Introduction
Platinum-associated mutagenesis has been observed in tumors directly treated with these agents and in secondary hematologic malignancies.1-6 However, data supporting a direct association of platinum exposure with genetic oncogenic events promoting secondary solid tumorigenesis are lacking. A 70-year-old woman was diagnosed with ovarian endometrioid adenocarcinoma, treated first with oophorectomy, hysterectomy, and chemotherapy (intravenous carboplatin, taxol, and topotecan), then with intraperitoneal and intravenous cisplatin and gemcitabine over 3 years, and finally with radiation therapy after recurrence. Eight years later, she was diagnosed with papillary thyroid carcinoma (PTC) and underwent total thyroidectomy and lymph node dissection. Histopathology revealed classic PTC with vascular invasion, extrathyroidal extension, and three perithyroidal nodal involvement. Despite high-dose radioactive iodine ablation and long-term thyroid-stimulating hormone suppression therapy, PTC recurred on the left lateral neck 4 and 8 years later, treated surgically on both occasions (Fig 1A). Tissue samples from ovarian tumor (n = 1), primary PTC (n = 10), and two recurrent PTC excisions (n = 1 each) were submitted for molecular profiling (Methods). The locations of tissue samples from primary PTC thyroidectomy, as documented in pathology reports with approximate relative positions and temporal annotations, are depicted in Figure 2A.
FIG 1.
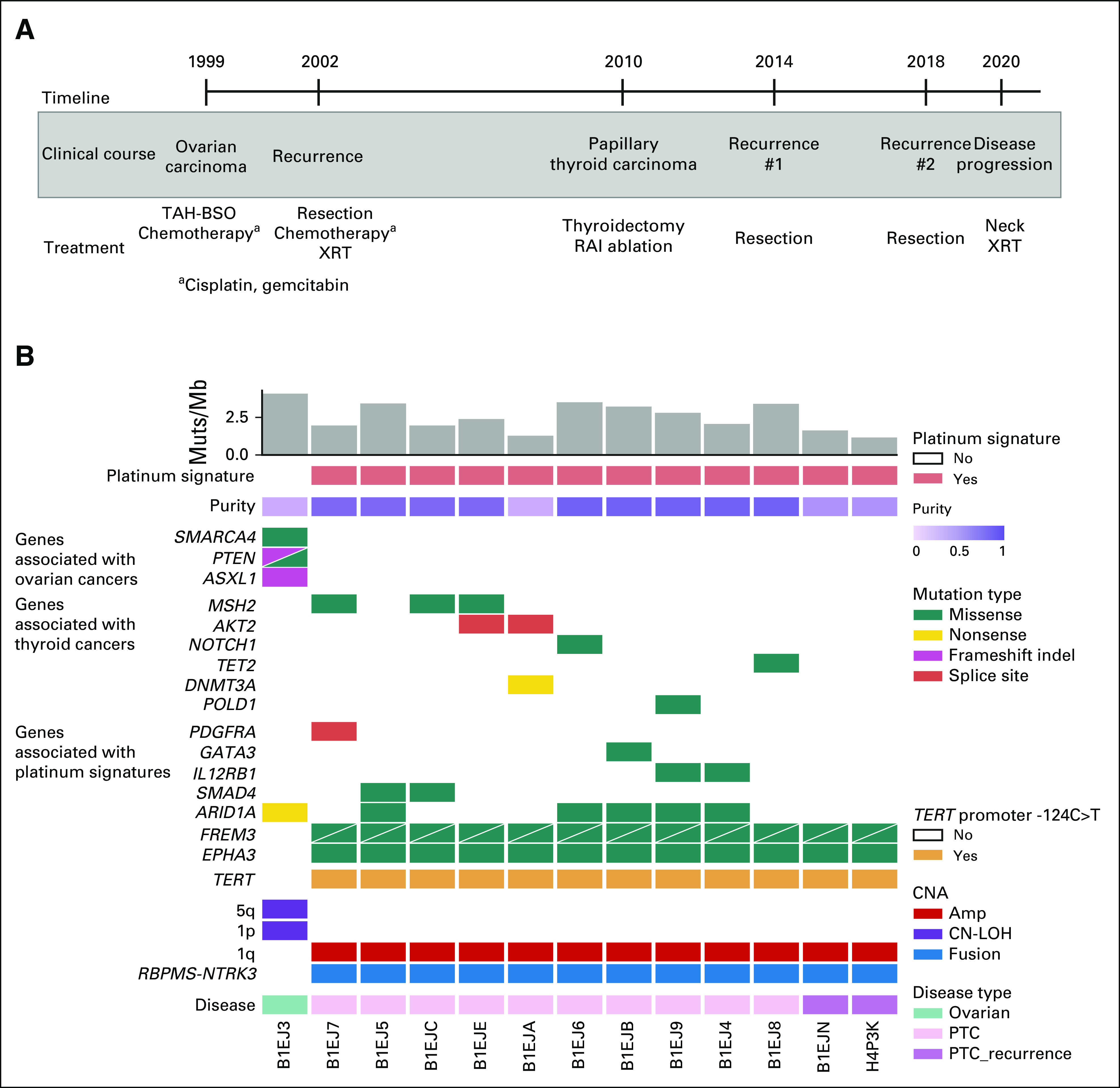
Genomic landscape of the patient's ovarian and PTC. (A) Timeline of sample collection, the patient's clinical course, and treatment history identified in ovarian (1999-2002) and thyroid cancer (2010-2020). (B) The CoMut7 plot illustrates select single nucleotide and insertion/deletion events, as well as CNA, RBPMS-NTRK3 RNA fusion, and TERT promoter status. The plot also includes the mutation burden, presence of the platinum signature determined by deconstructSigs and SigProfiler, and tumor purity as determined by ABSOLUTE. Each row represents the mutation or copy-number status for the indicated gene, and each column represents a unique tumor sample (ovarian or PTC samples). Two mutations in the same gene are represented by triangles. Amp, amplification; CN-LOH, copy-neutral loss of heterozygosity; CNA, copy-number alteration; Muts, mutations; PTC, papillary thyroid carcinoma; RAI, radioactive iodine; TAH-BSO, total abdominal hysterectomy and bilateral salpingo-oophorectomy; XRT, radiation therapy.
FIG 2.
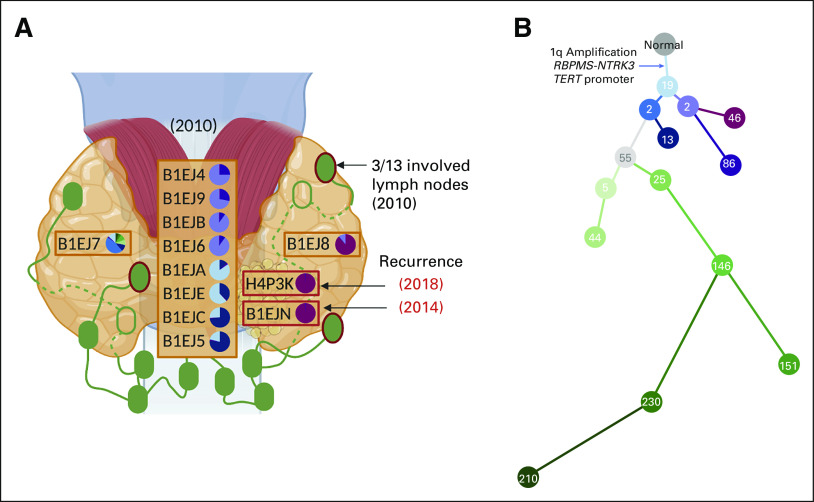
The spatial and temporal tumor heterogeneity in PTC. (A) A map of the PTC sample locations collected during the initial 2010 thyroidectomy (three orange boxes with 10 samples) and subsequent surgeries for locoregional recurrence in 2014 and 2018 (two red boxes with two samples). Three lymph nodes (green ovals with red borders) were positive for the spread of the tumor. Green empty ovals represent two lymph nodes posterior to the thyroid. Pie charts represent the fraction of each subclone found in each sample with a common ancestor existing in all PTC samples. (B) A phylogenetic tree of each subclone represented in the pie charts of Figure 2A. Number in the circle indicates the number of variants assigned to each subclone. The clonal events shared by all PTC samples were annotated in the tree. PTC, papillary thyroid carcinoma.
Methods
Written informed consent was obtained from the patient under Dana-Farber Cancer Institute's Institutional Review Board 09-472. Molecular analysis was performed on formalin-fixed paraffin-embedded tumor samples and germline DNA obtained from a peripheral blood sample as previously described.8 Detailed analysis methods presented in the Data Supplement.
Results
Molecular origins and evolution of primary ovarian and secondary papillary thyroid cancers.
Prior intratumoral heterogeneity studies have revealed considerable variations in genetic makeup in tumors across anatomic locations and disease stages,9 which we hypothesized may inform molecular origins and evolution of this secondary PTC, given aggressive course and prior clinical context. We evaluated multiregional and multitemporal samples (12 thyroid and one ovarian) to interrogate genetic makeup of ovarian, primary, and recurrent PTC. Along with an ovarian cancer sample collected in 1999, 10 samples were collected from different locations in total thyroidectomy and two samples from PTC locoregional recurrences (Fig 1A). Germline analysis did not identify any known pathogenic genetic alteration associated with ovarian or thyroid cancer or cancer-related genetic syndrome. Comparison of somatic genomic features, including mutations and copy-number alterations of these samples, indicated that ovarian and thyroid tumor did not share common genetic alterations and originated from genetically distinct tumorigenic events (Fig 1B, Data Supplement). The ovarian cancer harbored canonical somatic mutations (eg, PTEN and SMARCA4 mutations), whereas PTC harbored driver events including RBPMS-NTRK3 fusion and a TERT promoter mutation, both associated with aggressive PTC behavior (Fig 1B, Data Supplement).10,11
To explore the evolutionary relationship between tumor foci in these multiregional and multitemporal thyroid samples, we clustered mutations to subclones and built a phylogenetic tree representing subclone relationships (Figs 2A and 2B: Methods). All PTC subclones shared a 1q amplification and canonical driver events implicated in PTC oncogenesis (RBPMS-NTRK3 fusion and TERT promoter mutation; Fig 2B, Data Supplement). We observed four distinct phylogenetic groups across all PTC samples with varying degrees of subclones. Eight samples fell into one of two phylogenetic branches on the basis of their clonal architecture: B1EJA, B1EJE, B1EJC, and B1EJ5 were dominated by the most recent common ancestor of all PTC subclones and a closely related descendant (Figs 2A and 2B: blue branches); B1EJ4, B1EJ9, B1EJB, and B1EJ6 samples were dominated by a different descendant of the most recent common ancestor (Figs 2A and 2B: purple branches). Anatomically, these samples were spatially near each other within the same phylogenetically defined groups (Figs 2A and 2B: matching color in pie chart and tree). One site (B1EJ7) had the most diverse subclones (Figs 2A and 2B: green branches). The two samples corresponding to locoregional recurrences (B1EJN and H4P3K) shared similar dominant subclones with total thyroidectomy PTC sample (B1EJ8). Sample B1EJ8 was from an area of tumor present at thyroidectomy resection margin and in closest spatial proximity to recurrences (Figs 2A and 2B: red branches), supporting the relationship between pathologic findings and molecular spatial patterns of recurrent thyroid carcinoma.
Cisplatin mutational signature was present in primary and recurrent PTC two decades after chemotherapy.
To examine sources of mutagenesis underlying mutation patterns observed in this PTC, we performed mutational signature analysis on ovarian cancer and PTC samples (Methods). Both ovarian and PTC tumors harbored the ubiquitous clock-like (SBS1 and SBS5) mutational signatures (Data Supplement). Only PTC samples, obtained 11-19 years after exposure to platinum chemotherapy, had evidence of platinum mutational signatures (SBS31 and SBS35; Data Supplement).
We then calculated the likelihood of observing a mutation in the specific trinucleotide context induced by a specific signature (Methods and Fig 3, Data Supplement). An example of a missense mutation mostly attributed to the clock-like (SBS1 and SBS5) signatures was an A[C>T]G alteration in NOTCH1 (Figs 3A and 3B). Additional mutations related to the platinum chemotherapy signatures across PTC samples were identified in genes such as EPHA3, SMAD4, and GATA3 (Figs 1B, 3A and 3B). The clonal c.-124C>T TERT promoter mutation is an established driver of aggressive PTC (along with the c.-146C>T mutation)11-15 and was found in a mutational context, C[C>T]T, characteristic of the platinum chemotherapy signature (Methods and Figs 3A and 3B, Data Supplement). This mutation was present in all PTC tumor samples (Figs 1B and Data Supplement) and thus links an event driving thyroid cancer pathogenesis to treatment exposure occurring decades previously.1,2
FIG 3.
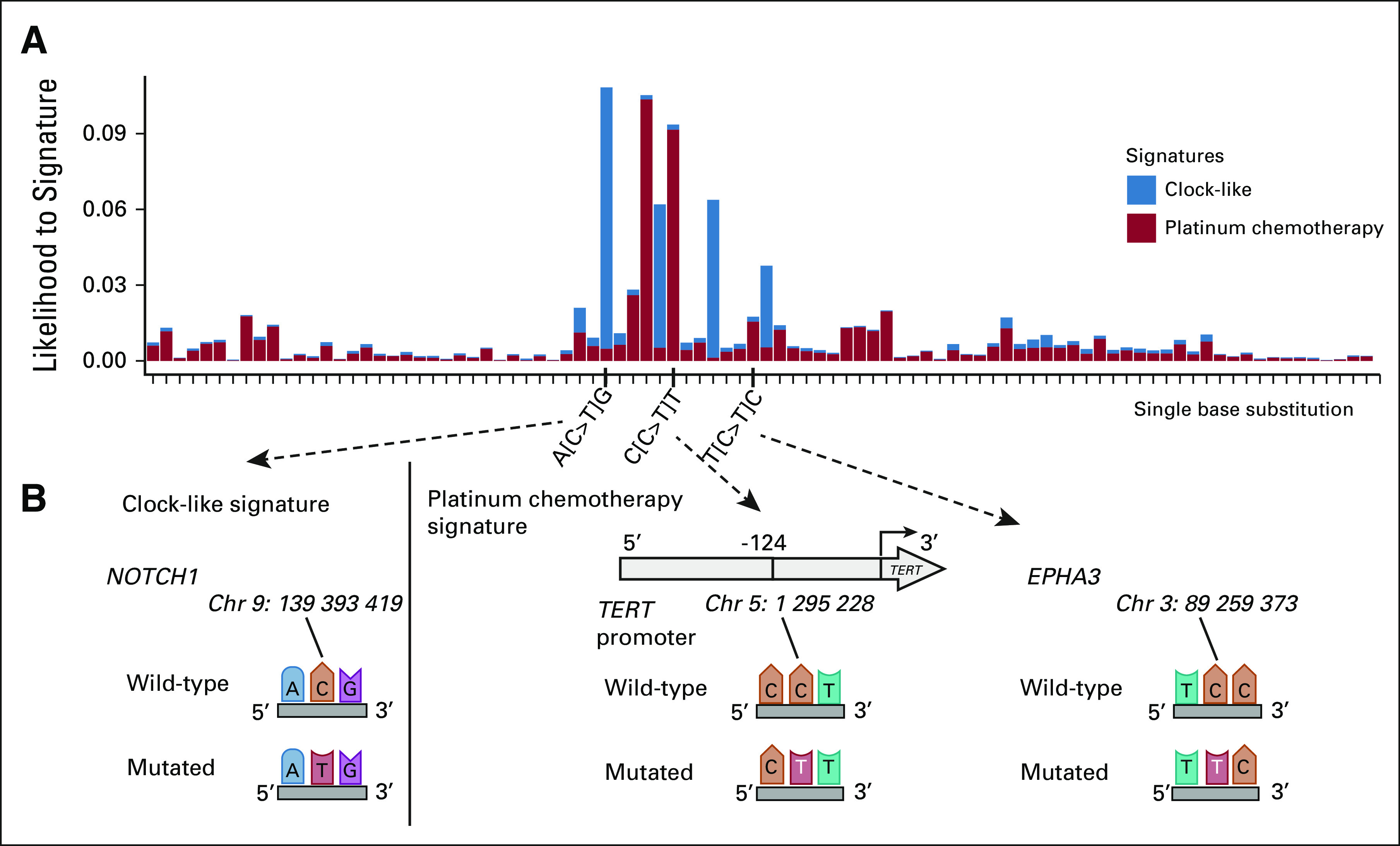
Single base substitution attribution to platinum chemotherapy signature. (A) The y-axis indicates the likelihood of observing the single base substitution induced by a specific signature using SigProfiler. The x-axis indicates single base substitution in 96 trinucleotide contexts. SBS1 and SBS5 were summarized as clock-like signatures. SBS31 and SBS35 were indicated as platinum chemotherapy signatures. (B) Nucleotide context with single base substitution changes in NOTCH1, TERT promoter, and EPHA3 genes.
Discussion
Recent studies discussed the chemotherapy mutational signatures that have been detected in metastatic tumors or in secondary hematologic malignancies.1-6 However, molecular origins and evolution of secondary solid tumors in patients previously exposed to platinum-based chemotherapy remain to be elucidated, although there are paradigms of chemotherapy-induced mutagenesis leading to drug-resistant clones and affecting clinical outcomes.1,2 Here, PTC molecular profiling at different time points and locations revealed significant intratumoral heterogeneity and genetic alterations distinct from an ovarian cancer observed in the same patient (Fig 1 and Data Supplement). Thyroid cancer is typically considered largely homogeneous at the molecular level, with the The Cancer Genome Atlas study proposing two major classifications: BRAF-like and RAS-like.16 Literature supports the presence of concomitant mutations, heterogeneous presence of driver mutations (such as BRAFV600E), and discordant profile of primary and metastatic PTC.17-20 By exploring evolutionary relationships across multiregional and multitemporal samples from the same patient, we illustrated the relationships between different subclones, and branches of samples with distinct subclones associated with the anatomical locations of collected samples (Fig 2 and Data Supplement). As the pathology report noted positive margins on excision of the patient's tumor during total thyroidectomy, it is possible that locoregional recurrences arose from remnant cancer cells escaping radioactive iodine ablation. No BRAF or RAS mutation was identified in primary and recurrent PTC samples. Instead, all PTC samples harbored RBPMS-NTRK3 fusion, TERT promoter c.-124C>T mutation, and 1q amplification, suggesting that these genetic alterations are closely linked with this PTC's pathogenesis and aggressive features (Figs 1 and 2). NTRK-altered PTC is rare, comprising under 2% of cases, and is characterized by multinodular growth, prominent fibrosis, extensive lymphovascular spread, and high risk of recurrence and metastatic disease,10 consistent with this patient's tumor pathology and behavior. Similarly, TERT promoter mutation, enriched in poorly differentiated and anaplastic thyroid carcinomas,11-15 and 1q amplification are associated with higher disease stage.16
Platinum chemotherapy mutational signatures were observed in all PTC samples, a footprint present 19 years after chemotherapy exposure (Data Supplement). Tumor location and primary versus recurrent site did not affect the degree of platinum mutational signatures observed; however, we demonstrated that c.-124C>T TERT promoter single base substitution in C[C>T]T context was mostly attributed to a platinum-associated mutational signature (Fig 3 and Data Supplement). Although ionizing radiation is a well-established risk factor for PTC, and gene fusions (in particular RET-PTC rearrangements) and copy-number alterations are enriched in radiation-induced PTC,21,22 no such link has been recognized for chemotherapy. Yet, in a study of 12,547 childhood cancer survivors, treatment with alkylating agents was associated with increased PTC risk, beyond the relative risk attributable to prior ionizing radiation therapy.23 This PTC case harbored uncommon genetic patterns with NTRK fusion and 1q amplification. Taken together with prior knowledge of how chemotherapy may induce DNA damage and breakpoints,24,25 these findings provide a mechanistic hypothesis for how platinum mutagenesis might have induced a TERT promoter mutation and contributed to the aggressive course of this patient's PTC.
This study may offer a mechanistic explanation for elevated thyroid cancer risk in patients after platinum chemotherapy exposure,2,23 who may benefit from increased awareness and lower threshold to screen for secondary PTC. For those exposed to platinum chemotherapy who later develop thyroid cancers, assessing for rare but prognostically significant driver events may be informative. The American Thyroid Association guidelines26 do not specifically address thyroid cancer screening in chemotherapy-treated patients, and there are limited data on the impact of chemotherapy on thyroid tumorigenesis. Future studies of a larger cohort of thyroid cancer patients with exposure to chemotherapy for a previous cancer will be necessary to determine whether there is a larger pattern of platinum chemotherapy-induced driver mutations explaining increased incidence of thyroid cancer seen in this population.
Theodora Pappa
Employment: Replimune (I), Deciphera (I)
Stock and Other Ownership Interests: AbbVie (I), Abbott Laboratories (I), Deciphera (I), Replimune (I)
Jake Conway
Employment: PATHAI
Consulting or Advisory Role: Tango Therapeutics
Travel, Accommodations, Expenses: PATHAI
Brendan Reardon
Patents, Royalties, Other Intellectual Property: Institutional patents filed on methods for clinical Interpretation
Darren Stanizzi
Employment: Vor Biopharma
Stock and Other Ownership Interests: Vor Biopharma
Meng Xiao He
Employment: Genentech/Roche
Stock and Other Ownership Interests: Genentech/Roche
Consulting or Advisory Role: Ikena Oncology, Amplify Medicines, Janssen
Jochen H. Lorch
Consulting or Advisory Role: Bayer, Eisai, Genentech, Novartis
Research Funding: Novartis (Inst), Bayer (Inst), Bristol Myers Squibb (Inst), Millennium (Inst)
Eliezer M. Van Allen
Stock and Other Ownership Interests: Syapse, Tango Therapeutics, Genome Medical, Microsoft, ervaxx, Monte Rosa Therapeutics, Manifold Bio
Consulting or Advisory Role: Syapse, Roche, Third Rock Ventures, Takeda, Novartis, Genome Medical, InVitae, Illumina, Tango Therapeutics, Ervaxx, Janssen, Monte Rosa Therapeutics, Manifold Bio, Genomic Life
Speakers' Bureau: Illumina
Research Funding: Bristol Myers Squibb, Novartis, Sanofi (Inst)
Patents, Royalties, Other Intellectual Property: Patent on discovery of retained intron as source of cancer neoantigens (Inst), Patent on discovery of chromatin regulators as biomarkers of response to cancer immunotherapy (Inst), Patent on clinical interpretation algorithms using cancer molecular data (Inst)
Travel, Accommodations, Expenses: Roche/Genentech
No other potential conflicts of interest were reported.
SUPPORT
Supported by National Cancer Institute (F31CA239347; J.C.); National Institutes of Health (T32 GM008313; M.X.H.); National Science Foundation (GRFP DGE1144152; M.X.H.); National Institutes of Health (T32 5T32HL007609–33; T.P.); National Institutes of Health (R37 CA222574; J.P., E.M.V.A.); National Institutes of Health (R01 CA227388; J.P., E.M.V.A.); Damon Runyon Cancer Research Foundation, Clinical Investigator Award (E.M.V.A.); National Institutes of Health (K99 CA262152; F.D.); Claudia Adams Barr Program, Innovative Cancer Research (F.D.); AWS Cloud, AWS Cloud Credits for Research Program (F.D.); Mark Foundation For Cancer Research, ASPIRE Award (F.D., E.M.V.A.)
J.S. and T.P. contributed equally to this work. J.P., J.H.L., and E.M.V.A. are cosenior authors.
PREPRINT VERSION
Preprint version available on bioRxiv. https://www.biorxiv.org/content/10.1101/2022.03.14.484002v1
AUTHOR CONTRIBUTIONS
Conception and design: Julia Schiantarelli, Jake Conway, Julian Huang, Jihye Park, Jochen H. Lorch, Eliezer M. Van Allen
Financial support: Jochen H. Lorch, Eliezer M. Van Allen
Administrative support: Eliezer M. Van Allen
Provision of study materials or patients: Darren Stanizzi, Jochen H. Lorch, Eliezer M. Van Allen
Collection and assembly of data: Julia Schiantarelli, Julian Huang, Darren Stanizzi, Evan Carey, Justine A. Barletta, Jihye Park, Jochen H. Lorch, Eliezer M. Van Allen
Data analysis and interpretation: Julia Schiantarelli, Theodora Pappa, Jake Conway, Jett Crowdis, Brendan Reardon, Felix Dietlein, Julian Huang, Alice Bosma-Moody, Alma Imamovic, Seunghun Han, Sabrina Camp, Eric Kofman, Meng Xiao He, David Liu, Jihye Park, Jochen H. Lorch, Eliezer M. Van Allen
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Theodora Pappa
Employment: Replimune (I), Deciphera (I)
Stock and Other Ownership Interests: AbbVie (I), Abbott Laboratories (I), Deciphera (I), Replimune (I)
Jake Conway
Employment: PATHAI
Consulting or Advisory Role: Tango Therapeutics
Travel, Accommodations, Expenses: PATHAI
Brendan Reardon
Patents, Royalties, Other Intellectual Property: Institutional patents filed on methods for clinical Interpretation
Darren Stanizzi
Employment: Vor Biopharma
Stock and Other Ownership Interests: Vor Biopharma
Meng Xiao He
Employment: Genentech/Roche
Stock and Other Ownership Interests: Genentech/Roche
Consulting or Advisory Role: Ikena Oncology, Amplify Medicines, Janssen
Jochen H. Lorch
Consulting or Advisory Role: Bayer, Eisai, Genentech, Novartis
Research Funding: Novartis (Inst), Bayer (Inst), Bristol Myers Squibb (Inst), Millennium (Inst)
Eliezer M. Van Allen
Stock and Other Ownership Interests: Syapse, Tango Therapeutics, Genome Medical, Microsoft, ervaxx, Monte Rosa Therapeutics, Manifold Bio
Consulting or Advisory Role: Syapse, Roche, Third Rock Ventures, Takeda, Novartis, Genome Medical, InVitae, Illumina, Tango Therapeutics, Ervaxx, Janssen, Monte Rosa Therapeutics, Manifold Bio, Genomic Life
Speakers' Bureau: Illumina
Research Funding: Bristol Myers Squibb, Novartis, Sanofi (Inst)
Patents, Royalties, Other Intellectual Property: Patent on discovery of retained intron as source of cancer neoantigens (Inst), Patent on discovery of chromatin regulators as biomarkers of response to cancer immunotherapy (Inst), Patent on clinical interpretation algorithms using cancer molecular data (Inst)
Travel, Accommodations, Expenses: Roche/Genentech
No other potential conflicts of interest were reported.
REFERENCES
- 1. Boot A, Huang MN, Ng AWT, et al. In-depth characterization of the cisplatin mutational signature in human cell lines and in esophageal and liver tumors. Genome Res. 2018;28:654–665. doi: 10.1101/gr.230219.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brady SW, Gout AM, Zhang J. Therapeutic and prognostic insights from the analysis of cancer mutational signatures. Trends Genet. 2022;38:194–208. doi: 10.1016/j.tig.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pich O, Muiños F, Lolkema MP, et al. The mutational footprints of cancer therapies. Nat Genet. 2019;51:1732–1740. doi: 10.1038/s41588-019-0525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52:1219–1226. doi: 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pich O, Cortes-Bullich A, Muiños F, et al. The evolution of hematopoietic cells under cancer therapy. Nat Commun. 2021;12:4803. doi: 10.1038/s41467-021-24858-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu D, Abbosh P, Keliher D, et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat Commun. 2017;8:2193. doi: 10.1038/s41467-017-02320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crowdis J, He MX, Reardon B, et al. CoMut: Visualizing integrated molecular information with comutation plots. Bioinformatics. 2020;36:4348–4349. doi: 10.1093/bioinformatics/btaa554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He MX, Cuoco MS, Crowdis J, et al. Transcriptional mediators of treatment resistance in lethal prostate cancer. Nat Med. 2021;27:426–433. doi: 10.1038/s41591-021-01244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turajlic S, Xu H, Litchfield K, et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 2018;173:581–594.e12. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu Y-H, Dias-Santagata D, Farahani AA, et al. Clinicopathologic and molecular characterization of NTRK-rearranged thyroid carcinoma (NRTC) Mod Pathol. 2020;33:2186–2197. doi: 10.1038/s41379-020-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Landa I, Ibrahimpasic T, Boucai L, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X, Bishop J, Shan Y, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20:603–610. doi: 10.1530/ERC-13-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melo M, da Rocha AG, Vinagre J, et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J Clin Endocrinol Metab. 2014;99:E754–E765. doi: 10.1210/jc.2013-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKelvey BA, Umbricht CB, Zeiger MA. Telomerase reverse transcriptase (TERT) regulation in thyroid cancer: A review. Front Endocrinol. 2020;11:485. doi: 10.3389/fendo.2020.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu R, Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer. 2016;23:R143–R155. doi: 10.1530/ERC-15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gawin M, Kurczyk A, Stobiecka E, et al. Molecular heterogeneity of papillary thyroid cancer: Comparison of primary tumors and synchronous metastases in regional lymph nodes by mass spectrometry imaging. Endocr Pathol. 2019;30:250–261. doi: 10.1007/s12022-019-09593-2. [DOI] [PubMed] [Google Scholar]
- 18. Masoodi T, Siraj AK, Siraj S, et al. Evolution and impact of subclonal mutations in papillary thyroid cancer. Am J Hum Genet. 2019;105:959–973. doi: 10.1016/j.ajhg.2019.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Biase D, Cesari V, Visani M, et al. High-sensitivity BRAF mutation analysis: BRAFV600E is acquired early during tumor development but is heterogeneously distributed in a subset of papillary thyroid carcinomas. J Clin Endocrinol Metab. 2014;99:E1530–E1538. doi: 10.1210/jc.2013-4389. [DOI] [PubMed] [Google Scholar]
- 20. Masoodi T, Siraj AK, Siraj S, et al. Whole-exome sequencing of matched primary and metastatic papillary thyroid cancer. Thyroid. 2020;30:42–56. doi: 10.1089/thy.2019.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yeager M, Machiela MJ, Kothiyal P, et al. Lack of transgenerational effects of ionizing radiation exposure from the Chernobyl accident. Science. 2021;372:725–729. doi: 10.1126/science.abg2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hess J, Newbern DK, Beebe KL, et al. High prevalence of gene fusions and copy number alterations in pediatric radiation therapy-induced papillary and follicular thyroid carcinomas. Thyroid. 2022;32:411–420. doi: 10.1089/thy.2021.0217. [DOI] [PubMed] [Google Scholar]
- 23. Veiga LHS, Bhatti P, Ronckers CM, et al. Chemotherapy and thyroid cancer risk: A report from the childhood cancer survivor study. Cancer Epidemiol Biomarkers Prev. 2012;21:92–101. doi: 10.1158/1055-9965.EPI-11-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Juul N, Wang Y, Kim J, et al. A genomic-profile derived summary measure of chromosomal breakpoints predicts response to treatment with the DNA-damaging agent cisplatin Cancer Res 69 111 111 2009. 19117993 [Google Scholar]
- 25. Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 26. Haugen BR, Alexander EK, Bible KC, et al. 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]