Abstract
The nonallosteric and allosteric l-lactate dehydrogenases of Lactobacillus pentosus and L. casei, respectively, exhibited broad substrate specificities, giving virtually the same maximal reaction velocity and substrate Km values for pyruvate and oxaloacetate. Replacement of Pro101 with Asn reduced the activity of the L. pentosus enzyme toward these alternative substrates to a greater extent than the activity toward pyruvate.
l-Lactate dehydrogenase (l-LDH; EC 1.1.1.27) and l-malate dehydrogenase (l-MDH; EC 1.1.1.37) are similar with respect to both protein structure and catalytic machinery, and they catalyze oxidation-reduction of common 2-ketoacids and l-2-hydroxyacids with NAD as the coenzyme (1, 19, 26). Nevertheless, unless artificially modified, most l-LDHs and l-MDHs strictly discriminate their own substrates, pyruvate (l-lactate) and oxaloacetate (l-malate), respectively (15), although a duck ɛ-crystallin (37) and the Bifidobacterium longum (11) l-LDHs are known to exhibit relatively high l-MDH activities that are only 25- and 44-fold, respectively, lower than their own l-LDH activities.
In lactic acid bacteria such as lactobacilli, l-LDHs show a great variety of catalytic properties and play key roles in the fermentation of lactic acid, acting in the last step of the anaerobic glycolysis pathway by converting pyruvate and NADH to l-lactate and NAD+ (19). While vertebrate cells possess nonallosteric l-LDH isozymes, depending on the tissue (19), many bacterial cells possess allosteric types of l-LDHs, which usually require fructose 1,6-bisphosphate [Fru(1,6)P2] for activity (13). We are carrying out a comparative study of the nonallosteric and allosteric types of l-LDHs from Lactobacillus pentosus, previously called L. plantarum, and L. casei, respectively, to understand their structure-function relationships (28–30). In the course of this study, we found that these two enzymes exhibit marked activities toward oxaloacetate that are comparable to their activities for pyruvate.
The cultivation of Escherichia coli cells harboring expression plasmids for the genes encoding the l-LDHs of L. pentosus JCM1558 (=ATCC 8041) and L. casei IAM 12473 (=ATCC 393) and purification of the enzymes were performed essentially as described previously (29, 30). Protein concentrations were determined with Bio-Rad protein assay reagent by the Bradford method (4), using bovine serum albumin as the standard. All enzyme assays were performed at 30°C. The 2-ketoacid reduction by L. pentosus l-LDH was assayed in 50 mM sodium MES (morpholineethanesulfonic acid) buffer (pH 6.0) containing 0.1 mM NADH and various concentrations of sodium pyruvate, oxaloacetate, or another 2-ketoacid. The reduction of pyruvate and oxaloacetate by the L. casei enzymes was assayed in 50 mM sodium acetate buffer (pH 5.0) and sodium MOPS (morpholinepropanesulfonic acid) buffer (pH 7.0), respectively. The assay mixtures for oxaloacetate were freshly prepared before each use, using newly purchased oxaloacetic acid (Wako Fine Chemicals, Osaka, Japan). One unit was defined as the catalytic rate of the conversion of 1 μmol of substrate per min. Since the catalytic reactions by the L. pentosus and L. casei enzymes in the presence of Fru(1,6)P2 were markedly inhibited by high concentrations of substrates, kinetic parameters such as Km and maximal velocity (Vmax) were estimated using only data obtained at substrate concentrations sufficiently low and noninhibitory to give linear lines on double-reciprocal plots. For reactions that showed significant cooperativity, the Hill equation (6) was used for sigmoidal curve fitting to obtain kinetic parameters such as Hill coefficient (h) and half-saturating substrate concentration (S0.5).
Oligodeoxynucleotide 5′-GTT GTG ATT ACC GCT GGC GCC AAT CAA AAG CCT GGC GAA TCA CG-3′ was purchased from Takara Shuzo to obtain a mutant L. pentosus l-LDH (P101N). Site-directed mutagenesis was performed with a GeneEditor in vitro mutagenesis kit (Promega).
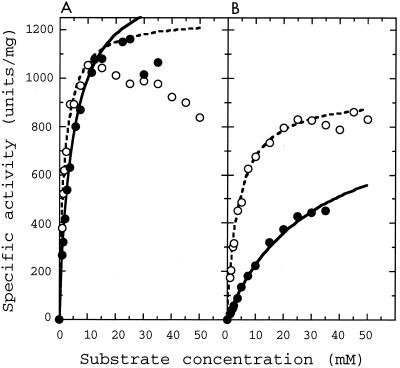
When oxaloacetate was used as the substrate, L. pentosus l-LDH exhibited high catalytic activity that was comparable to the activity toward pyruvate (Fig. 1A), exhibiting Km and Vmax values slightly higher than those for pyruvate (Table 1). The less than threefold differences in the oxaloacetate and pyruvate Km values indicate that the observed l-MDH activities arise mostly from oxaloacetate in the assay mixture, not from contaminant pyruvate. In addition to oxaloacetate, hydroxypyruvate was a good substrate for the enzyme reaction, exhibiting Km and Vmax values virtually the same as those for oxaloacetate. While the enzyme was relatively active toward 2-ketobutylate and phenylpyruvate, it was less active toward 2-ketoglutarate and 2-ketoacid substrates with long aliphatic chains, such as 2-ketovalerate, 2-ketocaproate, and 2-ketoisocaproate (Table 1), which are favorable substrates for the l-2-hydroxyisocaproate dehydrogenase from L. confusus (27).
FIG. 1.
Saturation curves for pyruvate and oxaloacetate for the wild-type and mutant L. pentosus l-LDHs. Reaction velocities for the wild-type (A) and P101N mutant (B) L. pentosus l-LDHs were measured in the presence of the indicated concentrations of pyruvate (open symbols) or oxaloacetate (closed symbols). Dashed and solid lines indicate the calculated saturation curves for pyruvate and oxaloacetate, respectively, by kinetic parameters shown in Table 1.
TABLE 1.
Kinetic parameters for various substrates for L. pentosus l-LDH
| Substrate | Wild type
|
P101N
|
||||
|---|---|---|---|---|---|---|
| Km (mM) | Vmax (U/mg) | Vmax/Km (min−1 · mg−1) | Km (mM) | Vmax (U/mg) | Vmax/Km (min−1 · mg−1) | |
| Pyruvate | 1.8 | 1,250 | 690 | 4 | 940 | 235 |
| Oxaloacetate | 4.6 | 1,450 | 315 | 30 | 890 | 30 |
| Hydroxypyruvate | 4.1 | 1,450 | 350 | 20 | 840 | 42 |
| 2-Ketobutylate | 32 | 830 | 26 | 27 | 180 | 6.7 |
| Phenylpyruvate | 15 | 480 | 32 | 50 | 110 | 2.2 |
| 2-Ketovalerate | 23 | 75 | 3.3 | 75 | 22 | 0.29 |
| 2-Ketocaproate | 29 | 110 | 3.8 | 54 | 30 | 0.56 |
| 2-Ketoisocaproate | 44 | 4 | 0.1 | NDa | ND | ND |
| 2-Ketoglutarate | 20 | 8 | 0.4 | 100 | 4 | 0.04 |
ND, not determined (enzyme activity too weak for determination of exact values).
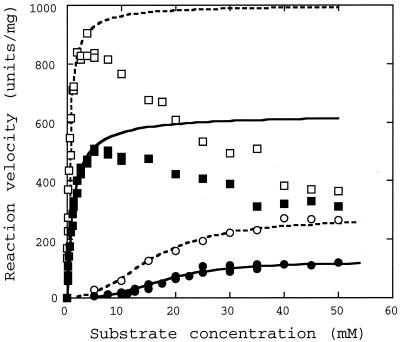
Unlike the L. pentosus enzyme, the L. casei enzyme is a representative allosteric l-LDH that requires Fru(1,6)P2 to exhibit its activity (14, 18, 20). Under acidic conditions such as pH 5.0, the L. casei enzyme shows marked catalytic activity even in the absence of Fru(1,6)P2, giving a sigmoidal saturation curve for substrate pyruvate but a hyperbolic saturation curve in the presence of Fru(1,6)P2. When oxaloacetate was used as the substrate at pH 5.0, the L. casei enzyme also exhibited high catalytic activity toward oxaloacetate in both the absence and presence of Fru(1,6)P2 (Fig. 2). In the absence of Fru(1,6)P2, for oxaloacetate a sigmoidal saturation curve was obtained for the enzyme reaction; as for pyruvate, an h value slightly higher than that for pyruvate was observed (Table 2). Under these conditions, we observed for oxaloacetate only an about twofold lower Vmax and slightly higher S0.5 for the enzyme reaction than for pyruvate. In the presence of 5 mM Fru(1,6)P2, on the other hand, the L. casei enzyme gave a hyperbolic saturation curve for oxaloacetate, as for pyruvate (Fig. 2), and slightly higher Km and lower Vmax values for oxaloacetate than for pyruvate (Table 2).
FIG. 2.
Saturation curves for pyruvate and oxaloacetate for L. casei l-LDH at pH 5.0. Reaction velocities were measured in the presence of the indicated concentrations of pyruvate (open symbols) or oxaloacetate (closed symbols), without (circles) or with (squares) 5 mM Fru(1,6)P2. Dashed and solid lines indicate the calculated saturation curves for pyruvate and oxaloacetate, respectively, by kinetic parameters shown in Table 2.
TABLE 2.
Kinetic parameters for pyruvate and oxaloacetate for L. casei l-LDH
| Condition | S0.5 (mM) | Vmax (U/mg) | Vmax/S0.5 (min−1 · mg−1) | h |
|---|---|---|---|---|
| pH 5.0 | ||||
| No Fru(1,6)P2 | ||||
| Pyruvate | 16.0 | 270 | 17 | 2.4 |
| Oxaloacetate | 18.0 | 120 | 7 | 3.1 |
| 5 mM Fru(1,6)P2 | ||||
| Pyruvate | 0.4 | 1,000 | 2,500 | 1 |
| Oxaloacetate | 1.1 | 625 | 570 | 1 |
| pH 7.0 | ||||
| 5 mM Fru(1,6)P2 | ||||
| Pyruvate | 30.0 | 560 | 19 | 2.4 |
| 5 mM Fru(1,6)P2 + 10 mM MnSO4 | ||||
| Pyruvate | 4.7 | 1,000 | 210 | 1 |
| Oxaloacetatea | 3.4 | 825 | 240 | 1 |
Kinetic parameters were calculated using the data for <10 mM oxaloacetate, since oxaloacetate was insoluble above 20 mM under these conditions.
Under neutral conditions such as pH 7.0, on the other hand, it is known that the L. casei enzyme absolutely requires Fru(1,6)P2 for activity, and the activation function of Fru(1,6)P2 is markedly improved in the presence of certain divalent cations such as Mn2+ (14, 18, 20). At pH 7.0, the L. casei enzyme exhibited no apparent catalytic activity toward oxaloacetate in the absence or in the presence of a nonsaturating concentration (5 mM) of Fru(1,6)P2, where pyruvate gives a sigmoidal saturation curve for the enzyme reaction (14, 18). Like the activity toward pyruvate, however, the activity toward oxaloacetate was also greatly enhanced by addition of 10 mM MnSO4 in the presence of 5 mM Fru(1,6)P2, giving a hyperbolic saturation curve with about 1.4- and 1.2-fold reduced Km and Vmax values, respectively, compared with those for pyruvate (Table 2).
Since the two Lactobacillus enzymes were essentially randomly chosen from the allosteric and nonallosteric types of Lactobacillus l-LDHs, respectively, it is likely that Lactobacillus l-LDHs generally exhibit such a broad substrate specificity regardless of their allosteric properties. Significant l-MDH activity has been observed in cell extracts of L. plantarum, L. casei, L. acidophilus, and L. helveticus (10, 25). These four lactobacilli also exhibit marked activities of citrate synthase, aconitase, citrate lyase, fumarase, and fumarate reductase but possibly lack isocitrate dehydrogenase, 2-ketoglutarate dehydrogenase, and succinate dehydrogenase activities, indicating that lactobacilli are deficient in biosynthetic and bioenergetic functions required for the complete citric acid cycle (25). This suggests that Lactobacillus l-LDHs do not physiologically catalyze the oxidation of l-malate, but can effect the reduction of oxaloacetate to l-malate in such an incomplete citric acid cycle, though L. plantarum cells may possess another protein that can catalyze oxaloacetate reduction with NADH (10).
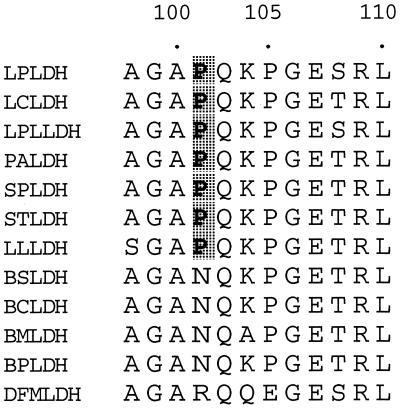
Although it has been shown that Gln102, Asp197, and Thr246 (positions are numbered according to Eventoff et al. 8) directly participate in substrate discrimination by Bacillus stearothermophilus l-LDH (35), all of these amino acids are highly conserved in the Lactobacillus enzymes, as in the case of other usual l-LDHs. On the other hand, it is also known that the active-site loop comprising positions 98 to 110 of l-LDH is essentially involved in the catalytic reaction (5) and substrate recognition (22, 35, 36). It is notable that Bacillus and Lactobacillus l-LDHs have different amino acids, i.e., Asn and Pro, respectively, at position 101 in the active-site loop (Fig. 3), although the two types of l-LDH are relatively close to each other evolutionarily (16), suggesting that l-LDHs from aerobic and anaerobic gram-positive bacteria may be distinguished by whether there is Asn or Pro at this position. The ternary complex structure of the B. stearothermophilus enzyme indicates that the side chain of Asn101 forms a hydrogen bond with the Gln102 main chain and thereby stabilizes the turn structure of the active-site loop (34). It is possible that the Asn101-to-Pro change affects the structure or flexibility of the active-site loop, since Pro limits the dihedral angles of the linked peptide bonds, besides impairing the ability to form the corresponding hydrogen bond. For L. pentosus l-LDH, the replacement of Pro101 with Asn significantly reduced activities toward pyruvate and, to a much greater extent, oxaloacetate (Fig. 1). The substitution increased the oxaloacetate Km (6.5-fold) more than the pyruvate Km (2.2-fold) (Table 1), and increased the activity toward pyruvate/activity toward oxaloacetate ratio from 2.2 to 7.8, when enzyme activity was evaluated on the basis of Vmax/Km. Like the activity toward oxaloacetate, enzyme activities toward other alternative 2-ketoacid substrates were reduced by the Pro101-to-Asn change more than the activity toward pyruvate, with an apparently similar trend. These results indicate that Pro101 contributes to widening of the substrate specificity of the Lactobacillus enzyme, although it is unlikely that only Pro101 is critical for the broad substrate specificity of the L. pentosus enzyme. It is known that in most l-LDHs the guanidino group of Arg171 forms a bidental, ionic hydrogen bond with the carboxyl group of substrate pyruvate and allows pyruvate to be strongly bound and properly oriented in the catalytic site (17). Recently, we determined the three-dimensional structure of the L. pentosus holoenzyme at 2.3-Å resolution, where the guanidino group undergoes an unusual intersubunit interaction across the Q-axis subunit interface (H. Uchikoba et al., unpublished results). Through crystallography and site-directed mutagenesis, we are further investigating the unusual substrate recognition of Lactobacillus l-LDHs.
FIG. 3.
Alignment of amino acid sequences of the active-site loops of l-LDHs of bacilli and lactic acid bacteria. LPLDH, L. pentosus (28); LCLDH, L. casei (23); LPLLDH, L. plantarum (9); PALDH, Pediococcus acidilactici (12); SPLDH, Streptococcus mutans (30); STLDH, S. thermophilus (31); LLLDH, Lactococcus lactis (16, 24); BSLDH, B. stearothermophilus (2); BCLDH, B. caldotenax (3); BMLDH, B. megaterium (33); BPLDH, B. psychrosaccharolyticus (32); DFMLDH, dogfish muscle (31). Proline residues conserved in l-LDHs of lactic acid bacteria are shaded.
Acknowledgments
This work was supported by a Grant-in-Aid for Science Research to H.T. from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Banaszak L J, Bradshaw R A. Malate dehydrogenase. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 11. New York, N.Y: Academic Press; 1975. pp. 369–397. [Google Scholar]
- 2.Barstow D A, Clarke A R, Chia W N, Wigley D, Sharman A F, Holbrook J J, Atkinson T, Minton N P. Cloning, expression, and complete nucleotide sequence of the Bacillus stearothermophilusl-lactate dehydrogenase gene. Gene. 1986;46:47–55. doi: 10.1016/0378-1119(86)90165-4. [DOI] [PubMed] [Google Scholar]
- 3.Barstow D A, Murphy J P, Sharman A F, Clarke A R, Holbrook J J, Atkinson T. Amino acid sequence of l-lactate dehydrogenase of Bacillus caldotenax deduced from the nucleotide sequence of the cloned gene. Eur J Biochem. 1987;165:581–596. doi: 10.1111/j.1432-1033.1987.tb11479.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Clarke A R, Wigley D B, Chia W N, Barstow D A, Atkinson T, Holbrook J J. Site-directed mutagenesis reveals role of mobile arginine residue in lactate dehydrogenase catalysis. Nature. 1986;324:699–702. doi: 10.1038/324699a0. [DOI] [PubMed] [Google Scholar]
- 6.Dixon M, Webb E C. Enzyme. 3rd ed. London, United Kingdom: Longman; 1979. pp. 400–402. [Google Scholar]
- 7.Duncan M J, Hillman J D. DNA sequence and in vitro mutagenesis of the gene encoding the fructose-1,6-diphosphate-dependent l-(+)-lactate dehydrogenase of Streptococcus mutans. Infect Immun. 1991;59:3930–3934. doi: 10.1128/iai.59.11.3930-3934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eventoff W, Rossmann M G, Taylor S S, Torff H-J, Meyer H, Keil W, Kiltz H-H. Structural adaptations of lactate dehydrogenase isozymes. Proc Natl Acad Sci USA. 1977;74:2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferain T, Garmyn D, Bernard N, Hols P, Delcour J. Lactobacillus plantarum ldhL gene: overexpression and deletion. J Bacteriol. 1994;176:596–601. doi: 10.1128/jb.176.3.596-601.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferain T, Schanck A N, Delcour J. 13C nuclear magnetic resonance analysis of glucose and citrate end products in an ldhL-ldhD double-knockout strain of Lactobacillus plantarum. J Bacteriol. 1996;178:7311–7315. doi: 10.1128/jb.178.24.7311-7315.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fushinobu S, Ohta T, Matsuzawa H. Homotropic activation via the subunit interaction and allosteric symmetry revealed on analysis of hybrid enzymes of l-lactate dehydrogenase. J Biol Chem. 1998;273:2971–2976. doi: 10.1074/jbc.273.5.2971. [DOI] [PubMed] [Google Scholar]
- 12.Garmyn D, Ferain T, Bernard N, Hols P, Delcour J. Cloning, nucleotide sequence, and transcriptional analysis of the Pediococcus acidilacticil-(+)-lactate dehydrogenase gene. Appl Environ Microbiol. 1995;61:266–272. doi: 10.1128/aem.61.1.266-272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garvie E I. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon G L, Doelle H W. Purification, properties and immunological relationship of l-(+)-lactate dehydrogenase from Lactobacillus casei. Eur J Biochem. 1976;67:543–555. doi: 10.1111/j.1432-1033.1976.tb10720.x. [DOI] [PubMed] [Google Scholar]
- 15.Goward C R, Nicholls D J. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 1994;3:1883–1888. doi: 10.1002/pro.5560031027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin H G, Swindell S R, Gasson M J. Cloning and sequence analysis of the gene encoding l-lactate dehydrogenase from Lactococcus lactis: evolutionary relationships between 21 different LDH enzymes. Gene. 1992;122:193–197. doi: 10.1016/0378-1119(92)90049-u. [DOI] [PubMed] [Google Scholar]
- 17.Hart K W, Clarke A R, Wigley D B, Waldman A D, Chia W N, Barstow D A, Atkinson T, Jones J B, Holbrook J J. A strong carboxylate-arginine interaction is important in substrate orientation and recognition in lactate dehydrogenase. Biochim Biophys Acta. 1986;914:294–298. doi: 10.1016/0167-4838(87)90289-5. [DOI] [PubMed] [Google Scholar]
- 18.Hensel R, Mayr U, Stetter K O, Kandler O. Comparative studies of lactic acid dehydrogenases in lactic acid bacteria. 1. Purification and kinetics of the allosteric l-lactic acid dehydrogenase from Lactobacillus casei ssp. casei and Lactobacillus curvatus. Arch Microbiol. 1977;112:81–93. doi: 10.1007/BF00446658. [DOI] [PubMed] [Google Scholar]
- 19.Holbrook J J, Liljas A, Steindel S J, Rossmann M G. Lactate dehydrogenase. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 11. New York, N.Y: Academic Press; 1975. pp. 191–292. [Google Scholar]
- 20.Holland R, Pritchard G G. Regulation of the l-lactate dehydrogenase from Lactobacillus casei by fructose-1,6-bisphosphate and metal ions. J Bacteriol. 1975;121:777–784. doi: 10.1128/jb.121.3.777-784.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito Y, Sasaki T. Cloning and nucleotide sequence of l-lactate dehydrogenase gene from Streptococcus thermophilus M-192. Biosci Biotechnol Biochem. 1994;58:1569–1573. doi: 10.1271/bbb.58.1569. [DOI] [PubMed] [Google Scholar]
- 22.Kallwass H K W, Luyten M A, Parris W, Gold M, Kay C M, Jones J B. Effects of Gln102Arg and Cys97Gly mutations of the structural specificity and stereospecificity of the l-lactate dehydrogenase from Bacillus stearothermophilus. J Am Chem Soc. 1992;114:4551–4557. [Google Scholar]
- 23.Kim S F, Baek S J, Pack M Y. Cloning and nucleotide sequence of the Lactobacillus casei lactate dehydrogenase gene. Appl Environ Microbiol. 1991;57:2413–2417. doi: 10.1128/aem.57.8.2413-2417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llanos R M, Hillier A J, Davidson B E. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding l-(+)-lactate dehydrogenase, from Lactococcus lactis. J Bacteriol. 1992;174:6956–6964. doi: 10.1128/jb.174.21.6956-6964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morishita T, Yajima M. Incomplete operation of biosynthetic and bioenergenic functions of the citric acid cycle in multiple autotrophic Lactobacilli. Biosci Biotechnol Biochem. 1995;59:251–255. [Google Scholar]
- 26.Rossmann M G, Liljas A, Brändén C-I, Banaszak L J. Evolutionary and structural relationships among dehydrogenases. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 11. New York, N.Y: Academic Press; 1975. pp. 61–102. [Google Scholar]
- 27.Schütte H, Hummel W, Kula M-R. l-2-Hydroxyisocaproate dehydrogenase. A new enzyme from Lactobacillus confusus for the stereospecific reduction of 2-ketocarboxylic acid. Appl Microbiol Biotechnol. 1984;19:167–176. [Google Scholar]
- 28.Taguchi H, Ohta T. d-Lactate dehydrogenase is a member of d-isomer-specific 2-hydroxyacid dehydrogenase family. J Biol Chem. 1991;266:12588–12594. [PubMed] [Google Scholar]
- 29.Taguchi H, Ohta T. Unusual amino acid substitution in the anion-binding site of Lactobacillus plantarum non-allosteric l-lactate dehydrogenase. Eur J Biochem. 1992;209:993–998. doi: 10.1111/j.1432-1033.1992.tb17373.x. [DOI] [PubMed] [Google Scholar]
- 30.Taguchi H, Ohta T. Role of histidine 188 in fructose 1,6-bisphosphate- and divalent cation-regulated l-lactate dehydrogenase of Lactobacillus casei. Biosci Biotechnol Biochem. 1995;59:451–458. doi: 10.1271/bbb.59.451. [DOI] [PubMed] [Google Scholar]
- 31.Taylor S S. Amino acid sequence of dogfish muscle lactate dehydrogenase. J Biol Chem. 1977;252:1799–1806. [PubMed] [Google Scholar]
- 32.Vckovski V, Schlatter D, Zuber H. Structure and function of l-lactate dehydrogenase from thermophilic and mesophilic bacteria. IX. Identification, isolation and nucleotide sequence of two l-lactate dehydrogenase genes of the psychrophilic bacterium Bacillus psychrosaccharolyticus. Biol Chem Hoppe-Seyler. 1990;371:103–110. doi: 10.1515/bchm3.1990.371.1.103. [DOI] [PubMed] [Google Scholar]
- 33.Waldvogel S, Weber H, Zuber H. Structure and function of l-lactate dehydrogenase from thermophilic and mesophilic bacteria. VII. Nucleotide sequence of the lactate dehydrogenase gene from the mesophilic bacterium Bacillus megaterium. Preparation and properties of a hybrid lactate dehydrogenase comprising moieties of the B. megaterium and B. stearothermophilus enzymes. Biol Chem Hoppe-Seyler. 1987;368:1391–1399. doi: 10.1515/bchm3.1987.368.2.1391. [DOI] [PubMed] [Google Scholar]
- 34.Wigley D B, Gamblin S J, Turkenburg J P, Dodson E J, Piontek K, Muirhead H, Holbrook J J. Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 Å resolution. J Mol Biol. 1992;223:317–335. doi: 10.1016/0022-2836(92)90733-z. [DOI] [PubMed] [Google Scholar]
- 35.Wilks H M, Hart K W, Feeney R, Dunn C R, Muirhead H, Chia W N, Barstow D A, Atkinson T, Clarke A R, Holbrook J J. A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science. 1988;242:1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- 36.Wilks H M, Halsall D J, Atkinson T, Chia W N, Clarke A R, Holbrook J J. Designs for a broad substrate specificity ketoacid dehydrogenase. Biochemistry. 1990;29:8587–8591. doi: 10.1021/bi00489a013. [DOI] [PubMed] [Google Scholar]
- 37.Wu C-Y, Chen S-T, Chiou S-H, Wang K-T. Kinetic analysis of duck ɛ-crystallin with l-lactate dehydrogenase activity: determination of kinetic constants and comparison of substrate specificity. Biochem Biophys Res Commun. 1992;186:874–880. doi: 10.1016/0006-291x(92)90827-8. [DOI] [PubMed] [Google Scholar]