Abstract
BACKGROUND:
Hepatitis C virus (HCV) genotype distribution is different in various regions. A variety of strategies could be used to detect HCV genotypes and subtypes. The aim of the present study was to introduce a genotyping method by an in-house protocol that could be used to determine HCV drug-resistant variants and phylogeny studies.
METHODS:
Samples from 91 patients with thalassemia were used for HCV genotyping by Cobas 4800 platform, and 50 cases of 1a, 1b, and 3a genotypes underwent amplification and sequencing of NS5A and NS5B by using consensus primers via conventional reverse transcription-polymerase chain reaction (RT-PCR) method. An ABI 3730xl system used for direct sequencing. Raw sequences were analyzed by popular bioinformatics software MEGA6 and CLC workbench 5. Phylogenetic construction was drawn using 1000 replicates bootstrap by the neighbor-joining method. Multiple sequence alignment (MSA) was performed for mutation detection.
RESULTS:
Sequencing results of 50 HCV isolates subtypes 1a (31/45), 3a (15/22) and 1b (4/8) NS5A and NS5B genes showed there were 72 NS5A and 105 NS5B mutations. Moreover, 8 resistant associated substitutions (RASs) were identified in nine thalassemia cases by multiple sequence alignment (MSA) protein analysis. The phylogenetic tree construct drew confirmed by the Cobas HCV genotyping results.
CONCLUSION:
The phylogenetic analysis could be a useful tool for HCV genotyping in case of determining the drug-resistant substitutions; however, it is time-consuming and needs expert analysis and interpretation. This preliminary study in Iranian patients with thalassemia introduces specific conventional RT-PCR to find RASs to direct acting antivirals (DAAs) and subtype determination at the same time.
Keywords: Hepatitis C virus (HCV), Genotyping, Phylogeny, Thalassemia
INTRODUCTION
Hepatitis C virus (HCV), as a worldwide health concern, has infected about 71 million people.1 In 2016, HCV mortality was estimated at 399,000, generally by developing cirrhosis and hepatocellular carcinoma. HCV is found worldwide. The most affected geographical regions in 2015 included WHO Eastern Mediterranean Region (2.3%), and the WHO European Region (1.5%). HCV is transmitted by blood-borne routes, especially in injection drug users.2,3 Globally, it is reported that there were 1.75 million new HCV infections in 2015 (23.7 per 100 000 people).3
HCV, as a member of Flaviviridae family Hepacivirus genus, has an enveloped, spherical and about 50 nm in diameter virion. Moreover, it has a monopartite, linear, ssRNA genome by almost 10 kb in size. It encodes structural (E1, E2, C, F, P7) and non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A, NS5B).1,4 HCV has seven genotypes with 30-35% genomic differences and 70 confirmed subtypes with less than 15% genomic differences.5 There are different locations for HCV genotyping included NS5, core, E1, and 5’ UTR regions, which could be utilized by a specific polymerase chain reaction (PCR), restriction fragment length polymorphisms, and line probe assays (LiPA).5
HCV genotype distribution around the world include 49.1% genotype 1, 17.9% genotype 3, 16.8% genotype 4, 11.0% genotype 2, 2.0% genotype 5, and 1.4% genotype 6 in which subtypes 1a, 1b, 2a, and 3a are the most prevalent.2,6 In Iran, HCV prevalence accounts for 0.5% of the general population, 7-13% among hemodialysis patients, 40.8% among patients with hemophilia, and 18% among patients with thalassemia. HCV subtype distribution in Iran includes 1a as the most common, which is followed by 3a and then 1b. Moreover, HCV genotype 1 is the most common among patients with thalassemia and hemophilia, and those undergoing hemodialysis, and also in solid organ recipients in Iran.7 The present study aimed to introduce a specific reverse transcription-polymerase chain reaction (RT-PCR) method to find HCV drug-resistant variants as well as HCV genotype and subtype determination by using NS5A, and NS5B partial sequencing in patients with thalassemia suffered from chronic HCV infection.
MATERIALS AND METHODS
Patients
Patients with thalassemia who suffered from chronic HCV and were referred to Firoozgar Hospital, affiliated to Iran University of Medical Sciences, Tehran, Iran, from October 2016 to March 2017 were enrolled in the present study. Most of the participants had a history of HCV antiviral therapy, and a small number of them were new cases. Inclusion criteria were having thalassemia major and intermediate by related data sheet in the hospital repository, being a known case of HCV carrier by related laboratory tests, and signing the written informed consent. Wrong sample collection, preparation or storage condition, leading to exclusion of specimen. Also, incomplete or insufficient datasheet related to the disease, and age less than 18 years, were excluded. Adequate laboratory tests and history of hematological deficiencies were reviewed by expert practitioners. Patients were receiving deferoxamine as a regular treatment. HCV detection results were reported by using UBI HCV EIA (Organon Teknika, Netherlands), and HCV detection One-step commercial kits (Genesig, Primerdesign, UK). All biochemical and hematological parameters tested for patients were obtained from medical history, which included alanine aminotransferase (ALT), aspartate aminotransferase (AST), white blood cell count (WBC), platelet (Plt) count, hemoglobin (Hb), viral load, HBV co-infection, and cirrhosis. A 10 cc whole blood was taken from each participant, and the serum was separated by appropriate centrifugation at 3000 g for 10 min and then kept at -70°c deep freezer.
Cobas HCV genotyping test
A Cobas 4800 system (Roche Molecular Diagnostics, Pleasanton, CA, USA) was used for HCV genotyping as a primary screening and genotyping test, which used a real-time PCR-based genotyping assay for use on the Cobas 4800 platform, according to the manufacturer’s instructions. Briefly, 400 µL of plasma was taken from each patient and separated in a vacuum tube containing EDTA, which was used for analysis on Cobas 4800 instrument.
RNA Extraction and Complementary DNA (cDNA) synthesis
Viral RNA was extracted from 200 μL serum using a High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Mannheim, Germany) with respect to protocols. RNA qualification and concentration were analyzed by a nanodrop spectrophotometer (Thermo Scientific, Wilmington, MA). They were kept at -70°c until further use. A cDNA kit for reverse transcriptase (RT)-PCR (MBI Fermentas, Toronto, Canada) was used for cDNA synthesis with regards to its protocol. Then, they were put in a deep freezer at -20°c.
Primer design
HCV subtypes 1a, 1b, and 3a were used for NS5A and NS5B specific primer designing according to a previous study.8 In order to investigate the drug-resistant subtypes, we targeted coding regions of the active site of the protein or the region by more identified mutation to DAAs.9-11
PCR Amplification
A conventional PCR was performed for amplification of specific regions for NS5A, and NS5B subtypes 1a, 1b, and 3a by a Bio-Rad (T100TM Thermal Cycler) instrument similar to a previous study.8 Briefly, a 50 µl reaction mixture was used. The heating protocol was as the previous study.8 Visualization of PCR products was performed by 1.5% agarose gel stained with SYBR Green (SG) safe stain against an ultra-violet (UV) transilluminator.
Nucleotide Sequencing
PCR products purification was performed via High Pure PCR Product Purification Kit (Roche Diagnostic, Mannheim, Germany) according to the protocol. An ABI 3730xl sequencer was used for both directions sequencing. Designed forward and reverse primers were used for sequencing. Bioinformatics software Clustal X program and MEGA software version 6 were used for multiple sequence alignment (MSA) after trimming raw data with CLC Genomics workbench 5 software (CLC bio, Aarhus, Denmark) against reference sequences NC_004102 for subtype 1a, EU781825 for 1b, and NC_009824 for 3a subtypes obtained from GeneBank (https://www.ncbi.nlm.nih.gov/).
Phylogenetic construction
The CLC Genomics workbench 5 software (CLC bio, Aarhus, Denmark) was used for raw sequences trimming. Consensus sequences were obtained by comparison of bidirectional Sanger sequencing raw data against reference sequences NC_004102 for subtype 1a, EU781825 for 1b, and NC_009824 for 3a subtypes. MEGA software version 6 was used for the alignment of sequences by MSA tool. A phylogenetic tree was drawn by using 1000 replicates bootstrap in a neighbor-joining method.
Sequence submission
BankIt online software (available at: https://www.ncbi.nlm.nih.gov/WebSub/) was used to sequence submission in GenBank database by using consensus sequences nucleotide and a translated frame. Feature of each protein was submitted to the datasheets. Primary released GenBank accession numbers for all subtypes NS5A were MT603183 to MT603232, and for NS5B were MT603233 to MT603282.
Ethical consideration
All ethical issues were in accordance with the Helsinki declaration (H Declaration - 1975). Research ethics was approved by the Ethics Committee of Iran University of Medical Sciences, Tehran, Iran (Code IR.IUMS.REC 1396.30299).
RESULTS
Of a total of 91 patients with thalassemia, 51 (56%) were male, and 40 (44%) were female. The mean ± SD of age (years) was 32 ± 6 years, Hb was 9.0 ± 2.0 (g/dl), ALT was 65 ± 48 (mg/dl), AST was 69 ± 47 (mg/dl), Plt was 431 ± 207 (× 10^3) (mm3), WBC was 10.0 ± 7.0 (× 10^3) (mm3), Viral load was 203 ± 1562 (× 10^5) (copy/dl). Of the patients, 74 had thalassemia major, and 17 had the intermediate type. HBV co-infection was found in two cases. Cirrhosis was reported in 37 patients.
HCV genotyping by Cobas HCV genotyping method showed that most of the isolates were 1a (49.4%) followed by 3a (24.1%) and 1b (8.7%). Figure 1 shows the HCV genotypes prevalence reported in our studied isolates.
Fig. 1:
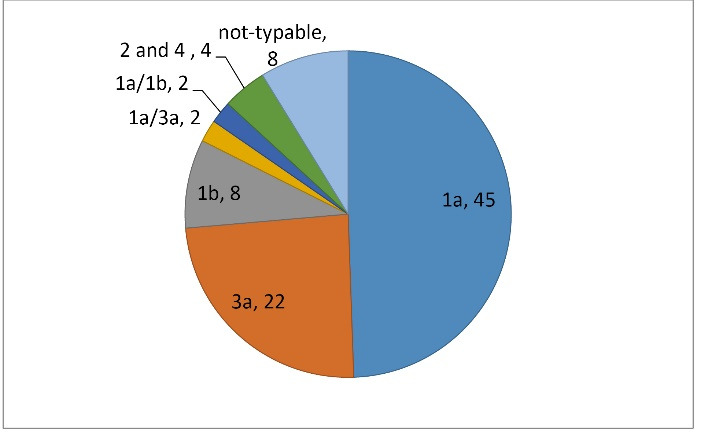
HCV genotyping assay by Cobas HCV genotyping method in 91 patients with thalassemia suffered from chronic HCV infection.
RT-PCR amplification
A total of 75 HCV subtypes 1a, 1b, and 3a NS5A and NS5B sequences were amplified using RT-PCR method. Figure 2 shows PCR products of each gene visualization on 1.5% agarose gel electrophoresis. High-quality PCR products were used for purification and nucleotide sequencing by the Sanger method.
Fig. 2:
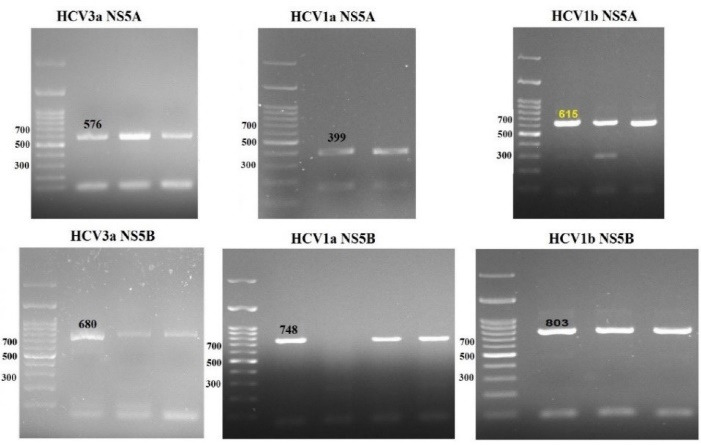
RT-PCR products of the patients with thalassemia infected with HCV.
Phylogenetic analysis
A total of 50 HCV subtypes 1a (31/45), 3a (15/22) and 1b (4/8) were used for amplification of NS5A and NS5B and phylogenetic analysis. Sequence analysis was performed by popular bioinformatics software and trimmed sequences used for phylogenetic tree construction. Figure 3 shows NS5A, and figure 4 shows NS5B tree construction.
Fig. 3:
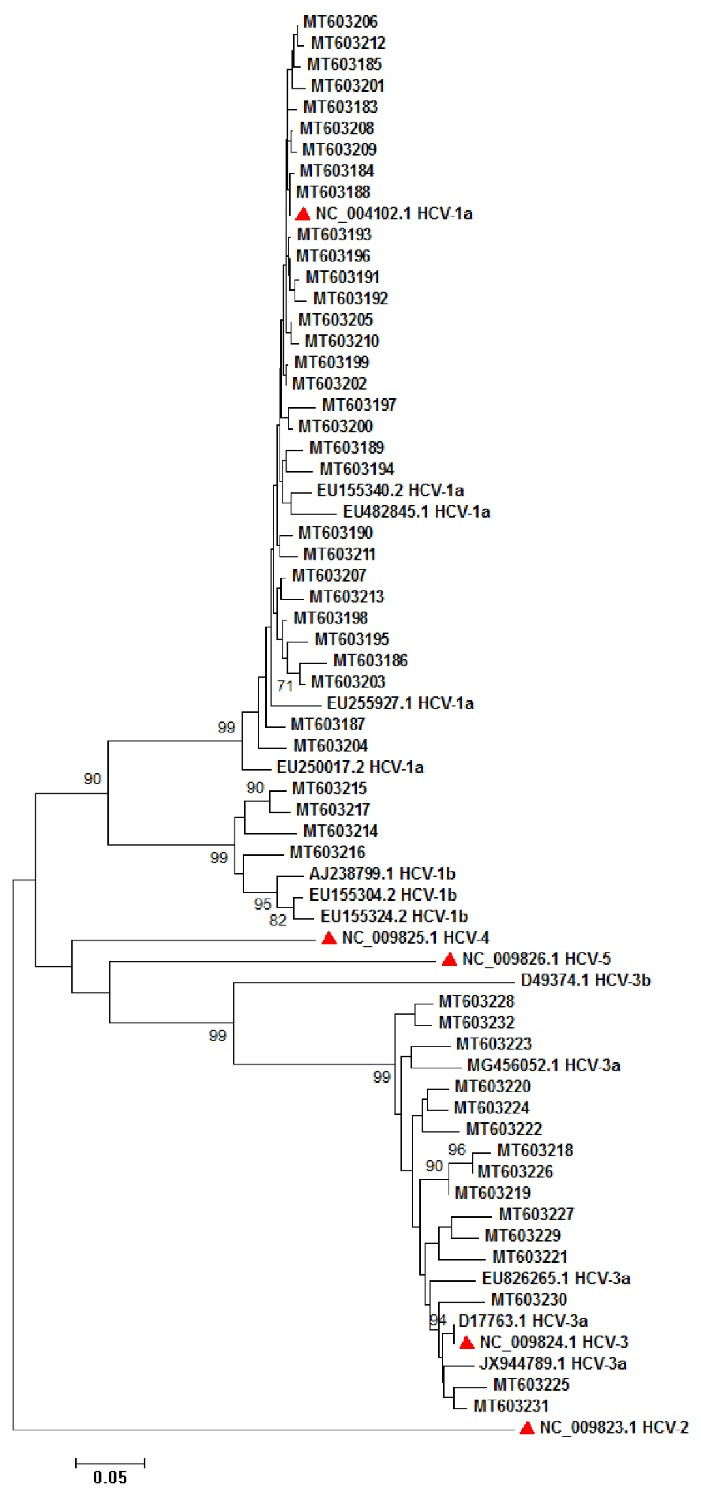
Phylogenetic analysis of 50 HCV NS5A sequences subtypes 1a, 1b, and 3a. Red triangles show the reference sequences for HCV subtypes 1a (NC_004102), 1b, 2 (NC_009823), 3 (NC_009824), 4 (NC_009825), 5 (NC_009826). The phylogenetic tree was drawn by using 1000 replicates bootstrap into a neighbor-joining method.
Fig. 4:
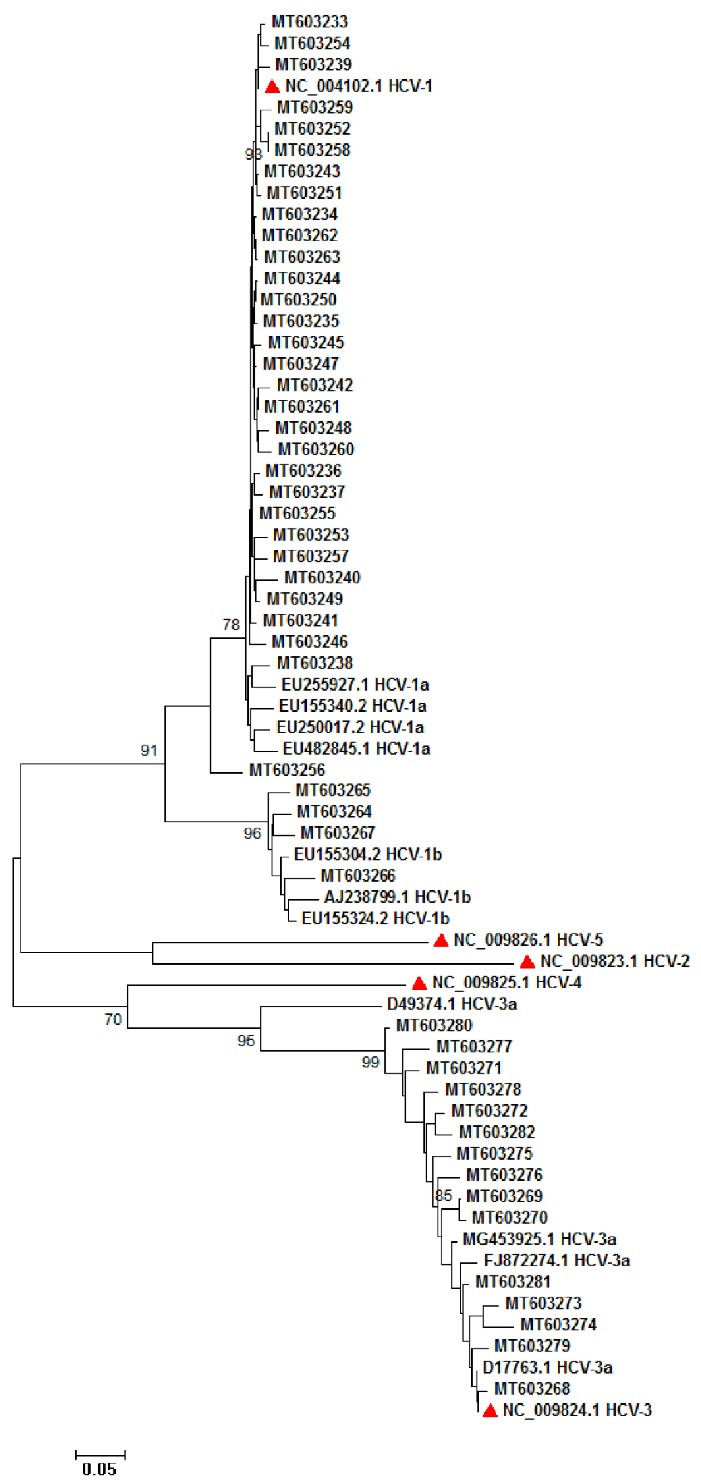
Phylogenetic analysis of 50 HCV NS5B sequences subtypes 1a, 1b, and 3a. Red triangles show the reference sequences for HCV subtypes 1a (NC_004102), 1b, 2 (NC_009823), 3 (NC_009824), 4 (NC_009825), 5 (NC_009826). The phylogenetic tree was drawn by using 1000 replicates bootstrap into a neighbor-joining method.
Mutation analysis
Sequences were aligned with reference sequences obtained from GeneBank database and found that there were totally 72 NS5A and 105 NS5B mutations and eight resistant-associated substitutions (RASs) in five subtypes 1a, three subtypes 1b, and one subtype 3a patients, which had been reported previously.8 Mutation analysis was performed based on the previous studies.10-15
DISCUSSION:
Global HCV subtypes prevalence in Iran included 1a as the predominant, followed by 3a.16 There are different strategies for HCV genotyping. NS3 region could be used as phylogenetic analysis and HCV subtype determination. Moreover, both core/E1 and NS5B regions could be used for phylogenetic analysis.17 Whole-genome sequencing or complete genome sequencing is another high-throughput method for HCV subtype determination. There are a few studies focused on phylogenetic analysis of HCV subtypes in Iran.16
The present study aimed at HCV genotyping and subtype determination as well as mutation analysis by NS5A and NS5B nucleotide sequencing. In the present study, we partially sequenced NS5A and NS5B of 50 HCV isolates obtained from Iranian patients with thalassemia. We have found that NS5A and NS5B could be used for HCV genotyping and subtype determination and found that there were 72 mutations in NS5A and 105 in NS5B amplified regions. There were eight RASs to DAAs in nine isolates obtained from patients with thalassemia, of which five were in 1a, 3 in 1b, and 1 in 3a subtypes based on previous studies.10-15
The phylogenetic analysis could be used as a main tool for the study of rapidly-evolving RNA viruses such as HCV. HCV genetic evolution could be monitored as a useful tool for global surveillance and to develop efficient preventive and therapeutic strategies.10,18 There are different studies that analyzed E1 and E2 sequences in various patients. A study of HCV in mixed cryoglobulinemia type 2 (MC2) showed no mutation in E1 and E2 sequences in these patients.19 A study of acute hepatitis C (AHC) by E1 and E2 phylogenetic analysis showed a common source of infection among them.20 In the study of E2 and NS5A sequence of HCV 3a subtypes, it was concluded that treatment response was not statistically significant among HIV-HCV co-infected patients.21 Recent studies use NS3, NS5A, and NS5B for phylogenetic purposes, but they have lower genetic variability than E1 and E2. Phylogeny of different genes could show their inconsistencies.10,22 NS5A respective 76 amino acid positions could be used for phylogenetic construction. Moreover, large-scale sequencing of HCV genome could be used as a valuable tool for clinical follow-up and drug-resistance testing.23 A study used NS5B for HCV genotyping and found 19 (76%) genotype 4d, two (8%) genotype 4a, one (4%) genotype 1b, and three (12%) genotype 2a.18
By the present study, we have introduced a suitable region for HCV genotyping as well as drug-resistant analysis. In this regard, we have designed primers for amplification of mutation-sensitive regions of NS5A and NS5B that were associated with drug resistance, especially DAAs.9-11 Phylogenetic construction showed the compliance of our results with Cobas HCV genotyping method.
There were some limitations to our study. The sample size that was restricted to patients with specific hematologic malignancies, including thalassemia major and intermediate, was the main limitation. Specific primers designed for each subtype enforced us to use three common subtypes 1a, 1b, and 3a. Time consumption and difficulty in interpretation and analysis for our staff could be another drawback.
In conclusion, although there are different methods for HCV genotyping and drug-resistant testing, our study showed a suitable region for both HCV genotyping and drug-resistant investigation at the same time by direct sequencing of the specific region of NS5A and NS5B of subtypes 1a, 1b, and 3a. Phylogenetic construction showed that there was great compliance with Cobas HCV genotyping method. HCV phylogenetic analysis in Iranian patients with thalassemia by NS5A and NS5B was studied for the first time in Iran.
Please cite this paper as: Safarnezhad Tameshkel F, Karbalaie Niya MH, Zamani F, Ajdarkosh H, Khoonsari MR, Faraji AH, Motamed N, Nikkhah M, Ameli M, Miri SM, Azarkeivan A, Sohrabi MR, Keyvani H. Simultaneous Hepatitis C Virus Genotyping and Variant Detection in Patients with Thalassemia: A Single-Center Phylogenetic Study. Middle East J Dig Dis 2022;14:124-130. doi: 10.34172/mejdd.2022.265.
Footnotes
FUNDING
This study was supported in part by grant 95-04-128-30299 from the Iran University of Medical Sciences, Tehran, Iran
ETHICAL APPROVAL
Ethics Committee of Iran University of Medical Sciences, Tehran, Iran, approved the study (Code: IR.IUMS.REC 1396.30299)
AUTHORS’ CONTRIBUTIONS
Study concept and design: M. H. K. N., H. K.; acquisition of data: F. S. T., F. Z.; analysis and interpretation of data: H. A., M. N.; drafting the manuscript: M K., N. M.; critical revision of the manuscript for important intellectual content: Amir H. F., S. M. M.; statistical analysis: M. A., S. M. M.; administrative, technical, and material support: A. A., M. R. S., F. Z.; study supervision: H. K., M. H. K. N.
CONFLICT OF INTEREST
All authors declare that they have no financial interests related to the material in the manuscript.
References
- 1.Goonawardane N, Yin C, Harris M. Phenotypic analysis of mutations at residue 146 provides insights into the relationship between NS5A hyperphosphorylation and hepatitis C virus genome replication. J Gen Virol. 2020;101:252–64. doi: 10.1099/jgv.0.001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahmud S, Chemaitelly HS, Kouyoumjian SP, Al Kanaani Z, Abu‐Raddad LJ. Key associations for hepatitis C virus genotypes in the Middle East and North Africa. J Med Virol. 2020;92:386–93. doi: 10.1002/jmv.25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmelzer J, Dugan E, Blach S, Coleman S, Cai Z, DePaola M. et al. Global prevalence of hepatitis C virus in children in 2018: a modelling study. Lancet Gastroenterol Hepatol. 2020;5:374–92. doi: 10.1016/S2468-1253(19)30385-1. [DOI] [PubMed] [Google Scholar]
- 4.Zayedi E, Makvandi M, Teimoori A, Samarbaf-Zadeh AR, Ghafari S, Seyedian SS. et al. Prevalence of hepatitis C virus among HIV-infected patients. Iran J Microbiol. 2020;12:156–63. doi: 10.18502/ijm.v12i2.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22:7824–40. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietschmann T, Brown RJ. Hepatitis C virus. Trends Microbiol. 2019;27:379–80. doi: 10.1016/j.tim.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Taherkhani R, Farshadpour F. Epidemiology of hepatitis C virus in Iran. World J Gastroenterol. 2015;21:10790–810. doi: 10.3748/wjg.v21.i38.10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tameshkel FS, Niya MHK, Zamani F, Motamed N, Ajdarkosh H, Vafaeimanesh J. et al. Resistance-associated substitutions (RASs) to HCV direct-acting antivirals (DAAs) at baseline of treatment in thalassemia patients: a referral center study. Arch Virol. 2020;165:2193–2203. doi: 10.1007/s00705-020-04728-x. [DOI] [PubMed] [Google Scholar]
- 9.Simicic P, Grgic I, Santak M, Vince A, Lepej SZ. Frequency of baseline NS5A resistance-associated substitutions in patients infected with genotype 1 of hepatitis C virus in Croatia. Microb Pathog. 2019;136:103694. doi: 10.1016/j.micpath.2019.103694. [DOI] [PubMed] [Google Scholar]
- 10.Bertoli A, Sorbo MC, Aragri M, Lenci I, Teti E, Polilli E. et al. Prevalence of single and multiple natural NS3, NS5A and NS5B resistance-associated substitutions in hepatitis C virus genotypes 1–4 in Italy. Sci Rep. 2018;8:8988. doi: 10.1038/s41598-018-26862-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones BR, Howe AY, Harrigan PR, Joy JB. The global origins of resistance-associated variants in the non-structural proteins 5A and 5B of the hepatitis C virus. Virus Evol. 2018;4:vex041. doi: 10.1093/ve/vex041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patiño-Galindo JÁ, Salvatierra K, González-Candelas F, López-Labrador FX. Comprehensive screening for naturally occurring hepatitis C virus resistance to direct-acting antivirals in the NS3, NS5A, and NS5B genes in worldwide isolates of viral genotypes 1 to 6. Antimicrob Agents Chemother. 2016;60:2402–16. doi: 10.1128/AAC.02776-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q, Perales C, Soria ME, García-Cehic D, Gregori J, Rodríguez-Frías F. et al. Deep-sequencing reveals broad subtype-specific HCV resistance mutations associated with treatment failure. Antiviral Res. 2020;174:104694. doi: 10.1016/j.antiviral.2019.104694. [DOI] [PubMed] [Google Scholar]
- 14.Sharafi H, Ghalamkari S, Hassanshahi A, Alavian SM. Pooled Prevalence of NS5A Resistance-Associated Substitutions in Chronic HCV Genotype 3 Infection: A Study Based on Deposited Sequences in GenBank. Microb Drug Resist. 2019;25:1072–9. doi: 10.1089/mdr.2018.0358. [DOI] [PubMed] [Google Scholar]
- 15.Uribe-Noguez LA, Mata-Marín JA, Ocaña-Mondragón A, Pompa-Mera EN, Ribas-Aparicio RM, Arroyo-Anduiza CI. et al. Comparison of direct sequencing of the NS5B region with the Versant HCV genotype 20 assay for genotyping of viral isolates in Mexico. J Infect Chemother. 2020;26:205–10. doi: 10.1016/j.jiac.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Salehi Moghadam F, Mohebbi SR, Hosseini SM, Romani S, Mirtalebi H, Azimzadeh P. et al. Phylogenetic analysis of hepatitis C virus strains and risk factors associated with infection and viral subtypes among Iranian patients. J Med Virol. 2014;86:1342–9. doi: 10.1002/jmv.23947. [DOI] [PubMed] [Google Scholar]
- 17.Paraskevis D, Stylianou DC, Hezka J, Stern Z, Oikonomopoulou M, Mamais I. et al. HCV phylogeography of the General population and High-Risk Groups in Cyprus Identifies the Island as a Global sink for and source of Infection. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-46552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciccozzi M, Presti AL, Ciccaglione AR, Zehender G, Ciotti M. Phylogeny and phylodinamic of Hepatitis C in Italy. BMC Infect Dis. 2012;12:1–6. doi: 10.1186/1471-2334-12-s2-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerotto M, Dal Pero F, Loffreda S, Bianchi FB, Alberti A, Lenzi M. A 385 insertion in the hypervariable region 1 of hepatitis C virus E2 envelope protein is found in some patients with mixed cryoglobulinemia type 2. Blood, J Am Society Hematol. 2001;98:2657–63. doi: 10.1182/blood.v98.9.2657. [DOI] [PubMed] [Google Scholar]
- 20.Larghi A, Zuin M, Crosignani A, Ribero ML, Pipia C, Battezzati PM. et al. Outcome of an outbreak of acute hepatitis C among healthy volunteers participating in pharmacokinetics studies. Hepatology. 2002;36:993–1000. doi: 10.1053/jhep.2002.36129. [DOI] [PubMed] [Google Scholar]
- 21.Bagaglio S, Bruno R, Lodrini S, De MM, Andreone P, Loggi E. et al. Genetic heterogeneity of hepatitis C virus (HCV) in clinical strains of HIV positive and HIV negative patients chronically infected with HCV genotype 3a. J Biol Regul Homeost Agents. 2003;17:153–61. [PubMed] [Google Scholar]
- 22.Cuypers L, Thijssen M, Shakibzadeh A, Deboutte W, Sarvari J, Sabahi F. et al. Signature of natural resistance in NS3 protease revealed by deep sequencing of HCV strains circulating in Iran. Infect Genet Evol. 2019;75:103966. doi: 10.1016/j.meegid.2019.103966. [DOI] [PubMed] [Google Scholar]
- 23.Cuypers L, Pérez AB, Chueca N, Aldamiz-Echevarría T, Alados JC, Martínez-Sapiña AM. et al. Relapse or reinfection after failing hepatitis C direct acting antiviral treatment: Unravelled by phylogenetic analysis. PloS One. 2018;13:e0201268. doi: 10.1371/journal.pone.0201268. [DOI] [PMC free article] [PubMed] [Google Scholar]


