Abstract
The objective of this study is to conduct a population pharmacokinetic (PK) model-based analysis on 10 warfarin metabolites [4’-, 6-, 7-, 8-, and 10-hydroxylated (OH)-S- and R- warfarin], when warfarin is administered alone or together with either fluconazole or rifampin. One or two compartment PK models expanded from target mediated drug disposition models developed previously for warfarin enantiomers were able to sufficiently characterize the PK profiles of 10 warfarin metabolites in plasma and urine under different conditions. Model-based analysis shows CYP2C9 mediated metabolic elimination pathways are more inhibitable by fluconazole [% formation clearance (CLf) of 6- and 7-OH-S-warfarin decrease: 73.2% and 74.8%] but less inducible by rifampin (% CLf of 6- and 7-OH-S-warfarin increase: 85% and 75%), compared with non-CYP2C9 mediated elimination pathways (% CLf of 10-OH-S-warfarin and renal clearance of S-warfarin decrease in the presence of fluconazole: 65.0% and 15.3%; % CLf of 4’-, 8-, and 10-OH-S-warfarin increase in the presence of rifampin: 260%, 127%, and 355%), which potentially explains the CYP2C9 genotype-dependent drug-drug interactions exhibited by S-warfarin, when warfarin is administrated together with fluconazole or rifampin. Additionally, for subjects with CYP2C9 *2 and *3 variants, a model-based analysis of warfarin metabolite profiles in subjects with various CYP2C9 genotypes demonstrates CYP2C9 mediated elimination is less important and non-CYP2C9 mediated elimination is more important, compared with subjects without these variants. To our knowledge, this is so far one of the most comprehensive population-based PK analyses of warfarin metabolites in subjects with various CYP2C9 genotypes under different comedications.
SIGNIFICANCE STATEMENT
The present study conducted population model-based analyses of 10 warfarin metabolites after racemic warfarin is administered either alone or together with metabolic inhibitors or inducers. The differential inhibition and induction of various elimination pathways of warfarin potentially explains the CYP2C9 genotype-dependent drug-drug interactions of S-warfarin. The analysis also facilitates a deeper understanding of warfarin disposition.
Introduction
Since being introduced into clinical practice in the 1950s, warfarin continues to be one of the most commonly prescribed anticoagulant drugs in the world nowadays (Shapiro, 1953; Asiimwe et al., 2021). Although highly effective in treating diseases such as atrial fibrillation and venous thromboembolism, the narrow therapeutic index, high interindividual variability, and potentially life-threatening and dose-limiting toxicities, such as intracranial hemorrhage, compromise the clinical use of warfarin (Smith et al., 1990; Takahashi and Echizen, 2001; Hart et al., 2007; Ansell et al., 2008).
After the oral administration of warfarin enantiomers, both R- and S-warfarin are absorbed rapidly and eliminated primarily through cytochrome P450 (P450) mediated hepatic metabolism to form multiple monohydroxylated metabolites (Kaminsky and Zhang, 1997; Ufer, 2005). S-warfarin, the pharmacologically more active enantiomer of warfarin, is primarily (>80%) metabolized by CYP2C9 to form either 7- or 6-hydroxy (OH)-S-warfarin, although 4’-, 8-, and 10-OH-S-warfarin can also be formed via catalysis by other P450s such as CYP2C19 and CYP3A4 (Rettie et al., 1992; Ufer, 2005; Pouncey et al., 2018). R-warfarin, in contrast, is metabolized by multiple P450 enzymes, such as CYP1A2, CYP2C19, and CYP3A4, to form 4’-, 6-, 7-, 8-, and 10-OH-R-warfarin (Zhang et al., 1995; Wienkers et al., 1996; Ufer, 2005; Rettie and Tai, 2006; Pouncey et al., 2018). After the formation of monohydroxylated metabolites, several warfarin metabolites, such as 4’-, 6-, 7-, and 8-hydroxy S- or R-warfarin, can undergo either urinary excretion or further metabolism, such as glucuronidation, although the relative importance in vivo of these conjugation pathways is unclear to date (Jansing et al., 1992; Takahashi et al., 1997; Ufer, 2005; Zielinska et al., 2008; Miller et al., 2009; Jones et al., 2010b; Pugh et al., 2014). Diastereomeric warfarin alcohols were also identified in human urine, the formation of which is P450-independent (Lewis and Trager, 1970; Moreland and Hewick, 1975; Hermans and Thijssen, 1989; Ufer, 2005).
Numerous previous studies suggested that CYP2C9 contributes significantly to the high interindividual variability (IIV) in S-warfarin exposure, complicating warfarin dosing (Rettie et al., 1994; Takahashi and Echizen, 2001; Hamberg et al., 2007; Flora et al., 2017; Xue et al., 2017). For enzymes encoded by the CYP2C9 gene with *2 (430C>T) and *3 (1075A>C) variants, in vitro studies suggest the intrinsic clearance (CL) of CYP2C9 mediated S-warfarin 7-hydroxylation is approximately 5.5-fold and 27-fold lower, respectively, compared with enzyme encoded by wild type CYP2C9 gene (Rettie et al., 1994; Haining et al., 1996; Steward et al., 1997; Rettie et al., 1999; Ufer, 2005). A population pharmacokinetic (PK) analysis demonstrated that subjects homozygous with respect to CYP2C9 *2 or *3 exhibit a 72% and 85% reduction in S-warfarin CL, respectively, compared with subjects homozygous for CYP2C9 *1/*1 (Hamberg et al., 2007). Additionally, a previous study showed that the CYP2C9 *1B haplotype (characterized by -3089G_A and -2663delTG) is in linkage disequilibrium with CYP2C19 *2, which can impact the autoinduction of phenytoin (Chaudhry et al., 2010). The presence of CYP2C9 *1B haplotype is also associated with the lower maintenance doses of phenytoin in patients with epilepsy but not associated with warfarin maintenance doses in a Chinese patient population (Chaudhry et al., 2010). However, whether these genetic variants are associated with the PK of warfarin metabolites is largely unclear.
Despite many studies being conducted with warfarin, a comprehensive model-based analysis of the metabolic profiles of the warfarin enantiomers and their metabolites, after the administration of either warfarin alone or together with a P450 inhibitor or inducer and utilizing a population PK modeling approach is still lacking. The study presented here is the second manuscript of a two-part companion series describing warfarin kinetics. In the first analysis, we found that CYP2C9 genotypes are associated with the magnitude of S-warfarin CL changes when warfarin is coadministrated with P450 inhibitors or P450 inducers, so called CYP2C9 genotype-dependent drug-drug interactions (DDIs) (Cheng et al., 2022). However, the mechanism behind it is poorly understood. Taking advantage of the plasma and urine PK profiles collected for 10 warfarin metabolites (4’-, 6-, 7-, 8-, and 10-OH-S- and R-warfarin), the goal of the herein presented work is to conduct a model-based analysis to provide more mechanistic insights behind the CYP2C9 genotype-dependent DDIs exhibited by S-warfarin shown in our companion analysis (Cheng et al., 2022). Interestingly, several unexpected findings were obtained in the current analysis of metabolite PK, such as the effect of CYP2C9 *1B on the magnitude of formation CL (CLf) changes of several metabolites after the administration of warfarin together with rifampin.
Methods
Data
Data used for developing the warfarin metabolite models were collected from an open-label, multiphase and crossover clinical pharmacogenetic study approved by the University of Minnesota’s Institutional Review Board. Details about the study population and study design are provided in the first manuscript of our companion manuscripts (Cheng et al., 2022). Briefly, 29 healthy subjects with CYP2C9 *1/*1 (n = 8), CYP2C9 *1B/*1B (n = 5), CYP2C9 *1/*3 (n = 9), CYP2C9 *2/*3 (n = 3), and CYP2C9 *3/*3 (n = 4) were enrolled in the study with written informed consent.
Each subject underwent three treatment periods. For the first period, each subject was treated with a single 10 mg oral dose of warfarin followed by an 11–15 day sampling phase and a 7-day washout phase before entering the second period. For the second period, subjects were randomized and administered either 400 mg fluconazole or 300 mg rifampin orally once daily for 7 consecutive days to allow the concentration of fluconazole or rifampin to reach steady state. Afterward, a single 10 mg oral dose of warfarin was administered for each subject followed by an 11–15 day sampling phase and a 7-day washout phase. Fluconazole or rifampin was continuously administered on a daily basis during the sampling phase. The design of the third period was the same as the second period with subjects crossing over to the alternative interacting drug.
Blood samples were taken at 2 hours, 6 hours, 24 hours, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 9 days, and 11 days after the dose of warfarin for each subject. For subjects with CYP2C9 *1/*3, an additional blood sample was taken at 13 days, and for subjects with CYP2C9 *2/*3 and *3/*3 genotypes, two more blood samples were taken at 13 days and 15 days, since these subjects were assumed to exhibit a longer warfarin elimination half-life. Urine samples were collected at 24-hour intervals for each subject at 1 day, 4 days, 7 days, and 10 days. Concentrations of 4’-, 6-, 7-, 8-, and 10-OH-S and R-warfarin in both plasma and urine were analyzed by liquid chromatography/mass spectrometry (LC/MS) with the methods described in the previous studies (Miller et al., 2009; Flora et al., 2017). The lower limit of quantifications (LLOQs) for 4’-, 6-, 7-, 8-, and 10-OH-warfarin metabolites were 0.75, 0.4, 0.67, 0.5, and 0.75 ng/ml, respectively (Flora et al., 2017).
PK Model Development
The PK models for S- and R-warfarin metabolites were built subsequently on the basis of the PK models developed for S- and R-warfarin parent compounds (Cheng et al., 2022). After the development of the S- and R-warfarin models with CYP2C9 genotype and drug interaction covariate effects, the Empirical Bayes Estimates of individual PK parameters were exported to the S- and R-warfarin metabolite data sets, respectively, to derive the metabolite PK profiles. The molecular weight difference between parent compounds (308 Da) and metabolites (324 Da) were adjusted during the model fitting process. The covariate effects of CYP2C9 genotypes and cotreatments were parameterized using eqs. 1 and 2 as shown below.
[TVP, typical values of parameters; TVPref, typical values of parameters in reference groups; P_Geno i: CYP2C9 genotype effect on parameters (i = 1, 2, 3, 4, 5 represent CYP2C9 *1/*1. *1B/*1B, *1/*3, *2/*3, *3/*3, respectively); P_TRT: cotreatment effect on parameters (TRT: Flu, fluconazole; Rif, rifampin)]
If an association between CYP2C9 genotypes and P_TRT is identified by visual inspection, CYP2C9 genotype effects were added on P_TRT as a covariate using eq. 3 as shown below.
[P_TRT_Geno i: cotreatment effect on parameters for subjects with genotype i (i = 1, 2, 3, 4, 5 represent CYP2C9 *1/*1. *1B/*1B, *1/*3, *2/*3, *3/*3, respectively)]
A covariate introducing a 3.84 decrease in objective function values with one degree of freedom at an α level of 0.05 is considered to be statistically significant.
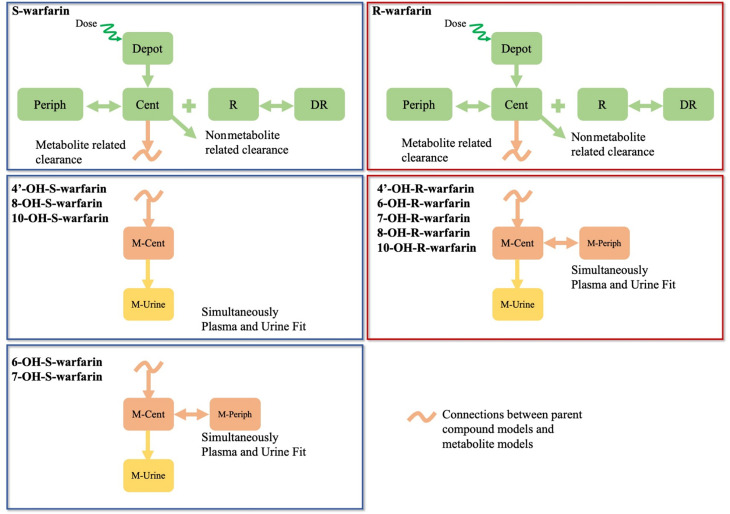
The schematic PK model structures for each warfarin metabolite are shown in Fig. 1. For 4’-, 8-, and 10-OH-S-warfarin, a one-compartment PK model with a urine compartment connected with the S-warfarin PK model with covariates was able to adequately describe the PK profiles for each metabolite in both plasma and urine under different conditions. For 6- and 7-OH-S-warfarin, and all the R-warfarin related metabolites, a two-compartment PK model with a urine compartment connected with either S- or R-warfarin PK model with covariates was able to adequately describe each metabolite’s PK profiles in both plasma and urine under different conditions.
Fig. 1.
PK model structures for warfarin parent and metabolites. Dark blue frames: S-warfarin parent compound and metabolites models; dark red frames: R-warfarin parent compound and metabolites models. Cent, central; DR, drug-receptor complexes; M-Cent, metabolite central compartment; M-periph, metabolite peripheral compartment; M-urine, metabolite urine compartment; Periph, peripheral; R, receptors.
Without studies conducted with each metabolite administrated by itself and prior knowledge about the fraction of parent compound converted to each metabolite, the volume of distribution (Vd) of each metabolite is theoretically not identifiable using only the metabolite plasma PK profiles after the administration of parent compound (Cosson et al., 2007). However, this can potentially be overcome if the plasma and urine PK profiles of each metabolite are fitted simultaneously, assuming the metabolite produced is 100% excreted through urine. The model fitting for each warfarin metabolite performed in this study was based on this assumption with plasma and urine measurements fitted simultaneously. Although this assumption allows the Vd of each metabolite model to become estimable, this may lead to underestimation of metabolite Vd, especially for metabolites that undergo extensive phase II metabolism, such as glucuronidation or sulphation. The underestimation of Vd can further lead to underestimation of CLf of each metabolite. Indeed, several warfarin metabolites, such as 4’-, 6-, 7-, and 8-OH warfarin, have been shown to undergo various extents of glucuronidation, and the overall percentages of warfarin metabolites that undergo glucuronidation vary from 14% to 59% between patients as shown by previous studies (Zielinska et al., 2008; Jones et al., 2010b). Thus, the Vd and CLf estimated in this study should be interpreted as the minimum possible values for each metabolite.
The baseline plasma concentrations for the warfarin only treatment period were assumed to be zero for all the metabolites during the entire modeling process. The baseline plasma concentrations in the second and third treatment periods were assumed to be nonzero for 4’-, 6-, 7-, and 8-OH-S- and 7- and 10-OH-R-warfarin, and parameters for baseline concentrations in the central and peripheral compartments were included for estimations. Additionally, the baseline plasma concentrations in the second and third treatment periods were assumed to be zero for 10-OH-S- and 4’-, 6-, and 8-OH-R-warfarin, since the baseline concentration parameters in the second and third treatment periods cannot be estimated with adequate precision for these metabolites.
IIV terms were parameterized by assuming a log-normal distribution. For each undesired IIV, a fixed 0.3% interindividual variability was assumed to facilitate the optimization efficiency of the expectation-maximization based method (Chigutsa et al., 2017). Proportional error models were used for modeling residual unexplained variabilities (RUVs). All IIVs were assumed to be independent of each other; thus, no off-diagonal matrix elements were estimated. Because of the existence of many plasma concentrations below the lower limit of quantification (LLOQ), the M3 method (Ahn et al., 2008; Bergstrand and Karlsson, 2009) proposed by Stuart Beal was used to account for the missing concentrations in the plasma PK profiles of each metabolite. NONMEM code for 7-OH-S-warfarin has been provided as an example in supplemental material (code for 7-OH-S-warfarin plasma).
Model Evaluations
The evaluations of model fitting were performed by visual prediction check (VPC) stratified by model covariates, such as CYP2C9 genotypes and comedications, with 200 simulations. Because of the existence of plasma concentrations below LLOQ, a two panel VPC procedure was followed to evaluate model fitting in plasma for each metabolite (Bergstrand and Karlsson, 2009). The precision of the parameter estimations was evaluated with the relative standard error (RSE) generated with the covariance steps and 95% confidence intervals (CIs) constructed after the sampling importance resampling (SIR) procedure (Dosne et al., 2016).
Model-Based Analysis
Percentage changes in CLf of each metabolite and percentage changes in renal CL (CLR) of each parent compound after the administration of fluconazole or rifampin were calculated with model parameters estimated by each metabolite model or parent compound model (Cheng et al., 2022) with eq. 4 as shown below.
[| CLf_TRT (or CLR_TRT) –100% | represent the absolute difference between cotreatment effects on CLf of each metabolite (or CLR of each parent compound) and 100% (TRT: Flu, fluconazole; Rif, rifampin)]
The 95% CIs were constructed with RSE estimated from the covariance steps by assuming a symmetrical normal distribution.
For model-based analysis of warfarin and its metabolites, S- and R-warfarin CL, CLR, and the covariate effects on them were extracted from our companion study (Cheng et al., 2022). The S- and R-warfarin CL, CLR, and CLf for each warfarin metabolite in subjects with various CYP2C9 genotypes under different cotreatments were first calculated with the corresponding covariates. The percentages of CLf of each metabolite or CLR of each parent compound with respect to S- or R-warfarin CL, in subjects with various CYP2C9 genotypes under different cotreatments, were then calculated using eq. 5 as shown below.
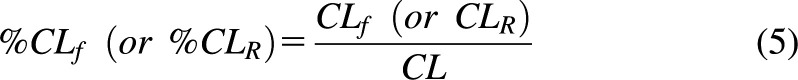 |
Data Analysis
Model fittings were implemented with the Importance Sampling algorithm with interaction using “AUTO=1” option and mu-reference within NONMEM 7.4 (ICON Development Solutions, Ellicott City, Maryland) (Bauer, 2015). Perl-speaks-NONMEM (PsN 4.9.0, Uppsala, Sweden) within Pirana interface (Keizer et al., 2011) was used to facilitate the implementation of VPC and SIR. R 3.6.3 (The R Foundation for Statistical Computing) and RStudio 1.1.453 (RStudio, lnc., Boston, Massachusetts) were used for data pre- and postprocessing and data visualization.
Results
Data
The number of plasma concentrations and urine samples included for developing each metabolite PK model are shown in Supplemental Table 1. The plasma measurements are categorized into baseline concentrations, nonbaseline concentrations above the LLOQ, and nonbaseline concentrations below LLOQ.
Parameters representing baseline plasma concentrations in the second and third treatment periods were initially included for 10-OH-S- and 4’-, 6-, and 10-OH-R-warfarin, but relatively imprecise estimations (large RSE) were observed. Thus, for these compounds, no parameters related to baseline plasma concentrations were included, and baseline plasma concentrations were assumed to be zero in all treatment periods. Additionally, poor fitting of the 10-OH-S-warfarin plasma PK profiles in three subjects was observed with all the structure models tested that resulted in an excessive influence on the overall model fitting and parameter estimations. Thus, the 10-OH-S-warfarin PK profiles for these three subjects were subsequently excluded during model fitting.
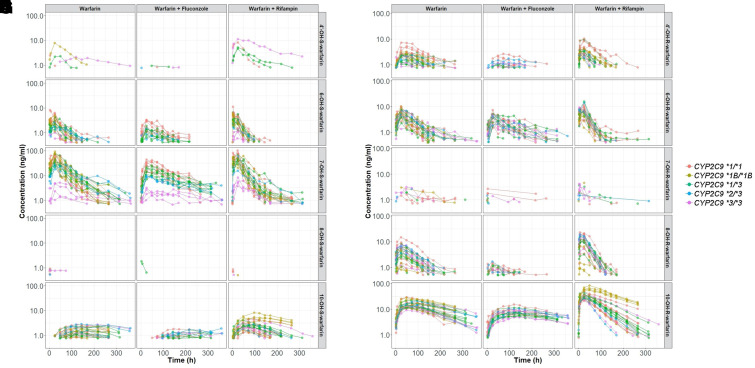
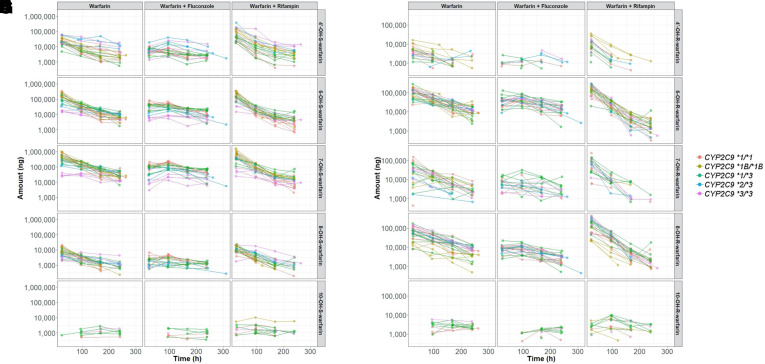
The plasma and urine PK profiles used for developing each metabolite model are shown in Figs. 2 and 3, respectively. In general, plasma concentrations were readily detectable and substantial for most of the warfarin metabolites except for 4’- and 8-OH-S- and 7-OH-R-warfarin. The large number of plasma concentrations below the LLOQ indicate an extremely low plasma exposure of these metabolites (Supplemental Table 1). In contrast, 4’- and 8-OH-S- and 7-OH-R-warfarin were readily detected in urine, suggesting that they are rapidly eliminated after their enzymatic generation. Additionally, despite substantial concentrations of 10-OH-S- and 10-OH-R-warfarin in plasma, these metabolites were only sparsely detected in urine.
Fig. 2.
Plasma PK profiles for S-warfarin metabolites [left, 4’-(A), 6-(C), 7-(E), 8-(G), and 10-(I) OH-S-warfarin] and R-warfarin metabolites [right, 4’-(B), 6-(D), 7-(F), 8-(H), and 10-(J) OH-R-warfarin]. Colors represent subjects’ CYP2C9 genotypes. Plots are on log scales.
Fig. 3.
Urine PK profiles for S-warfarin metabolites [left, 4’-(A), 6-(C), 7-(E), 8-(G), and 10-(I) OH-S-warfarin] and R-warfarin metabolites (right, 4’-(B), 6-(D), 7-(F), 8-(H), and 10-(J) OH-R-warfarin]. Colors represent subjects’ CYP2C9 genotypes. Plots are on log scales.
Model Parameters
The parameter estimations for 4’-, 6-, 7-, 8-, and 10-OH-S- and R-warfarin are shown in Supplemental Table 2–11, respectively. Model parameters were estimated with reasonable precision as indicated by the RSE estimated from the covariance steps and 95% CIs generated after the SIR procedure. The inhibitory and inducing effects of fluconazole and rifampin, respectively, on the CLf of the warfarin metabolites were variable and metabolite dependent. As would be expected, the CYP2C9 genotype impacted the CLf of 6- and 7-OH-S-warfarin, but, surprisingly, also 8-OH-S-warfarin.
For metabolites with sparse plasma PK profiles, such as 4’-OH-S-, 7-OH-R-, and 8-OH-S-warfarin, large RUV estimations (128%, 2640%, and 352%, respectively) for the plasma PK profile fits were observed (Supplemental Table 2, 7, and 8). In addition, for these metabolites, several excessively large IIV estimations for CL and central volume of distribution (VC) were also noted. It is suspected that the existence of the substantial number of plasma concentrations below the LLOQ contributed to the excessive amount of uncertainty (either IIV or RUV) estimated.
Model Evaluations
Again, the model parameters were estimated with reasonable precision based on the RSEs generated with the covariance steps and the 95% CIs calculated with SIR overall (Supplemental Table 2–11). The visual prediction checks (VPCs), which are stratified by CYP2C9 genotypes and comedications, suggested the developed models can predict the PK observations in both plasma and urine for each metabolite reasonably well (Supplemental Figure 1–10, A and C). Importantly, the observed fractions of concentrations below quantification limits in plasma PK profiles align well with the model predicted fractions of concentrations below quantification limits (Supplemental Figure 1–10, B), which indicate the appropriate use of the M3 method.
Model-Based Analysis
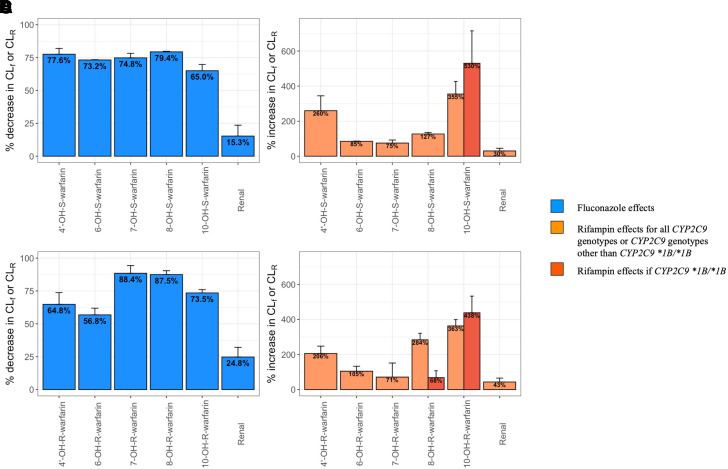
The estimated inhibitory effect of fluconazole and inducing effect of rifampin on the CLf of each metabolite or on the CLR of each parent compound are presented in Fig. 4. After the administration of fluconazole, the CLf of each metabolite and the CLR of each parent enantiomer decrease. Notably, the decrease in CLR is much less compared with the decrease in CLf of each metabolite after the administration of fluconazole. In contrast, after the administration of rifampin, the CLf of each metabolite and the CLR of each parent compound increase, with a much lesser increase in CLR compared with the increase in CLf of each metabolite. Interestingly, our model identified that subjects with the CYP2C9 *1B/*1B genotype exhibit a greater induction of CLf (higher CLf_Rif) for 10-OH-S- and 10-OH-R-warfarin but a lesser effect on CLf (lower CLf_Rif) for 8-OH-R-warfarin after the administration of rifampin (Fig. 4, B and D).
Fig. 4.
Percentage changes in CLf or CLR of S-warfarin metabolites (A and B) and R-warfarin metabolites (C and D) after the administration of fluconazole (A and C) or rifampin (B and D). Colors represent the fluconazole and rifampin introduced inhibition and induction, respectively, as shown in the figure legend. Error bars represent 95% confidence intervals constructed with relative standard error (RSE) estimated from covariance steps assuming a symmetrical normal distribution.
7- and 6-OH-S-warfarin are the most abundant metabolites of S-warfarin, the formation of which is mediated by CYP2C9 (Table 1). After the administration of fluconazole, the percentage decrease in CLf of 7- and 6-OH-S-warfarin was similar to the decrease in CLf of 4’- and 8-OH-S-warfarin (Fig. 4A). However, fluconazole had a lesser impact on 10-OH-S-warfarin CLf and a minimal impact on S-warfarin CLR (Fig. 4A). Rifampin administration, however, increased the CLf of 7- and 6-OH-S-warfarin to a lesser extent than any of the other S-warfarin metabolites (Fig. 4B).
TABLE 1.
Summary of the major P450 enzymes involved in the formation of each metabolite
The information of P450s is based on reference Ufer (2005) and Pouncey et al. (2018).
| S-Warfarin Metabolites | P450s | R-Warfarin Metabolites | P450s |
|---|---|---|---|
| 4’-OH-S-warfarin | 2C19, 3A4, 2C8 | 4’-OH-R-warfarin | 2C19, 3A4, 2C8 |
| 6-OH-S-warfarin | 2C9 (major), 2C19 (minor) | 6-OH-R-warfarin | 1A2 (major), 2C19 (minor) |
| 7-OH-S-warfarin | 2C9 (major), 2C19 (minor) | 7-OH-R-warfarin | 2C19, 1A2, 2C8 |
| 8-OH-S-warfarin | 2C19 | 8-OH-R-warfarin | 2C19 (major), 1A2 (minor) |
| 10-OH-S-warfarin | 3A4 | 10-OH-R-warfarin | 3A4 |
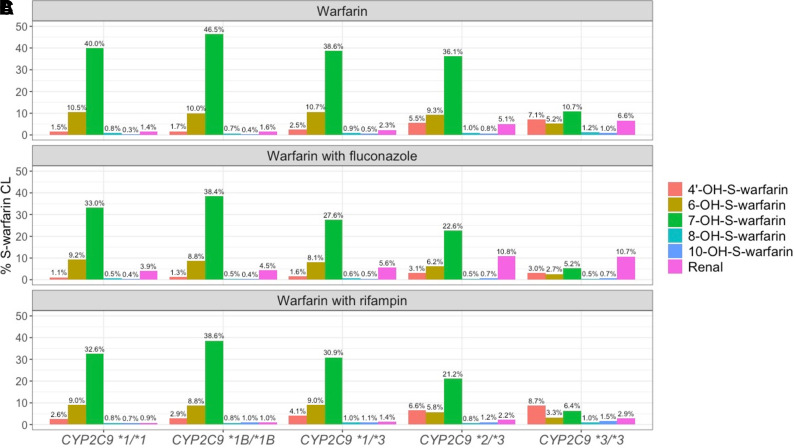
The metabolic profiles for S-warfarin (the percentages of CLf of each metabolite or CLR of each parent compound with respect to S-warfarin CL) in subjects with various CYP2C9 genotypes under different cotreatments are shown in Fig. 5. For subjects with the CYP2C9 *2 and *3 variants, the proportion of CYP2C9 mediated S-warfarin CL occurring through the formation of 6- and 7-OH-S-warfarin decreases, but the proportion of non-CYP2C9 mediated warfarin clearance increases. Interestingly, the CLR of S-warfarin contributes more toward S-warfarin CL in subjects possessing the CYP2C9 *2 and *3 variants compared with wild-type subjects (CYP2C9 *1/*1), regardless of comedications administered. Despite prior suggestions that the CLR of S-warfarin is generally inconsequential (Ufer, 2005), our results suggest that for subjects with the CYP2C9 *2/*3 and *3/*3 variants, the CLR of S-warfarin can constitute up to 5.1% and 6.6% of overall S-warfarin CL, respectively, when warfarin is administered alone. Furthermore, the CLR of S-warfarin can constitute up to 10.8% and 10.7% of overall S-warfarin CL when warfarin is administered together with fluconazole.
Fig. 5.
S-warfarin metabolic profiles in subjects with different CYP2C9 genotypes after the administration of warfarin alone (A) and warfarin together with fluconazole (B) or rifampin (C). Color represents different elimination pathway as shown in the figure legend.
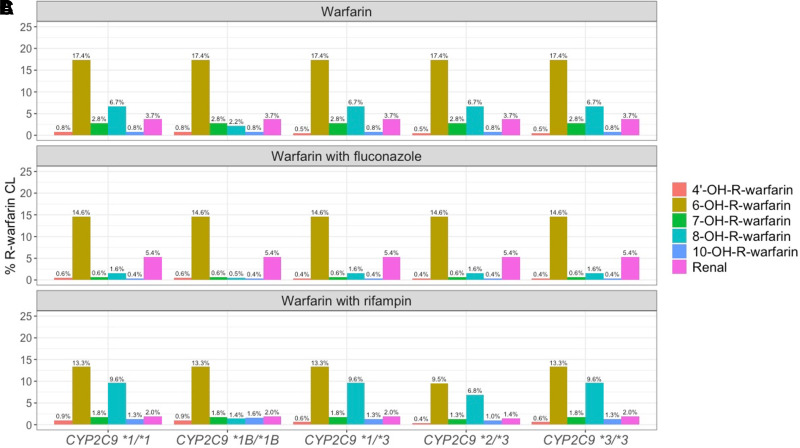
The percentages of CLf for each metabolite or CLR for each parent enantiomer with respect to R-warfarin CL in subjects with various CYP2C9 genotypes under different cotreatments are shown in Fig. 6. Unlike S-warfarin, no obvious CYP2C9 genotype-dependency is detected in the R-warfarin metabolic profiles. After the administration of fluconazole, CLR contributes more and CLf through R-warfarin metabolites contributes less toward overall CL of R-warfarin. After the coadministration of rifampin, CLR of R-warfarin contributes less toward overall CL. Interestingly, the formation of 4’-,8-, and 10-OH-R-warfarin play a more prominent role in R-warfarin elimination after the coadministration of rifampin compared with the administration of warfarin alone. In contrast, the formation of 6- and 7-OH-R-warfarin metabolites appears to play a lesser role in R-warfarin elimination after coadministration of rifampin.
Fig. 6.
R-warfarin metabolic profiles in subjects with different CYP2C9 genotypes after the administration of warfarin alone (A) and warfarin together with fluconazole (B) or rifampin (C). Color represents different elimination pathway as shown in the figure legend.
Discussion
In this study, we developed PK models for 10 warfarin metabolites on the basis of the S-and R-warfarin PK models built in our companion study (Cheng et al., 2022). Our models were able to characterize the plasma and urine PK profiles of 10 warfarin metabolites reasonably well with reasonable parameter estimation precisions. Importantly, the developed warfarin metabolite models can provide more mechanistic insights about warfarin dispositions after warfarin administered either alone or together with fluconazole or rifampin.
Many previous studies show 7- and 6-OH-S warfarin are the major metabolites for S-warfarin, the formation of which are mediated by CYP2C9 (Rettie et al., 1992; Kaminsky and Zhang, 1997; Ufer, 2005). As expected, our model-based analysis confirmed previous findings by showing 7- and 6-OH-S-warfarin are the most predominant metabolites for S-warfarin, the CLf of which is impacted by CYP2C9 (Fig. 5). Interestingly, our model detects a CYP2C9 genotype-dependent elimination clearance for 10-OH-S-warfarin. Subjects with more defective CYP2C9 exhibit a higher elimination clearance for 10-OH-S-warfarin. This potentially suggests further P450-dependent metabolism or conjugation, such as glucuronidation, may be involved in the elimination of 10-OH-S-warfarin. Indeed, the involvement of subsequent metabolism steps may also explain the relatively low urine level of 10-OH-S-warfarin (Fig. 3I). Interestingly, a previous study also suggests the exposure level of 10-OH-S-warfarin is related to CYP2C9 genotypes (Pouncey et al., 2018). However, this study concluded that subjects who are CYP2C9 extensive metabolizers may exhibit a lower concentration of 10-OH-S-warfarin, which is paradoxical with our analysis. Given that many studies show the formation of 10-OH-S-warfarin is mediated by CYP3A4 (Kaminsky and Zhang, 1997; Ngui et al., 2001; Pouncey et al., 2018), the impact of CYP2C9 on the elimination rather than the formation of 10-OH-S-warfarin demonstrated by our analysis provides additional insight about its disposition that warrants further investigation. The formation of 10-OH-R-warfarin is mediated by CYP3A4 as well (Ufer, 2005; Pouncey et al., 2018). The plasma concentration of 10-OH-R-warfarin appears to be the highest among the R-warfarin metabolites (Fig. 2), but its PK have heretofore not been extensively assessed. Here, our model identified a relatively low CLf and Vd for 10-OH-R-warfarin (Supplemental Table 11). The noncompartmental analysis reported in our previous study demonstrated that 10-OH-R-warfarin also has an uncommonly long terminal half-life (>100 hours) and exhibited elimination rate-limited kinetics (Flora et al., 2017). In addition, the urinary excretion of 10-OH-R-warfarin appears to be very limited (Fig. 3J). The mechanism behind the unusual PK behavior of 10-OH-R warfarin is unknown. However, as indicated in our previous study, we suspect that 10-OH-R-warfarin may undergo enterohepatic recirculation, fecal elimination, and sequential metabolism to another metabolite (Flora et al., 2017). Interestingly, a previous study has reported 10-OH-metabolites of warfarin can potently inhibit the S-warfarin CYP2C9 metabolic clearance in a competitive manner (Jones et al., 2010a). Thus, more studies focused on the disposition of 10-OH-warfarin may be valuable for better understanding of the interplay between warfarin metabolites and the disposition of S-warfarin, the pharmacologically more active component in warfarin.
Our model also discovered that subjects with the CYP2C9 *1B/*1B genotype exhibit a lower 8-OH-R-warfarin CLf compared with subjects with other CYP2C9 genotypes. Interestingly, a strong linkage disequilibrium between CYP2C9*1B and CYP2C19 *2 was previously reported (Chaudhry et al., 2010). In addition, CYP2C19 *2 is known to be the most common mutation encoding a defective CYP2C19 protein (de Morais et al., 1994; Dehbozorgi et al., 2018). Since the formation of 8-OH-R-warfarin is mediated mainly by CYP2C19 (Pouncey et al., 2018), the higher probability of the existence of the CYP2C19 *2 genotype in subjects with the CYP2C9 *1B/*1B genotype may explain the lower 8-OH-R-warfarin CLf in these subjects. Our companion study also suggests that the increase in S- and R-warfarin CL after the coadministration of rifampin is similar between subjects with the CYP2C9 *1/*1 and CYP2C9 *1B/*1B (Cheng et al., 2022). However, the current study demonstrates that subjects with the CYP2C9 *1B/*1B exhibit a more pronounced induction of the CLf for CYP3A4-mediated 10-hydroxylation of both S- and R-warfarin (Fig. 4, B and D; Table 1), but a less inducible CLf for CYP2C19-mediated 8-hydroxylation of R-warfarin (Fig. 4B; Table 1). Interestingly, Chaudhry et al. (2010) also demonstrated that subjects with the CYP2C9 *1B variant tend to exhibit a greater induction of CYP3A4 but a lesser induction of CYP2C19, although both associations were not statistically significant. The mechanism for the altered inducibility of CYP3A4 and CYP2C19 is speculated to be attributable to binding element changes in transcription factors, such as Yin Yang 1, introduced by CYP2C9 *1B (Chaudhry et al., 2010).
In our companion study, we successfully used a target mediated drug disposition model to characterize warfarin PK profiles in both plasma and urine when warfarin is administered either alone or together with fluconazole or rifampin. Our model-based analysis suggested the changes in S-warfarin CL after the administration of warfarin, together with fluconazole or rifampin, follow a CYP2C9 genotype-dependent manner (Cheng et al., 2022). As such, the study presented herein provides more mechanistic insights behind the CYP2C9 genotype-dependent DDIs exhibited by S-warfarin. Specifically, after the administration of fluconazole, the inhibitory effects on the CLf of CYP2C9 related metabolites, such as 7-and 6-OH-S-warfarin, are higher than some of the non-CYP2C9 related metabolites, such as 10-OH-S-warfarin, and are much higher than the CLR for S-warfarin (Fig. 4A). Since for subjects possessing the CYP2C9 *2 and *3 variants, CLf of CYP2C9 related metabolites represents a smaller proportion of the overall S-warfarin CL, compared with subjects with CYP2C9 *1/*1 (Fig. 5), a higher inhibitability of CYP2C9-mediated CLf compared with other elimination pathways makes the S-warfarin CL in subjects with CYP2C9 *2 and *3 variants less susceptible to fluconazole inhibition. In contrast, after the administration of rifampin, the inducing effects on CLf for the CYP2C9 related metabolites are less than the non-CYP2C9 related metabolites (Fig. 4B). Since for subjects with CYP2C9 *2 and *3 variants, CLf of non-CYP2C9 related metabolites represents a larger proportion of the overall S-warfarin CL compared with subjects with CYP2C9 *1/*1 (Fig. 5), the greater induction of non-CYP2C9 mediated CL makes subjects with the CYP2C9 *2 and *3 variants subject to higher CL changes after the coadministration of rifampin.
The present study confirmed some previous findings. For example, Heimark et al. (1987) found that in the presence of rifampin cotreatment, the CL of warfarin enantiomers and the CLf of 6-, 7-, and 8-OH metabolites increase. Heimark et al. (1987) also found that the percentages of R-warfarin dose excreted in the urine decrease as 6- and 7-OH metabolites, but increase as 4’- and 8-OH metabolites. Additionally, after the coadministration of fluconazole, (Black et al., 1996) found that the CL of warfarin enantiomers and the CLf of 6-, 7-, 8-, and 10-OH metabolites decrease. Fluconazole inhibits the formation of 6-, 7-, and 8-OH S-warfarin to a similar extent, by around 70% (Black et al., 1996). These findings are consistent with the analysis presented in this study (Figs. 4 and 6).
Limitations to our models are recognized. Firstly, for metabolites such as 4’-OH-S-, 7-OH-R-, and 8-OH-S-warfarin, the model predicted plasma PK profiles included a high proportion of concentrations below 0 (Supplemental Figure 1, 6, and 7, A). This occurred because the estimated RUVs for these metabolites are excessively large (>100%). In addition, several IIV terms in PK models were also uncommonly large (Supplemental Table 2, 7, and 8). We suspect that a significant proportion of the concentrations below LLOQ for these metabolites (Supplemental Table 1) leads to substantial inflation in the estimations of IIVs and RUVs. However, given the sufficient urine measurements collected, the model parameters were still estimated with reasonable precision (Supplemental Table 2, 7, and 8). In the future, more advanced analytical techniques with higher sensitivity for detection of these metabolites will be needed to better characterize their plasma PK profiles. Additionally, some systemic bias in model predictions was noticed. For example, the developed model for 4’-OH-S-warfarin appears to underpredict the urinary PK profiles in subjects with CYP2C9 *1B/*1B and inadequately characterize the urinary PK profiles in subjects with CYP2C9 *2/*3 when warfarin is administered together with fluconazole (Supplemental Figure 1C).
Secondly, without the administration of each metabolite alone or any prior knowledge about the fraction of warfarin enantiomers metabolized to a particular metabolite, the Vd of each metabolite is theoretically not estimable based on metabolite plasma PK profiles alone when only the parent compound is administered (Cosson et al., 2007). A common approach to address this is to either fix the fraction of metabolism based on literature (Vanobberghen et al., 2016; Mian et al., 2019) or to assume a metabolite Vd (for instance, equal to Vd of parent compound) (Patel et al., 2017). However, the Vd of each metabolite becomes identifiable if the metabolite plasma and urine PK profiles are fitted simultaneously and if all the metabolites generated are excreted in the urine and only renally eliminated. In fact, in vitro studies conducted by Zielinska et al. (2008) suggest that 4’-, 6-, 7-, and 8-hydroxywarfarin can undergo glucuronidation mediated by multiple UDP-glucuronosyltransferases (UGTs), such as UGT1A1 and UGT1A10. Human studies also suggest the overall percentages of glucuronidation of monohydroxylated warfarin metabolites can vary between 14%–59% (Miller et al., 2009; Jones et al., 2010b). This in vitro and in vivo evidence of phase II metabolism suggests that the metabolite PK models presented may potentially underestimate the actual Vd and CLf of each metabolite. Thus, the CLf estimated in our study should be considered as the minimum possible CLf for each metabolite. Nevertheless, despite the limitations discussed above, our collective analyses presented in Figs. 5 and 6 can still reasonably reflect the warfarin metabolic profiles in subject with various CYP2C9 under different cotreatments.
To our knowledge, our model-analysis of warfarin parent enantiomers and metabolites is the most comprehensive population PK analysis of warfarin disposition in subjects with different CYP2C9 to date. This study involved the development of population PK models for 10 warfarin metabolites and conducted a model-based analysis of the warfarin metabolic profiles. The differential effects of fluconazole inhibition and rifampin induction on the CLf of warfarin metabolites across CYP2C9 genotypes largely explains the CYP2C9 genotype-dependent effects on the warfarin enantiomers observed in our companion study (Cheng et al., 2022). In addition, the differential inhibitability and inducibility of several P450 enzymes elucidated by our study of warfarin metabolites is potentially useful in informing the predicted extent of DDIs of other drugs eliminated by similar metabolic pathways.
In conclusion, we developed population PK models for 10 warfarin metabolites and performed comprehensive model-based analysis. The model-based analysis provides mechanistic insights behind the CYP2C9 genotype-dependent DDIs exhibited by S-warfarin, as suggested in our companion study. In addition, our analysis provides a better understanding of warfarin disposition for subjects with various CYP2C9 genotypes after the administration of both inhibitor and inducer comedications.
Author Contributions
Participated in research design: Cheng, Flora, Rettie, Brundage, Tracy.
Conducted experiments: Flora, Rettie, Tracy.
Performed data analysis: Cheng, Brundage.
Wrote or contributed to the writing of the manuscript: Cheng, Flora, Rettie, Brundage, Tracy.
Abbreviations
- CI
confidence interval
- CL
clearance
- CLf
formation clearance
- CLR
renal clearance
- DDI
drug-drug interaction
- IIV
interindividual variability
- LLOQ
lower limit of quantification
- P450
cytochrome P450
- PK
pharmacokinetic
- RSE
relative standard error
- RUV
residual unexplained variability
- SIR
sampling importance resampling
- Vd
volume of distribution
- VPC
visual predictive check
Footnotes
This manuscript was funded by National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM069753] (T.S.T.) and [Grant P01-GM032165] (T.S.T.).
Current affiliation: Metrum Research Group, Tariffville, Connecticut.
Current affiliation: GRYT Health Inc., Rochester, New York.
Current affiliation: Tracy Consultants, Huntsville, Alabama.
This work is part of the Ph.D. thesis of S.C.
Citation of meeting abstracts: Cheng S., Flora D.R., Tracy T. S., Rettie A.E., Brundage R.C. Genotype-Dependent Changes in Warfarin Clearance upon Co-administration of an Inhibitor (fluconazole) and an inducer (rifampin): A Model-based Analysis. American Conference of Pharmacometrics (ACOP) 11
The authors declare no conflict of interest.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ahn JE, Karlsson MO, Dunne A, Ludden TM (2008) Likelihood based approaches to handling data below the quantification limit using NONMEM VI. J Pharmacokinet Pharmacodyn 35:401–421. [DOI] [PubMed] [Google Scholar]
- Ansell J, Hirsh J, Hylek E, Jacobson A, Crowther M, Palareti G (2008) Pharmacology and management of the vitamin K antagonists: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition). Chest 133:160S–198S. [DOI] [PubMed] [Google Scholar]
- Asiimwe IG, Zhang EJ, Osanlou R, Jorgensen AL, Pirmohamed M (2021) Warfarin dosing algorithms: a systematic review. Br J Clin Pharmacol 87:1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer RJ (2015) NONMEM Users Guide: Introduction to NONMEM 7.3.0, Gaithersburg, Maryland. [Google Scholar]
- Bergstrand M, Karlsson MO (2009) Handling data below the limit of quantification in mixed effect models. AAPS J 11:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DJ, Kunze KL, Wienkers LC, Gidal BE, Seaton TL, McDonnell ND, Evans JS, Bauwens JE, Trager WF (1996) Warfarin-fluconazole. II. A metabolically based drug interaction: in vivo studies. Drug Metab Dispos 24:422–428. [PubMed] [Google Scholar]
- Chaudhry ASUrban TJLamba JKBirnbaum AKRemmel RPSubramanian MStrom SYou JHKasperaviciute DCatarino CB, et al. (2010) CYP2C9*1B promoter polymorphisms, in linkage with CYP2C19*2, affect phenytoin autoinduction of clearance and maintenance dose. J Pharmacol Exp Ther 332:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Flora DR, Rettie AE, Brundage RC, Tracy TS (2022) Pharmacokinetic modeling of warfarin I – model-based analysis of warfarin enantiomers with a target mediated drug disposition model reveals CYP2C9 genotype-dependent drug-drug interactions of S-warfarin. Drug Metab Dispos DOI: 10.1124/dmd.122.000876 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigutsa E, Long AJ, Wallin JE (2017) Exposure-response analysis of necitumumab efficacy in squamous non-small cell lung cancer patients. CPT Pharmacometrics Syst Pharmacol 6:560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson V, Jorga K, Fuseau E (2007) Modeling of metabolite pharmacokinetics in a large pharmacokinetic data set: an application, in Pharmacometrics: The Science of Quantitative Pharmacology (Ette EI, Williams PJ, eds) pp 1107–1136, Wiley, Hoboken. [Google Scholar]
- de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA (1994) The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem 269:15419–15422. [PubMed] [Google Scholar]
- Dehbozorgi M, Kamalidehghan B, Hosseini I, Dehghanfard Z, Sangtarash MH, Firoozi M, Ahmadipour F, Meng GY, Houshmand M (2018) Prevalence of the CYP2C19*2 (681 G>A), *3 (636 G>A) and *17 (-806 C>T) alleles among an Iranian population of different ethnicities. Mol Med Rep 17:4195–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosne AG, Bergstrand M, Harling K, Karlsson MO (2016) Improving the estimation of parameter uncertainty distributions in nonlinear mixed effects models using sampling importance resampling. J Pharmacokinet Pharmacodyn 43:583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora DR, Rettie AE, Brundage RC, Tracy TS (2017) CYP2C9 genotype-dependent warfarin pharmacokinetics: impact of CYP2C9 genotype on R- and S-warfarin and their oxidative metabolites. J Clin Pharmacol 57:382–393. [DOI] [PubMed] [Google Scholar]
- Haining RL, Hunter AP, Veronese ME, Trager WF, Rettie AE (1996) Allelic variants of human cytochrome P450 2C9: baculovirus-mediated expression, purification, structural characterization, substrate stereoselectivity, and prochiral selectivity of the wild-type and I359L mutant forms. Arch Biochem Biophys 333:447–458. [DOI] [PubMed] [Google Scholar]
- Hamberg AK, Dahl ML, Barban M, Scordo MG, Wadelius M, Pengo V, Padrini R, Jonsson EN (2007) A PK-PD model for predicting the impact of age, CYP2C9, and VKORC1 genotype on individualization of warfarin therapy. Clin Pharmacol Ther 81:529–538. [DOI] [PubMed] [Google Scholar]
- Hart RG, Pearce LA, Aguilar MI (2007) Meta-analysis: antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann Intern Med 146:857–867. [DOI] [PubMed] [Google Scholar]
- Heimark LD, Gibaldi M, Trager WF, O’Reilly RA, Goulart DA (1987) The mechanism of the warfarin-rifampin drug interaction in humans. Clin Pharmacol Ther 42:388–394. [DOI] [PubMed] [Google Scholar]
- Hermans JJ, Thijssen HH (1989) The in vitro ketone reduction of warfarin and analogues. Substrate stereoselectivity, product stereoselectivity and species differences. Biochem Pharmacol 38:3365–3370. [DOI] [PubMed] [Google Scholar]
- Jansing RL, Chao ES, Kaminsky LS (1992) Phase II metabolism of warfarin in primary culture of adult rat hepatocytes. Mol Pharmacol 41:209–215. [PubMed] [Google Scholar]
- Jones DR, Kim SY, Guderyon M, Yun CH, Moran JH, Miller GP (2010a) Hydroxywarfarin metabolites potently inhibit CYP2C9 metabolism of S-warfarin. Chem Res Toxicol 23:939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DR, Moran JH, Miller GP (2010b) Warfarin and UDP-glucuronosyltransferases: writing a new chapter of metabolism. Drug Metab Rev 42:55–61. [DOI] [PubMed] [Google Scholar]
- Kaminsky LS, Zhang ZY (1997) Human P450 metabolism of warfarin. Pharmacol Ther 73:67–74. [DOI] [PubMed] [Google Scholar]
- Keizer RJ, van Benten M, Beijnen JH, Schellens JH, Huitema AD (2011) Piraña and PCluster: a modeling environment and cluster infrastructure for NONMEM. Comput Methods Programs Biomed 101:72–79. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Trager WF (1970) Warfarin metabolism in man: identification of metabolites in urine. J Clin Invest 49:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian P, Valkenburg AJ, Allegaert K, Koch BCP, Breatnach CV, Knibbe CAJ, Tibboel D, Krekels EHJ (2019) Population pharmacokinetic modeling of acetaminophen and metabolites in children after cardiac surgery with cardiopulmonary bypass. J Clin Pharmacol 59:847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GP, Jones DR, Sullivan SZ, Mazur A, Owen SN, Mitchell NC, Radominska-Pandya A, Moran JH (2009) Assessing cytochrome P450 and UDP-glucuronosyltransferase contributions to warfarin metabolism in humans. Chem Res Toxicol 22:1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland TA, Hewick DS (1975) Studies on a ketone reductase in human and rat liver and kidney soluble fraction using warfarin as a substrate. Biochem Pharmacol 24:1953–1957. [DOI] [PubMed] [Google Scholar]
- Ngui JS, Chen Q, Shou M, Wang RW, Stearns RA, Baillie TA, Tang W (2001) In vitro stimulation of warfarin metabolism by quinidine: increases in the formation of 4′- and 10-hydroxywarfarin. Drug Metab Dispos 29:877–886. [PubMed] [Google Scholar]
- Patel YT, Daryani VM, Patel P, Zhou D, Fangusaro J, Carlile DJ, Martin PD, Aarons L, Stewart CF (2017) Population pharmacokinetics of selumetinib and its metabolite N-desmethyl-selumetinib in adult patients with advanced solid tumors and children with low-grade gliomas. CPT Pharmacometrics Syst Pharmacol 6:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouncey DL, Hartman JH, Moore PC, Dillinger DJ, Dickerson KW, Sappington DR, Smith ES 3rd, Boysen G, Miller GP (2018) Novel isomeric metabolite profiles correlate with warfarin metabolism phenotype during maintenance dosing in a pilot study of 29 patients. Blood Coagul Fibrinolysis 29:602–612. [DOI] [PubMed] [Google Scholar]
- Pugh CP, Pouncey DL, Hartman JH, Nshimiyimana R, Desrochers LP, Goodwin TE, Boysen G, Miller GP (2014) Multiple UDP-glucuronosyltransferases in human liver microsomes glucuronidate both R- and S-7-hydroxywarfarin into two metabolites. Arch Biochem Biophys 564:244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettie AE, Haining RL, Bajpai M, Levy RH (1999) A common genetic basis for idiosyncratic toxicity of warfarin and phenytoin. Epilepsy Res 35:253–255. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Korzekwa KR, Kunze KL, Lawrence RF, Eddy AC, Aoyama T, Gelboin HV, Gonzalez FJ, Trager WF (1992) Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol 5:54–59. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Tai G (2006) The pharmocogenomics of warfarin: closing in on personalized medicine. Mol Interv 6:223–227. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR (1994) Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 4:39–42. [DOI] [PubMed] [Google Scholar]
- Shapiro S (1953) Warfarin sodium derivative: (coumadin sodium); an intravenous hypoprothrombinemia-inducing agent. Angiology 4:380–390. [DOI] [PubMed] [Google Scholar]
- Smith P, Arnesen H, Holme I (1990) The effect of warfarin on mortality and reinfarction after myocardial infarction. N Engl J Med 323:147–152. [DOI] [PubMed] [Google Scholar]
- Steward DJ, Haining RL, Henne KR, Davis G, Rushmore TH, Trager WF, Rettie AE (1997) Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics 7:361–367. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Echizen H (2001) Pharmacogenetics of warfarin elimination and its clinical implications. Clin Pharmacokinet 40:587–603. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kashima T, Kimura S, Muramoto N, Nakahata H, Kubo S, Shimoyama Y, Kajiwara M, Echizen H (1997) Determination of unbound warfarin enantiomers in human plasma and 7-hydroxywarfarin in human urine by chiral stationary-phase liquid chromatography with ultraviolet or fluorescence and on-line circular dichroism detection. J Chromatogr B Biomed Sci Appl 701:71–80. [DOI] [PubMed] [Google Scholar]
- Ufer M (2005) Comparative pharmacokinetics of vitamin K antagonists: warfarin, phenprocoumon and acenocoumarol. Clin Pharmacokinet 44:1227–1246. [DOI] [PubMed] [Google Scholar]
- Vanobberghen F, Penny MA, Duthaler U, Odermatt P, Sayasone S, Keiser J, Tarning J (2016) Population pharmacokinetic modeling of tribendimidine metabolites in opisthorchis viverrini-infected adults. Antimicrob Agents Chemother 60:5695–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienkers LC, Wurden CJ, Storch E, Kunze KL, Rettie AE, Trager WF (1996) Formation of (R)-8-hydroxywarfarin in human liver microsomes. A new metabolic marker for the (S)-mephenytoin hydroxylase, P4502C19. Drug Metab Dispos 24:610–614. [PubMed] [Google Scholar]
- Xue LHolford NDing XLShen ZYHuang CRZhang HZhang JJGuo ZNXie CZhou L, et al. (2017) Theory-based pharmacokinetics and pharmacodynamics of S- and R-warfarin and effects on international normalized ratio: influence of body size, composition and genotype in cardiac surgery patients. Br J Clin Pharmacol 83:823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Fasco MJ, Huang Z, Guengerich FP, Kaminsky LS (1995) Human cytochromes P4501A1 and P4501A2: R-warfarin metabolism as a probe. Drug Metab Dispos 23:1339–1346. [PubMed] [Google Scholar]
- Zielinska A, Lichti CF, Bratton S, Mitchell NC, Gallus-Zawada A, Le VH, Finel M, Miller GP, Radominska-Pandya A, Moran JH (2008) Glucuronidation of monohydroxylated warfarin metabolites by human liver microsomes and human recombinant UDP-glucuronosyltransferases. J Pharmacol Exp Ther 324:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]