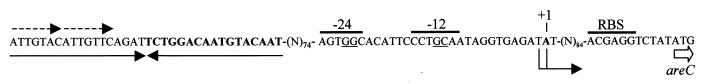Abstract
The areCBA genes in Acinetobacter sp. strain ADP1, determining growth on benzyl alkanoates, are shown to be transcribed as a single operon and regulated by areR, which encodes a regulatory protein of the NtrC/XylR family. Assays of the Are enzymes and of two insertions of lacZ as a reporter gene have shown that the operon is induced by benzyl acetate, benzyl alcohol, and benzaldehyde, as well as 2- and 4-hydroxybenzyl acetates and benzyl propionate and butyrate. Two adjacent sites of transcriptional initiation were 97 and 96 bp upstream of the start codon for areC, near a ς54-dependent −12, −24 promoter. Inactivation of areR and rpoN (for RNA polymerase ς54) drastically reduced growth rates on the Are substrates and induction of the operon.
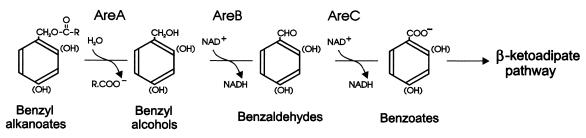
The areC, -B, and -A genes in Acinetobacter sp. strain ADP1 are located (9, 10) in a 30-kbp supraoperonic cluster of catabolic genes which includes the ben and cat genes encoding the enzymes of the catechol branch of the β-ketoadipate pathway (5, 6, 7, 17, 19). The are genes (Fig. 1) encode an esterase and two dehydrogenases, responsible for the sequential catabolism of benzyl alkanoates to benzoate, salicylate, or 4-hydroxybenzoate, respectively (Fig. 2), which feed into the β-ketoadipate pathway (9, 10). Sequence analysis of areC, -B, and -A and the promoter region suggests that they may be cotranscribed and expressed as an operon (9), upstream of which is a gene designated areR (homologous to genes encoding transcriptional activators of the NtrC/XylR family [16, 18] and a possible regulator of the putative operon).
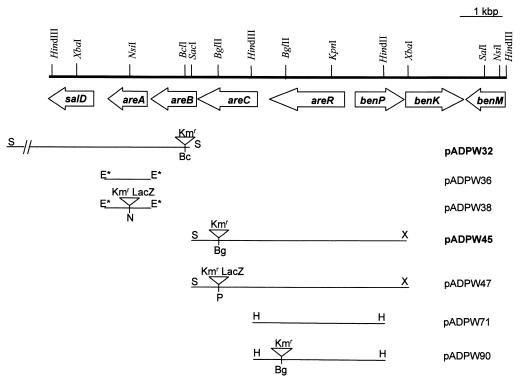
FIG. 1.
Physical map of the DNA adjacent to the ben genes in ADP1. Physical map of the areCBA genes and their locations relative to one end of the supraoperonic ben-cat cluster. The various inserts of the plasmids produced from cloning genomic DNA into vectors are specified in Table 1. Plasmids named in boldface contain inserts that were cloned directly from genomic DNA. All other plasmids were produced by PCR from genomic DNA or by subcloning from plasmids containing genomic DNA. Sites at the termini of the inserts marked with an asterisk were incorporated via PCR primers. The Km cassette or lacZ-Km cassette insertions are not to scale. The abbreviations for the restriction sites are Bc, BclI; Bg, BglII; E, EcoRI; H, HindIII; N, NsiI; P, PstI; S, SacI; X, XbaI.
FIG. 2.
Proposed pathway for catabolism of benzyl alkanoates by Acinetobacter sp. strain ADP1.
Table 1 lists the plasmids and bacterial strains used in this study. The latter were cultivated as previously described (9). Aromatic substrates were obtained as previously described (9). Standard methods for DNA manipulations and plasmid preparation (20) or previously described methods (9) were used. Growth of cells was monitored, and preparation of cell extracts and assays of Are enzymes was conducted, as described previously (9). β-Galactosidase in lysed cells was determined using the method described by Miller (15). The promoterless lacZ-Km cassette was removed from pKOK6.1 (12) and inserted into the NsiI site within areA on pADPW36 to create pADPW38 (Fig. 1). For its insertion into areC, genomic DNA from ADPW56 (areC::Km) (Table 1) was digested with XbaI and SacI and ligated into pUC18, selecting for Apr Kmr transformants: the plasmid obtained (pADPW45) contained a 6.2-kb insert with the whole of areR and an areC::Km insert (Fig. 1). The XbaI-SacI fragment from pADPW45 was then cloned into pUC18NP to create pADPW46 (Table 1). The Km cassette in areC was then replaced by the lacZ-Km on a PstI fragment to create pADPW47 (Fig. 1). In both pADPW38 and pADPW47, lacZ was transcribed in the same direction as areA and areC, respectively. Both plasmids were linearized and separately transformed into ADP1 to create ADPW61 (areA::lacZ-Km) and ADPW63 (areC::lacZ-Km), respectively, and integration into the chromosomes was confirmed by Southern blot analysis (data not shown).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotype | Reference or source |
|---|---|---|
| Acinetobacter strains | ||
| ADP1 (BD413) | Wild type | 11 |
| ADPW56 | areC::Km; transformation of ADP1 with pADPW22 | 9 |
| ADPW61 | areA::lacZ-Km; transformation of ADP1 with pADPW38 | This study |
| ADPW63 | areC::lacZ-Km; transformation of ADP1 with pADPW47 | This study |
| ADPW79 | areR::Km; transformation of ADP1 with pADPW90 | This study |
| ACN274 | rpoN::Km derivative of ADP1 | E. L. Neidle |
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Gibco BRL |
| XL1-Blue MRF' | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173′ endA1′ supE44′ thi-1′ recA1′ gyrA96′ relA1′ lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pUC18 | Apr, cloning vector | 28 |
| pUC18NP | pUC18 without PstI in the MCS | 27 |
| pUC4K | Apr Kmr; source plasmid for Kmr cassette | 26 |
| pKOK6.1 | Apr KmrlacZ; source plasmid for promoterless lacZ-Km cassette | 12 |
| pADPW32 | 8.0-kbp SacI fragment cloned from ADPW57 containing Kmr cassette and the whole of areA in pUC18 | This study |
| pADPW36 | 1.2-kbp EcoRIa fragment containing areA in pUC18 | This study |
| pADPW38 | pADPW36 with lacZ-Km cassette from pKOK6.1 cloned into NsiI site in areA | This study |
| pADPW45 | 6.2-kbp HindIII fragment cloned from ADPW56 containing areR and areC::Km in pUC18 | This study |
| pADPW46 | 6.2-kbp HindIII fragment cloned from ADPW56 containing areR and areC::Km in pUC18NP | This study |
| pADPW47 | pADPW46 with lacZ-Km cassette from pKOK6.1 cloned into PstI of the Kmr cassette | This study |
| pADPW71 | 3.5-kbp HindIII fragment from pADPW45 in pUC18 | This study |
| pADPW90 | pADPW71 with Kmr cassette from pUC4K cloned into the BglII site in areR | This study |
∗, restriction site added by PCR.
The Km cassette from pUC4K (26) was inserted into the sole BglII restriction site in the areR gene of pADPW71, a HindIII subclone of pADPW45 (Fig. 1), to form pADPW90 (Fig. 1). It was linearized and transformed into ADP1 to create ADPW79 (areR::Km), as was confirmed by Southern blot analysis.
Total RNA was prepared from Acinetobacter sp. using an RNeasy kit (Qiagen). The primer, for primer extension reactions (5′-ATCAAGTAATGTCATATAGACCTCGTA-3′) complementary to nucleotides on either side of the areC translational start (boldface), was end labeled with [γ-32P]dATP using T4 polynucleotide kinase and was annealed to the total RNA at 45°C for 3 h. Reactions were carried out with avian myeloblastosis virus reverse transcriptase (RT) (Promega, Madison, Wis.) at 37°C for 1 h. No sequence ladder was obtained with the same primer, but one was created instead from the cloning vector M13mp18 (28) using the M13 forward primer.
RT-PCR was carried out as described previously (10) by using the following primer pairs: across the intergenic regions between areC and areB, forward (primer CBf) 5′-TCAAAGCGCGTGTAATCGAAAAGGTCAAAC-3′ and reverse (primer CBr) 5′-ATGCCCATCTGGATCTCCACCACTGAAGT-3′; for the intergenic region between areB and areA, forward (primer BAf) 5′-CAGGCGGTGGTGTAAAGTTTGCTCTTGAAT-3′ and reverse (primer BAr) 5′-ATTGCCCCCTGCGCTGTCTCCTG-3′. Sequence determinations and analysis were performed as described previously (9, 10).
Induction of the areCBA genes in ADP1.
Induced AreA, AreB, and AreC activities were found in extracts of cells grown on all three pathway substrates that were compared with uninduced (succinate-grown) cells (Table 2), indicating that growth on each induces all three are genes.
TABLE 2.
Specific activities of the Are enzymes in extracts of ADP1 and its derivatives
| Strain and growth substrate | Sp
acta (U/mg of protein) against benzyl substrates
|
||
|---|---|---|---|
| Benzaldehyde dehydrogenase | Benzyl alcohol dehydrogenase | Benzyl esterase | |
| ADP1 | |||
| Succinate | <0.04 | <0.04 | <0.04 |
| Benzaldehyde | 0.5 | 0.6 | 11 |
| Benzyl alcohol | 0.4 | 0.9 | 9 |
| Benzyl acetate | 0.26 | 0.7 | 8 |
| Succinate + benzyl alcohol | 0.24 | 0.33 | 6 |
| ADPW79 | |||
| Succinate + benzyl alcohol | <0.04 | <0.04 | 0.14 |
| ACN274 | |||
| Succinate + benzyl alcohol | <0.04 | 0.15 | 0.6 |
Specific activities were determined from three determinations at two protein concentrations on each of two cell extracts prepared from independent cultures.
Induction of areA and areC expression by are operon substrates in strains ADPW61 and ADPW63.
β-Galactosidase activities of whole cells of ADPW61 (areA::lacZ-Km) and ADPW63 (areC::lacZ-Km) cultures were determined after growth to late log phase on succinate in the presence of aromatic inducers (15).
Benzyl acetate, benzyl alcohol, and benzaldehyde induced transcription of β-galactosidase in ADPW61 and ADPW63 (Table 3), and in the latter, the activities were approximately 8- to 10-fold higher than in ADPW61. Benzyl acetate, 2-hydroxybenzyl (salicyl) acetate, and 4-hydroxybenzyl acetate were equally effective as inducers, but the level of induction decreased as the length of the aliphatic chain of benzyl esters increased from acetate through propionate to butyrate (Table 3).
TABLE 3.
Induction by benzyl acetate and its metabolites of β-galactosidase activity expressed from are::lacZ chromosomal fusions
| Inducerb | β-Galactosidase
activitya (Miller units)
|
|
|---|---|---|
| ADPW61 (areA::lacZ-Km) | ADPW63 (areC::lacZ-Km) | |
| None | 44 (±20) | 660 (±220) |
| Benzaldehyde | 1,050 (±145) | 9,200 (±2,200) |
| Benzyl alcohol | 950 (±125) | 8,450 (±1,900) |
| Benzyl acetate | 1,000 (±150) | 8,200 (±1,600) |
| Benzyl propionate | 730 (±65) | 4,150 (±700) |
| Benzyl butyrate | 380 (±50) | 2,100 (±350) |
| 2-Hydroxybenzyl acetate | 1,000 (±50) | 8,040 (±820) |
| 4-Hydroxybenzyl acetate | 1,100 (±200) | 8,400 (±1,000) |
All values are the average of three independent duplicated experiments. Figures in parentheses are the standard deviations of the means.
Inducers were added at 1 mM.
The transcription initiation site of the are genes.
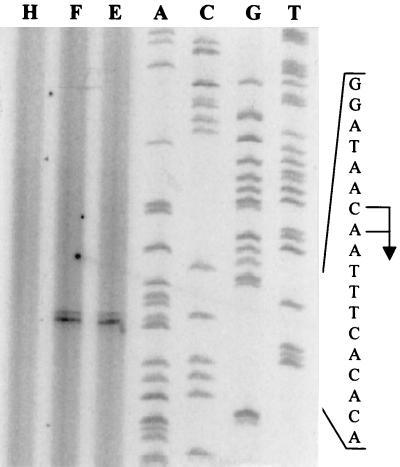
Primer extension reactions using RNA purified from induced ADP1 cells grown on either benzyl acetate or benzyl alcohol (Fig. 3) showed two products. The strongest corresponded to 97 bases upstream of the areC ATG, showing the transcription start site at an A residue at position +1 (Fig. 4), but there was also a second, weaker extension product corresponding to the adjacent T residue at position −1. No primer extension products were found in succinate-grown cells. Upstream of the start of transcription is a −12, −24 consensus element for a ς54 promoter binding site. The genetic organization of the areR-to-areC intragenic region, including the start codon and the putative ribosome binding site of areC, can be deduced (Fig. 4).
FIG. 3.
Primer extension of mRNA from Acinetobacter sp. strain ADP1. The experiment was performed with total RNA and a primer which overlaps the putative start codon of areC. Lanes A, C, G, and T show the respective products from M13mp18 that were sequenced using the M13 forward primer to size the extension products. The bases on the left are from the corresponding M13 sequence. Lanes E, F, and H represent the signal obtained from the experiment using 10 μg of total RNA from cells of ADP1 grown with benzyl acetate, benzyl alcohol, or succinate, respectively, as the sole carbon sources. The locations corresponding to both transcriptional starts are designated by the arrow and are positions 112 and 111 from the 5′ end of the universal primer. The corresponding bases on the ADP1 sequence are shown in Fig. 4.
FIG. 4.
Regulatory sequences upstream of areC. The start codon of areC is indicated, and the putative ribosome binding site (Shine-Dalgarno) of areC is indicated above the sequence (RBS). The nucleotides corresponding to both primer extension signals (Fig. 3) are indicated by the double-headed arrow with the strongest numbered as the +1 site. The putative −12, −24 promoter elements are indicated. Arrows below the sequence mark an inverted repeat that is hypothesized to act as the binding site for the regulator protein. Dashed arrows above the sequence mark a direct repeat.
RT-PCR of induced ADP1 total RNA.
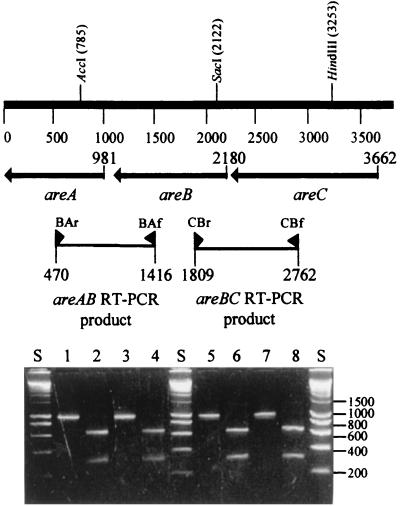
RT-PCR conducted on RNA that was purified from induced ADP1 cells amplified products across the boundaries between areC to areB (953 bp) and areB to areA (946 bp), which had both the expected sizes and restriction sites in the expected positions (Fig. 5). No RT-PCR products using primers whose products would span the areR-to-areC and areA-to-salD intergenic regions were obtained from the same RNA preparations, indicating the absence of transcription across these regions (data not shown).
FIG. 5.
Agarose gel electrophoresis of RT-PCR products amplified by primers from ADP1 grown on benzyl acetate and benzyl alcohol. The positions of the primers for spanning the areAB intergenic region (BAr, BAf) and the areCB intragenic region (CBr, CBf) are shown relative to the gene organization of areCBA. The values for molecular size markers (in base pairs) in lanes S (HyperLadder I; Bioline, London, United Kingdom) are indicated on the right side of the gel. Lanes: 1, areCB, benzyl acetate-grown cells (expected size, 953 bp); 2, areCB, benzyl acetate-grown cells digested with SacI (640 and 313 bp); 3, areCB, benzyl alcohol-grown cells (expected size, 953 bp); 4, areCB, benzyl alcohol-grown cells digested with SacI (640 and 313 bp); 5, areBA, benzyl acetate-grown cells (expected size, 946 bp); 6, areBA, benzyl acetate-grown cells digested with AccI (631 and 315 bp); 7, areBA, benzyl alcohol-grown cells (expected size, 946 bp); 8, areBA, benzyl alcohol-grown cells digested with AccI (631 and 315 bp). No detectable products were obtained in control reactions with each pair of primers, from which RT had been omitted, or in reactions carried out on succinate-grown cells (data not shown).
Analysis of ADPW79 (areR::Km) growth phenotype.
Growth of ADPW79 on minimal medium plates containing benzyl acetate, benzyl alcohol, and benzaldehyde was drastically reduced, taking 2 to 3 days to grow to the same colony size that ADP1 attained overnight. Thus, as with the areCBA gene disruptions (9), insertional inactivation of the gene did not result in a complete loss of ability to grow on the substrates and is probably due to low levels of other enzymes with overlapping substrate specificities (9).
Induction of areCBA in ADPW79 and ACN274.
By comparison with ADP1, AreA, -B, and -C in extracts of ADPW79 (areR::Km) grown on minimal media containing succinate plus benzyl alcohol (Table 2) exhibited no induced activities. In the rpoN mutant ACN274, which has the same reduced growth on the Are substrates as does ADPW79, the activity levels of AreA and AreC were partially elevated by growth on succinate plus benzyl alcohol above those in uninduced ADP1 but were still significantly below those in induced cells of ADP1.
All the evidence presented indicates that areCBA is an operon which is coordinately induced by AreR. The three genes are transcribed in the same direction, with only short intragenic regions. At the mRNA level, we have shown by RT-PCR that transcription is continuous across both intragenic regions areC to areB and areB to areA but that continuity of transcription does not extend to the two flanking genes, areR and salD. The only hint of an additional regulatory element is the presence of a small, inverted repeat within the areB-to-areA intragenic region, which might act as a partial transcription terminator and could account for the difference in activities beween ADPW61 and ADPW63. Downstream, 11 bp from the stop codon of areA, is a 17-bp perfect inverted repeat sequence, 5′-AATTAAAAAGGTTCTTAATAAGAACCTTTTTAATT-3′, which is the likely candidate to be a transcriptional terminator for the operon.
Biochemical analysis also points to AreA, -B, and -C being coordinately induced. We demonstrated that AreB and AreC activities were induced by growth of ADP1 on benzyl acetate and benzyl alcohol (9) and have now extended the assays of ADP1 to include the esterase (AreA). Growth on benzyl acetate, benzyl alcohol, or benzaldehyde causes induction of all three enzymes (Table 2). This indicates coordinate rather than sequential expression, since in the case of growth on benzyl alcohol and benzaldehyde, the esterase AreA (and also the alcohol dehydrogenase AreB for growth on benzaldehyde) is not required for growth and yet is induced. It cannot be deduced from these data for ADP1 which compound or compounds can induce, since active metabolism is occurring. However, the use of a promoterless lacZ as a reporter gene to monitor are gene expression clarifies this (Table 3), since the lacZ-Km cassette has its two genes in opposite orientations with a central transcriptional terminator and therefore exerts a polar effect on downstream genes (12). With lacZ inserted into areA (ADPW61), β-galactosidase activity was induced by benzyl acetate, confirming that benzyl acetate must be an inducer, since it cannot be metabolized because of the areA insertional inactivation. Additionally in ADPW63, with lacZ in areC, thus blocking transcription of all three genes, β-galactosidase was induced by all three aromatic metabolites, indicating nonspecificity in the control of expression. In addition, a variety of esters with modifications in the aromatic alcohol moiety (2- and 4-hydroxybenzyl acetates) and in the alkanoic acid moieties (benzyl propionate and butyrate), all of which act as substrates, also induce β-galactosidase in both ADPW61 and -63.
The regulator protein for areCBA appears to be AreR. Upstream, 74 bp from the −12, −24 promoter is an 18-bp inverted repeat with only two mismatches. Each half of the inverted repeat contains two direct repeats of 7 bp (Fig. 4), which could provide the binding site for a regulator protein, since an inverted repeat sequence has also been postulated to act as the regulator binding site in the related DmpR system (22). It could also serve as a transcription terminator for areR, since it is located only 10 bp downstream of its termination codon.
The sequence of areR has all the characteristics of the ς54-dependent NtrC/XylR family of regulatory proteins (16, 18, 21). The two most homologous proteins in the databanks are the AcoR proteins from Clostridium magnum (13) and Pseudomonas putida (GenBank accession no. CAA72019). Alignment of amino acid sequences of AreR with sequences of these proteins (data not shown), as well as with the two related regulators of aromatic catabolism in Pseudomonas, DmpR (24) and XylR (8), shows the three domains characteristic of the bacterial enhancer binding protein family (16, 18). The amino-terminal A domain, which is responsive to the inducers (23), is the least conserved and the most variable in length, but the sequence from residues 159 and 174 of AreR (PVFNGQGKILGALDIT) is completely conserved between AreR and both AcoR proteins. The central C domain, carrying the ATPase activity, is well conserved between the five aligned sequences, and the lengths of the C domains of the genes are very similar.
In addition to the sequence analysis implicating AreR as the regulator of areCBA, we have confirmed its physiological role by insertion of a Km cassette into areR. This not only drastically reduces the growth rate on the are substrates but also knocks out the ability of benzyl alcohol to induce all three Are protein activities (Table 2). Induction of the Are enzymes is also much reduced in an rpoN mutant of ADP1 lacking the ς54 subunit of RNA polymerase, completing the circle of evidence implicating the regulatory protein, promoter, and sigma factor in the induction of areCBA.
Acknowledgments
This research was funded by a BBSRC research grant (5/P12776).
We thank Helen Wing, University of Birmingham, Birmingham, United Kingdom, for help with the primer extension experiment.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Bauchop T, Elsden S R. The growth of microorganisms in relation to energy supply. J Gen Microbiol. 1960;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- 4.Buck M, Cannon W. Specific binding of the transcriptional factor sigma-54 to promoter DNA. Nature. 1992;358:422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- 5.Collier L S, Gaines III G L, Neidle E L. Regulation of benzoate degradation in Acinetobactersp. strain ADP1 by BenM, a LysR-type transcriptional activator. J Bacteriol. 1998;180:2493–2501. doi: 10.1128/jb.180.9.2493-2501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobactersp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 8.Inouye S, Nakazawa A, Nakazawa T. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene. 1988;66:301–306. doi: 10.1016/0378-1119(88)90366-6. [DOI] [PubMed] [Google Scholar]
- 9.Jones R M, Collier L S, Neidle E L, Williams P A. areABC genes determine the catabolism of aryl esters in Acinetobactersp. strain ADP1. J Bacteriol. 1999;181:4568–4575. doi: 10.1128/jb.181.15.4568-4575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones R M, Pagmantidis V, Williams P A. sal genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobactersp. strain ADP1. J Bacteriol. 2000;182:2018–2025. doi: 10.1128/jb.182.7.2018-2025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juni E, Janik A. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 13.Krüger N, Oppermann F B, Lorenzl H, Steinbüchel A. Biochemical and molecular characterization of the Clostridium magnumacetoin dehydrogenase enzyme system. J Bacteriol. 1994;176:3614–3630. doi: 10.1128/jb.176.12.3614-3630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick M J. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N) Mol Microbiol. 1993;5:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 16.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidle E L, Shapiro M K, Ornston L N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticusgenes for benzoate degradation. J Bacteriol. 1987;169:5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.North A K, Klose K E, Steadman K M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ornston L N, Neidle E L. Evolution of genes for the β-ketoadipate pathway in Acinetobacter calcoaceticus. In: Towner K, Bergogne-Berezin E, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 201–237. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Shingler V. Signal sensing by sigma54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 22.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shingler V, Moore T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonassp. strain CF600. J Bacteriol. 1994;176:1555–1560. doi: 10.1128/jb.176.6.1555-1560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonassp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weights, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 27.Williams P A, Shaw L M, Pitt C W, Vrecl M. xylUW, two genes at the start of the upper pathway operon of TOL plasmid pWWO, appear to play no essential part in determining its catabolic phenotype. Microbiology. 1997;143:101–107. doi: 10.1099/00221287-143-1-101. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]