was attributed to a reduced binding of HNF1α, HNF3, and RXR:RAR to Slco1b2 promoter (Li and Klaassen, 2004). The increased expression of the cytokine tumor necrosis factor alpha (TNF-α) was reported to be a mechanism underlying the reduced expression of Slco1a1 and Slco1a4 in the mice with liver injury, which was probably through an HNF1-dependent signaling (Geier et al., 2003). In HNF1α-null mice, the mRNA expression of Slco1a1, Slco1a4, Slco1a5, Slco1b2, and Slco2b1 in the liver; Slco1a1, Slco1a6, Slco2b1, Slco3a1, and Slco4c1 in the kidney; and Slco2a1 in the duodenum were altered (Maher et al., 2006). There is also in vivo evidence in human subjects suggesting the regulation of SLCO1B1 expression by HNF1α (He et al., 2008). Ursodeoxycholic acid (UDCA) is an inhibitor of HNF1α. In healthy subjects, treatment with UDCA could cause significant alteration in the pharmacokinetics of rosuvastatin, which is a well characterized OATP1B1 substrate. The regulation of SLCO transcription by HNFs could involve other NRs. For example, HNF1α and HNF3 could partner with RXR:RAR at the Slco1b2 promoter (Li and Klaassen, 2004). The downregulation of SLCO1B3 expression in hepatocellular carcinoma (HCC) has been related to the high expression of HNF3β (Vavricka et al., 2004). Later, it was found that FXR, HNF1α, and HNF3β simultaneously regulated the transcription of SLCO1B3 (Ohtsuka et al., 2006). In addition, HNF4α has also been found to promote SLCO1B3 transcription through the coactivation of β-catenin in HCC (Kitao et al., 2018).
Nrf2 is a nuclear transcription factor that regulates about 250 genes involved in cell homeostasis, including antioxidant proteins, detoxifying enzymes, drug transporters, and many cell-protective proteins (Tonelli et al., 2018). In rodents, the activation of Nrf2 could downregulate the transcriptional expression of Slco1a1 but upregulate those of Slco2b1 and Slco1b2 (X Cheng et al., 2005; Q Cheng et al., 2011; Aleksunes and Klaassen, 2012; Wu et al., 2012). In the liver of Nrf2 knockout mice, the mRNA expression of Slco1a6, Slco2b1 (Anwar-Mohamed et al., 2011), and Slco1b2 (Tanaka et al., 2009) were significantly decreased.
There are additional NRs whose role in the transcription of SLCOs has been explored. In human hepatocytes, all-trans retinoic acid (atRA) could activate the formation of RXR:RAR heterodimers, which could downregulate the mRNA and protein expression of OATP1B1 and OATP2B1 (Le Vee et al., 2013). The effects of atRA treatment on SLCO expression could be counteracted by knockdown of RXR and RAR. VDR, an NR that in complex with hormonally active vitamin D [1,25(OH)2D3] regulates the expression of a wide array of physiologically important genes. In small intestinal epithelial cells, VDR has been shown to be involved in the transcriptional activation of SLCO1A2 (Eloranta et al., 2012). In addition, PPARα has also been reported to play a vital role in the downregulation of Slco1a1, Slco1a4, and Slco1b2 in mouse livers by the environmental toxic chemicals perfluorooctanoic acid and perfluorodecanoic acid (Cheng and Klaassen, 2008). In wild-type mice, treatment with PPARα ligands such as clofibrate, ciprofibrate, and diethylhexylphthalate could significantly decrease the mRNA levels of Slco1a1, Slco1b2, Slco2a1, and Slco2b1 in the liver (Cheng et al., 2005). However, as recently reviewed elsewhere (Rodrigues et al., 2020; Zamek-Gliszczynski et al., 2021), we would like to point out that the direct clinical evidence remains very limited to support an importance of alteration in OATP transcriptional regulation in determining drug disposition and response. Further clinical studies are therefore needed to characterize the clinical relevance for these various regulatory mechanisms of OATP transcription.
Transcriptional Regulation of OATs
Epigenetic mechanisms are involved in the transcriptional regulation of OATs. Both DNA methylation and histone modification may play a vital role. By identifying the TSS of SLC22A8 gene, the Sugiyama group found that the promoter region of SLC22A8 has multiple CpG dinucleotides as the targets of DNA methylation (Kikuchi et al., 2006). In their follow up studies, DNA methylation was found to repress the activity of the SLC22A8 promoter and decrease the expression of SLC22A8, whereas transcription could be resumed when using methyltransferase inhibitors (Kikuchi et al., 2006). DNA methylation is also highly related to the tissue-specific expression of OAT1 and OAT3 (Jin et al., 2012). Compared with the renal cortex, most of the CpG dinucleotides around the TSSs of hepatic Slc22a6 and Slc22a8 were highly methylated, consistent with their high expression levels in the kidney but low levels in the liver. Recently, another study showed that histone hypoacetylation rather than DNA hypermethylation was involved in the transcriptional suppression of SLC22A7 in HCC cell lines (Wang et al., 2021b). When histone hypoacetylation was mitigated with a histone deacetylase inhibitor, the expression of SLC22A7 was increased.
Earlier findings have revealed the transcriptional regulation of OATs by transcriptional factors and particularly various NRs. As early as 2003, studies have found that the promoter regions of SLC22A6 and SLC22A8 have the binding sites for several transcription factors, including PAX1, PBX, WT1, and HNF1 (Eraly et al., 2003). There is considerable evidence supporting that transcription factors function in transcription of OATs. For example, in reporter gene assays, expression of the transcription factor B-cell chronic lymphocytic leukemia (CLL)/lymphoma 6 (BCL6) led to increased activities of the reporter luciferase that was driven by the promoters of Slc22a6 and Slc22a8 (Wegner et al., 2012). However, BCL6 seemed to not directly activate the promoter of SLC22A6. It indirectly enhanced the transcription by increasing the protein expression of HNF1α (Wegner et al., 2014). In addition, estrogen receptor alpha (ER α) can indirectly regulate the transcription of SLC22A6 as well (Euteneuer et al., 2019). There was no direct binding of ERα to the SLC22A6 promoter. Instead, the ligand-activated ERα could activate the CCAAT-box–binding transcription factor (CBF) and heterogeneous nuclear ribonucleic protein K (HNRNPK), both of which could bind to the promoter of SLC22A6 and enhance its transcription. On the other hand, the ligand-activated LXRs could reduce SLC22A6 expression in the renal S2 cells, which in turn resulted in a decreased transport of OAT1 substrates in these cells (Kittayaruksakul et al., 2012). However, LXRs may have a cell-specific effect on the expression of different OATs. In the hepatocytes isolated from the rats with hypercholesterolemia, the expression of both LXRα and Oat2 were found to be reduced. In follow up experiments with HepG2 cells treated with hypercholesterolemic serum, LXRα activation could rescue the reduced expression of SLC22A7 (Liu et al., 2016a).
Studies have also shown the regulation of OAT transcription by xenobiotic responsive NRs. Ligand-activated CAR and AhR could downregulate SLC22A7 expression (Jigorel et al., 2006; Le Vee et al., 2015). The indoxyl sulfate was reported to induce the expression of SLC22A6 via AhR-EGFR (epidermal growth factor receptor) signaling in the kidney, which could be suppressed by AhR antagonism (Jansen et al., 2019). Methotrexate is a well characterized substrate of OATs. Interestingly, treatment with methotrexate could decrease the expression of Slc22a6 and Slc22a8 in rat kidneys (Shibayama et al., 2006). As the decrease was accompanied with downregulation of CAR and PXR expression, it is likely that these two NRs were involved in the reduction of Slc22a6 and Slc22a8 expression. Moreover, the activation of Nrf2 was reported to cause a significant decrease in Slc22a7 mRNA expression (Wu et al., 2012). In addition, FXR could regulate the transcription of SLC22A7 either directly or indirectly (Popowski et al., 2005). FXR could directly bind to and inhibit the transactivation of SLC22A7 promoter. FXR activation could also lead to an increased expression of the gene encoding the SHP protein (Lee et al., 2000), which is a corepressor of HNF4α. The increased SHP would therefore decrease the nuclear binding activity of HNF4α, ultimately leading to a decrease in the expression of SLC22A7 (Popowski et al., 2005).
The role of cellular signaling with HNFs in regulation of OAT transcription has been widely studied. HNF1α and HNF4α may regulate the transcription of many OAT genes in developing kidneys because there exist a number of HNF1α/HNF4α binding elements at the proximal promoter regions of OAT genes (Gallegos et al., 2012; Martovetsky et al., 2013; Nigam et al., 2015). Motif analysis of cis-regulatory enhancers in the developing proximal tubules of rodents has suggested that Hnf4a and Hnf1a were the main transcriptional regulators of multiple SLC drug transporters, which include Slc22a6, Slc22a8, Slc22a1, Slc22a2, and Slc47a1 (Martovetsky et al., 2013). Human HNF1α/β was also found to bind with the HNF1-motif that was located near the TSS of SLC22A6, SLC22A8, SLC22A11, and SLC22A12 genes, thereby enhancing the promoter activity and activating the transcription of these transporter genes (Kikuchi et al., 2006, 2007; Saji et al., 2008; Jin et al., 2012). Of note, mutations in the HNF1-motif could fully abolish the enhanced promoter activity that was associated with HNF1α/β binding. Besides HNF1α/β, HNF4α also regulates the transcription of OAT genes in a similar manner (Ogasawara et al., 2007; Thiagarajan et al., 2011). The involvement of HNF1α in OAT gene transcription has also been evidenced in genetic mouse models (Maher et al., 2006; Kikuchi et al., 2007). In HNF1α-null mice, the mRNA expression of Slc22a6, Slc22a8, and Slc22a12 was significantly decreased in the kidney and that of Slc22a7 was reduced in both liver and kidney. However, the expression of Slc22a8 was increased in the duodenum. OAT2, OAT5, and OAT7 are three OATs in human liver. Although SLC22A7 gene is subject to the transactivation by HNF4α (Popowski et al., 2005), the experimental evidence from genetic manipulation and reporter gene assays as well as with expression analysis in surgical liver tissues have indicated that HNF1α is a critical transcriptional factor in regulation of SLC22A10 and SLC22A9 expression (Klein et al., 2010).
Transcriptional Regulation of OCTs
The impact of genetic variants and epigenetic regulation of OCTs in general has been briefly summarized recently (Kölz et al., 2021). Herein we select to review the findings related to transcriptional regulation. Among epigenetic mechanisms, DNA methylation is likely the most extensively investigated for the transcriptional regulation of OCT genes. OCT1 is highly expressed in human livers. Based on microarray analyses with patient liver tissues, Schaeffeler et al. (2011) found that SLC22A1 expression was lower in hepatocellular carcinoma compared with normal liver tissues, which was accompanied with DNA hypermethylation at the promoter of SLC22A1. In addition, the level of SLC22A1 mRNA in cisplatin-resistant human esophageal cancer cells was found to be markedly reduced compared with that in the sensitive cells (Lin et al., 2013). Consistently, the promoter of SLC22A1 was highly methylated in the cisplatin-resistant cells. As cisplatin is a moderate substrate of OCT1, DNA methylation could be a mechanism accounting for cisplatin chemoresistance in some cancer patients.
Human OCT2 is a rather kidney-specific transporter. DNA methylation was found to be associated with the tissue specificity of human OCT2. The CpG sites in the proximal promoter of SLC22A2 gene were reported to be hypermethylated in the liver but hypomethylated in the kidney, which is consistent with a relatively low hepatic expression but a much higher renal expression of SLC22A2 (Aoki et al., 2008). The hypomethylation of SLC22A2 promoter occurred particularly at the CpG site in the enhancer box (E-box) that is the binding site of the basal transcription factor called upstream stimulating factor (USF)1. Increased methylation of SLC22A2 proximal promoter could dramatically reduce the binding of USF1 to the E-box, leading to a reduced SLC22A2 transcription. Renal cell carcinoma (RCC) is characterized by multidrug resistance. The Zeng group has identified the repressed expression of SLC22A2 as a mechanism for oxaliplatin resistance in RCC (Liu et al., 2016b). They revealed that the repressed SLC22A2 expression was characterized by hypermethylation in SLC22A2 promoter, which blocked the interaction between the transcription factor MYC and the E-box motif and prevented MYC from recruiting the histone methylase mixed-lineage leukemia 1 (MLL1) to catalyze the trimethylation of lysine 4 on histone 3 (H3K4me3), eventually leading to the repressed SLC22A2 transcription. Importantly, the epigenetic activation of SLC22A2 by the DNA methylation inhibitor decitabine could sensitize RCC cells to oxaliplatin treatment in the animal model. They further found that histone acetylation also regulates SLC22A2 expression in RCC (Zhu et al., 2019). In normal kidney tissues, a dynamic balance of histone acetylation is maintained by histone acetyltransferases (HATs) and histone deacetylases (HDACs). In RCC cells, the interaction between HDAC7 and MYC was disrupted, leading to high abundance of HDAC7 and low levels of histone acetylation at H3K18ac and H3K27ac around SLC22A2 promoter, which contributed to the repression of SLC22A2 transcription. In addition, Saito et al. (2011) reported that the placental expression of SLC22A2 was likely subject to epigenetic regulation as well. The mRNA expression of SLC22A2 was higher in the biallelic placental samples than that in monoallelic samples, which was associated with an increased H3 acetylation but decreased trimethylation of lysine 9 on histone H3 (H3K9me3) around SLC22A2 promoter.
For the widely expressed human OCT3, the proximal promoter has been reported to be located within a large CpG island which hypermethylation might cause a reduced expression of SLC22A3 gene (Chen et al., 2013). Compared with normal tissues, the hypermethylation of SLC22A3 promoter was associated with reduced SLC22A3 transcript levels in prostate tumors. However, the hypermethylation of SLC22A3 promoter appeared to be prostate tumor–specific, as it was not detected in other tumor tissues (Chen et al., 2013). In addition, metformin, which is a first-line therapy for type 2 diabetes, has been reported to reduce DNA methylation at the genes encoding metformin transporters, including SLC22A3 (García-Calzón et al., 2017). Although the exact mechanism has not been revealed, these epigenetic changes seem to be associated with metformin antidiabetic efficacy.
The role of NRs in regulation of OCT gene transcription has also been explored. At present, the regulatory role of PXR in SLC22A1 expression remains controversial. It has been suggested that doxorubicin treatment could suppress SLC22A1 expression by increasing the expression of PXR in renal tubular epithelial cells (Nagai et al., 2019). The transcriptional activity of SLC22A1 promoter and consequently the mRNA expression of SLC22A1 were reported to be subject to the suppression by ligand-activated PXR in human hepatocytes (Hyrsova et al., 2016). The effect by a PXR agonist has been ascribed to its competitive sequestration of the coactivator steroid receptor coactivator 1 (SRC-1) from the HNF4α response element and E-box in the SLC22A1 promoter, reducing the transactivation of SLC22A1 gene by HNF4α and USF transcription factors. However, in chronic myeloid leukemia (CML) cell lines and primary CML cells, the expression of SLC22A1 was upregulated by the agonists of PXR, RAR, and RXR (Austin et al., 2015). The PXR agonist rifampin was reported to increase the mRNA expression of SLC22A1 in peripheral blood cells and enhance the glucose-lowering effect of metformin in healthy human subjects (Cho et al., 2011). In addition, overexpression of PXR was found to increase the mRNA expression of Slc22a1 in rat hepatocytes, whereas treatment with the PXR agonist PCN induced the expression of Slc22a1 in the liver and that of Slc22a2 in the kidney in rats (Maeda et al., 2007). Further studies are needed to clarify the role of PXR in OCT gene transcription in different cells and tissues and the in vivo effects.
It has been reported that an increased level of CDCA, which could be induced by cholestasis, downregulates the mRNA expression of SLC22A1 and SLC22A3 in the liver (Saborowski et al., 2006; Nies et al., 2009). As CDCA is a ligand of FXR, the downregulation has been attributed to the inhibition of HNF4α-mediated transactivation that results from CDCA-induced expression of the transcriptional repressor SHP via FXR signaling. In agreement with this, the expression of Slc22a1 was significantly downregulated in rats with obstructive cholestasis (Denk et al., 2004). In addition, by treating human renal proximal tubular cells (RPTEC/TERT1 cells) and OCT2-CHO-K1 cells with CDCA and FXR antagonist Z-guggulsterone, a recent study has suggested that FXR also plays a role in regulation of renal SLC22A2 expression (Wongwan et al., 2020).
Other NRs are also involved in transcriptional regulation of OCT genes. By measuring the promoter activity in rat renal epithelial cell line LLC-PK1, Asaka et al. (2006) have demonstrated that testosterone treatment could stimulate the promoter activity of rat Slc22a2 via the androgen receptor–mediated signaling pathway. In the kidney of PPARα-null mice, the expression of Slc22a2 was decreased, suggesting that PPARα, which is highly expressed in the liver and kidney, may regulate the expression of Slc22a2 (Freitas-Lima et al., 2020). As the uptake of cisplatin into the proximal tubules is significantly mediated by OCTs, deletion of PPARα was protective of cisplatin-induced nephrotoxicity in mice (Freitas-Lima et al., 2020). PPARα was also found to mediate the circadian expression of Slc22a2 in mice (Oda et al., 2014). There are two HNF4α response elements in human SLC22A1 promoter that were involved in HNF4α-mediated transactivation of SLC22A1 transcription (Saborowski et al., 2006). HNF4α could enhance SLC22A1 promoter activity by synergistic interaction with the transcription factor USF1 or USF2 (Kajiwara et al., 2008). Last but not least, the activation of glucocorticoid receptor could indirectly increase the expression of SLC22A1 by upregulating HNF4α expression in human primary hepatocytes (Rulcova et al., 2013).
Transcriptional Regulation of OCTNs
There is evidence supporting a role of DNA methylation in regulation of OCTN gene transcription. Qu et al. (2013) identified a region (from −354 to +85) in SLC22A5 promoter that was critical to the transcription of SLC22A5 gene. The methylation of CpG sites in this region was found to be negatively correlated with the expression of SLC22A5 in cancer cells. Treatment with the demethylating agent decitabine could significantly reduce the hypermethylation at the SLC22A5 promoter, which enhanced the expression of SLC22A5 and consequently the transporter activities in HepG2 and LS174T cancer cells. Scalise et al. (2012) have also indicated that DNA methylation is an important mechanism underlying the downregulation of SLC22A5 expression in epithelial cancer cells. After treatment with 5-aza-cytidine, which is a demethylating agent, the mRNA expression of SLC22A5 could increase by 10 times in these cancer cells. Future studies are needed to determine the details of epigenetic regulation on OCTN transcription.
PPARs are among the major transcriptional factors characterized in the transcriptional regulation of OCTNs. The regulation of OCTNs by PPAR has been found to widely exist in humans, rodents, cattle, pigs, and other animals. With multiple approaches to activation of PPARα, including treatment with PPARα agonists such as clofibrate and WY-14,643, oxidized fat diet feeding, and food restriction, the Eder group have demonstrated that PPARα activation can lead to the upregulation of Slc22a5 in mice, rats, and pigs (Luci et al., 2006, 2007, 2008; Koch et al., 2007, 2008; Ringseis et al., 2007a,b,c, 2008a,b, 2009). In line with this, the expression of Slc22a5 is significantly reduced in PPARα-null mice (van Vlies et al., 2007; Koch et al., 2008). Moreover, Maeda et al. (2008) have analyzed the 5′-flanking promoter region of rat Slc22a5 and identified several putative peroxisome proliferator response elements (PPREs). After treatment with the PPARα agonist fenofibrate, there was an enhanced binding of PPARα to the proximal PPREs of Slc22a5. However, the increase of Slc22a5 promoter activity after PPARα activation was less than what was expected from that of Slc22a5 mRNA levels, suggesting existence of additional PPREs that might be located outside the proximal promoter region of Slc22a5 gene. In fact, a subsequent study by Eder group revealed that there were functional PPREs in the first intron of Slc22a5, which were highly responsive to both exogenous PPARα/RXR expression and agonists (Wen et al., 2010). The sequence of intron 1 containing the PPREs showed a high degree of similarity between mice and rats. Functional PPREs have also been identified in the intron 1 of porcine, bovine, and human SLC22A5 genes (Luo et al., 2014), suggesting that the regulation of SLC22A5 by PPARα is highly conserved across species. Besides OCTN2, the transcription of OCTN3 seems to also be subject to PPARα regulation. The expression of Octn3 gene was lower in the kidney and small intestine of PPARα-null mice compared with wild-type mice (van Vlies et al., 2007; Koch et al., 2008). PPARα activation increased the expression of Octn3 and its transport function in rat astrocytes as well (Januszewicz et al., 2009).
By using different PPAR-null models and specific PPAR inhibitors, D’Argenio et al. (2010) have demonstrated that PPARγ could also regulate the expression of Slc22a5 gene in colon via binding to the PPRE at the first intron as a heterodimer with RXRα. The expression of SLC22A5 and the corresponding transport function could be increased by the PPARγ agonists thiazolidinediones and luteolin in mice and colon cells (D’Argenio et al., 2010; Qu et al., 2014). Notably, the colon Slc22a5 expression in PPARα-null mice was significantly upregulated by overexpression of a constitutively active PPARγ mutant, indicating an independent regulation by PPARγ (D’Argenio et al., 2010). Additional transcriptional factors may be involved in regulation of OCTN expression. For example, SLC22A5 has been reported to be an estrogen-dependent gene via an intronic estrogen-responsive element (ERE) in breast cancer cells (Wang et al., 2012a). In addition, as a line of clinical evidence, the mRNA level of SLC22A5 has been reported to be decreased in liver biopsies of those patients who received the PXR agonist carbamazepine (Oscarson et al., 2006). However, the number of transcriptional factors in understanding of OCTN transcription are still limited.
Transcriptional Regulation of PEPTs
The evidence on the transcriptional regulation of PEPT transporters is not as much as that of the SLC drug transporters described above. Most related studies have been focused on PEPT1. There has been little understanding of epigenetic regulation of PEPTs until very recently. Wang et al. (2021a) reported a suppressed expression of SLC15A1 (PEPT1) in colon cancer due to DNA methylation and histone deacetylation. DNA methyltransferase 1 was characterized as the primary determinant of the hypermethylation of SLC15A1 proximal promoter. In addition, the absence of CBP/p300-mediated H3K18/27Ac combined with the histone deacetylase–mediated histone hypoacetylation around the SLC15A1 promoter also accounted for the repressed expression of SLC15A1.
The transcriptional regulation of PEPTs by NRs has not been well investigated either. Among a few studies, cholate-induced FXR/PPARα activation/inhibition has been reported to play a role in SLC15A1 transcription (Okamura et al., 2014; Liang et al., 2020). In the intestine of diabetic rats, the level of CDCA was significantly increased. As a natural ligand of FXR, CDCA could downregulate the expression of rat Slc15a1, which could be reversed by treatment with the FXR inhibitor glycine-β-muricholic acid or by FXR knockdown (Liang et al., 2020). On the other hand, treatment with the PPARα agonist WY-14643 increased the mRNA level of SLC15A1 and its transport function in rat intestine and human intestinal Caco-2 cells (Shimakura et al., 2006a). The increase in free fatty acids, which are endogenous ligands for PPARα, could also significantly increase the expression of Slc15a1 mRNA in rodents (Shimakura et al., 2006a). Two nucleotide sequences homologous to PPRE have been identified within 2.0 kilobases (kb) upstream to the TSS of the mouse and human SLC15A1 gene (Okamura et al., 2014). The bile acids accumulated in intestinal epithelial cells after eating could interfere with the recruitment of the cotranscriptional activator CBP/p300 to the promoter region of SLC15A1 gene, thereby inhibiting PPARα-mediated transactivation of SLC15A1. However, the regulatory effects of PPARα on SLC15A1 expression have not been fully confirmed in other studies (Hirai et al., 2007; Saito et al., 2008).
A few other transcription factors, including Sp1, Cdx2, and Nrf2, have also been shown to regulate the transcriptional activity of SLC15A1 promoter (Shimakura et al., 2005, 2006b; Geillinger et al., 2014). The −172 ∼ −35 base pair (bp) region of SLC15A1 promoter lacks TATA-box; however, it contains guanine-cytosine (GC)-rich sites that can bind with the transcription factor Sp1 to recruit the TATA-binding protein and initiate transcription (Shimakura et al., 2005). The mutation of the Sp1 sites resulted in a reduced transcriptional activity of human SLC15A1 promoter. Moreover, although coexpression of Cdx2 and Sp1 has been found to synergistically activate the SLC15A1 promoter, the mutation of Sp1 sites reduced the effect of Cdx2 (Shimakura et al., 2006b). In addition, an increased Nrf2 activity was found to upregulate the expression of SLC15A1 via binding with the antioxidant response elements (AREs), which are close to the start codon in human SLC15A1 gene (Geillinger et al., 2014).
Transcriptional Regulation of MATEs
Only limited findings are available regarding transcriptional regulation on MATE transporters. A CpG island located in the 27 kb upstream of SLC47A1 gene has been identified as an enhancer (Tanaka et al., 2018). The methylation levels of this CpG island were negatively correlated with the mRNA expression of SLC47A1 in the liver (Tanaka et al., 2018). However, the detail regulation of SLC47A1 expression through DNA methylation is currently unclear. As for OCT2 described above, the expression of another MATE transporter, MATE2, has been reported to be suppressed in RCC (Yu et al., 2017). The suppression was related to histone modification. Specifically, the protein scaffold adjacent to the SLC47A2 gene was found to be enriched with histone H3K4me3 and lysine 27 trimethylation (H3K27me3) in normal renal tissues. In RCC, the binding between the histone methylase mixed-lineage leukemia 1 (MLL1) and the promoter region was lost, reducing H3K4me3 enrichment and the transcriptional activity of SLC47A2 promoter. In addition, there was deacetylation of H3K27 in RCC, which prevented the enrichment of H3K4me3 as well.
Several studies have explored the role of transcriptional factors in transcription of MATEs. The Giacomini group has characterized the transcriptional activity of the basal promoter of human SLC47A1 gene. They identified polymorphic sequences in the promoter region that could bind to two transcriptional factors, activating protein-1 (AP-1) and activating protein-2 repressor (AP-2rep), in regulation of the transcription of SLC47A1 gene (Ha Choi et al., 2009). By using different NR pharmacological activators, a study by Lickteig et al. (2008) has suggested that AhR, CAR, PXR, Nrf2, and PPARα do not play a critical role in the transcription of Slc47a1 and Slc47a2 in mouse livers. However, the mRNA expression of Slc47a1 in the kidney of PPARα-null mice was increased (Freitas-Lima et al., 2020) and dramatically decreased in the liver of HNF4α-null mice (Lu et al., 2010), suggesting a role of these two NRs in tissue-specific MATE transcription. In addition, the mRNA expression of Slc47a1 was significantly increased in the liver of NADPH–cytochrome P450 oxidoreductase (Cpr)-null mice where Nrf2 was activated (Cheng et al., 2014). Moreover, treatment with the Nrf2 activator bardoxolone methyl was found to significantly increase the mRNA expression of SLC47A1 in human renal proximal tubular epithelial cells (Atilano-Roque et al., 2016). Interestingly, by using microfluidic culture of human proximal tubules, Fukuda et al. (2017) have demonstrated that fluid shear stress stimulates MATE2-K expression via Nrf2 pathway activation. Last but not least, FXR may be involved in the transcriptional regulation of human MATEs because treatment with its ligand CDCA could significantly increase the mRNA expression of SLC47A1 and SLC47A2 in human renal proximal tubular cells (Wongwan et al., 2020). Further mechanistic studies are needed to clarify the regulatory role of these NRs in transcription of MATE genes.
Closing Remarks
SLC drug transporters are present in tissues with varying abundance. It would be a prerequisite to clarify the accurate localization and abundance of these transporters before elucidating their role in the absorption, distribution, metabolism, excretion, and consequently the pharmacological and toxicological effects of clinical drugs. The transcriptional regulation is the major determinant of tissue distribution and abundance for these drug transporters. We have provided an overview and collected information on transcriptional regulation of major SLC drug transporters in this review. In consideration of space limit, however, we are unable to discuss the molecular mechanisms in detail for the transcriptional regulation of these transporters, nor did we include all SLC families whose members sometimes facilitate the movement of clinically used drugs across the membrane. In addition, there are progresses in terms of preclinical evidence and clinical implication that deserve a specific review under individual topics. These limitations have inevitably compromised the comprehensiveness of this review.
Although the importance of SLC transporters in drug disposition and response is widely appreciated, understanding the role of epigenetic- and NR-mediated transcriptional regulation has just begun. There are progresses suggesting clinical implication of such regulation for SLC drug transporter genes. For example, by targeting the epigenetic mechanism of OCT2 transcription, a sequential combination therapy using the DNA methylation inhibitor decitabine has been proposed to overcome the resistance of RCC to oxaliplatin chemotherapy (Liu et al., 2016b). However, overall, the clinical evidence remains very limited. It is challenging to ascertain the effects by an alteration in transporter transcription on drug disposition and response in animal models and particularly in human subjects in vivo. Although one mechanism or one NR may govern the transcription of multiple SLC drug transporter genes, an individual transporter can be regulated via multiple mechanisms as well. The lack of selective probe substrates, inhibitors, and regulatory molecules for specific drug transporters has been bringing another layer of complexity in interpretation of in vivo data. Current effort in characterizing drug-drug interactions (DDIs) for an increasing number of compounds will bring us much-needed probe drugs to assess the function of SLC drug transporters and their regulation. Moreover, investigating the impact of SLC transcriptional changes on drug disposition and response with appropriate probe compounds in genetic animal models and genetically polymorphic human populations may yield invaluable preclinical and clinical evidence soon. Of note, there are also different levels of regulation on transporter function. For example, posttranslational modification on transporter proteins has been widely reported to affect SLC drug transporter function (Murray and Zhou, 2017; Xu and You, 2017; Czuba et al., 2018; Lee et al., 2020). A novel kinase-dependent posttranslational regulation of OATP1B1 activity has been recently characterized as a mechanism underlying DDIs between broad OATP1B1 substrates and the commonly prescribed tyrosine kinase inhibitors (TKIs) (Hayden et al., 2021). We have also identified a mechanism regulating the membrane translocation of OCT2 and consequently the nephrotoxicity of cisplatin, a widely prescribed anticancer drug (Yang et al., 2020). Our understanding of a full range of regulatory mechanisms for drug transporters will eventually have an impact on optimization of pharmacotherapy regimens to improve drug efficacy and avoid unnecessary toxicity.
Abbreviations
- AhR
aryl hydrocarbon receptor
- atRA
all-trans retinoic acid
- CAR
constitutive androgen receptor
- CDCA
chenodeoxycholic acid
- CpG
cytosine-phosphate-guanine
- CRA
9-cis retinoic acid
- DDI
drug-drug interaction
- ERα
estrogen receptor alpha
- E-box
enhancer box
- FXR
farnesoid X receptor
- HAT
histone acetylase
- HCC
hepatocellular carcinoma
- HDAC
histone deacetylase
- HIF-1α
hypoxia-inducible factor 1 alpha
- H3K4me3
trimethylation of lysine 4 on histone 3
- HNF
hepatocyte nuclear factor
- LPS
lipopolysaccharide
- LXR
liver X receptor
- MATE
multidrug and toxin extrusion protein
- NR
nuclear receptor
- Nrf2
nuclear factor erythroid 2-related factor 2
- OAT
organic anion transporter
- OATP
organic anion-transporting polypeptide
- OCT
organic cation transporter
- OCTN
organic cation/carnitine transporter
- PCN
pregnenolone 16α-carbonitrile
- PEPT
peptide transporter
- PPARα
peroxisome proliferator-activated receptor alpha
- PPRE
peroxisome proliferator response element
- PXR
pregnane X receptor
- RAR
retinoic acid receptor
- RCC
renal cell carcinoma
- RXR
retinoid X receptor
- SHP
small heterodimer partner 1
- SLC
solute carrier
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TSS
transcription start site
- UDCA
ursodeoxycholic acid
- USF
upstream stimulating factor
- VDR
vitamin D receptor
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Zhou, Shu.
Footnotes
This work was supported by National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM099742] (Y.S.).
No author has an actual or perceived conflict of interest with the contents of this article.
References
- Aleksunes LM, Klaassen CD (2012) Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos 40:1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amacher DE (2016) The regulation of human hepatic drug transporter expression by activation of xenobiotic-sensing nuclear receptors. Expert Opin Drug Metab Toxicol 12:1463–1477. [DOI] [PubMed] [Google Scholar]
- Anderson CM, Thwaites DT (2010) Hijacking solute carriers for proton-coupled drug transport. Physiology (Bethesda) 25:364–377. [DOI] [PubMed] [Google Scholar]
- Anwar-Mohamed A, Degenhardt OS, El Gendy MA, Seubert JM, Kleeberger SR, El-Kadi AO (2011) The effect of Nrf2 knockout on the constitutive expression of drug metabolizing enzymes and transporters in C57Bl/6 mice livers. Toxicol In Vitro 25:785–795. [DOI] [PubMed] [Google Scholar]
- Aoki M, Terada T, Kajiwara M, Ogasawara K, Ikai I, Ogawa O, Katsura T, Inui K (2008) Kidney-specific expression of human organic cation transporter 2 (OCT2/SLC22A2) is regulated by DNA methylation. Am J Physiol Renal Physiol 295:F165–F170. [DOI] [PubMed] [Google Scholar]
- Asaka J, Terada T, Okuda M, Katsura T, Inui K (2006) Androgen receptor is responsible for rat organic cation transporter 2 gene regulation but not for rOCT1 and rOCT3. Pharm Res 23:697–704. [DOI] [PubMed] [Google Scholar]
- Atilano-Roque A, Aleksunes LM, Joy MS (2016) Bardoxolone methyl modulates efflux transporter and detoxifying enzyme expression in cisplatin-induced kidney cell injury. Toxicol Lett 259:52–59. [DOI] [PubMed] [Google Scholar]
- Austin G, Holcroft A, Rinne N, Wang L, Clark RE (2015) Evidence that the pregnane X and retinoid receptors PXR, RAR and RXR may regulate transcription of the transporter hOCT1 in chronic myeloid leukaemia cells. Eur J Haematol 94:74–78. [DOI] [PubMed] [Google Scholar]
- Ayka A, Şehirli AO (2020) The role of the SLC transporters protein in the neurodegenerative disorders. Clin Psychopharmacol Neurosci 18:174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarczyk D, Sanghvi MV (2020) Organic anion transporting polypeptide 2B1 (OATP2B1), an expanded substrate profile, does it align with OATP2B1's hypothesized function? Xenobiotica 50:1128–1137. [DOI] [PubMed] [Google Scholar]
- Benson EA, Eadon MT, Desta Z, Liu Y, Lin H, Burgess KS, Segar MW, Gaedigk A, Skaar TC (2016) Rifampin regulation of drug transporters gene expression and the association of microRNAs in human hepatocytes. Front Pharmacol 7:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutia YD, Babu E, Ramachandran S, Yang S, Thangaraju M, Ganapathy V (2016) SLC transporters as a novel class of tumour suppressors: identity, function and molecular mechanisms. Biochem J 473:1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkhem-Bergman L, Bergström H, Johansson M, Parini P, Eriksson M, Rane A, Ekström L (2013) Atorvastatin treatment induces uptake and efflux transporters in human liver. Drug Metab Dispos 41:1610–1615. [DOI] [PubMed] [Google Scholar]
- César-Razquin ASnijder BFrappier-Brinton TIsserlin RGyimesi GBai XReithmeier RAHepworth DHediger MAEdwards AM, et al. (2015) A call for systematic research on solute carriers. Cell 162:478–487. [DOI] [PubMed] [Google Scholar]
- Chen LHong CChen ECYee SWXu LAlmof EUWen CFujii KJohns SJStryke D, et al. (2013) Genetic and epigenetic regulation of the organic cation transporter 3, SLC22A3. Pharmacogenomics J 13:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Taguchi K, Aleksunes LM, Manautou JE, Cherrington NJ, Yamamoto M, Slitt AL (2011) Constitutive activation of nuclear factor-E2-related factor 2 induces biotransformation enzyme and transporter expression in livers of mice with hepatocyte-specific deletion of Kelch-like ECH-associated protein 1. J Biochem Mol Toxicol 25:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Gu J, Klaassen CD (2014) Adaptive hepatic and intestinal alterations in mice after deletion of NADPH-cytochrome P450 oxidoreductase (Cpr) in hepatocytes. Drug Metab Dispos 42:1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD (2008) Critical role of PPAR-alpha in perfluorooctanoic acid- and perfluorodecanoic acid-induced downregulation of Oatp uptake transporters in mouse livers. Toxicol Sci 106:37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Maher J, Dieter MZ, Klaassen CD (2005) Regulation of mouse organic anion-transporting polypeptides (Oatps) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab Dispos 33:1276–1282. [DOI] [PubMed] [Google Scholar]
- Cho SK, Yoon JS, Lee MG, Lee DH, Lim LA, Park K, Park MS, Chung JY (2011) Rifampin enhances the glucose-lowering effect of metformin and increases OCT1 mRNA levels in healthy participants. Clin Pharmacol Ther 89:416–421. [DOI] [PubMed] [Google Scholar]
- Czuba LC, Hillgren KM, Swaan PW (2018) Post-translational modifications of transporters. Pharmacol Ther 192:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio G, Petillo O, Margarucci S, Torpedine A, Calarco A, Koverech A, Boccia A, Paolella G, Peluso G (2010) Colon OCTN2 gene expression is up-regulated by peroxisome proliferator-activated receptor gamma in humans and mice and contributes to local and systemic carnitine homeostasis. J Biol Chem 285:27078–27087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk GU, Soroka CJ, Mennone A, Koepsell H, Beuers U, Boyer JL (2004) Down-regulation of the organic cation transporter 1 of rat liver in obstructive cholestasis. Hepatology 39:1382–1389. [DOI] [PubMed] [Google Scholar]
- Eloranta JJ, Hiller C, Jüttner M, Kullak-Ublick GA (2012) The SLCO1A2 gene, encoding human organic anion-transporting polypeptide 1A2, is transactivated by the vitamin D receptor. Mol Pharmacol 82:37–46. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Hamilton BA, Nigam SK (2003) Organic anion and cation transporters occur in pairs of similar and similarly expressed genes. Biochem Biophys Res Commun 300:333–342. [DOI] [PubMed] [Google Scholar]
- Euteneuer AM, Seeger-Nukpezah T, Nolte H, Henjakovic M (2019) Estrogen receptor α (ERα) indirectly induces transcription of human renal organic anion transporter 1 (OAT1). Physiol Rep 7:e14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Lima LC, Budu A, Arruda AC, Perilhão MS, Barrera-Chimal J, Araujo RC, Estrela GR (2020) PPAR-α deletion attenuates cisplatin nephrotoxicity by modulating renal organic transporters MATE-1 and OCT-2. Int J Mol Sci 21:7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Kaishima M, Ohnishi T, Tohyama K, Chisaki I, Nakayama Y, Ogasawara-Shimizu M, Kawamata Y (2017) Fluid shear stress stimulates MATE2-K expression via Nrf2 pathway activation. Biochem Biophys Res Commun 484:358–364. [DOI] [PubMed] [Google Scholar]
- Gallegos TF, Martovetsky G, Kouznetsova V, Bush KT, Nigam SK (2012) Organic anion and cation SLC22 “drug” transporter (Oat1, Oat3, and Oct1) regulation during development and maturation of the kidney proximal tubule. PLoS One 7:e40796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Calzón S, Perfilyev A, Männistö V, de Mello VD, Nilsson E, Pihlajamäki J, Ling C (2017) Diabetes medication associates with DNA methylation of metformin transporter genes in the human liver. Clin Epigenetics 9:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier A, Dietrich CG, Voigt S, Kim SK, Gerloff T, Kullak-Ublick GA, Lorenzen J, Matern S, Gartung C (2003) Effects of proinflammatory cytokines on rat organic anion transporters during toxic liver injury and cholestasis. Hepatology 38:345–354. [DOI] [PubMed] [Google Scholar]
- Geier A, Wagner M, Dietrich CG, Trauner M (2007) Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim Biophys Acta 1773:283–308. [DOI] [PubMed] [Google Scholar]
- Geillinger KE, Kipp AP, Schink K, Röder PV, Spanier B, Daniel H (2014) Nrf2 regulates the expression of the peptide transporter PEPT1 in the human colon carcinoma cell line Caco-2. Biochim Biophys Acta 1840:1747–1754. [DOI] [PubMed] [Google Scholar]
- Glastras SJ, Wong MG, Chen H, Zhang J, Zaky A, Pollock CA, Saad S (2015) FXR expression is associated with dysregulated glucose and lipid levels in the offspring kidney induced by maternal obesity. Nutr Metab (Lond) 12:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy PHewitt NJAlbrecht UAndersen MEAnsari NBhattacharya SBode JGBolleyn JBorner CBöttger J, et al. (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87:1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo GL, Choudhuri S, Klaassen CD (2002a) Induction profile of rat organic anion transporting polypeptide 2 (oatp2) by prototypical drug-metabolizing enzyme inducers that activate gene expression through ligand-activated transcription factor pathways. J Pharmacol Exp Ther 300:206–212. [DOI] [PubMed] [Google Scholar]
- Guo GL, Staudinger J, Ogura K, Klaassen CD (2002b) Induction of rat organic anion transporting polypeptide 2 by pregnenolone-16alpha-carbonitrile is via interaction with pregnane X receptor. Mol Pharmacol 61:832–839. [DOI] [PubMed] [Google Scholar]
- Ha Choi JWah Yee SKim MJNguyen LHo Lee JKang JOHesselson SCastro RAStryke DJohns SJ, et al. (2009) Identification and characterization of novel polymorphisms in the basal promoter of the human transporter, MATE1. Pharmacogenet Genomics 19:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch N, Reichel C, Stieger B, Cattori V, Fattinger KE, Landmann L, Meier PJ, Kullak-Ublick GA (2001) Effect of phenobarbital on the expression of bile salt and organic anion transporters of rat liver. J Hepatol 34:881–887. [DOI] [PubMed] [Google Scholar]
- Han S, Kim K, Thakkar N, Kim D, Lee W (2013) Role of hypoxia inducible factor-1α in the regulation of the cancer-specific variant of organic anion transporting polypeptide 1B3 (OATP1B3), in colon and pancreatic cancer. Biochem Pharmacol 86:816–823. [DOI] [PubMed] [Google Scholar]
- Hayden ERChen MPasquariello KZGibson AAPetti JJShen SQu JOng SSChen TJin Y, et al. (2021) Regulation of OATP1B1 function by tyrosine kinase-mediated phosphorylation. Clin Cancer Res 27:4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YJZhang WTu JHKirchheiner JChen YGuo DLi QLi ZYChen HHu DL, et al. (2008) Hepatic nuclear factor 1alpha inhibitor ursodeoxycholic acid influences pharmacokinetics of the organic anion transporting polypeptide 1B1 substrate rosuvastatin and bilirubin. Drug Metab Dispos 36:1453–1456. [DOI] [PubMed] [Google Scholar]
- Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA (2004) The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch 447:465–468. [DOI] [PubMed] [Google Scholar]
- Hirai T, Fukui Y, Motojima K (2007) PPARalpha agonists positively and negatively regulate the expression of several nutrient/drug transporters in mouse small intestine. Biol Pharm Bull 30:2185–2190. [DOI] [PubMed] [Google Scholar]
- Holla VR, Backlund MG, Yang P, Newman RA, DuBois RN (2008) Regulation of prostaglandin transporters in colorectal neoplasia. Cancer Prev Res (Phila) 1:93–99. [DOI] [PubMed] [Google Scholar]
- Hota SK, Bruneau BG (2016) ATP-dependent chromatin remodeling during mammalian development. Development 143:2882–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CS, Chen HW, Lin TY, Lin AH, Lii CK (2018) Shikonin upregulates the expression of drug-metabolizing enzymes and drug transporters in primary rat hepatocytes. J Ethnopharmacol 216:18–25. [DOI] [PubMed] [Google Scholar]
- Huang KM, Uddin ME, DiGiacomo D, Lustberg MB, Hu S, Sparreboom A (2020) Role of SLC transporters in toxicity induced by anticancer drugs. Expert Opin Drug Metab Toxicol 16:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrsova L, Smutny T, Carazo A, Moravcik S, Mandikova J, Trejtnar F, Gerbal-Chaloin S, Pavek P (2016) The pregnane X receptor down-regulates organic cation transporter 1 (SLC22A1) in human hepatocytes by competing for (“squelching”) SRC-1 coactivator. Br J Pharmacol 173:1703–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara S, Kikuchi R, Kusuhara H, Imai S, Maeda K, Sugiyama Y (2010) DNA methylation profiles of organic anion transporting polypeptide 1B3 in cancer cell lines. Pharm Res 27:510–516. [DOI] [PubMed] [Google Scholar]
- Imai S, Kikuchi R, Kusuhara H, Sugiyama Y (2013a) DNA methylation and histone modification profiles of mouse organic anion transporting polypeptides. Drug Metab Dispos 41:72–78. [DOI] [PubMed] [Google Scholar]
- Imai S, Kikuchi R, Kusuhara H, Yagi S, Shiota K, Sugiyama Y (2009) Analysis of DNA methylation and histone modification profiles of liver-specific transporters. Mol Pharmacol 75:568–576. [DOI] [PubMed] [Google Scholar]
- Imai S, Kikuchi R, Tsuruya Y, Naoi S, Nishida S, Kusuhara H, Sugiyama Y (2013b) Epigenetic regulation of organic anion transporting polypeptide 1B3 in cancer cell lines. Pharm Res 30:2880–2890. [DOI] [PubMed] [Google Scholar]
- International Transporter Consortium, Giacomini KMHuang SMTweedie DJBenet LZBrouwer KLChu XDahlin AEvers RFischer V, et al. (2010) Membrane transporters in drug development. Nat Rev Drug Discov 9:215–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JJansen KNeven EPoesen ROthman Avan Mil ASluijter JSastre Torano JZaal EABerkers CR, et al. (2019) Remote sensing and signaling in kidney proximal tubules stimulates gut microbiome-derived organic anion secretion. Proc Natl Acad Sci USA 116:16105–16110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszewicz E, Pajak B, Gajkowska B, Samluk L, Djavadian RL, Hinton BT, Nałecz KA (2009) Organic cation/carnitine transporter OCTN3 is present in astrocytes and is up-regulated by peroxisome proliferators-activator receptor agonist. Int J Biochem Cell Biol 41:2599–2609. [DOI] [PubMed] [Google Scholar]
- Jigorel E, Le Vee M, Boursier-Neyret C, Parmentier Y, Fardel O (2006) Differential regulation of sinusoidal and canalicular hepatic drug transporter expression by xenobiotics activating drug-sensing receptors in primary human hepatocytes. Drug Metab Dispos 34:1756–1763. [DOI] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD (2011) DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2:607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Kikuchi R, Saji T, Kusuhara H, Sugiyama Y (2012) Regulation of tissue-specific expression of renal organic anion transporters by hepatocyte nuclear factor 1 α/β and DNA methylation. J Pharmacol Exp Ther 340:648–655. [DOI] [PubMed] [Google Scholar]
- Jung D, Hagenbuch B, Gresh L, Pontoglio M, Meier PJ, Kullak-Ublick GA (2001) Characterization of the human OATP-C (SLC21A6) gene promoter and regulation of liver-specific OATP genes by hepatocyte nuclear factor 1 alpha. J Biol Chem 276:37206–37214. [DOI] [PubMed] [Google Scholar]
- Jung D, Podvinec M, Meyer UA, Mangelsdorf DJ, Fried M, Meier PJ, Kullak-Ublick GA (2002) Human organic anion transporting polypeptide 8 promoter is transactivated by the farnesoid X receptor/bile acid receptor. Gastroenterology 122:1954–1966. [DOI] [PubMed] [Google Scholar]
- Kajiwara M, Terada T, Asaka J, Aoki M, Katsura T, Ikai I, Inui K (2008) Regulation of basal core promoter activity of human organic cation transporter 1 (OCT1/SLC22A1). Am J Physiol Gastrointest Liver Physiol 295:G1211–G1216. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M (1999) Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol 19:6318–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Stienstra R (2017) The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie 136:75–84. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kusuhara H, Hattori N, Kim I, Shiota K, Gonzalez FJ, Sugiyama Y (2007) Regulation of tissue-specific expression of the human and mouse urate transporter 1 gene by hepatocyte nuclear factor 1 alpha/beta and DNA methylation. Mol Pharmacol 72:1619–1625. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kusuhara H, Hattori N, Shiota K, Kim I, Gonzalez FJ, Sugiyama Y (2006) Regulation of the expression of human organic anion transporter 3 by hepatocyte nuclear factor 1alpha/beta and DNA methylation. Mol Pharmacol 70:887–896. [DOI] [PubMed] [Google Scholar]
- Kitao AMatsui OYoneda NKozaka KKobayashi SKoda WMinami TInoue DYoshida KYamashita T, et al. (2018) Gadoxetic acid-enhanced magnetic resonance imaging reflects co-activation of β-catenin and hepatocyte nuclear factor 4α in hepatocellular carcinoma. Hepatol Res 48:205–216. [DOI] [PubMed] [Google Scholar]
- Kittayaruksakul S, Soodvilai S, Asavapanumas N, Muanprasat C, Chatsudthipong V (2012) Liver X receptor activation downregulates organic anion transporter 1 (OAT1) in the renal proximal tubule. Am J Physiol Renal Physiol 302:F552–F560. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM (2010) Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev 62:1–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K, Jüngst C, Mwinyi J, Stieger B, Krempler F, Patsch W, Eloranta JJ, Kullak-Ublick GA (2010) The human organic anion transporter genes OAT5 and OAT7 are transactivated by hepatocyte nuclear factor-1α (HNF-1α). Mol Pharmacol 78:1079–1087. [DOI] [PubMed] [Google Scholar]
- Koch A, König B, Luci S, Stangl GI, Eder K (2007) Dietary oxidised fat up regulates the expression of organic cation transporters in liver and small intestine and alters carnitine concentrations in liver, muscle and plasma of rats. Br J Nutr 98:882–889. [DOI] [PubMed] [Google Scholar]
- Koch A, König B, Stangl GI, Eder K (2008) PPAR alpha mediates transcriptional upregulation of novel organic cation transporters-2 and -3 and enzymes involved in hepatic carnitine synthesis. Exp Biol Med (Maywood) 233:356–365. [DOI] [PubMed] [Google Scholar]
- Koepsell H (2020) Organic cation transporters in health and disease. Pharmacol Rev 72:253–319. [DOI] [PubMed] [Google Scholar]
- Kölz C, Schaeffeler E, Schwab M, Nies AT (2021) Genetic and epigenetic regulation of organic cation transporters. Handb Exp Pharmacol 266:81–100. [DOI] [PubMed] [Google Scholar]
- Kullak-Ublick GA, Stieger B, Meier PJ (2004) Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology 126:322–342. [DOI] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT (2018) The human transcription factors. Cell 172:650–665. [DOI] [PubMed] [Google Scholar]
- Lau HH, Ng NHJ, Loo LSW, Jasmen JB, Teo AKK (2018) The molecular functions of hepatocyte nuclear factors - in and beyond the liver. J Hepatol 68:1033–1048. [DOI] [PubMed] [Google Scholar]
- Le Vee M, Jouan E, Stieger B, Fardel O (2013) Differential regulation of drug transporter expression by all-trans retinoic acid in hepatoma HepaRG cells and human hepatocytes. Eur J Pharm Sci 48:767–774. [DOI] [PubMed] [Google Scholar]
- Le Vee M, Jouan E, Stieger B, Lecureur V, Fardel O (2015) Regulation of human hepatic drug transporter activity and expression by diesel exhaust particle extract. PLoS One 10:e0121232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Ha JM, Sugiyama Y (2020) Post-translational regulation of the major drug transporters in the families of organic anion transporters and organic anion-transporting polypeptides. J Biol Chem 295:17349–17364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD (2000) The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol 20:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Klaassen CD (2004) Role of liver-enriched transcription factors in the down-regulation of organic anion transporting polypeptide 4 (oatp4; oatplb2; slc21a10) by lipopolysaccharide. Mol Pharmacol 66:694–701. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang W, Song J, Zhang X, Ran L, He Y (2019) SLCO4C1 promoter methylation is a potential biomarker for prognosis associated with biochemical recurrence-free survival after radical prostatectomy. Clin Epigenetics 11:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang LM, Zhou JJ, Xu F, Liu PH, Qin L, Liu L, Liu XD (2020) Diabetes downregulates peptide transporter 1 in the rat jejunum: possible involvement of cholate-induced FXR activation. Acta Pharmacol Sin 41:1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickteig AJ, Cheng X, Augustine LM, Klaassen CD, Cherrington NJ (2008) Tissue distribution, ontogeny and induction of the transporters Multidrug and toxin extrusion (MATE) 1 and MATE2 mRNA expression levels in mice. Life Sci 83:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Yee SW, Kim RB, Giacomini KM (2015) SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov 14:543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Li X, Li J, Zhang L, Xu F, Chu Y, Li J (2013) Long-term cisplatin exposure promotes methylation of the OCT1 gene in human esophageal cancer cells. Dig Dis Sci 58:694–698. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhu D, Wen J, Ding W, Huang S, Xia C, Zhang H, Xiong Y (2020) Berberine promotes OATP1B1 expression and rosuvastatin uptake by inducing nuclear translocation of FXR and LXRα. Front Pharmacol 11:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X (2019) SLC family transporters. Adv Exp Med Biol 1141:101–202. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pu QH, Wu MJ, Yu C (2016a) Proteomic analysis for the impact of hypercholesterolemia on expressions of hepatic drug transporters and metabolizing enzymes. Xenobiotica 46:940–947. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng X, Yu Q, Wang H, Tan F, Zhu Q, Yuan L, Jiang H, Yu L, Zeng S (2016b) Epigenetic activation of the drug transporter OCT2 sensitizes renal cell carcinoma to oxaliplatin. Sci Transl Med 8:348ra97. [DOI] [PubMed] [Google Scholar]
- Lončar J, Popović M, Krznar P, Zaja R, Smital T (2016) The first characterization of multidrug and toxin extrusion (MATE/SLC47) proteins in zebrafish (Danio rerio). Sci Rep 6:28937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Kornberg RD (2017) Chromatin-remodeling for transcription. Q Rev Biophys 50:e5. [DOI] [PubMed] [Google Scholar]
- Lu H, Gonzalez FJ, Klaassen C (2010) Alterations in hepatic mRNA expression of phase II enzymes and xenobiotic transporters after targeted disruption of hepatocyte nuclear factor 4 alpha. Toxicol Sci 118:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci S, Geissler S, König B, Koch A, Stangl GI, Hirche F, Eder K (2006) PPARalpha agonists up-regulate organic cation transporters in rat liver cells. Biochem Biophys Res Commun 350:704–708. [DOI] [PubMed] [Google Scholar]
- Luci S, Giemsa B, Kluge H, Eder K (2007) Clofibrate causes an upregulation of PPAR-alpha target genes but does not alter expression of SREBP target genes in liver and adipose tissue of pigs. Am J Physiol Regul Integr Comp Physiol 293:R70–R77. [DOI] [PubMed] [Google Scholar]
- Luci S, Hirche F, Eder K (2008) Fasting and caloric restriction increases mRNA concentrations of novel organic cation transporter-2 and carnitine concentrations in rat tissues. Ann Nutr Metab 52:58–67. [DOI] [PubMed] [Google Scholar]
- Luo HZhang YGuo HZhang LLi XRingseis RWen GHui DLiang AEder K, et al. (2014) Transcriptional regulation of the human, porcine and bovine OCTN2 gene by PPARα via a conserved PPRE located in intron 1. BMC Genet 15:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Oyabu M, Yotsumoto T, Higashi R, Nagata K, Yamazoe Y, Tamai I (2007) Effect of pregnane X receptor ligand on pharmacokinetics of substrates of organic cation transporter Oct1 in rats. Drug Metab Dispos 35:1580–1586. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wakasawa T, Funabashi M, Fukushi A, Fujita M, Motojima K, Tamai I (2008) Regulation of Octn2 transporter (SLC22A5) by peroxisome proliferator activated receptor alpha. Biol Pharm Bull 31:1230–1236. [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Callaghan TN, Cheng X, Cheung C, Gonzalez FJ, Klaassen CD (2006) Alterations in transporter expression in liver, kidney, and duodenum after targeted disruption of the transcription factor HNF1alpha. Biochem Pharmacol 72:512–522. [DOI] [PubMed] [Google Scholar]
- Malagnino V, Hussner J, Issa A, Midzic A, Meyer Zu Schwabedissen HE (2019) OATP1B3-1B7, a novel organic anion transporting polypeptide, is modulated by FXR ligands and transports bile acids. Am J Physiol Gastrointest Liver Physiol 317:G751–G762. [DOI] [PubMed] [Google Scholar]
- Martovetsky G, Tee JB, Nigam SK (2013) Hepatocyte nuclear factors 4α and 1α regulate kidney developmental expression of drug-metabolizing enzymes and drug transporters. Mol Pharmacol 84:808–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzolini C, Tirona RG, Gervasini G, Poonkuzhali B, Assem M, Lee W, Leake BF, Schuetz JD, Schuetz EG, Kim RB (2007) A common polymorphism in the bile acid receptor farnesoid X receptor is associated with decreased hepatic target gene expression. Mol Endocrinol 21:1769–1780. [DOI] [PubMed] [Google Scholar]
- Masuyama H, Suwaki N, Tateishi Y, Nakatsukasa H, Segawa T, Hiramatsu Y (2005) The pregnane X receptor regulates gene expression in a ligand- and promoter-selective fashion. Mol Endocrinol 19:1170–1180. [DOI] [PubMed] [Google Scholar]
- Meyer Zu Schwabedissen HE, Böttcher K, Chaudhry A, Kroemer HK, Schuetz EG, Kim RB (2010) Liver X receptor α and farnesoid X receptor are major transcriptional regulators of OATP1B1. Hepatology 52:1797–1807. [DOI] [PubMed] [Google Scholar]
- Meyer zu Schwabedissen HE, Kim RB (2009) Hepatic OATP1B transporters and nuclear receptors PXR and CAR: interplay, regulation of drug disposition genes, and single nucleotide polymorphisms. Mol Pharm 6:1644–1661. [DOI] [PubMed] [Google Scholar]
- Meyer zu Schwabedissen HE, Tirona RG, Yip CS, Ho RH, Kim RB (2008) Interplay between the nuclear receptor pregnane X receptor and the uptake transporter organic anion transporter polypeptide 1A2 selectively enhances estrogen effects in breast cancer. Cancer Res 68:9338–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Suzuki T, Kitada K, Yabuki N, Shibuya R, Moriya T, Ishida T, Ohuchi N, Blumberg B, Sasano H (2006) Expression of the steroid and xenobiotic receptor and its possible target gene, organic anion transporting polypeptide-A, in human breast carcinoma. Cancer Res 66:535–542. [DOI] [PubMed] [Google Scholar]
- Miller JL, Grant PA (2013) The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell Biochem 61:289–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Patterson AD, Perdew GH (2014) Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer 14:801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M, Zhou F (2017) Trafficking and other regulatory mechanisms for organic anion transporting polypeptides and organic anion transporters that modulate cellular drug and xenobiotic influx and that are dysregulated in disease. Br J Pharmacol 174:1908–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Fukuno S, Yamamoto K, Omotani S, Hatsuda Y, Myotoku M, Konishi H (2019) Downregulation of organic cation transporter 1 and breast cancer resistance protein with the induction of pregnane X receptor in rat kidney impaired by doxorubicin. Pharmazie 74:744–746. [DOI] [PubMed] [Google Scholar]
- Niemi M (2010) Transporter pharmacogenetics and statin toxicity. Clin Pharmacol Ther 87:130–133. [DOI] [PubMed] [Google Scholar]
- Nies AT, Koepsell H, Winter S, Burk O, Klein K, Kerb R, Zanger UM, Keppler D, Schwab M, Schaeffeler E (2009) Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology 50:1227–1240. [DOI] [PubMed] [Google Scholar]
- Nigam SK (2018) The SLC22 transporter family: a paradigm for the impact of drug transporters on metabolic pathways, signaling, and disease. Annu Rev Pharmacol Toxicol 58:663–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK, Bush KT, Martovetsky G, Ahn SY, Liu HC, Richard E, Bhatnagar V, Wu W (2015) The organic anion transporter (OAT) family: a systems biology perspective. Physiol Rev 95:83–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C, Wang Y, Zhao X, Tep S, Murakami E, Subramanian R, Smith B, Lai Y (2019) Organic anion-transporting polypeptide genes are not induced by the pregnane X receptor activator rifampin: studies in hepatocytes in vitro and in monkeys in vivo. Drug Metab Dispos 47:1433–1442. [DOI] [PubMed] [Google Scholar]
- Oda M, Koyanagi S, Tsurudome Y, Kanemitsu T, Matsunaga N, Ohdo S (2014) Renal circadian clock regulates the dosing-time dependency of cisplatin-induced nephrotoxicity in mice. Mol Pharmacol 85:715–722. [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Terada T, Asaka J, Katsura T, Inui K (2007) Hepatocyte nuclear factor-4alpha regulates the human organic anion transporter 1 gene in the kidney. Am J Physiol Renal Physiol 292:F1819–F1826. [DOI] [PubMed] [Google Scholar]
- Ohtsuka HAbe TOnogawa TKondo NSato TOshio HMizutamari HMikkaichi TOikawa MRikiyama T, et al. (2006) Farnesoid X receptor, hepatocyte nuclear factors 1alpha and 3beta are essential for transcriptional activation of the liver-specific organic anion transporter-2 gene. J Gastroenterol 41:369–377. [DOI] [PubMed] [Google Scholar]
- Okamura A, Koyanagi S, Dilxiat A, Kusunose N, Chen JJ, Matsunaga N, Shibata S, Ohdo S (2014) Bile acid-regulated peroxisome proliferator-activated receptor-α (PPARα) activity underlies circadian expression of intestinal peptide absorption transporter PepT1/Slc15a1. J Biol Chem 289:25296–25305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscarson M, Zanger UM, Rifki OF, Klein K, Eichelbaum M, Meyer UA (2006) Transcriptional profiling of genes induced in the livers of patients treated with carbamazepine. Clin Pharmacol Ther 80:440–456. [DOI] [PubMed] [Google Scholar]
- Pácha J, Balounová K, Soták M (2021) Circadian regulation of transporter expression and implications for drug disposition. Expert Opin Drug Metab Toxicol 17:425–439. [DOI] [PubMed] [Google Scholar]
- Perland E, Fredriksson R (2017) Classification systems of secondary active transporters. Trends Pharmacol Sci 38:305–315. [DOI] [PubMed] [Google Scholar]
- Pizzagalli MD, Bensimon A, Superti-Furga G (2021) A guide to plasma membrane solute carrier proteins. FEBS J 288:2784–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowski K, Eloranta JJ, Saborowski M, Fried M, Meier PJ, Kullak-Ublick GA (2005) The human organic anion transporter 2 gene is transactivated by hepatocyte nuclear factor-4 alpha and suppressed by bile acids. Mol Pharmacol 67:1629–1638. [DOI] [PubMed] [Google Scholar]
- Qu Q, Qu J, Guo Y, Zhou BT, Zhou HH (2014) Luteolin potentiates the sensitivity of colorectal cancer cell lines to oxaliplatin through the PPARγ/OCTN2 pathway. Anticancer Drugs 25:1016–1027. [DOI] [PubMed] [Google Scholar]
- Qu Q, Qu J, Zhan M, Wu LX, Zhang YW, Lou XY, Fu LJ, Zhou HH (2013) Different involvement of promoter methylation in the expression of organic cation/carnitine transporter 2 (OCTN2) in cancer cell lines. PLoS One 8:e76474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran ABetts GBhana SHelme GBlick CMoller-Levet CSaunders EValentine HPepper SMiller CJ, et al. (2013) An in vivo hypoxia metagene identifies the novel hypoxia inducible factor target gene SLCO1B3. Eur J Cancer 49:1741–1751. [DOI] [PubMed] [Google Scholar]
- Rawłuszko-Wieczorek AA, Horst N, Horbacka K, Bandura AS, Świderska M, Krokowicz P, Jagodziński PP (2015) Effect of DNA methylation profile on OATP3A1 and OATP4A1 transcript levels in colorectal cancer. Biomed Pharmacother 74:233–242. [DOI] [PubMed] [Google Scholar]
- Ringseis R, Gutgesell A, Dathe C, Brandsch C, Eder K (2007a) Feeding oxidized fat during pregnancy up-regulates expression of PPARalpha-responsive genes in the liver of rat fetuses. Lipids Health Dis 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringseis R, Luci S, Spielmann J, Kluge H, Fischer M, Geissler S, Wen G, Hirche F, Eder K (2008a) Clofibrate treatment up-regulates novel organic cation transporter (OCTN)-2 in tissues of pigs as a model of non-proliferating species. Eur J Pharmacol 583:11–17. [DOI] [PubMed] [Google Scholar]
- Ringseis R, Lüdi S, Hirche F, Eder K (2008b) Treatment with pharmacological peroxisome proliferator-activated receptor alpha agonist clofibrate increases intestinal carnitine absorption in rats. Pharmacol Res 58:58–64. [DOI] [PubMed] [Google Scholar]
- Ringseis R, Muschick A, Eder K (2007b) Dietary oxidized fat prevents ethanol-induced triacylglycerol accumulation and increases expression of PPARalpha target genes in rat liver. J Nutr 137:77–83. [DOI] [PubMed] [Google Scholar]
- Ringseis R, Pösel S, Hirche F, Eder K (2007c) Treatment with pharmacological peroxisome proliferator-activated receptor alpha agonist clofibrate causes upregulation of organic cation transporter 2 in liver and small intestine of rats. Pharmacol Res 56:175–183. [DOI] [PubMed] [Google Scholar]
- Ringseis R, Wege N, Wen G, Rauer C, Hirche F, Kluge H, Eder K (2009) Carnitine synthesis and uptake into cells are stimulated by fasting in pigs as a model of nonproliferating species. J Nutr Biochem 20:840–847. [DOI] [PubMed] [Google Scholar]
- Rives ML, Javitch JA, Wickenden AD (2017) Potentiating SLC transporter activity: emerging drug discovery opportunities. Biochem Pharmacol 135:1–11. [DOI] [PubMed] [Google Scholar]
- Rodrigues AD, Lai Y, Shen H, Varma MVS, Rowland A, Oswald S (2020) Induction of human intestinal and hepatic organic anion transporting polypeptides: where is the evidence for its relevance in drug-drug interactions? Drug Metab Dispos 48:205–216. [DOI] [PubMed] [Google Scholar]
- Roth M, Obaidat A, Hagenbuch B (2012) OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. Br J Pharmacol 165:1260–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulcova A, Krausova L, Smutny T, Vrzal R, Dvorak Z, Jover R, Pavek P (2013) Glucocorticoid receptor regulates organic cation transporter 1 (OCT1, SLC22A1) expression via HNF4α upregulation in primary human hepatocytes. Pharmacol Rep 65:1322–1335. [DOI] [PubMed] [Google Scholar]
- Saborowski M, Kullak-Ublick GA, Eloranta JJ (2006) The human organic cation transporter-1 gene is transactivated by hepatocyte nuclear factor-4alpha. J Pharmacol Exp Ther 317:778–785. [DOI] [PubMed] [Google Scholar]
- Saito H, Terada T, Shimakura J, Katsura T, Inui K (2008) Regulatory mechanism governing the diurnal rhythm of intestinal H+/peptide cotransporter 1 (PEPT1). Am J Physiol Gastrointest Liver Physiol 295:G395–G402. [DOI] [PubMed] [Google Scholar]
- Saito J, Hirota T, Kikunaga N, Otsubo K, Ieiri I (2011) Interindividual differences in placental expression of the SLC22A2 (OCT2) gene: relationship to epigenetic variations in the 5′-upstream regulatory region. J Pharm Sci 100:3875–3883. [DOI] [PubMed] [Google Scholar]
- Saji T, Kikuchi R, Kusuhara H, Kim I, Gonzalez FJ, Sugiyama Y (2008) Transcriptional regulation of human and mouse organic anion transporter 1 by hepatocyte nuclear factor 1 alpha/beta. J Pharmacol Exp Ther 324:784–790. [DOI] [PubMed] [Google Scholar]
- Scalise M, Galluccio M, Accardi R, Cornet I, Tommasino M, Indiveri C (2012) Human OCTN2 (SLC22A5) is down-regulated in virus- and nonvirus-mediated cancer. Cell Biochem Funct 30:419–425. [DOI] [PubMed] [Google Scholar]
- Schaeffeler E, Hellerbrand C, Nies AT, Winter S, Kruck S, Hofmann U, van der Kuip H, Zanger UM, Koepsell H, Schwab M (2011) DNA methylation is associated with downregulation of the organic cation transporter OCT1 (SLC22A1) in human hepatocellular carcinoma. Genome Med 3:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger A, Welch MA, van Vlijmen H, Korzekwa K, Swaan PW, Matsson P (2018) Molecular modeling of drug-transporter interactions-an international transporter consortium perspective. Clin Pharmacol Ther 104:818–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CWu JBChu GCLi QWang RZhang CZhang YKim HLWang JZhau HE, et al. (2014) Heptamethine carbocyanine dye-mediated near-infrared imaging of canine and human cancers through the HIF-1α/OATPs signaling axis. Oncotarget 5:10114–10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama Y, Ushinohama K, Ikeda R, Yoshikawa Y, Motoya T, Takeda Y, Yamada K (2006) Effect of methotrexate treatment on expression levels of multidrug resistance protein 2, breast cancer resistance protein and organic anion transporters Oat1, Oat2 and Oat3 in rats. Cancer Sci 97:1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakura J, Terada T, Katsura T, Inui K (2005) Characterization of the human peptide transporter PEPT1 promoter: Sp1 functions as a basal transcriptional regulator of human PEPT1. Am J Physiol Gastrointest Liver Physiol 289:G471–G477. [DOI] [PubMed] [Google Scholar]
- Shimakura J, Terada T, Saito H, Katsura T, Inui K (2006a) Induction of intestinal peptide transporter 1 expression during fasting is mediated via peroxisome proliferator-activated receptor alpha. Am J Physiol Gastrointest Liver Physiol 291:G851–G856. [DOI] [PubMed] [Google Scholar]
- Shimakura J, Terada T, Shimada Y, Katsura T, Inui K (2006b) The transcription factor Cdx2 regulates the intestine-specific expression of human peptide transporter 1 through functional interaction with Sp1. Biochem Pharmacol 71:1581–1588. [DOI] [PubMed] [Google Scholar]
- Sperling S (2007) Transcriptional regulation at a glance. BMC Bioinformatics 8 (Suppl 6):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Woody S, Sun M, Cui W (2013) Nuclear-receptor-mediated regulation of drug- and bile-acid-transporter proteins in gut and liver. Drug Metab Rev 45:48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieger B, Geier A (2011) Genetic variations of bile salt transporters as predisposing factors for drug-induced cholestasis, intrahepatic cholestasis of pregnancy and therapeutic response of viral hepatitis. Expert Opin Drug Metab Toxicol 7:411–425. [DOI] [PubMed] [Google Scholar]
- Stieger B, Hagenbuch B (2014) Organic anion-transporting polypeptides. Curr Top Membr 73:205–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Toyohara T, Akiyama Y, Takeuchi Y, Mishima E, Suzuki C, Ito S, Soga T, Abe T (2011) Transcriptional regulation of organic anion transporting polypeptide SLCO4C1 as a new therapeutic modality to prevent chronic kidney disease. J Pharm Sci 100:3696–3707. [DOI] [PubMed] [Google Scholar]
- Svoboda M, Riha J, Wlcek K, Jaeger W, Thalhammer T (2011) Organic anion transporting polypeptides (OATPs): regulation of expression and function. Curr Drug Metab 12:139–153. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Hirota T, Ieiri I (2018) Relationship between DNA methylation in the 5′ CpG island of the SLC47A1 (multidrug and toxin extrusion protein MATE1) gene and interindividual variability in MATE1 expression in the human liver. Mol Pharmacol 93:1–7. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Cui YJ, Klaassen CD (2009) ANIT-induced intrahepatic cholestasis alters hepatobiliary transporter expression via Nrf2-dependent and independent signaling. Toxicol Sci 108:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan RD, Georgas KM, Rumballe BA, Lesieur E, Chiu HS, Taylor D, Tang DT, Grimmond SM, Little MH (2011) Identification of anchor genes during kidney development defines ontological relationships, molecular subcompartments and regulatory pathways. PLoS One 6:e17286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli C, Chio IIC, Tuveson DA (2018) Transcriptional regulation by Nrf2. Antioxid Redox Signal 29:1727–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyohara TSuzuki TMorimoto RAkiyama YSouma TShiwaku HOTakeuchi YMishima EAbe MTanemoto M, et al. (2009) SLCO4C1 transporter eliminates uremic toxins and attenuates hypertension and renal inflammation. J Am Soc Nephrol 20:2546–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vlies N, Ferdinandusse S, Turkenburg M, Wanders RJ, Vaz FM (2007) PPAR alpha-activation results in enhanced carnitine biosynthesis and OCTN2-mediated hepatic carnitine accumulation. Biochim Biophys Acta 1767:1134–1142. [DOI] [PubMed] [Google Scholar]
- Vavricka SR, Jung D, Fried M, Grützner U, Meier PJ, Kullak-Ublick GA (2004) The human organic anion transporting polypeptide 8 (SLCO1B3) gene is transcriptionally repressed by hepatocyte nuclear factor 3beta in hepatocellular carcinoma. J Hepatol 40:212–218. [DOI] [PubMed] [Google Scholar]
- Wang C, Uray IP, Mazumdar A, Mayer JA, Brown PH (2012a) SLC22A5/OCTN2 expression in breast cancer is induced by estrogen via a novel intronic estrogen-response element (ERE). Breast Cancer Res Treat 134:101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xin M, Li Y, Zhang P, Zhang M (2016) The functions of histone modification enzymes in cancer. Curr Protein Pept Sci 17:438–445. [DOI] [PubMed] [Google Scholar]
- Wang Y, Viscarra J, Kim SJ, Sul HS (2015) Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell Biol 16:678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Yang L, Qiu L, Hua Y, Wu S, Zeng S, Yu L, Zheng X (2021a) Epigenetic regulation of intestinal peptide transporter PEPT1 as a potential strategy for colorectal cancer sensitization. Cell Death Dis 12:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu Q, Hu H, Zhu H, Yang B, He Q, Yu L, Zeng S (2021b) Upregulation of histone acetylation reverses organic anion transporter 2 repression and enhances 5-fluorouracil sensitivity in hepatocellular carcinoma. Biochem Pharmacol 188:114546. [DOI] [PubMed] [Google Scholar]
- Wang YM, Chai SC, Brewer CT, Chen T (2014) Pregnane X receptor and drug-induced liver injury. Expert Opin Drug Metab Toxicol 10:1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Ong SS, Chai SC, Chen T (2012b) Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol 8:803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner W, Burckhardt BC, Burckhardt G, Henjakovic M (2012) Male-dominant activation of rat renal organic anion transporter 1 (Oat1) and 3 (Oat3) expression by transcription factor BCL6. PLoS One 7:e35556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner W, Burckhardt G, Henjakovic M (2014) Transcriptional regulation of human organic anion transporter 1 by B-cell CLL/lymphoma 6. Am J Physiol Renal Physiol 307:F1283–F1291. [DOI] [PubMed] [Google Scholar]
- Wen G, Ringseis R, Eder K (2010) Mouse OCTN2 is directly regulated by peroxisome proliferator-activated receptor alpha (PPARalpha) via a PPRE located in the first intron. Biochem Pharmacol 79:768–776. [DOI] [PubMed] [Google Scholar]
- Wongwan T, Chatsudthipong V, Soodvilai S (2020) Farnesoid X receptor activation stimulates organic cations transport in human renal proximal tubular cells. Int J Mol Sci 21:6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Cui JY, Klaassen CD (2012) Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS One 7:e39006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, You G (2017) Loops and layers of post-translational modifications of drug transporters. Adv Drug Deliv Rev 116:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shu Y (2021) Role of membrane transporters in drug disposition, in Fundamentals of Drug Delivery (Benson HAE, Roberts MS, Williams AC, Liang X, eds) pp 193–230, John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- Yang H, Tang J, Guo D, Zhao Q, Wen J, Zhang Y, Obianom ON, Zhou S, Zhang W, Shu Y (2020) Cadmium exposure enhances organic cation transporter 2 trafficking to the kidney membrane and exacerbates cisplatin nephrotoxicity. Kidney Int 97:765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XH, Liu SY, Xing AY (2014) Molecular regulation of organic anion transporting polypeptide 1A2 (OATP1A2) by taurocholic acid in Bewo Cells. Cell Mol Biol 60:22–26. [PubMed] [Google Scholar]
- Yu QLiu YZheng XZhu QShen ZWang HHe HLin NJiang HYu L, et al. (2017) Histone H3 lysine 4 trimethylation, lysine 27 trimethylation, and lysine 27 acetylation contribute to the transcriptional repression of solute carrier family 47 member 2 in renal cell carcinoma. Drug Metab Dispos 45:109–117. [DOI] [PubMed] [Google Scholar]
- Zamek-Gliszczynski MJPatel MYang XLutz JDChu XBrouwer KLRLai YLee CANeuhoff SPaine MF, et al. (2021) Intestinal P-gp and putative hepatic OATP1B induction: International Transporter Consortium perspective on drug development implications. Clin Pharmacol Ther 109:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun Z, Jia J, Du T, Zhang N, Tang Y, Fang Y, Fang D (2021) Overview of histone modification. Adv Exp Med Biol 1283:1–16. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Yu L, Qin Z, Chen L, Hu H, Zheng X, Zeng S (2019) Regulation of OCT2 transcriptional repression by histone acetylation in renal cell carcinoma. Epigenetics 14:791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziello JE, Jovin IS, Huang Y (2007) Hypoxia-inducible factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J Biol Med 80:51–60. [PMC free article] [PubMed] [Google Scholar]
- Zolk O, Schnepf R, Muschler M, Fromm MF, Wendler O, Traxdorf M, Iro H, Zenk J (2013) Transporter gene expression in human head and neck squamous cell carcinoma and associated epigenetic regulatory mechanisms. Am J Pathol 182:234–243. [DOI] [PubMed] [Google Scholar]