PURPOSE
The prospective Neoadjuvant Breast Registry Symphony Trial compared the 80-gene molecular subtyping signature with clinical assessment by immunohistochemistry and/or fluorescence in situ hybridization in predicting pathologic complete response (pCR) and 5-year outcomes in patients with early-stage breast cancer.
METHODS
Standard-of-care neoadjuvant chemotherapy combined with trastuzumab or trastuzumab plus pertuzumab was given to patients with human epidermal growth factor receptor 2 (HER2)–positive tumors (n = 295). pCR was the primary end point, with secondary end points of distant metastasis-free survival and overall survival at 5 years.
RESULTS
Among clinically defined HER2-positive (cHER2) tumors, the 80-gene assay identified 29.5% (87 of 295) as Luminal-Type (cHER2/gLuminal), 14.9% (44 of 295) as Basal-Type (cHER2/gBasal), and 55.6% (164 of 295) as HER2-Type (cHER2/genomically classified as HER2 [gHER2]). Patients with cHER2/gHER2 tumors had a higher pCR rate (61.6%) compared with non-gHER2 tumors (26.7%; P < .001). Dual targeting for cHER2/gHER2 tumors yielded a higher pCR rate (75%) compared with those treated with single HER2-targeted therapy (54%; P = .006). For cHER2/gBasal tumors, the 42.9% pCR rate observed with dual targeting was not different from that with trastuzumab alone (46.4%; P = .830). Among those with cHER2/gBasal tumors, 5-year distant metastasis-free survival (68.6%; 95% CI, 49.1 to 81.9) was significantly worse than in patients with cHER2/gLuminal tumors (88.9%; 95% CI, 78.0 to 94.6) and cHER2/gHER2 tumors (87.4%; 95% CI, 80.2 to 92.2; P = .010), with similar corresponding overall survival differences.
CONCLUSION
The 80-gene assay identified meaningful genomic diversity in patients with cHER2 disease. Patients with cHER2/gHER2 tumors, who benefitted most from dual HER2-targeted therapy, accounted for approximately half of the cHER2 cohort. Genomically Luminal tumors had low pCR rates but good 5-year outcomes. cHER2/gBasal tumors derived no benefit from dual therapy and had significantly worse 5-year prognosis; these patients merit special consideration in future trials.
INTRODUCTION
Patients with histologically assessed human epidermal growth factor receptor 2 (HER2)–positive (HER2+) disease receive HER2-targeted therapy, such as trastuzumab, which results in prolonged survival.1-3 Pertuzumab, another HER2-targeted agent, provides dual targeting when combined with trastuzumab. The addition of pertuzumab to trastuzumab and chemotherapy improved pathologic complete response (pCR) rates and overall survival (OS) in patients with HER2+ disease.4,5 However, dual HER2-targeted therapy has additional financial costs and toxicities, and the benefit is not uniform across all patients.6 Benefit from HER2-targeted treatment may vary as a result of clinically relevant genomic diversity within HER2+ tumors, which is not identified by standard pathology.7,8 HER2 positivity is clinically assessed by immunohistochemistry (IHC) and/or fluorescence in situ hybridization (FISH). Yet, not all clinically defined HER2+ (cHER2) tumors have functional HER2 gene expression signatures,8-10 which may affect response to HER2-targeted therapy. Genomic information may be helpful in clinical management by clarifying the extent to which patients may benefit from single or dual HER2 targeting. Moreover, cHER2 tumors with discordant genomic signatures may not respond as favorably to HER2-directed treatment, suggesting a need for subtype-specific treatment.
CONTEXT
Key Objective
The 80-gene and 70-gene signatures (80-GS, 70-GS) further classify clinically assessed tumors, which more precisely corresponds with chemotherapy responses. In this study, the association between the combined 80-GS and 70-GS, chemotherapy response, and 5-year outcomes were evaluated in patients with histologically confirmed human epidermal growth factor receptor 2–positive (HER2+) breast cancer.
Knowledge Generated
Among HER2+ tumors, 80-GS identified 44.4% as genomically Luminal-Type or Basal-Type. Genomically HER2-Type tumors had significantly higher rates of pathologic complete response compared with clinically defined HER2+ tumors. Independent of single or dual HER2 therapy, patients with genomically Luminal-Type tumors had excellent 5-year outcomes, whereas genomically Basal-Type tumors had poor outcomes. By contrast, patients with genomically HER2-Type tumors benefitted most from dual therapy.
Relevance
Genomic characterization of clinically defined HER2+ tumors identifies patients who benefit from dual therapy and a substantial subset of patients with genomically non–HER2-Type tumors that may need alternative treatment. Understanding the underlying tumor biology may help tailor appropriate treatment options.
The Neoadjuvant Breast Registry Symphony Trial (NBRST) enrolled 1,091 patients with early-stage breast cancer and evaluated the prediction of response rates to neoadjuvant therapy by comparing the combined 70-gene risk classification (MammaPrint) and 80-gene molecular subtyping (BluePrint) signatures with traditional IHC/FISH assessment.11,12 MammaPrint is a gene expression–based assay that classifies tumors as having a Low Risk or High Risk of distant recurrence.13 BluePrint is an 80-gene molecular subtyping assay that stratifies tumors as Luminal-Type, HER2-Type, or Basal-Type.14 In NBRST, BluePrint and MammaPrint further classified 22% of clinically assessed tumors as a different subtype, which correlated more accurately with observed chemotherapy responses than IHC-/FISH-defined tumors.12 Importantly, IHC-/FISH-defined HER2+ tumors were the largest clinical subtype further classified by BluePrint in NBRST, with a substantial proportion classified as non–HER2-Type.11 During the enrollment period of NBRST, pertuzumab was approved as a neoadjuvant therapy for HER2+ disease, which provided the opportunity to evaluate two HER2-targeted treatment arms: trastuzumab versus trastuzumab plus pertuzumab. Here, we report pCR rates and 5-year outcomes in the cHER2 cohort from NBRST, stratified by BluePrint and treatment regimen, consisting of either single or dual HER2-targeted therapy.
METHODS
Study Cohort
Patients with early-stage breast cancer (N = 1,091) were enrolled into the prospective registry, NBRST (ClinicalTrial.gov identifier: NCT01479101), across 67 US institutions between 2011 and 2014. This study was conducted in accordance with the ethical standards established in the Declaration of Helsinki. Institutional Review Boards at all participating institutions approved the protocol, and informed consent for trial participation, which included tissue and clinical data collection, was obtained from all participants. Patients aged 18-90 years without metastatic disease and scheduled to receive neoadjuvant therapy were eligible for inclusion, and of the 1,069 eligible patients, 1,024 had available clinical data (Data Supplement). The current study focuses on 295 of 1,091 (27.0%) patients with histologically/FISH-confirmed HER2+ tumors. Patients with HER2+ tumors received chemotherapy with either single (trastuzumab) or dual (trastuzumab and pertuzumab) HER2-targeted therapy. Neoadjuvant and adjuvant therapies were given at the physician's discretion, in agreement with National Comprehensive Cancer Network guidelines and standard of care. Case report forms were used for clinical data collection, recurrence, and death events, and clinical data were collected 6 weeks after receiving MammaPrint and BluePrint results, 4 weeks postsurgery, 2-3 years postsurgery, and 5 years postsurgery.
Clinical and Molecular Subtyping
Preoperative core biopsies were used to assess hormone receptor (HR) and HER2 status by IHC or IHC/FISH, respectively, per ASCO/College of American Pathologists guidelines.15,16 IHC/FISH classified tumors as (1) estrogen receptor–positive and/or progesterone receptor–positive, and HER2-positive (HR+HER2+), or (2) estrogen receptor–negative and progesterone receptor–negative, and HER2-positive (HR–HER2+).
RNA from the same core biopsies was isolated for MammaPrint and BluePrint assays, which was performed as standard of care by Agendia (Irvine, CA). Tumors were classified with MammaPrint as Low Risk (MammaPrint index > 0.000) or High Risk (MammaPrint index ≤ 0.000).17 BluePrint categorizes tumors as Luminal-Type, HER2-Type, or Basal-Type.14
Objectives and End Points
The primary end point for NBRST was pCR, which is defined as the absence of invasive carcinoma (breast and axilla) on microscopic examination of the surgically resected tissue, irrespective of any carcinoma in situ (ypT0/TisN0).11 Distant metastasis-free survival (DMFS) and OS were end points for long-term follow-up. DMFS is defined as the time from the date of diagnosis to the date of first distant metastasis, death of any cause, or censoring at the last follow-up date. OS is defined as the date of diagnosis to death from any cause or censoring at the last follow-up date.
Statistical Analysis
Clinical characteristics were summarized using descriptive statistics. Differences in age were evaluated by one-way analysis of variance. Other clinical characteristics were assessed by chi-square or Fisher's exact test. Statistical significance was defined by two-sided P < .05 for all tests. Therapeutic response (pCR) rates for each subgroup were calculated. The pCR of each genomic subtype was compared with the pCR rate of the remaining HER2+ population using a two-tailed proportional z-test. For survival analyses, 5-year DMFS and OS survival curves were estimated by the Kaplan-Meier method and the log-rank test was used to determine survival differences. All statistical analyses were conducted using Stata, version 16 (Stata Corp, College Station, TX).
RESULTS
Clinical Characteristics
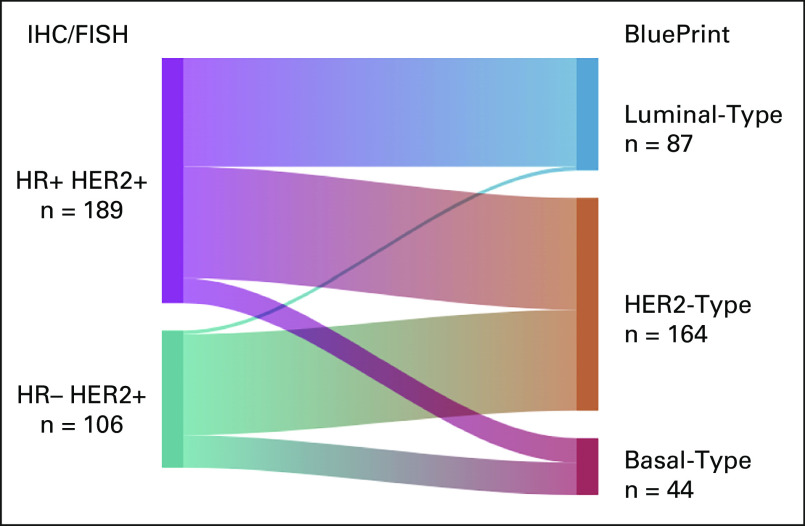
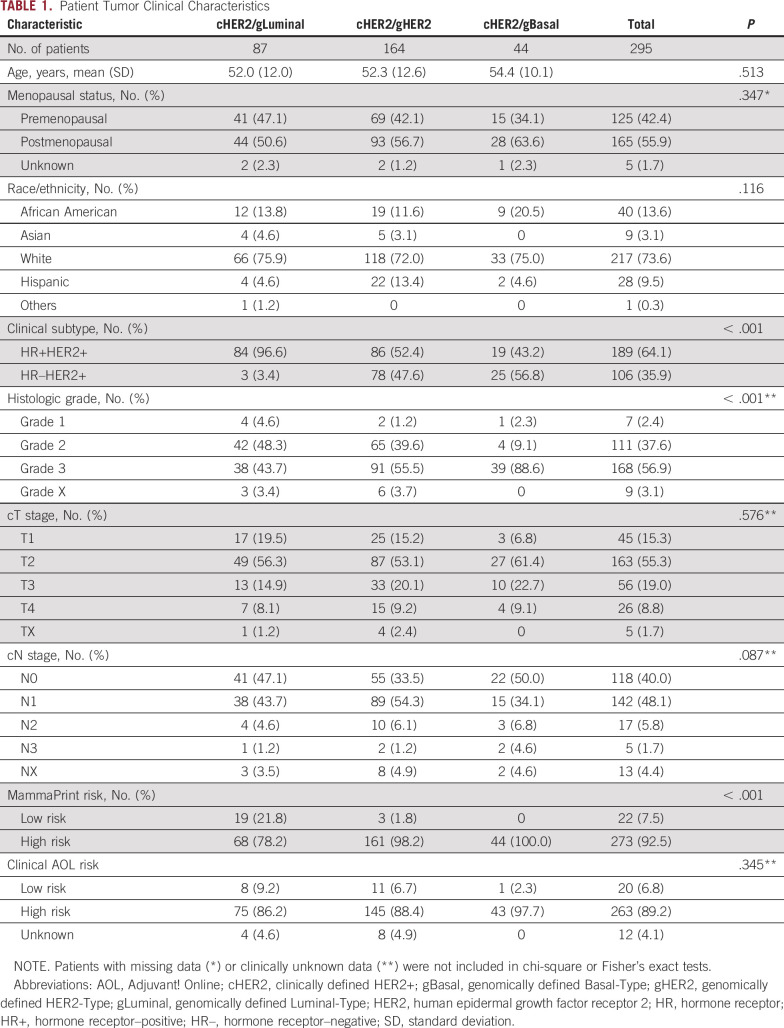
Overall, 295 eligible patients within NBRST were clinically defined as HER2+ by IHC/FISH (cHER2), of which 55.6% (164 of 295) were genomically classified as HER2-Type (cHER2/gHER2) and 44.4% as non–HER2-Type by BluePrint (29.5% [n = 87 of 295] Luminal-Type [cHER2/gLuminal] and 14.9% [n = 44 of 295] Basal-Type [cHER2/gBasal]; Fig 1). Of the 295 cHER2 patients, 92.5% were classified as High Risk by MammaPrint (Table 1).
FIG 1.
Further classification of IHC-/FISH-defined HER2+ patients with BluePrint. Sankey diagram depicting (Left) 295 IHC-/FISH-defined HER2+ patients sorted by (Right) genomic subtype using BluePrint into HER2-Type (cHER2/gHER2) or further classified as Luminal-Type (cHER2/gLuminal) or Basal-Type (cHER2/gBasal). cHER2, clinically defined HER2+; FISH, fluorescence in situ hybridization; gHER2, genomically classified as HER2; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; IHC, immunohistochemistry.
TABLE 1.
Patient Tumor Clinical Characteristics
Patient clinical characteristics within cHER2/gLuminal, cHER2/gHER2, and cHER2/gBasal tumors are summarized in Table 1. Median age (52.0-54.4 years) and menopausal status were comparable across BluePrint subtypes (P = .513 and P = .347, respectively). In total, 64.0% of patients had HR+ tumors (n = 189) and 36.0% had HR– tumors (n = 106). As is typical for neoadjuvant therapy candidates, most patients had grade 2-3 tumors (94.6%), larger tumors (T2-4; 83.1%), and positive lymph nodes (55.6%). The clinical adjuvant online risk, calculated per MINDACT guidelines,13 classified 89.2% of IHC-/FISH-defined HER2+ patients as high risk.
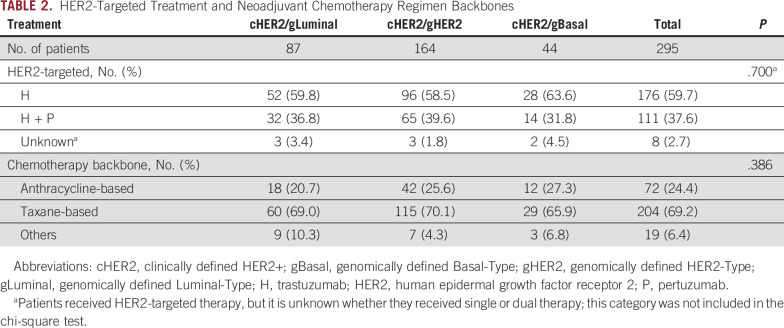
A similar distribution of patients who received single or dual HER2-targeted therapy was observed between BluePrint subtypes (Table 2; P = .700). In addition to standard-of-care HER2-targeted therapy (single or dual), 24.4% (72 of 295) of cHER2 patients received anthracycline-based chemotherapy followed by taxane chemotherapy, whereas 69.2% (204 of 295) received taxane-based chemotherapy (Table 2). Postoperative therapy followed standard HER2-targeted regimens.
TABLE 2.
HER2-Targeted Treatment and Neoadjuvant Chemotherapy Regimen Backbones
Therapeutic Response and 5-Year Outcome With BluePrint
The pCR rate among cHER2 patients was 46.1% (136 of 295). When further classified by BluePrint, the molecular subtypes were associated with more precise pCR rates (Fig 2A). To evaluate the pCR rates within BluePrint subtypes, the pCR rate of each subgroup (cHER2/gLuminal, cHER2/gHER2, or cHER2/gBasal) was compared with the pCR rate of the remaining cHER2 patients of different BluePrint subtypes. Patients with cHER2/gLuminal tumors had the lowest pCR rate of 17.2% (n = 15 of 87) relative to the remaining cHER2 patients (58.2%; n = 121 of 208; P < .001). Patients with cHER2/gHER2 tumors had a significantly higher pCR rate (61.6%; n = 101 of 164) relative to patients with non-gHER2 tumors (26.7%; n = 35 of 131; P < .001). Patients with cHER2/gBasal tumors had a 45.5% (n = 20 of 44) pCR rate although this was not significantly different when compared with the remaining cHER2 tumors (46.2%; n = 116 of 251; P = .926).
FIG 2.
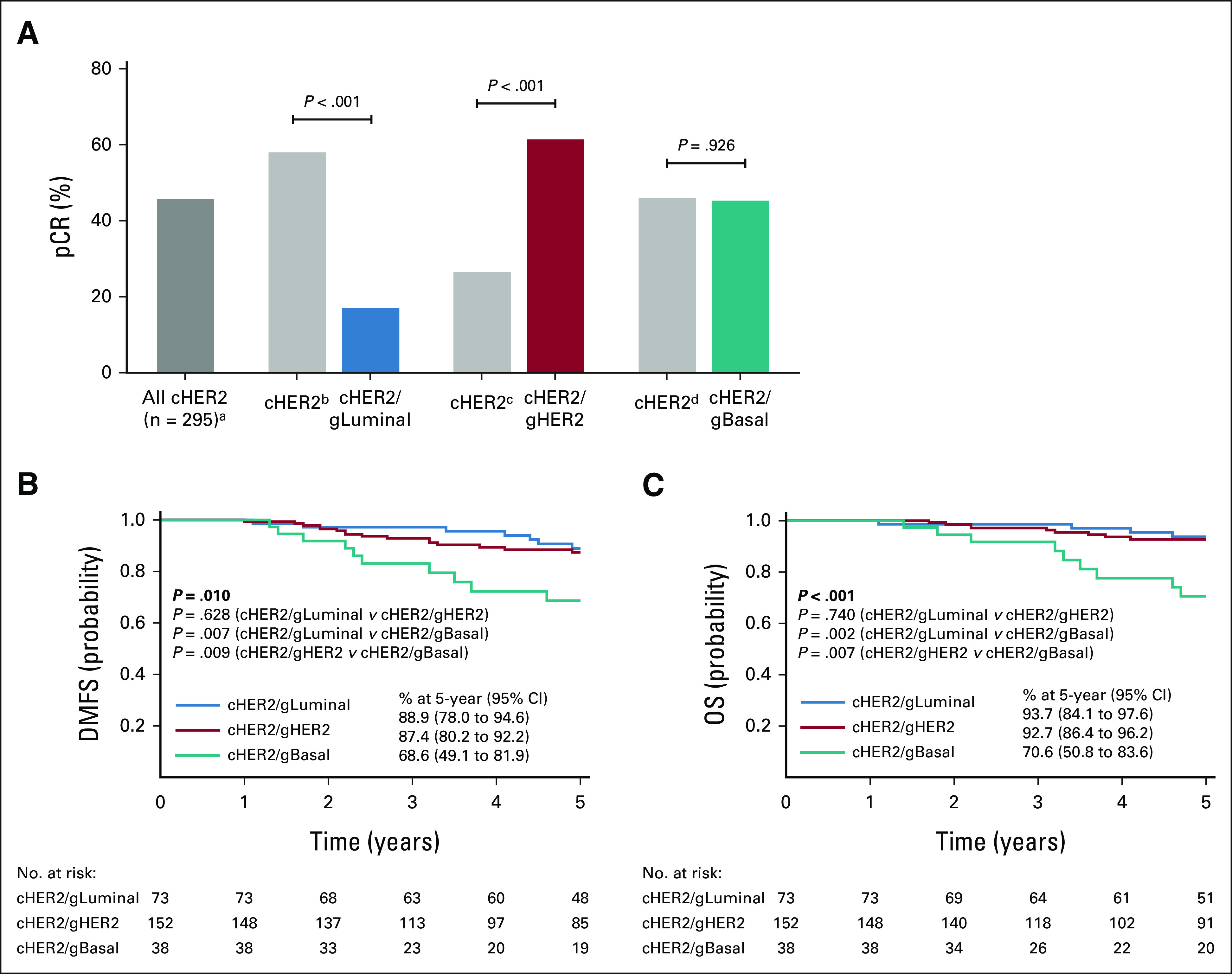
Therapeutic response and 5-year outcomes by BluePrint. (A) The pCR rate of all 295 cHER2 patients (adark gray bar) compared with pCR rates when stratified by BluePrint. The pCR rates of each BluePrint molecular subtype are shown (blue, cHER2/gLuminal; red, cHER2/gHER2; teal, cHER2/gBasal) with the remaining proportion of immunohistochemistry-/fluorescence in situ hybridization–defined HER2+ patients shown for each comparison in light gray bars: bcHER2/gHER2 and cHER2/gBasal, ccHER2/gLuminal and cHER2/gBasal, and dcHER2/gLuminal and cHER2/gHER2. Significance was evaluated using a two-tailed proportional z-test. Five-year probabilities of (B) DMFS and (C) OS for each BluePrint subgroup were assessed by the log-rank test. cHER2, clinically defined HER2+; DMFS, distant metastasis-free survival; gBasal, genomically defined Basal-Type; gHER2, genomically defined HER2-Type; gLuminal, genomically defined Luminal-Type; OS, overall survival; pCR, pathologic complete response.
Next, we evaluated the corresponding 5-year DMFS and OS rates for each BluePrint subtype. The median follow-up time was 5.4 years (range 0.3-7.3) for both DMFS and OS (n = 263/295). Five-year DMFS and OS probabilities in cHER2/gLuminal tumors were not significantly different from those in cHER2/gHER2 tumors (P = .628 and .740, respectively). However, patients with cHER2/gBasal tumors had significantly worse 5-year DMFS and OS outcomes compared with patients with cHER2/gHER2 (P = .009 and P = .007, respectively) and cHER2/gLuminal tumors (P = .007 and P = .002, respectively; Figs 2B and 2C). Patients with cHER2/gBasal tumors had a 5-year DMFS of 68.6% (95% CI, 49.1 to 81.9), which was significantly worse than those with cHER2/gHER2 (87.4%; 95% CI, 80.2 to 92.2) and cHER2/gLuminal tumors (88.9%; 95% CI, 78.0 to 94.6; P = .010; Fig 2B). OS was in line with DMFS outcomes, with the 5-year OS probability of 70.6% (95% CI, 50.8 to 83.6) in patients with cHER2/gBasal tumors, 92.7% (95% CI, 86.4 to 96.2) in patients with cHER2/gHER2, and 93.7% (95% CI, 84.1 to 97.6) in patients with cHER2/gLuminal tumors (P < .001; Fig 2C).
Association Between Therapeutic Response and 5-Year Outcomes
We assessed the association of pCR and 5-year outcomes in BluePrint subtypes, independent of therapy. The 5-year DMFS in patients with cHER2/gLuminal tumors was 100% with pCR versus 86.3% (95% CI, 73.2 to 93.2) with residual disease (Fig 3A; P = .185), and the OS probability was 100% with pCR versus 92.3% (95% CI, 80.6 to 97.1) with residual disease (Fig 3B; P = .329). In patients with cHER2/gHER2 tumors, the probability of DMFS was 9.3% higher in those who achieved a pCR (91.0%; 95% CI, 81.9 to 95.6) compared with those with residual disease (81.7%; 95% CI, 67.4 to 90.1; P = .086; Fig 3C). OS in patients with cHER2/gHER2 tumors was not significantly different between patients who achieved a pCR (93.2%; 95% CI, 84.4 to 97.1) versus those who did not (92.1%; 95% CI, 80.1 to 97.0; Fig 3D; P = .598). The probability of DMFS in patients with cHER2/gBasal tumors was substantially higher in those who achieved a pCR (84.9%; 95% CI, 51.2 to 96.0) compared with those who had residual disease (56.3%; 95% CI, 30.7 to 75.6; P = .066; Fig 3E). In addition, OS in patients with cHER2/gBasal tumors was significantly greater in patients who achieved a pCR (91.7%; 95% CI, 53.9 to 98.8) compared with those who did not (54.8%; 95% CI, 29.2 to 74.5; Fig 3F; P = .023).
FIG 3.
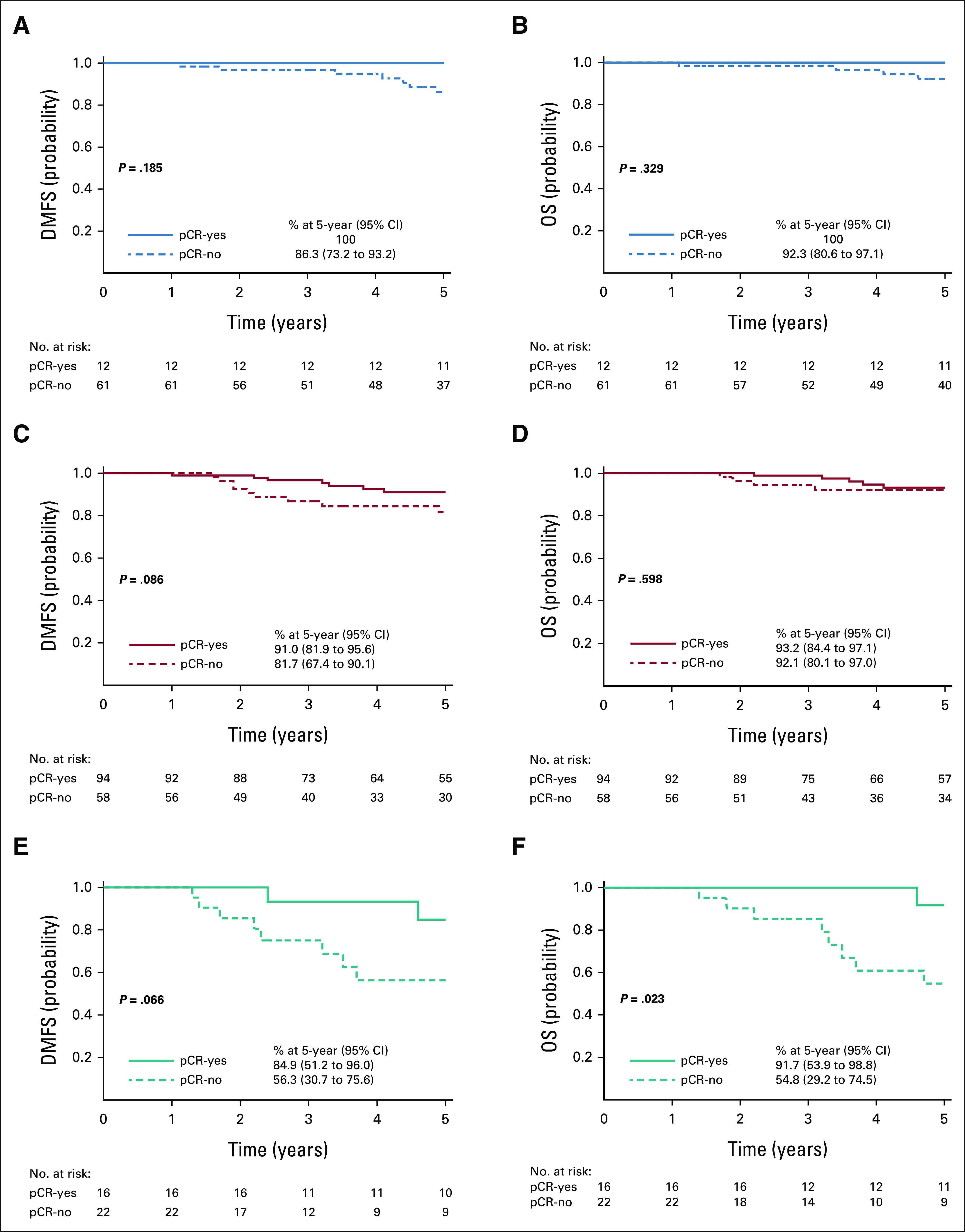
Association between therapeutic response and the 5-year outcome. Five-year probabilities of (A, C, and E) DMFS and (B, D, and F) OS stratified by pCR were assessed by the log-rank test for each BluePrint subtype: (A and B) cHER2/gLuminal, (C and D) cHER2/gHER2, and (E and F) cHER2/gBasal. cHER2, clinically defined HER2+; DMFS, distant metastasis-free survival; gBasal, genomically defined Basal-Type; gHER2, genomically defined HER2-Type; gLuminal, genomically defined Luminal-Type; HER2, human epidermal growth factor receptor 2; OS, overall survival; pCR, pathologic complete response.
Therapeutic Response and 5-Year Outcomes by Treatment Arm
To determine if a subset of patients with cHER2 tumors have improved outcomes with dual anti-HER2 therapy, we evaluated therapeutic response and outcomes stratified by HER2-targeted therapy and BluePrint molecular subtype. Of the 295 patients, 176 (59.7%) received single HER2-targeted therapy and 111 (37.6%) received dual targeted therapy, which was equally represented across BluePrint molecular subtypes (Fig 4A). Overall, patients treated with dual therapy had higher pCR rates (59.5%) versus those treated with single targeted therapy (39.2%; Fig 4B; P < .001). When stratified by BluePrint, however, patients with cHER2/gLuminal or cHER2/gHER2 tumors treated with dual therapy achieved significantly higher pCR rates (34.4% and 75.4%, respectively) compared with patients treated with single targeted therapy (7.7% and 54.2%; P = .002 and P = .006, respectively). By contrast, we observed no significant improvement in pCR rate in patients with cHER2/gBasal tumors with the addition of pertuzumab (42.9%) versus trastuzumab alone (46.4%; Fig 4C; P = .830).
FIG 4.
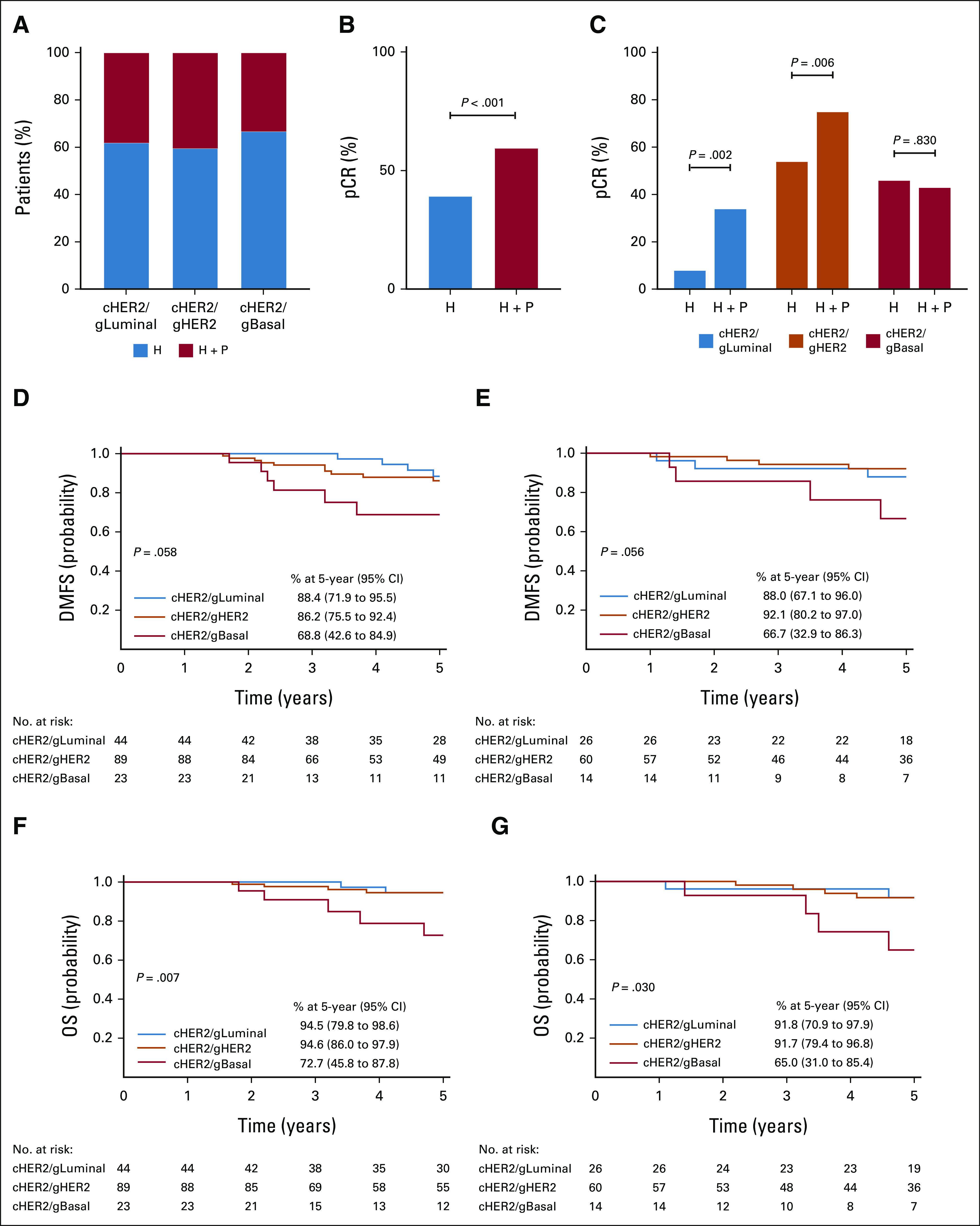
Therapeutic response and 5-year outcome by treatment arm. (A) The proportion of each BluePrint molecular subtype (cHER2/gLuminal, cHER2/gHER2, and cHER2/gBasal) treated with single HER2-targeted therapy (H; blue bars) versus dual HER2-targeted therapy (H + P; red bars). The pCR rate of (B) all immunohistochemistry-/fluorescence in situ hybridization–defined HER2+ patients and (C) BluePrint molecular subtypes (blue, cHER2/gLuminal; orange, cHER2/gHER2; and red, cHER2/gBasal), treated with H versus H + P. Significance was evaluated using a two-tailed proportional z-test. Five-year probabilities of (D and E) DMFS and (F and G) OS stratified by BluePrint molecular subtype and (D and F) treatment with H or (E and G) treatment with H + P. cHER2, clinically defined HER2+; DMFS, distant metastasis-free survival; gBasal, genomically defined Basal-Type; gHER2, genomically defined HER2-Type; gLuminal, genomically defined Luminal-Type; H, trastuzumab; HER2, human epidermal growth factor receptor 2; OS, overall survival; P, pertuzumab; pCR, pathologic complete response.
The 5-year DMFS probability differed by 19.6%-25.4% between BluePrint subtypes treated with single (P = .058) versus dual HER2-targeted therapy (P = .056; Figs 4D and 4E). We observed a similar range of 21.9%-26.7% in 5-year OS between BluePrint subtypes treated with single (P = .007) versus dual HER2-targeted therapy (Figs 4F and 4G; P = .030). Patients with cHER2/gLuminal tumors had comparable 5-year DMFS outcomes with single (88.4%; 95% CI, 71.9 to 95.5) or dual targeted therapy (88.0%; 95% CI, 67.1 to 96.0; Figs 4D and 4E and Data Supplement; P = .810). In addition, patients with cHER2/gLuminal tumors had excellent 5-year OS outcomes regardless of single (94.5%; 95% CI, 79.8 to 98.6) or dual targeted therapy (91.8%; 95% CI, 70.9 to 97.9; Figs 4F and 4G, Data Supplement; P = .650). The 5-year DMFS in patients with cHER2/gHER2 tumors given trastuzumab was 86.2% (95% CI, 75.5 to 92.4) compared with 92.1% (95% CI, 80.2 to 97.0) with trastuzumab plus pertuzumab (Figs 4D and 4E and Data Supplement; P = .355). The OS in patients with cHER2/gHER2 tumors was 94.6% (95% CI, 86.0 to 97.9) with single targeted therapy versus 91.7% (95% CI, 79.4 to 96.8) with dual targeted therapy (Figs 4F and 4G and Data Supplement; P = .597). Patients with cHER2/gBasal tumors had the worst 5-year outcomes regardless of the addition of pertuzumab, with 68.8% (95% CI, 42.6 to 84.9) DMFS probability with trastuzumab versus 66.7% (95% CI, 32.9 to 86.3) with trastuzumab and pertuzumab (Figs 4D and 4E and Data Supplement; P = .895) and similar OS (72.7%; 95% CI, 45.8 to 87.8 v 65.0%; 95% CI, 31.0 to 85.4, respectively; Figs 4F and 4G and Data Supplement; P = .684). Despite small numbers, when BluePrint subtypes were stratified by HR status, patients with HR+HER2+, gHER2 tumors had the greatest 5-year DMFS benefit with pertuzumab (12.3%; P = .084) compared with gLuminal tumors (–2.1%; P = .670) and gBasal tumors (8.7%; P = .705), albeit not significant (Data Supplement).
DISCUSSION
Among patients with IHC-/FISH-defined HER2+ tumors, those genomically classified as HER2-Type by BluePrint had the highest pCR rates compared with cHER2/gLuminal and cHER2/gBasal tumors, which also translated into excellent 5-year DMFS and OS outcomes, in line with studies using the PAM50 genomic classifer.18,19 Patients with cHER2/gHER2 tumors who achieved a pCR had a clinical benefit (9.3% increase in 5-year DMFS) compared with patients with residual disease. In addition, patients with \cHER2/gHER2 tumors treated with dual therapy exhibited the highest pCR rates, and although they did not have statistically significant 5-year DMFS benefit compared with single HER2-targeted therapy, the magnitude was 5.9% greater with dual therapy. Results from this NBRST study identified a larger clinical benefit in patients with HER2-Type tumors than that observed in the APHINITY trial, which reported a 0.9% benefit with the addition of pertuzumab to 3-year invasive disease-free survival (IDFS) and 3% benefit at 6 years.20,21 Molecular subtyping was performed using BluePrint on patients with HER2+ breast cancer enrolled in the APHINITY trial, which demonstrated patients with genomically HER2-Type tumors benefited most from the addition of pertuzumab to trastuzumab (hazard ratio 0.56; 95% CI, 0.27 to 1.15) compared with patients with cHER2/gLuminal (hazard ratio 0.93; 95% CI, 0.61 to 1.41) or cHER2/gBasal tumors (hazard ratio 0.89; 95% CI, 0.44 to 1.79).22 In the current study, patients with cHER2/gHER2 tumors had good 5-year OS, regardless of therapy. However, as concluded in the 6-year interim analysis of APHINITY, longer follow-up may be needed to assess OS, which did not reach statistical significance in either study.21 Together, these data suggest that the population of patients who benefit most from dual HER2-targeted therapy has genomically defined HER2-Type breast cancer.
Biologic diversity of HER2+ breast cancers has been described using BluePrint and other genomic assays such as PAM50, which consistently show a population of IHC/FISH-defined HER2+ patients with functionally activated Luminal or Basal pathways.8,23-25 Importantly, within this cHER2 NBRST cohort, BluePrint further classified 44.4% as either Luminal-Type or Basal-Type. BluePrint was performed on preoperative core biopsies, suggesting that this genomic diversity is not evident by IHC/FISH methods. Consistent with recent studies, even patients with cHER2/gLuminal tumors had the lowest pCR rates relative to cHER2/gHER2 or cHER2/gBasal tumors and they had good 5-year DMFS and OS outcomes, independent of the HER2-targeted therapy regimen or pCR rate, suggesting that these patients may derive the most significant benefit from endocrine therapy.19 Patients with cHER2/gBasal tumors had poorer 5-year DMFS and OS probabilities compared with those with cHER2/gLuminal and cHER2/gHER2 tumors, regardless of the addition of pertuzumab. Dual HER2-targeted therapy did not increase the proportion of patients with cHER2/gBasal tumors who achieved a pCR, and those with residual disease had significantly worse outcomes. These results indicate that treatment responses observed in patients with cHER2/gBasal tumors may be due to the chemotherapy backbone, rather than the HER2-targeted therapy, and these patients did not appear to benefit from the addition of pertuzumab to trastuzumab and chemotherapy. These data highlight the importance of identifying the genomic subtype, as cHER2/gBasal tumors represent a distinct subset within IHC-/FISH-defined HER2+ tumors requiring further investigation in light of their unique clinical behavior and therapy response.
As treatment recommendations and available targeted therapies for HER2+ disease rapidly change, it is important to understand the population of patients who will benefit most from all forms of HER2-targeted therapy. An interim analysis from the KATHERINE trial demonstrated that patients with residual disease after neoadjuvant therapy, most of whom were HR+HER2+, who were given adjuvant trastuzumab emtansine (T-DM1) had better 3-year IDFS compared with standard trastuzumab therapy.26 On the basis of data presented here, it is reasonable to speculate as to whether genomic subtyping could further identify a subpopulation of patients within the KATHERINE trial who benefitted most from T-DM1. Specifically, would patients with genomically Luminal tumors (cHER2/gLuminal) with residual disease benefit with T-DM1 as much as patients with genomically HER2 tumors? Another antibody-drug conjugate (ADC), trastuzumab deruxtecan, has not only exhibited benefit in patients with HER2+ breast cancer who had previously been treated with HER2-targeted therapies, but it has also demonstrated activity in non-HER2 subtypes (HER2 low, 1-2+ by IHC).27,28 Considering that patients with HR-HER2+ disease from the KATHERINE trial had improved IDFS with T-DM1,26 benefit from ADCs in patients with cHER2/gBasal tumors may be possible since they express the HER2 receptor and harbor an underlying activated Basal biology that may be more responsive to the targeted chemotherapy payload. However, this would need to be evaluated in trials comparing an ADC candidate with standard-of-care HER2-targeted therapy, such as the DESTINY-Breast02 or KRISTINE trials. Given the disappointingly small benefit that pertuzumab added in the APHINITY trial, which changed treatment guidelines, further genomic classification may be useful in finding those who benefit most.29 In this study, we show the genomic diversity that occurs in a cHER2 population and identify patients with the greatest magnitude of benefit from pertuzumab.
Because of the nature of observational registry trials, some limitations of the NBRST study include missing or unknown data and the limited number of patients for the study of non-HER2 BluePrint subtypes within the cHER2 subset. As a result, although we did evaluate cHER2 patients by BluePrint and HR status because of the distinct outcomes that have been described between HR+HER2+ and HR–HER2+ patients, the results were not statistically significant.6,8,24 Second, the long-term follow-up analyses were not the primary objective of NBRST, and the study was not powered to evaluate outcomes. Thus, although we see a substantial increase in 5-year DMFS benefit among patients with cHER2/gHER2 tumors treated with dual targeted therapy compared with the APHINITY trial, it was not statistically significant. Nevertheless, the 5-year DMFS and OS outcomes varied among the cHER2 patients, and BluePrint was able to better predict outcomes than by histology.
In conclusion, in this study, we identified a subgroup of patients with cHER2/gLuminal tumors who had good 5-year prognosis regardless of pCR rate, which may help inform adjuvant therapy decisions. We also identified a subgroup of patients, cHER2/gBasal, whose outcomes were not improved by dual HER2-targeted therapy and for whom alternative treatment strategies and treatment escalation may be necessary. In addition, we identified the subgroup of HER2+ patients with BluePrint, cHER2/gHER2, who are most likely to benefit from dual HER2-targeted therapy, which was approximately half of the cHER2 population. Genomic subtyping with BluePrint identifies biologic diversity within cHER2 breast cancers, providing further classification and precision in correlating subtypes with therapeutic response and long-term outcomes beyond traditional IHC/FISH subtyping.
ACKNOWLEDGMENT
We are grateful to all the patients who participated in this study, in addition to all the investigators, surgeons, pathologists, and research nurses.
APPENDIX 1. Institutional Investigators
Appendix 1.
Institutional Investigators
Pat W. Whitworth
Employment: Integra LifeSciences (I)
Leadership: Integra LifeSciences (I)
Stock and Other Ownership Interests: Targeted Medical Education Inc, Integra LifeSciences (I)
Honoraria: Puma Biotechnology
Consulting or Advisory Role: ImpediMed, Prelude Therapeutics, Becton Dickinson
Research Funding: Prelude Therapeutics, Agendia, Medneon
Travel, Accommodations, Expenses: Targeted Medical Education Inc
Peter D. Beitsch
Employment: Invitae
Leadership: Targeted Medical Education Inc
Stock and Other Ownership Interests: Targeted Medical Education Inc, Invitae
Research Funding: Invitae
Expert Testimony: Dune Medical Devices, ImpediMed
Uncompensated Relationships: Medneon
Paul D. Richards
Stock and Other Ownership Interests: NanoViricides
Research Funding: Carrick Therapeutics (Inst)
James V. Pellicane
Stock and Other Ownership Interests: PreludeDx
Honoraria: Agendia, PreludeDx
Speakers' Bureau: Agendia, PreludeDx
Beth B. Dupree
Leadership: Caliber Medical
Stock and Other Ownership Interests: Videra Surgical
Honoraria: Medtronic, Perimeter Medical
William Dooley
Leadership: Shaga Medical LLC
Stock and Other Ownership Interests: Shaga Medical
Research Funding: Agendia, Xoft
Patents, Royalties, Other Intellectual Property: patent pending—microendoscopy system
Shiyu Wang
Employment: Agendia
Patricia Dauer
Employment: Agendia
Stock and Other Ownership Interests: Agendia
Travel, Accommodations, Expenses: Agendia
Andrea R. Menicucci
Employment: Agendia
Erin B. Yoder
Employment: Agendia
Stock and Other Ownership Interests: Agendia
Travel, Accommodations, Expenses: Agendia
Lisa E. Blumencranz
Employment: Agendia
William Audeh
Employment: Agendia
Leadership: Agendia
Stock and Other Ownership Interests: Agendia
Consulting or Advisory Role: Celanese, Private Health
Research Funding: Agendia
Travel, Accommodations, Expenses: Agendia
No other potential conflicts of interest were reported.
SUPPORT
Supported by Agendia Inc.
CLINICAL TRIAL INFORMATION
The prospective NBRST registry trial (NCT01479101)
DATA SHARING STATEMENT
The raw data and clinical data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
AUTHOR CONTRIBUTIONS
Conception and design: Pat W. Whitworth, Paul L. Baron, Laura A. Lee, William Dooley, Patricia Dauer, Andrea R. Menicucci, Erin B. Yoder, Lisa E. Blumencranz, William Audeh
Administrative support: Pat W. Whitworth, William Dooley, Erin B. Yoder
Provision of study materials or patients: Pat W. Whitworth, Angela Mislowsky, Carrie L. Dul, Paul L. Baron, Rakhshanda Layeequr Rahman, Laura A. Lee, Beth B. Dupree, Pond R. Kelemen, William Dooley
Collection and assembly of data: Pat W. Whitworth, Peter D. Beitsch, Mary K. Murray, Angela Mislowsky, Carrie L. Dul, James V. Pellicane, Rakhshanda Layeequr Rahman, Laura A. Lee, Beth B. Dupree, Andrew Y. Ashikari, Raye J. Budway, Cristina Lopez-Penalver William Dooley, Erin B. Yoder, Christine Finn, Lisa E. Blumencranz, William Audeh
Data analysis and interpretation: Pat W. Whitworth, Paul D. Richards, Carrie L. Dul, Paul L. Baron, Rakhshanda Layeequr Rahman, Pond R. Kelemen, William Dooley, Shiyu Wang, Patricia Dauer, Andrea R. Menicucci, Erin B. Yoder, Lisa E. Blumencranz, William Audeh
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Pat W. Whitworth
Employment: Integra LifeSciences (I)
Leadership: Integra LifeSciences (I)
Stock and Other Ownership Interests: Targeted Medical Education Inc, Integra LifeSciences (I)
Honoraria: Puma Biotechnology
Consulting or Advisory Role: ImpediMed, Prelude Therapeutics, Becton Dickinson
Research Funding: Prelude Therapeutics, Agendia, Medneon
Travel, Accommodations, Expenses: Targeted Medical Education Inc
Peter D. Beitsch
Employment: Invitae
Leadership: Targeted Medical Education Inc
Stock and Other Ownership Interests: Targeted Medical Education Inc, Invitae
Research Funding: Invitae
Expert Testimony: Dune Medical Devices, ImpediMed
Uncompensated Relationships: Medneon
Paul D. Richards
Stock and Other Ownership Interests: NanoViricides
Research Funding: Carrick Therapeutics (Inst)
James V. Pellicane
Stock and Other Ownership Interests: PreludeDx
Honoraria: Agendia, PreludeDx
Speakers' Bureau: Agendia, PreludeDx
Beth B. Dupree
Leadership: Caliber Medical
Stock and Other Ownership Interests: Videra Surgical
Honoraria: Medtronic, Perimeter Medical
William Dooley
Leadership: Shaga Medical LLC
Stock and Other Ownership Interests: Shaga Medical
Research Funding: Agendia, Xoft
Patents, Royalties, Other Intellectual Property: patent pending—microendoscopy system
Shiyu Wang
Employment: Agendia
Patricia Dauer
Employment: Agendia
Stock and Other Ownership Interests: Agendia
Travel, Accommodations, Expenses: Agendia
Andrea R. Menicucci
Employment: Agendia
Erin B. Yoder
Employment: Agendia
Stock and Other Ownership Interests: Agendia
Travel, Accommodations, Expenses: Agendia
Lisa E. Blumencranz
Employment: Agendia
William Audeh
Employment: Agendia
Leadership: Agendia
Stock and Other Ownership Interests: Agendia
Consulting or Advisory Role: Celanese, Private Health
Research Funding: Agendia
Travel, Accommodations, Expenses: Agendia
No other potential conflicts of interest were reported.
REFERENCES
- 1. Perez EA, Romond EH, Suman VJ, et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: Planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J Clin Oncol. 2014;32:3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 3. Early Breast Cancer Trialists' Collaborative Group Trastuzumab for early-stage, HER2-positive breast cancer: A meta-analysis of 13 864 women in seven randomised trials. Lancet Oncol. 2021;22:1139–1150. doi: 10.1016/S1470-2045(21)00288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swain SM, Baselga J, Kim SB, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372:724–734. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 6. Goutsouliak K, Veeraraghavan J, Sethunath V, et al. Towards personalized treatment for early stage HER2-positive breast cancer. Nat Rev Clin Oncol. 2020;17:233–250. doi: 10.1038/s41571-019-0299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brunelli M, Manfrin E, Martignoni G, et al. Genotypic intratumoral heterogeneity in breast carcinoma with HER2/neu amplification: Evaluation according to ASCO/CAP criteria. Am J Clin Pathol. 2009;131:678–682. doi: 10.1309/AJCP09VUTZWZXBMJ. [DOI] [PubMed] [Google Scholar]
- 8. Carey LA, Berry DA, Cirrincione CT, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34:542–549. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daemen A, Manning G. HER2 is not a cancer subtype but rather a pan-cancer event and is highly enriched in AR-driven breast tumors. Breast Cancer Res. 2018;20:8. doi: 10.1186/s13058-018-0933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beitsch P, Whitworth P, Baron P, et al. Pertuzumab/trastuzumab/CT versus trastuzumab/CT therapy for HER2+ breast cancer: Results from the Prospective Neoadjuvant Breast Registry Symphony Trial (NBRST) Ann Surg Oncol. 2017;24:2539–2546. doi: 10.1245/s10434-017-5863-x. [DOI] [PubMed] [Google Scholar]
- 12. Whitworth P, Stork-Sloots L, de Snoo FA, et al. Chemosensitivity predicted by BluePrint 80-gene functional subtype and MammaPrint in the Prospective Neoadjuvant Breast Registry Symphony Trial (NBRST) Ann Surg Oncol. 2014;21:3261–3267. doi: 10.1245/s10434-014-3908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cardoso F, van't Veer LJ, Bogaerts J, et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 14. Krijgsman O, Roepman P, Zwart W, et al. A diagnostic gene profile for molecular subtyping of breast cancer associated with treatment response. Breast Cancer Res Treat. 2012;133:37–47. doi: 10.1007/s10549-011-1683-z. [DOI] [PubMed] [Google Scholar]
- 15. Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 16. Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 17. Van 't Veer L, Dal H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 18. Schettini F, Pascual T, Conte B, et al. HER2-enriched subtype and pathological complete response in HER2-positive breast cancer: A systematic review and meta-analysis. Cancer Treat Rev. 2020;84:101965. doi: 10.1016/j.ctrv.2020.101965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fernandez-Martinez A, Krop IE, Hillman DW, et al. Survival, pathologic response, and genomics in CALGB 40601 (Alliance), a neoadjuvant phase III trial of paclitaxel-trastuzumab with or without lapatinib in HER2-positive breast cancer. J Clin Oncol. 2020;38:4184–4193. doi: 10.1200/JCO.20.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. von Minckwitz G, Procter M, de Azambuja E, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N Engl J Med. 2017;377:122–131. doi: 10.1056/NEJMoa1703643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piccart M, Procter M, Fumagalli D, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer in the APHINITY trial: 6 years' follow-up. J Clin Oncol. 2021;39:1448–1457. doi: 10.1200/JCO.20.01204. [DOI] [PubMed] [Google Scholar]
- 22. Krop I, Mittempergher L, Paulson J, et al. BluePrint performance in predicting pertuzumab benefit in genomically HER2-positive patients: A biomarker analysis of the APHINITY trial. Cancer Res. 2021;81 abstr PD3-01. [Google Scholar]
- 23. Whitworth P, Beitsch P, Pellicane J, et al. Distinct neoadjuvant chemotherapy response and 5-year outcome in patients with ERpositive, HER2-negative breast tumors that reclassify as Basal-Type by the 80-gene signature. JCO Precis Oncol. 2022;6:e2100463. doi: 10.1200/PO.21.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prat A, Carey LA, Adamo B, et al. Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer. J Natl Cancer Inst. 2014;106:dju152. doi: 10.1093/jnci/dju152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brufsky A, Crozier JA, Chuba PJ, et al. Adding precision to 2018 ASCO/CAP HER2 testing guidelines in breast cancer with genomic profiling. J Clin Oncol. 2020;38:3570–3570. [Google Scholar]
- 26. von Minckwitz G, Huang CS, Mano MS, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. 2019;380:617–628. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 27. Modi S, Park H, Murthy RK, et al. Antitumor activity and safety of trastuzumab deruxtecan in patients with HER2-low-expressing advanced breast cancer: Results from a phase Ib study. J Clin Oncol. 2020;38:1887–1896. doi: 10.1200/JCO.19.02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Modi S, Saura C, Yamashita T, et al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med. 2020;382:610–621. doi: 10.1056/NEJMoa1914510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller KD. Questioning our APHINITY for more. N Engl J Med. 2017;377:186–187. doi: 10.1056/NEJMe1706150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data and clinical data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.