Abstract
cis-polyprenyl diphosphate synthases are involved in the biosynthesis of the glycosyl carrier lipid in most organisms. However, only little is known about this enzyme of archaea. In this report, we isolated the gene of cis-polyprenyl diphosphate synthase from a thermoacidophilic archaeon, Sulfolobus acidocaldarius, and characterized the recombinant enzyme.
In bacteria and eucarya, a specific prenyltransferase plays a key role during some of the glycosylation processes. The enyme, cis-polyprenyl diphosphate synthase (CPDS), catalyzes consecutive Z-type condensations of isopentenyl diphosphate (IPP) with allylic primer substrates, and the product diphosphates are utilized as the precursor of glycosyl carrier lipids, i.e., undecaprenyl monophosphate in bacteria and dolichyl monophosphate in eucarya. The genes of both bacterial and eucaryal CPDSs were recently cloned, and the high homology among them was revealed (1, 14, 15).
Glycosylation of proteins and membrane lipids also occurs ubiquitously in archaea (10, 11). Based on the sensitivity of the antibiotics such as bacitracin, the archaeal glycosylation process is thought to involve a glycosyl carrier lipid (11, 16). Previous studies using halophilic archaea revealed that the lipid has a polyprenyl chain of 11 or 12 isoprene units (8, 10). However, little is known about the CPDS involved in the biosynthesis of the archaeal glycosyl carrier lipid. To elucidate properties of the enzyme and obtain information on insights into its biological role in archaea, we isolated the gene encoding CPDS from a thermophilic archaeon, Sulfolobus acidocaldarius, and characterized the enzyme expressed in Escherichia coli cells.
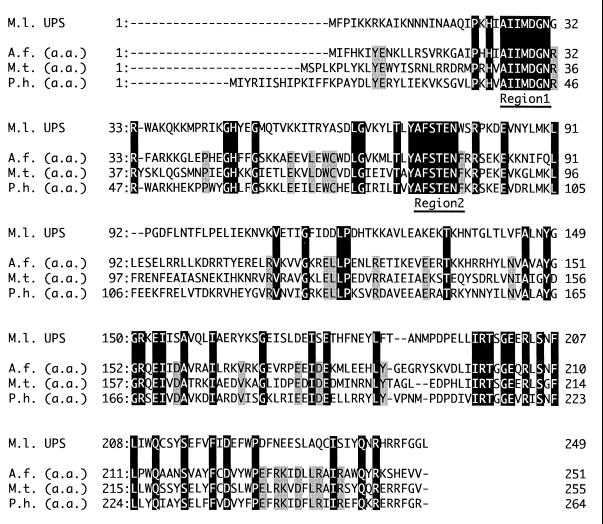
From the DNA databases in which whole genome sequences of several archaea, e.g., Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum, and Pyrococcus horikoshii, have been registered, we found the genes that encode the homologue of the known CPDSs by using BLAST, a homology search program. The alignment of amino acid sequences of these archaeal homologues shows the existence of several highly conserved regions that could be observed also in bacterial and eucaryal CPDSs (Fig. 1). Based on the nucleotide sequences of the regions, we designed degenerated PCR primers: FW1, 5′-GC(A/C/G)AT(A/C)AT(A/C/T)ATGGA(C/T)GG(A/T)AA-3′, and RV2, 5′-(C/G)(A/C)A(A/G)TT(C/T)TC(A/G/T)(C/G)T(A/T)(C/G)(A/T)(A/G)AA(G/T)GC-3′. As a consequence of PCR using these primers, KOD DNA polymerase (TOYOBO), and S. acidocaldarius genome as a template, a 200-bp fragment was amplified. Colony hybridization using this fragment as a probe yielded two positive clones from ca. 7,500 colonies of S. acidocaldarius genomic library. The plasmid p3d, which was one of the positive clones, was sequenced and proved to contain an open reading frame of 786 bp (here termed cpds) which encodes a protein of 262 amino acids (DDBJ/GenBank/EMBL accesion number AB048249). The encoded protein shows high sequence similarity to known CPDSs, 36% identity with both undecaprenyl diphosphate synthase of Micrococcus luteus B-P 26 and dehydrodolichyl diphosphate synthase of Saccharomyces cerevisiae, and shares the highly conserved regions proposed by Koyama et al. (7) and Apfel et al. (1)
FIG. 1.
Conserved regions in archaeal putative CPDSs. The hypothetical open reading frames which show high homology to previously cloned CPDSs, e.g., M. luteus B-P 26 undecaprenyl diphosphate synthase (M.1. UPS), were searched for among the whole genome sequences of three archaea, A. fulgidus (A.f.), M. thermoautotrophicum (M.t.), and P. horikoshii (P.h.), and their amino acid (a.a.) sequences are multiply aligned. Residues conserved among the bacterial enzyme and the three archaeal putative homologues are shown in black boxes, while shadowed areas denote the residues conserved only among the archaeal enzymes. Two highly conserved regions utilized for the design of degenerated PCR primers are underlined (regions 1 and 2).
Because we could not detect thermostable prenyltransferase activity in the crude extract of the transformant cells harboring the plasmid p3d, the full-length cpds gene was amplified using the PCR primers 5′-TGAGACCATGGCCAAAGATGTGATAACTAG-3′ and 5′-AGAATGGATCCTTAAGCCCCAAAGTTTC-3′. The restriction sites newly introduced in the primers, the NcoI site and BamHI site, are indicated by underlines. The amplified fragment was digested with NcoI and BamHI and then ligated into NcoI-BamHI sites of pET22b(+) vector. The E. coli BL21(DE3) cells were transformed with the resultant plasmid, designated pET-CPDS, and the transformant cells were grown in M9 minimal medium supplemented with yeast extract (2 g per liter), glycerol (2 g per liter), and ampicillin (50 mg per liter). When the optical density at 600 nm of the culture reached 0.5, 1.0 mM (final concentration) isopropyl 1-thio-β-d-galactoside was added to the medium and this was followed by overnight cultivation. The cells were harvested and disrupted by sonication in 50 mM Tris-Cl buffer, pH 7.7, containing 10 mM 2-mercaptoethanol and 1 mM EDTA. The homogenate was centrifuged at 3,000 × g for 20 min, and the supernatant was recovered as a crude extract. Prenyltransferase activity was assayed as follows. The assay mixture contained, in a final volume of 200 μl, 0.1 nmol of [1-14C]IPP (2.0 GBq/mmol), 1 nmol of the indicated allylic substrate (dimethylallyl diphosphate [DMAPP], geranyl diphosphate [GPP], [all-E] farnesyl diphosphate [FPP], and [all-E] geranylgeranyl diphosphate [GGPP]), 0.1 μmol of MgCl2, 2 μmol of phosphate buffer (pH 6.0), 0.1% Triton X-100, and a suitable amount of enzyme. This mixture was incubated at 55°C for 1 h, and the reaction was stopped by chilling the reaction mixture in an ice bath. The mixture was shaken with 600 μl of 1-butanol saturated with H2O. The butanol layer was washed with water saturated with NaCl, and radioactivity in the butanol layer was measured.
The crude extract of the BL21(DE3) strain carrying pET-CPDS showed much higher prenyltransferase activity than that of E. coli carrying the parental plasmid (data not shown). To confirm the expression of the archaeal thermostable CPDS, the crude extract was heated at 55°C for 7 h and then centrifuged. However, the enzyme activity was found to be coprecipitated with denatured proteins of E. coli during the heat treatment. This problem was circumvented by the addition of 1.5% Triton X-100 (final concentration) to the crude extract before the heat treatment, which allowed the enzyme activity to remain soluble in the supernatant. The solubilized activity was subjected to DEAE-Toyopearl column chromatography and was eluted as a single peak with a gradient of 0 to 0.85 M NaCl. The active fractions were collected and used for further characterization.
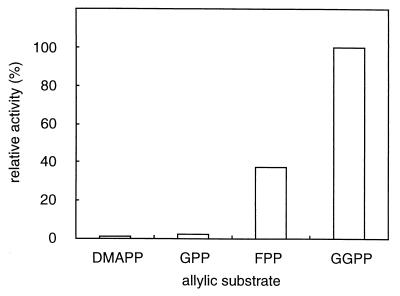
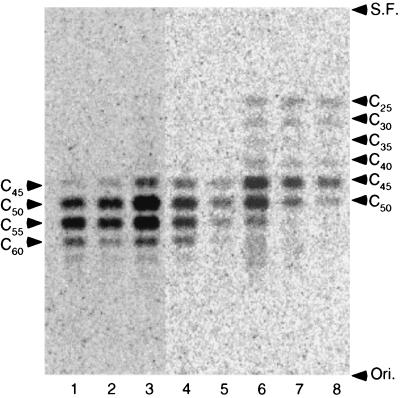
The partially purified enzyme was characterized in terms of its substrate and product specificities, stability, metal ion requirement, and effects of detergents on activity. As shown in Fig. 2, the archaeal CPDS prefers GGPP to FPP as the allylic substrate. On the other hand, DMAPP and GPP scarcely reacted. Thus, we used GGPP as the allylic substrate for subsequent characterization. The optimal reaction pH was 6.0, and the optimal reaction temperature was about 60°C, although more than 90% of the enzyme activity was retained after heat treatment at 70°C for 1 h. The Mg2+ ion was required for the enzyme activity, and its optimal concentration was 0.5 to 2 mM. Other divalent cations, such as Mn2+ and Ca2+, showed no ability to substitute for Mg2+. Triton X-100 activated the enzyme (sevenfold) at concentrations of 0.05 to 0.1%; however, at high concentrations, an inhibitory effect reminiscent of a surface dilution effect was observed (3, 4). To determine the conclusive product of the archaeal CPDS, enzyme reactions were made according to the assay conditions described above, except that 5 μM GGPP and various concentrations of IPP were added to the reaction mixture. The products were extracted with 1-butanol and then treated with acid phosphatase according to the method of Fujii et al. (5). The hydrolysates were extracted with pentane and analyzed by reversed-phase thin-layer chromatography using a precoated plate, LKC-18F, developed with acetone-H2O (9:1). Authentic standard alcohols were visualized with iodine vapor, and the distribution of radioactivity was analyzed by a Molecular Imager (Bio-Rad). As shown in Fig. 3, the chain lengths of synthesized polyprenyl diphosphates depend on the concentration of IPP. Decreasing the concentration of IPP resulted in increasing amounts of intermediates with shorter chain lengths. This should not arise from the depletion of IPP, because the majority of the substrate remained unreacted after termination of the reaction. In general, long-chain polyprenyl diphosphate synthases require an excess amount of IPP compared to the allylic primer substrate to synthesize the conclusive product (9, 12). When an amount of IPP that was 20 times larger than that of GGPP was added to the reaction mixture, the archaeal enzyme mainly produced C50 and C55 polyprenyl diphosphates along with much smaller amounts of C60, in a molar ratio, C50/C55/C60, of 3.5:5:1.5. When FPP was used as an allylic substrate, the chain length of the conclusive product remained unchanged (data not shown). Because the names of prenyltransferases are traditionally determined in terms of their conclusive products, we regarded the enzyme as undecaprenyl diphosphate synthase. However, it must be noted that the divalent cation and the detergent in the reaction mixture might affect the conclusive product in its length as reported on dehydrodolichyl diphosphate synthase of rat liver (9).
FIG. 2.
Substrate specificity of S. acidocaldarius CPDS. The allylic substrate preference of S. acidocaldarius CPDS was determined by using DMAPP, GPP, FPP, or GGPP. One of the allylic substrates and IPP were added to the reaction mixture at a concentration of 5 μM and 0.5 μM, respectively. The other assay conditions were as described in the text. “Relative activity” means the activity normalized to the activity with GGPP as the allylic substrate, which is set at 100%.
FIG. 3.
Effect of substrate concentration on product distribution of S. acidocaldarius CPDS. Each reaction mixture contained various concentrations of [14C]IPP and 5 μM GGPP. The concentration (and specific radioactivity) of [14C]IPP in each reaction mixture is as follows: lane 1, 100 μM (1 Ci/mol); lane 2, 50 μM (1 Ci/mol); lane 3, 25 μM (5 Ci/mol); lane 4, 10 μM (5 Ci/mol); lane 5, 5 μM (5 Ci/mol); lane 6, 2.5 μM (55 Ci/mol); lane 7, 1 μM (55 Ci/mol); lane 8, 0.5 μM (55 Ci/mol). The conditions of enzyme reaction and product analysis are described in the text. The carbon numbers of the products are indicated on both sides. Ori., origin; S.F., solvent front.
In most bacteria and eucarya, which generally contain FPP synthases, FPP acts as the primer substrate for the biosynthesis of glycosyl carrier lipids (6). Because FPP is also the substrate of many enzymes, e.g., medium- and long-chain trans-prenyl diphosphate synthases which supply the precursors of the side chains of respiratory quinones, it is regarded as the central intermediate of isoprenoid biosynthesis in these organisms. However, GGPP synthase, not FPP synthase, is the only short-chain (all-E) prenyl diphosphate synthase that supplies the primer substrate to CPDS in most archaea. Chen et al. (2) and Ohnuma et al. (13) proposed that the archaeal prenyltransferase would be a bifunctional GGPP or FPP synthase, based on the consideration that archaea also utilize FPP as the precursor of isoprenoid compounds. However, the substrate specificity of S. acidocaldarius CPDS strongly suggests that the allylic primer substrate for the biosynthesis of the glycosyl carrier lipid in the thermoacidophilic archaeon is GGPP, not FPP. To date it is not clear whether the enzymes that require FPP as a substrate exist in S. acidocaldarius, but it is conceivable that GGPP is the branch point of isoprenoid biosynthesis in this archaeon and that the metabolic specificity is generally in Archaea in contrast to the other domains of life.
Acknowledgments
This work was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan.
We are grateful to K. Ogura and T. Koyama, Tohoku University, for providing prenyl diphosphates. We thank Naoto Shimizu and Kazutake Hirooka for participating in helpful discussions.
REFERENCES
- 1.Apfel C M, Takacs B, Fountoulakis M, Stieger M, Keck W. Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: cloning, expression, and characterization of the essential uppS gene. J Bacteriol. 1999;181:483–492. doi: 10.1128/jb.181.2.483-492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen A, Poulter C D. Purification and characterization of farnesyl diphosphate/geranylgeranyl diphosphate synthase. A thermostable bifunctional enzyme from Methanobacterium thermoautotrophicum. J Biol Chem. 1993;268:11002–11007. [PubMed] [Google Scholar]
- 3.Dennis E A. Kinetic dependence of phospholipase A 2 activity on the detergent Triton X-100. J Lipid Res. 1973;14:152–159. [PubMed] [Google Scholar]
- 4.Dennis E A. Phospholipase A2 activity towards phosphatidylcholine in mixed micelles: surface dilution kinetics and the effect of thermotropic phase transitions. Arch Biochem Biophys. 1973;158:485–493. doi: 10.1016/0003-9861(73)90540-7. [DOI] [PubMed] [Google Scholar]
- 5.Fujii H, Koyama T, Ogura K. Efficient enzymatic hydrolysis of polyprenyl pyrophosphates. Biochim Biophys Acta. 1982;712:716–718. [PubMed] [Google Scholar]
- 6.Hemming F W. Biosynthesis of dolichols and related compounds. In: Porter J W, Spurgeon S L, editors. Biosynthesis of isoprenoid compounds. Vol. 2. New York, N.Y: John Wiley & Sons; 1983. pp. 305–354. [Google Scholar]
- 7.Koyama T, Shimizu N, Ogura K. Structure and function of polyisoprenoid chain elongation enzymes. In: Steinbuchel A, editor. Biochemistry principles and mechanisms of biosynthesis and biodegradation of polymers. Proceedings of an international symposium. Münster, Germany: Wiley-VCH; 1998. [Google Scholar]
- 8.Lechner J, Wieland F, Sumper M. Biosynthesis of sulfated saccharides N-glycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria. J Biol Chem. 1985;260:860–866. [PubMed] [Google Scholar]
- 9.Matsuoka S, Sagami H, Kurisaki A, Ogura K. Variable product specificity of microsomal dehydrodolichyl diphosphate synthase from rat liver. J Biol Chem. 1991;266:3464–3468. [PubMed] [Google Scholar]
- 10.Mescher M F, Hansen U, Strominger J L. Formation of lipid-linked sugar compounds in Halobacterium salinarium. Presumed intermediates in glycoprotein synthesis. J Biol Chem. 1976;251:7289–7294. [PubMed] [Google Scholar]
- 11.Mescher M F, Strominger J L. Glycosylation of the surface glycoprotein of Halobacterium salinarium via a cyclic pathway of lipid-linked intermediates. FEBS Lett. 1978;89:37–41. doi: 10.1016/0014-5793(78)80517-1. [DOI] [PubMed] [Google Scholar]
- 12.Ohnuma S, Koyama T, Ogura K. Chain length distribution of the products formed in solanesyl diphosphate synthase reaction. J Biochem. 1992;112:743–749. doi: 10.1093/oxfordjournals.jbchem.a123969. [DOI] [PubMed] [Google Scholar]
- 13.Ohnuma S-I, Suzuki M, Nishino T. Archaebacterial ether-linked lipid biosynthetic gene. Expression cloning, sequencing, and characterization of geranylgeranyl-diphosphate synthase. J Biol Chem. 1994;269:14792–14797. [PubMed] [Google Scholar]
- 14.Sato M, Sato K, Nishikawa S, Hirata A, Kato J, Nakano A. The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol Cell Biol. 1999;19:471–483. doi: 10.1128/mcb.19.1.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu N, Koyama T, Ogura K. Molecular cloning, expression, and purification of undecaprenyl diphosphate synthase. No sequence similarity between E- and Z-prenyl diphosphate synthases. J Biol Chem. 1998;273:19476–19481. doi: 10.1074/jbc.273.31.19476. [DOI] [PubMed] [Google Scholar]
- 16.Zhu B C, Drake R R, Schweingruber H, Laine R A. Inhibition of glycosylation by amphomycin and sugar nucleotide analogs PP36 and PP55 indicates that Haloferax volcanii β-glucosylates both glycoproteins and glycolipids through lipid-linked sugar intermediates: evidence for three novel glycoproteins and a novel sulfated dihexosyl-archaeol glycolipid. Arch Biochem Biophys. 1995;319:355–364. doi: 10.1006/abbi.1995.1305. [DOI] [PubMed] [Google Scholar]