PURPOSE
Guidelines recommend somatic and germline testing for men with advanced prostate cancer (PCa). Barriers to widespread implementation result in underutilization of germline testing. Somatic testing alone risks missing pathogenic germline variants (PGVs). We sought to determine whether the addition of germline testing to tumor-only sequencing improves detection of PGVs in men with advanced PCa. Secondarily, we sought to define the added value of combining somatic and germline testing to optimize detection of clinically actionable alterations.
PATIENTS AND METHODS
We analyzed results of independent germline testing and tumor-only sequencing from 100 men with advanced PCa from a prospective clinical trial (ClinicalTrials.gov identifier: NCT03328091). The primary outcome was the proportion of PGVs not reported with tumor-only sequencing. The secondary outcome was the association of locus-specific loss of heterozygosity for PGVs in homologous recombination genes with clinical-genomic features.
RESULTS
In the 100 men who underwent germline testing and tumor-only sequencing, 24 PGVs were identified, 17 of which were clinically actionable, in 23 patients. Tumor-only sequencing failed to report four (17%) of the PGVs. One additional PGV (4.2%) had variant allele frequency on tumor-sequencing below the threshold for follow-up germline testing. When integrating tumor-only sequencing with germling testing results, 33% of patients harbored clinically actionable alterations. Rates of locus-specific loss of heterozygosity were higher for BRCA2 PGVs in castration-resistant PCa than PGVs in other homologous recombination genes in hormone-sensitive PCa (P = .029).
CONCLUSION
Tumor-only sequencing failed to report more than 20% of PGVs in men with advanced PCa. These findings strongly support guideline recommendations for universal germline and somatic testing in this population. Combining tumor and germline sequencing doubled the chance of detecting a clinically actionable alteration.
INTRODUCTION
Molecular testing has transformed clinical management of advanced prostate cancer (PCa). Somatic tumor sequencing identifies clinically actionable alterations in 20%-25% of men with metastatic PCa, approximately half of which are pathogenic germline variants (PGVs).1,2 Consequently, the National Comprehensive Cancer Network (NCCN) and European Society of Medical Oncology (ESMO) PCa guidelines recommend germline and somatic testing for men with metastatic PCa.3,4 Germline testing identifies heritable mutations associated with increased cancer risk. Somatic testing identifies nonheritable acquired alterations as well as some, but not all, germline variants. Several limitations of tumor-only sequencing can result in under-reporting of PGVs, including (1) filtering of germline variants on the basis of population frequency, (2) somatic deletion of a gene harboring a germline variant, (3) incomplete coverage of genes that may harbor a germline variant, and (4) limited ability to detect certain germline variants, such as exon-level copy-number alterations and variants in high-homology regions.5-7 Furthermore, without a paired normal sample, tumor-only sequencing cannot definitively classify variants as germline versus somatic. Accurate reporting of PGVs is important to identify inherited cancer risk for patients and their family members.8
CONTEXT
Key Objective
Despite guideline recommendations to perform both somatic and germline testing in all men with advanced prostate cancer, germline testing remains underutilized. The implications of relying on tumor-only sequencing, and the added value of germline testing to tumor-only sequencing, are not well-defined.
Knowledge Generated
Tumor-only sequencing failed to identify one in five pathogenic germline variants in men with advanced prostate cancer. Obtaining paired somatic and germline sequencing increased the likelihood of detecting a clinically actionable alteration by two-fold.
Relevance
Herein, we highlight the strengths and limitations of tumor-only and germline-only sequencing in men with advanced prostate cancer. These findings strongly support the guideline recommendation that clinicians obtain both somatic and germline testing for all men with advanced prostate cancer. This approach maximizes detection of clinically relevant alterations to inform therapeutic decision making and facilitate cascade testing to identify affected family members of patients with germline alterations.
Despite guideline recommendations, germline testing is underutilized in PCa. Barriers to widespread implementation include a shortage of genetic counselors, inefficient workflows, gaps in insurance, insufficient educational materials, and concerns about genetic discrimination.8,9 Differences in access and mistrust of the health care system compound underutilization in minority and underserved populations.10,11 Provider education is critical as well. A recent survey reported that nearly 40% of academic genitourinary oncologists do not offer universal germline testing for men with metastatic PCa.9 Further data are needed to demonstrate the value of complementing tumor-only sequencing with germline testing in men with advanced PCa.
In this study, we analyzed a subset of subjects enrolled in ProGen (ClinicalTrials.gov identifier: NCT03328091), a prospective randomized clinical trial that evaluated pretest video education with post-test genetic counseling compared with in-person pretest genetic counseling in men with advanced PCa.12 ProGen demonstrated that video education resulted in similar uptake in germline testing as in-person genetic counseling. All consenting subjects underwent germline testing as part of the ProGen trial; a subset also underwent independent tumor-only sequencing during routine care. This cohort provided an opportunity to evaluate the added value of germline testing to tumor-only sequencing in men with advanced PCa. We hypothesized that the addition of germline testing to tumor-only sequencing would improve detection of PGVs. Secondarily, we assessed the complementary value of germline and somatic testing to identify clinically actionable alterations.
PATIENTS AND METHODS
ProGen Trial Enrollment and Selection of Patients for Study
Patients analyzed herein are a subset of those enrolled in ProGen.12 Only patients enrolled and treated at the Dana-Farber Cancer Institute (DFCI)/Brigham and Women's Hospital (BWH) were included. Eligible patients underwent tumor-only sequencing using the DFCI/BWH OncoPanel test as part of routine care,13 and had either localized unfavorable intermediate- or high-risk hormone-sensitive prostate cancer (HSPC) by NCCN criteria,3 (2) metastatic HSPC (mHSPC), or (3) metastatic castration-resistant prostate cancer (mCRPC; Appendix Fig A1). Written informed consent was obtained for all study participants. This study was approved by the DFCI IRB (Protocol 17-409).
Germline and Tumor Sequencing and Variant Curation
Germline DNA was isolated from buffy coat using the QIAGEN DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). Germline DNA was sequenced using a next-generation sequencing panel in a laboratory with Clinical Laboratory Improvement Amendments certification (CancerNext Expanded 67-gene panel, Ambry Genetics, Aliso Viejo, CA). Germline variants were annotated by Ambry Genetics and classified in accordance with the American College of Medical Genetics and Genomics and the Association for Molecular Pathology as pathogenic (P), likely pathogenic (LP), variant of unknown significance, likely benign, or benign.14 Only P/LP variants were included in this analysis.
Tumor samples were sequenced by the Center for Advanced Molecular Diagnostics, a Clinical Laboratory Improvement Amendments–certified clinical laboratory at BWH, using OncoPanel, a custom hybrid-capture sequencing assay.13 Sequence variants, copy-number alterations, and structural variants were identified as previously described.15,16 The lower limit of detection for sequence variants was 10% allelic fraction at 50× coverage. Likely polymorphisms and artifacts were filtered by comparing variant calls to in-batch normal controls, a panel of normal samples, and variants found in gnomAD databases at > 0.1% frequency in any subpopulation and/or the NHLBI Exome Sequencing Project. Variants flagged for filtering that were present in the COSMIC database at least twice were subsequently rescued. Three versions of OncoPanel were used during the study period, namely POPv1 (275 genes), POPv2 (300 genes), and POPv3 (447 genes). Only the 49 genes common to all three versions of OncoPanel and the Ambry CancerNext Expanded germline panel were included in this analysis (Appendix Fig A2).
For purposes of this analysis, mutations from OncoPanel were considered to be pathogenic if they were nonsense, splice site, small insertions or deletions, or missense substitutions predicted to be probably damaging by Polyphen-2 or deleterious by SIFT.17,18 Among the 49 genes included in the analysis, the following alterations were considered clinically actionable in PCa (Appendix Fig A2): deleterious alterations in homologous recombination (HR) genes included in the FDA approval for olaparib in mCRPC (ATM, BRCA1, BRCA2, BRIP1, CHEK2, and PALB2)19 and deleterious alterations in mismatch repair (MMR) genes (MLH1, MSH2, MSH6, and PMS2), which predict response to pembrolizumab across solid tumors.20
Clinical Outcomes
Patients in the cohort treated with olaparib for mCRPC were identified through chart review. Best prostate-specific antigen (PSA) response to olaparib was measured as the lowest on-treatment PSA relative to baseline PSA. Radiographic progression-free survival on olaparib was defined as time from treatment initiation to radiographic progression or death determined by a clinician blinded to patients' genomic status. For time-to-event analysis, patients without events were censored at the date of last imaging or death.
Statistical Analysis
Differences between groups were compared using Fisher's exact test for categorical variables and Mann-Whitney test for continuous variables. All P values were two-sided, with P < .05 considered statistically significant. The distribution of time-to-event was estimated using the Kaplan-Meier method.
RESULTS
Patient Characteristics
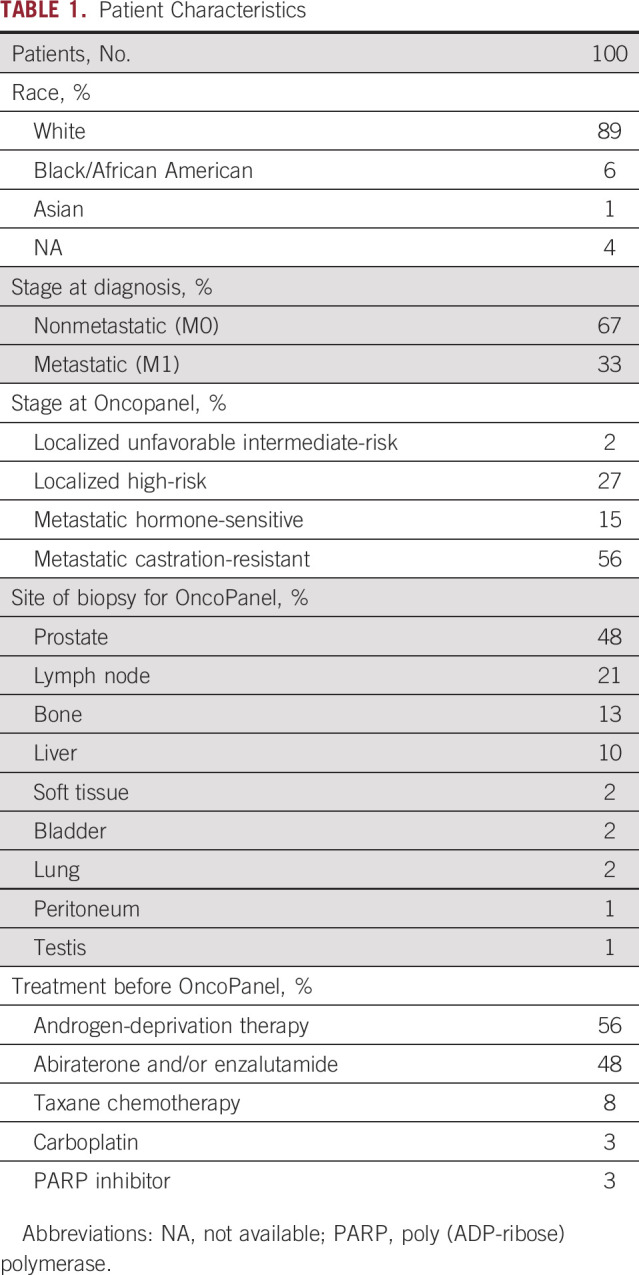
Among the 662 patients enrolled in the ProGen clinical trial, 100 met all eligibility criteria for inclusion in this analysis (Appendix Fig A1). Patient characteristics are listed in Table 1. At the time of Oncopanel, 29% had localized HSPC, 15% had mHSPC, and 56% had mCRPC. Therapy before OncoPanel included abiraterone and/or enzalutamide (48%), taxane chemotherapy (8%), and carboplatin or poly (ADP-ribose) polymerase (PARP) inhibitor (6%).
TABLE 1.
Patient Characteristics
Prevalence of Germline and Somatic Alterations
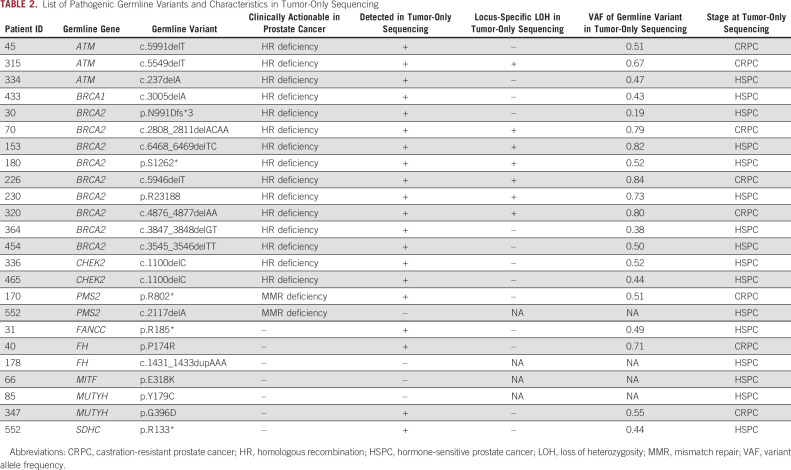
In the 100 patients who underwent germline testing, 23% harbored P/LP germline variants (Table 2). The most frequently altered gene was BRCA2 (9%). Additional variants were present in ATM (3%), CHEK2, FH, MUTYH, and PMS2 (2% each), and BRCA1, FANCC, MITF, and SDHC (1% each). One patient harbored variants in PMS2 and SDHC. All variants were heterozygous. Consistent with prior reports, clinically actionable P/LP germline variants were present in 17% of patients—15% with HR alterations and 2% with MMR alterations.21,22
TABLE 2.
List of Pathogenic Germline Variants and Characteristics in Tumor-Only Sequencing
OncoPanel identified a similar spectrum of somatic alterations to prior reports (Fig 1A; Appendix Fig A2).1,23 CRPC tumors harbored a significantly higher number of clinically actionable somatic alterations than HSPC tumors (P = .0017; Fig 1B). CRPC tumors were also significantly more likely than HSPC tumors to harbor a clinically actionable somatic alteration (38% v 9%; P = .0011; Fig 1C). Overall, clinically actionable somatic alterations were present in 19% of patients, including 16% with HR alterations, 2% with MMR alterations, and 1% with both HR and MMR alterations (Fig 1D). The most frequent clinically actionable somatic HR alterations were in ATM (9%) and BRCA2 (8%). Additional alterations were present in BRCA1 (2%), PALB2 (2%), and CHEK2 (1%). Clinically actionable somatic MMR alterations were present in MSH6 (2%) and PMS2 (1%). Integrating the germline-only and tumor-only data identified 33% of patients with clinically actionable P/LP germline variants and/or somatic alterations, including 29% with HR alterations and 5% with MMR mutations (Fig 1D).
FIG 1.
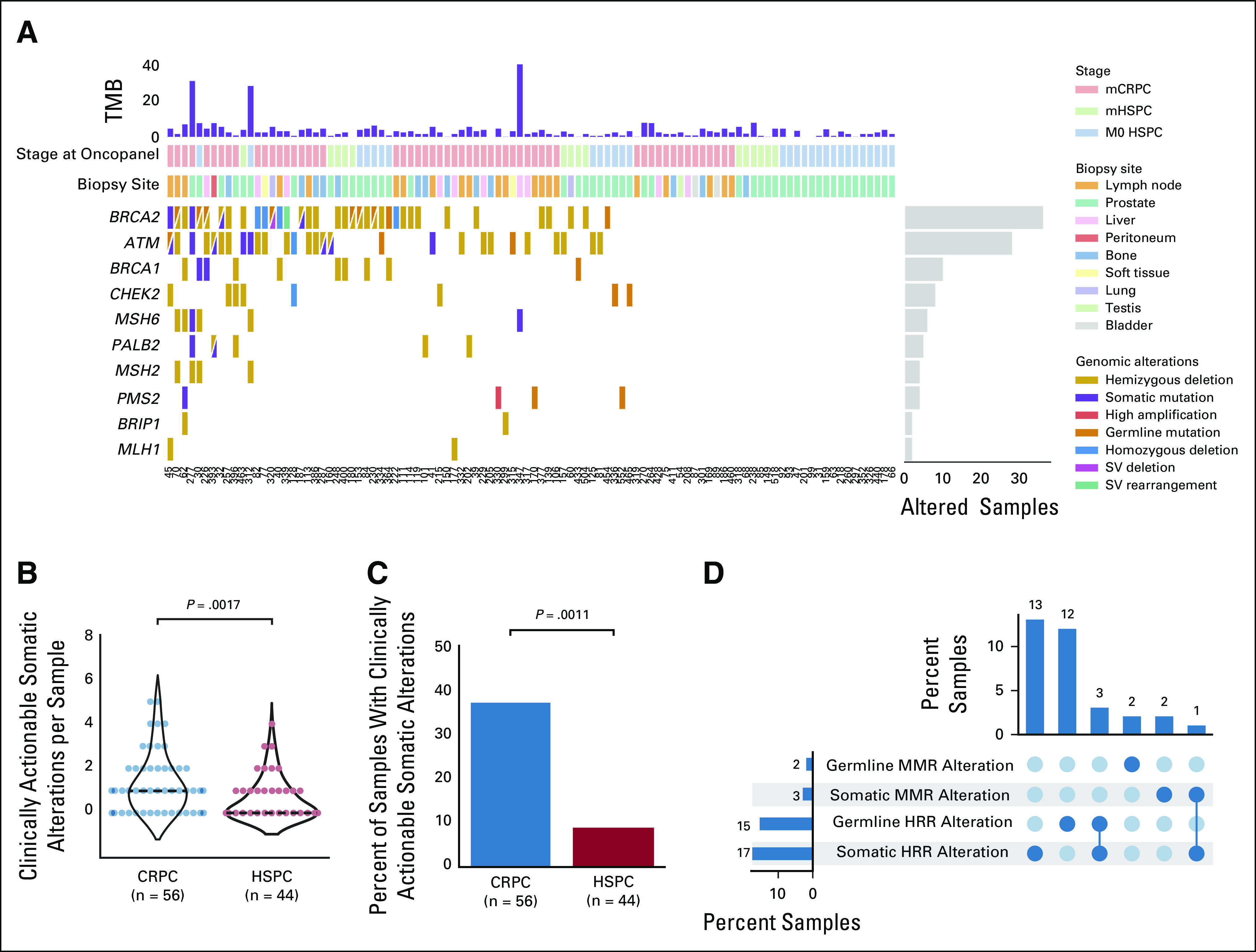
Landscape of clinically actionable germline and somatic alterations in 100 men with advanced prostate cancer. (A) CoMut plot of clinically actionable alterations in prostate cancer integrating germline testing and OncoPanel tumor-only sequencing data. Columns represent individual patients, ordered by number of alterations. Rows represent specific clinically actionable genes in prostate cancer, ordered by frequency. Mutations per Mb are shown in the upper histogram, and incidence of alterations in the cohort is in the right histogram. Cases with multiple alterations in a gene are represented by split colors. (B) Violin plot showing the number of clinically actionable somatic alterations per sample for CRPC versus HSPC tumors. Dashed black line indicates the median. (C) Bar graph showing the proportion of samples harboring clinically actionable somatic alterations in CRPC versus HSPC tumors. (D) Upset plot showing the proportion of samples harboring specific clinically actionable alterations across the entire cohort. CRPC, castration-resistant prostate cancer; HRR, homologous recombinational repair; HSPC, hormone-sensitive prostate cancer; mCRPC, metastatic castration-resistant prostate cancer; mHSPC, metastatic hormone-sensitive prostate cancer; MMR, mismatch repair; SV, structural variant; TMB, tumor mutational burden.
Limitations of Tumor-Only Sequencing to Report Germline Variants
Limitations of tumor-only sequencing can result in under-reporting of PGVs. To evaluate the accuracy of tumor-only sequencing in PCa, we assessed the number of bona fide P/LP germline variants that were reported in OncoPanel results. Of the 24 P/LP germline variants, four (17%) were not reported, including one (5.9%) of 17 clinically actionable variants (Table 2). Review of these four cases demonstrated that all four germline variants were present in the raw binary version of the sequencing alignment/map files. PMS2 c.2117delA, a clinically actionable germline variant in PCa, was not reported because of insufficient sequencing depth—the 32× coverage at this locus was below the validated assay coverage threshold of > 50× for accurate variant calling. The other three P/LP germline variants in FH, MITF, and MUTYH were not reported because of a filter that removes suspected germline variants; this is standard for tumor-only sequencing and analysis approaches to convert the binary version of the sequencing alignment/map files into processed variants. Although these genes are not clinically actionable in PCa, not reporting these variants would result in missed opportunities for cascade testing to identify affected family members.
We next evaluated whether the origin of an alteration—germline versus somatic—can be accurately inferred using variant allele frequency (VAF) from tumor-only sequencing. The OncoPanel VAF was significantly higher for germline than somatic variants (median 52%; interquartile range [IQR] 47%-72%] v 34% [IQR 17%-48%]; P = 7.2 × 10−5; Fig 2A). However, OncoPanel VAF had limitations for accurately predicting whether variants were germline or somatic. The ESMO Precision Medicine Working Group recommends that a variant on tumor-only sequencing be considered for germline-focused analysis if VAF is > 30% for single nucleotide variants or > 20% for indels.24 These cutoffs demonstrated high sensitivity for identifying variants of germline origin as 95% (n = 19/20) of P/LP germline variants had a VAF above these thresholds (Fig 2B). However, these cutoffs lacked specificity. Only 27% (n = 19/71) of variants with a VAF above the ESMO thresholds were of germline origin (Fig 2C). Although the majority (97%; n = 32/33) of alterations below the ESMO thresholds were somatic, there was one germline variant. The patient with a germline mutation below the specified thresholds harbored a germline BRCA2 frameshift mutation with a VAF of 19% on tumor sequencing despite 60% tumor content. Review of this case revealed a somatic heterozygous deletion of the allele inferred to carry the germline variant.
FIG 2.
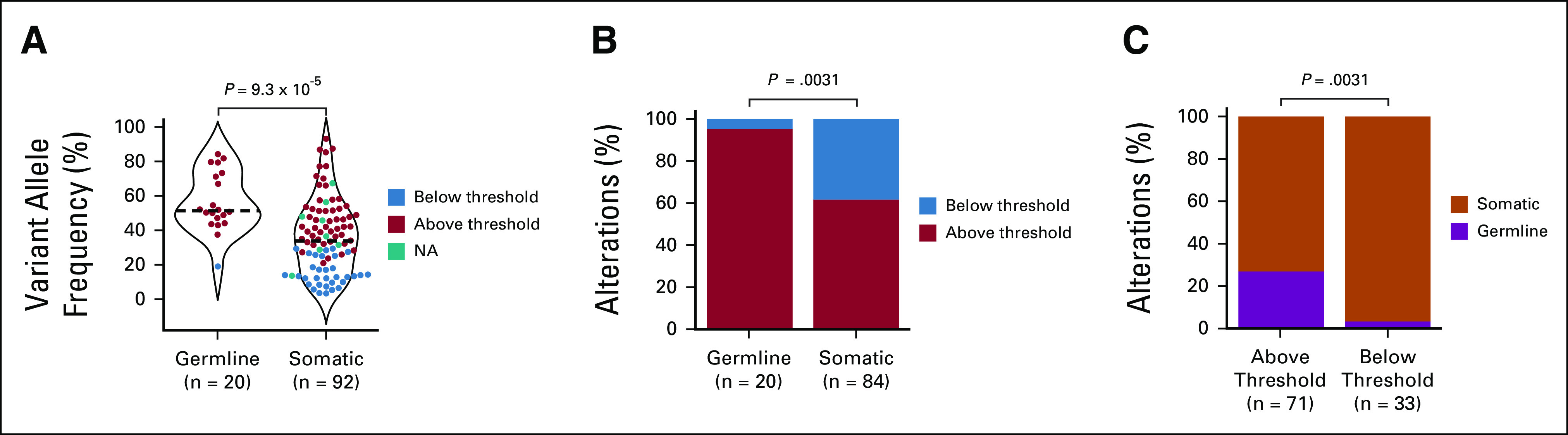
Limitations of tumor-only sequencing to report germline variants. (A) Violin plot showing the variant allele frequency for pathogenic germline and somatic variants. The color scheme indicates whether variants in the tumor‐only sequencing were above or below the ESMO Precision Medicine Working Group recommended thresholds for follow-up germline testing: >30% for SNVs or >20% for small indels.24 Splice variants are labeled as not applicable (N/A) given the absence of specific recommendations regarding a variant allele frequency threshold for follow-up germline testing. Dashed black line indicates the median. (B) Proportion of pathogenic germline and somatic variants that are above versus below the ESMO Precision Medicine Working Group thresholds. (C) Proportion of pathogenic variants that are above or below the ESMO Precision Medicine Working Group thresholds that are of germline versus somatic origin.
Locus-Specific Loss of Heterozygosity Associates With CRPC and BRCA2
Tumor suppressor genes, including most clinically actionable genes in PCa, canonically require inactivation of both alleles to affect phenotypic change. Given the association of deleterious alterations in BRCA2 with disease aggressiveness and development of mCRPC, we hypothesized that CRPC (compared with HSPC) and alterations in BRCA2 (compared with other HR genes) would be associated with higher rates of locus-specific loss of heterozygosity (LOH) for PGVs ie, somatically acquired inactivation of the second allele resulting in biallelic loss.25,26 Of the 15 clinically actionable PGVs in HR genes, 47% (n = 7) demonstrated locus-specific LOH on OncoPanel. VAF was significantly higher for mutations with versus without locus-specific LOH (median VAF 0.79 [IQR 0.70-0.81] v 0.46 [0.42-0.51]; P = .0018; Fig 3A). VAF > 55% had 100% specificity and 86% sensitivity to identify variants with locus-specific LOH. There was a trend toward higher rates of locus-specific LOH in CRPC than HSPC (80% v 30%; P = .12) and BRCA2 than other HR genes (67% v 17%; P = .12), but these were not statistically significant in the setting of small sample size (Fig 3B). We observed locus-specific LOH in 100% of BRCA2 mutations from CRPC tumors (n = 3/3), 50% of mutations in other HR genes from CRPC tumors (n = 1/2), 50% of BRCA2 mutations from HSPC tumors (n = 3/6), and 0% of mutations in other HR genes from HSPC tumors (n = 0/4; Fig 3B). BRCA2 mutations in CRPC tumors were significantly more likely to demonstrate locus-specific LOH than mutations in other HR genes in HSPC tumors (P = .029).
FIG 3.
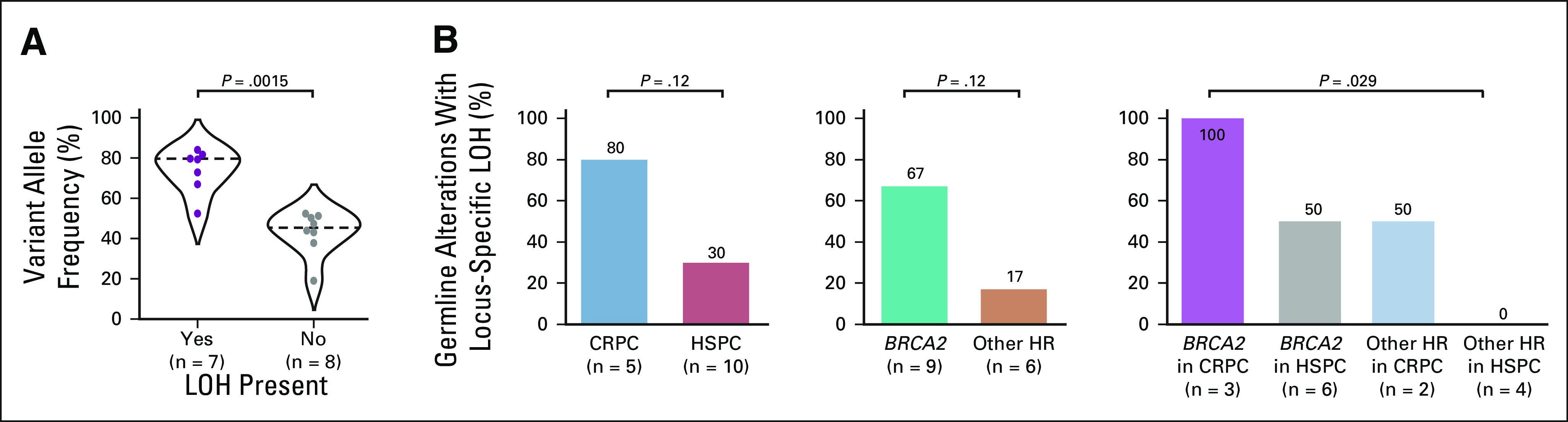
Locus-specific LOH associates with CRPC and BRCA2. (A) Violin plot showing the variant allele frequency for pathogenic germline and somatic variants with versus without LOH. (B) Bar graph showing the proportion of variants with LOH broken down by CRPC versus HSPC and BRCA2 versus other HR genes. CRPC, castration-resistant prostate cancer; HR, homologous recombination; HSPC, hormone-sensitive prostate cancer; LOH, loss of heterozygosity.
Response to Olaparib in mCRPC Strongly Associates With HR Deficiency
The PARP inhibitor, olaparib, is FDA-approved for men with mCRPC harboring deleterious HR alterations.19 Of the 14 patients in this cohort treated with olaparib for mCRPC, nine had clinically actionable HR alterations (HR-deficient)—eight with BRCA2 alterations and one with both ATM and PALB2 alterations—all of which were homozygous deletions or deleterious mutations with locus-specific LOH. The remaining five patients had no clinically actionable HR alterations (HR-proficient). Consistent with its known activity in biomarker-selected patients, 100% of HR-deficient patients achieved > 50% decrease in PSA (PSA50) on olaparib compared with 20% of HR-proficient patients (P = .0050; Figs 4A and 4B). Men with HR-deficient mCRPC had significantly longer radiographic progression-free survival to olaparib compared with those with HR-proficient disease (median 23.6 v 3.7 months; hazard ratio = 0.14; 95% CI, 0.033 to 0.60; P = .0036; Fig 4C).
FIG 4.
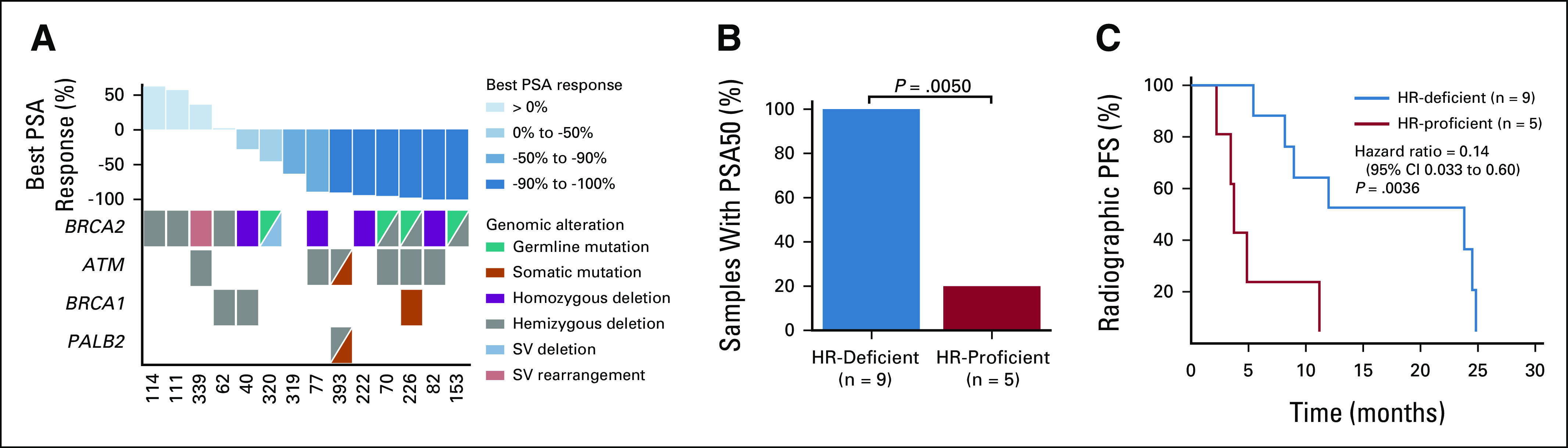
Response to olaparib in mCRPC strongly associates with HR deficiency. (A) Best PSA response to olaparib in 14 men with mCRPC. Columns represent individual patients, ordered by best PSA response. Rows represent specific genes, ordered by frequency. (B) Bar graph showing the proportion of HR-deficient versus HR‐proficient patients who achieved a decrease in PSA of 50% or more (PSA50) on olaparib. (C) Kaplan-Meier curve showing the radiographic PFS for HR‐deficient versus HR-proficient patients treated with olaparib. HR, homologous recombination; mCRPC, metastatic castration-resistant prostate cancer; PFS, progression-free survival; PSA, prostate-specific antigen; SV, structural variant.
DISCUSSION
We report the results from 100 men with advanced PCa who underwent independent germline testing and tumor-only sequencing. Several findings have clinical relevance for management of patients with advanced PCa. First, one third of men harbored clinically actionable alterations with similar contribution from germline and somatic alterations. Second, tumor-only sequencing failed to report several PGVs. Third, analyzing VAF in tumor-only sequencing lacked accuracy for predicting whether variants were of germline versus somatic origin. Fourth, locus-specific LOH was more commonly observed in BRCA2 than other HR genes and in CRPC than HSPC. Finally, men with HR-deficient mCRPC achieved significantly better response to olaparib than those with HR-proficient tumors. These findings highlight the importance of paired germline and somatic testing in PCa.
Approximately 11%-17% of men with advanced PCa harbor PGVs, many of which are clinically actionable.2,21 Despite NCCN and ESMO guideline recommendations for germline testing for all men with metastatic PCa,3 widespread implementation is suboptimal, especially for minority populations.10 We observed that 17% of PGVs, including one clinically actionable germline variant in this patient set, were not reported from tumor-only sequencing. This is consistent with a recent analysis of > 21,000 patients with solid tumors from MSK-IMPACT, in which 9% of PGVs were not reported on tumor-only sequencing.5 In our study, this was largely because of filtering on the basis of population frequency; tumor-only sequencing assays rely on computational algorithms to filter out presumed germline variants and, for some assays, subsequently rescue well-documented PGVs for reporting. This is an imperfect system, with significant interlaboratory variability, and limitations to this approach can result in PGVs not being reported, which is especially problematic in non-White patient populations because of lack of sufficient representation in databases used for in silico filtering.27 We also found that PGVs can have low VAF on tumor sequencing because of somatic deletion of the affected allele. This was exemplified by a patient with a pathogenic germline BRCA2 mutation with a VAF of 19% on OncoPanel, which is below the ESMO Precision Medicine Working Group cutoff for follow-up germline testing.24 This was observed in 2% of germline variants in the MSK-IMPACT cohort.5 These observations highlight the limitations of tumor-only sequencing in PCa. The decision to order germline testing based solely on finding presumed germline variants in tumor-only sequencing results would miss more than 20% of PGVs in our cohort, depriving such patients the opportunity for informed cancer screening and cascade testing to identify affected family members. These findings strongly reinforce the importance of germline testing in PCa.
Deleterious alterations in HR genes are a predictive biomarker of response to PARP inhibitors in men with advanced PCa.19 However, there is significant heterogeneity in response depending on which gene is altered and the type of alteration. This study provides four insights into our evolving understanding of the diversity of HR alterations as predictive biomarkers in PCa. First, we observed a higher rate of locus-specific LOH for pathogenic BRCA2 germline variants than those in other HR genes (67% v 17%). Genomic analysis of tumors from men with HR-deficient mCRPC found that two-copy loss—homozygous deletion or deleterious mutation with locus-specific LOH—was associated with higher response rates to olaparib than deleterious mutation without LOH.28 This observation may partially explain the clinical observation that, of all HR genes, alterations in BRCA2 most strongly associate with response to olaparib.19 Second, we observed a higher rate of LOH for PGVs in CRPC than HSPC tumors (80% v 30%). Several ongoing clinical trials are investigating PARP inhibitors in combination with ADT and an AR pathway inhibitor in men with HR-deficient mHSPC and as neoadjuvant therapy in men with localized high-risk PCa. Higher response rates to PARP inhibition in patients with locus-specific LOH and the lower rate of locus-specific LOH in HSPC than CRPC suggest that PARP inhibitor activity may be attenuated in earlier disease settings.28 Third, consistent with published data, we observed significantly greater benefit with olaparib in men with HR-deficient mCRPC, yet a minority of men with HR-proficient mCRPC also responded.29 Further characterization of occult mechanisms of HR deficiency and PARP inhibitor sensitivity, such as DNA methylation,30 is needed to improve precision care for men with PCa. Fourth, paired germline and somatic sequencing is required to optimize detection of clinically actionable HR alterations in men with advanced PCa. Germline-only and somatic-only testing identified clinically actionable HR alterations in 15% and 17% of men, respectively. When germline and somatic results were integrated, 28% of men harbored a clinically actionable HR alteration.
Our study has several limitations. First, our study was restricted to the subset of patients from the ProGen study who underwent tumor sequencing, raising the possibility for selection bias. Second, our analysis was limited to 49 genes common to the tumor and germline sequencing panels. Although this included most clinically actionable alterations in PCa, expanded molecular analyses may reveal additional complexities regarding these issues.31,32 Third, the sample size precluded definitive conclusions about associations of LOH with clinical-genomic features. Because all patients in this cohort with HR-deficient tumors had two-copy loss, we were unable to evaluate response on the basis of the presence or absence of locus-specific LOH. Finally, our cohort was composed largely of White men of European ancestry. This lack of diversity in race and ancestral origin may limit the generalizability of our results. Furthermore, this highlights the importance of including diverse populations in future clinical research efforts. Despite these limitations, this study provides insights into the complementary value of germline testing with tumor-only sequencing for men with advanced PCa.
In summary, we demonstrate that tumor-only sequencing with follow-up germline testing on the basis of VAF would miss more than 20% of PGVs in men with advanced PCa. We encourage clinicians to be cognizant of the limitations of tumor-only sequencing and obtain germline testing to maximize detection of PGVs and facilitate cascade testing to identify affected family members.
APPENDIX
FIG A1.
Study flow diagram. mCRPC, metastatic castration-resistant prostate cancer; mHSPC, metastatic hormone-sensitive prostate cancer.
FIG A2.
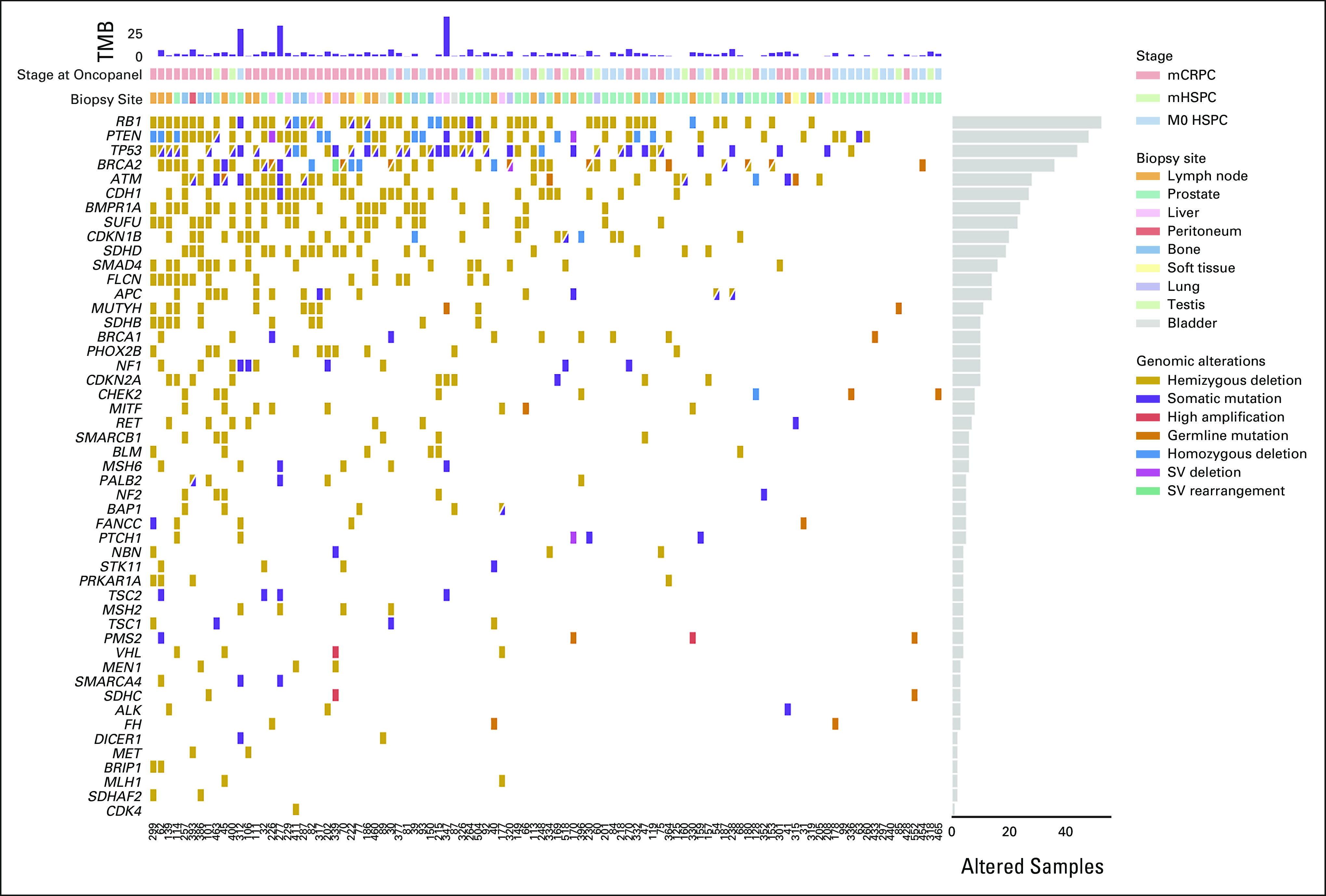
CoMut plot integrating germline testing and OncoPanel tumor-only sequencing data for the 49 genes common to all three versions of OncoPanel and the Ambry CancerNext Expanded germline panel. HSPC, hormone-sensitive prostate cancer; mCRPC, metastatic castration-resistant prostate cancer; mHSPC, metastatic hormone-sensitive prostate cancer; SV, structural variant; TMB, tumor mutational burden.
Jacob E. Berchuck
Employment: Cityblock Health
Stock and Other Ownership Interests: Genome Medical, Cityblock Health
Honoraria: UroToday
Consulting or Advisory Role: Genome Medical
Patents, Royalties, Other Intellectual Property: Institutional patent filed on methods for detecting prostate cancer resistance using cell-free DNA
Travel, Accommodations, Expenses: UroToday
Daniel Boiarsky
Stock and Other Ownership Interests: Tectonic Therapeutics
Rebecca Silver
Research Funding: Bayer
Harrison K. Tsai
Consulting or Advisory Role: Vertex
Alok K. Tewari
Stock and Other Ownership Interests: Moderna Therapeutics, Teladoc
Honoraria: UroToday
Consulting or Advisory Role: Best Doctors, Inc, B.A.I. Technologies
Travel, Accommodations, Expenses: AbbVie
Jonathan A. Nowak
Research Funding: NanoString Technologies, Illumina
Huma Q. Rana
Research Funding: Ambry Genetics, InVitae
Atish D. Choudhury
Employment: LeMaitre Vascular
Honoraria: Journal of Clinical Pathways/Oncology Learning Network, OncLive, Bayer, Targeted Oncology, Aptitude Health, Journal of Clinical Pathways/Oncology Learning Network, Cancer Network, Clinical Care Options, Great Debates and Updates, Pfizer
Consulting or Advisory Role: MedaCorp, Clovis Oncology, Dendreon, Bayer, Lilly, Blackstone, Astellas Pharma, AstraZeneca, Bayer, Blue Earth Diagnostics, Janssen Oncology, Tolmar
Research Funding: Janssen (Inst), Bayer
Travel, Accommodations, Expenses: Genentech
Mark M. Pomerantz
Honoraria: Bayer
Matthew L. Freedman
Stock and Other Ownership Interests: Precede
Consulting or Advisory Role: Precede
Patents, Royalties, Other Intellectual Property: A pending patent has been filed for detecting neuroendocrine prostate cancer using DNA methylation
Eliezer M. Van Allen
Stock and Other Ownership Interests: Syapse, Tango Therapeutics, Genome Medical, Microsoft, ervaxx, Monte Rosa Therapeutics, Manifold Bio, Genomic Life
Consulting or Advisory Role: Syapse, Roche, Third Rock Ventures, Takeda, Novartis, Genome Medical, InVitae, Illumina, Tango Therapeutics, ervaxx, Janssen, Monte Rosa Therapeutics, Manifold Bio, Genomic Life
Speakers' Bureau: Illumina
Research Funding: Bristol Myers Squibb, Novartis, Sanofi (Inst)
Patents, Royalties, Other Intellectual Property: Patent on discovery of retained intron as source of cancer neoantigens (Inst), Patent on discovery of chromatin regulators as biomarkers of response to cancer immunotherapy (Inst), Patent on clinical interpretation algorithms using cancer molecular data (Inst)
Travel, Accommodations, Expenses: Roche/Genentech
Mary-Ellen Taplin
Honoraria: Janssen-Ortho, Clovis Oncology, UpToDate, Research to Practice, Pfizer, AstraZeneca, Roivant, AbbVie, Arcus Biosciences, Constellation Pharmaceuticals, Epizyme, Targeted Oncology, Arvinas, Blue Earth Diagnostics, Hengrui Therapeutics
Consulting or Advisory Role: Janssen-Ortho, Bayer, Best Doctors, Inc, UpToDate, Clovis Oncology, Research to Practice, Myovant Sciences, Pfizer, AstraZeneca, Arcus Ventures
Research Funding: Janssen-Ortho (Inst)
Travel, Accommodations, Expenses: APCCC meeting
No other potential conflicts of interest were reported.
SUPPORT
J.E.B. is supported by the Department of Defense (W81XWH-20-1-0118). A.D.C. is supported by a Department of Defense Translational Science Award (W81XWH-20-1-0057). M.L.F. is supported by the Movember-Distinguished Gentleman's Ride-PCF Challenge Award, the H.L. Snyder Medical Research Foundation, the Cutler Family Fund for Prevention and Early Detection, the Donahue Family Fund, the National Cancer Institute (R01 CA251555), and the Department of Defense (W81XWH-19-1-0565 and W81XWH-21-1-0234). E.M.V.A. is supported by a PCF-V Foundation Challenge Award and a Mark Foundation Emerging Leader Award. M.-E.T. is supported by the Fairweather Family Fund and Pan-Mass Challenge Team IMAGINE, the Prostate Cancer Clinical Trial Consortium, a Prostate Cancer Foundation Challenge Award (16CHAL03), and the Dana-Farber/Harvard Cancer Center Prostate Cancer SPORE (NCI P50 CA090381). Financial assistance for germline testing for subjects enrolled in the ProGen study was provided by Ambry Genetics. The sponsors played no direct role in this study.
J.E.B., D.B., E.M.V.A., M.-E.T. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Jacob E. Berchuck, Daniel Boiarsky, Rebecca Silver, Rajitha Sunkara, Huma Q. Rana, Mary-Ellen Taplin
Administrative support: Neal I. Lindeman, Eliezer M. Van Allen
Provision of study materials or patients: Rajitha Sunkara, Neal I. Lindeman, Huma Q. Rana, Atish D. Choudhury
Collection and assembly of data: Jacob E. Berchuck, Daniel Boiarsky, Rebecca Silver, Rajitha Sunkara, Heather M. McClure, Neal I. Lindeman, Huma Q. Rana, Atish D. Choudhury, Mark M. Pomerantz, Mary-Ellen Taplin
Data analysis and interpretation: Jacob E. Berchuck, Daniel Boiarsky, Harrison K. Tsai, Stephanie Siegmund, Alok K. Tewari, Jonathan A. Nowak, Neal I. Lindeman, Huma Q. Rana, Atish D. Choudhury, Mark M. Pomerantz, Matthew L. Freedman, Eliezer M. Van Allen, Mary-Ellen Taplin
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jacob E. Berchuck
Employment: Cityblock Health
Stock and Other Ownership Interests: Genome Medical, Cityblock Health
Honoraria: UroToday
Consulting or Advisory Role: Genome Medical
Patents, Royalties, Other Intellectual Property: Institutional patent filed on methods for detecting prostate cancer resistance using cell-free DNA
Travel, Accommodations, Expenses: UroToday
Daniel Boiarsky
Stock and Other Ownership Interests: Tectonic Therapeutics
Rebecca Silver
Research Funding: Bayer
Harrison K. Tsai
Consulting or Advisory Role: Vertex
Alok K. Tewari
Stock and Other Ownership Interests: Moderna Therapeutics, Teladoc
Honoraria: UroToday
Consulting or Advisory Role: Best Doctors, Inc, B.A.I. Technologies
Travel, Accommodations, Expenses: AbbVie
Jonathan A. Nowak
Research Funding: NanoString Technologies, Illumina
Huma Q. Rana
Research Funding: Ambry Genetics, InVitae
Atish D. Choudhury
Employment: LeMaitre Vascular
Honoraria: Journal of Clinical Pathways/Oncology Learning Network, OncLive, Bayer, Targeted Oncology, Aptitude Health, Journal of Clinical Pathways/Oncology Learning Network, Cancer Network, Clinical Care Options, Great Debates and Updates, Pfizer
Consulting or Advisory Role: MedaCorp, Clovis Oncology, Dendreon, Bayer, Lilly, Blackstone, Astellas Pharma, AstraZeneca, Bayer, Blue Earth Diagnostics, Janssen Oncology, Tolmar
Research Funding: Janssen (Inst), Bayer
Travel, Accommodations, Expenses: Genentech
Mark M. Pomerantz
Honoraria: Bayer
Matthew L. Freedman
Stock and Other Ownership Interests: Precede
Consulting or Advisory Role: Precede
Patents, Royalties, Other Intellectual Property: A pending patent has been filed for detecting neuroendocrine prostate cancer using DNA methylation
Eliezer M. Van Allen
Stock and Other Ownership Interests: Syapse, Tango Therapeutics, Genome Medical, Microsoft, ervaxx, Monte Rosa Therapeutics, Manifold Bio, Genomic Life
Consulting or Advisory Role: Syapse, Roche, Third Rock Ventures, Takeda, Novartis, Genome Medical, InVitae, Illumina, Tango Therapeutics, ervaxx, Janssen, Monte Rosa Therapeutics, Manifold Bio, Genomic Life
Speakers' Bureau: Illumina
Research Funding: Bristol Myers Squibb, Novartis, Sanofi (Inst)
Patents, Royalties, Other Intellectual Property: Patent on discovery of retained intron as source of cancer neoantigens (Inst), Patent on discovery of chromatin regulators as biomarkers of response to cancer immunotherapy (Inst), Patent on clinical interpretation algorithms using cancer molecular data (Inst)
Travel, Accommodations, Expenses: Roche/Genentech
Mary-Ellen Taplin
Honoraria: Janssen-Ortho, Clovis Oncology, UpToDate, Research to Practice, Pfizer, AstraZeneca, Roivant, AbbVie, Arcus Biosciences, Constellation Pharmaceuticals, Epizyme, Targeted Oncology, Arvinas, Blue Earth Diagnostics, Hengrui Therapeutics
Consulting or Advisory Role: Janssen-Ortho, Bayer, Best Doctors, Inc, UpToDate, Clovis Oncology, Research to Practice, Myovant Sciences, Pfizer, AstraZeneca, Arcus Ventures
Research Funding: Janssen-Ortho (Inst)
Travel, Accommodations, Expenses: APCCC meeting
No other potential conflicts of interest were reported.
REFERENCES
- 1. Robinson D, Van Allen EM, Wu Y-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Comprehensive Cancer Network (NCCN) Prostate Cancer Guidelines Version 3.2022. 2022. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf [Google Scholar]
- 4. Parker C, Castro E, Fizazi K, et al. Prostate cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31:1119–1134. doi: 10.1016/j.annonc.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 5. Terraf P, Pareja F, Brown DN, et al. Comprehensive assessment of germline pathogenic variant detection in tumor-only sequencing. Ann Oncol. 2022;33:426–433. doi: 10.1016/j.annonc.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schienda J, Church AJ, Corson LB, et al. Germline sequencing improves tumor-only sequencing interpretation in a precision genomic study of patients with pediatric solid tumor. JCO Precis Oncol. 2021;5:1840–1852. doi: 10.1200/PO.21.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceyhan-Birsoy, PhD O, Misyura M, Mandelker D. A clinical approach to detecting germline pathogenic variants from tumor-only sequencing. JNCI Cancer Spectr. 2020;4:pkaa019. doi: 10.1093/jncics/pkaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Szymaniak BM, Facchini LA, Giri VN, et al. Practical considerations and challenges for germline genetic testing in patients with prostate cancer: Recommendations from the Germline Genetics Working Group of the PCCTC. JCO Oncol Pract. 2020;16:811–819. doi: 10.1200/OP.20.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paller CJ, Antonarakis ES, Beer TM, et al. Germline genetic testing in advanced prostate cancer; practices and barriers: Survey results from the germline genetics Working Group of the Prostate Cancer Clinical Trials Consortium. Clin Genitourin Cancer. 2019;17:275–282.e1. doi: 10.1016/j.clgc.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hann KEJ, Freeman M, Fraser L, et al. Awareness, knowledge, perceptions, and attitudes towards genetic testing for cancer risk among ethnic minority groups: A systematic review. BMC Public Health. 2017;17:503. doi: 10.1186/s12889-017-4375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weise N, Shaya J, Javier-Desloges J, et al. Disparities in germline testing among racial minorities with prostate cancer. Prostate Cancer Prostatic Dis. 2022;25:403–410. doi: 10.1038/s41391-021-00469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rana HQ, Stopfer JE, Petrucelli N, et al. A randomized controlled trial of video-education or in-person genetic counseling for men with prostate cancer (ProGen) J Clin Oncol. 2020;38:1507–1507. [Google Scholar]
- 13. Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia EP, Minkovsky A, Jia Y, et al. Validation of OncoPanel: A targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141:751–758. doi: 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 16. Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1:e87062. doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 20. Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: Results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicolosi P, Ledet E, Yang S, et al. Prevalence of germline variants in prostate cancer and implications for current genetic testing guidelines. JAMA Oncol. 2019;5:523–528. doi: 10.1001/jamaoncol.2018.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berchuck JE, Zhang Z, Silver R, et al. Impact of pathogenic germline DNA damage repair alterations on response to intense neoadjuvant androgen deprivation therapy in high-risk localized prostate cancer. Eur Urol. 2021;80:295–303. doi: 10.1016/j.eururo.2021.03.031. [DOI] [PubMed] [Google Scholar]
- 23. Abida W, Cyrta J, Heller G, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mandelker D, Donoghue M, Talukdar S, et al. Germline-focussed analysis of tumour-only sequencing: Recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. 2019;30:1221–1231. doi: 10.1093/annonc/mdz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castro E, Romero-Laorden N, Del Pozo A, et al. PROREPAIR-B: A prospective cohort study of the impact of germline DNA repair mutations on the outcomes of patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2019;37:490–503. doi: 10.1200/JCO.18.00358. [DOI] [PubMed] [Google Scholar]
- 27. Garofalo A, Sholl L, Reardon B, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 2016;8:79. doi: 10.1186/s13073-016-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carreira S, Porta N, Arce-Gallego S, et al. Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B trial. Cancer Discov. 2021;11:2812–2827. doi: 10.1158/2159-8290.CD-21-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kondrashova O, Topp M, Nesic K, et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9:3970. doi: 10.1038/s41467-018-05564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reardon B, Moore ND, Moore NS, et al. Integrating molecular profiles into clinical frameworks through the Molecular Oncology Almanac to prospectively guide precision oncology. Nat Cancer. 2021;2:1102–1112. doi: 10.1038/s43018-021-00243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. AlDubayan SH, Conway JR, Camp SY, et al. Detection of pathogenic variants with germline genetic testing using deep learning vs standard methods in patients with prostate cancer and melanoma. JAMA. 2020;324:1957–1969. doi: 10.1001/jama.2020.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]