Abstract
Analysis of a Brucella suis 1330 gene fused to a gfp reporter, and identified as being induced in J774 murine macrophage-like cells, allowed the isolation of a gene homologous to nikA, the first gene of the Escherichia coli operon encoding the specific transport system for nickel. DNA sequence analysis of the corresponding B. suis nik locus showed that it was highly similar to that of E. coli except for localization of the nikR regulatory gene, which lies upstream from the structural nikABCDE genes and in the opposite orientation. Protein sequence comparisons suggested that the deduced nikABCDE gene products belong to a periplasmic binding protein-dependent transport system. The nikA promoter-gfp fusion was activated in vitro by low oxygen tension and metal ion deficiency and was repressed by NiCl2 excess. Insertional inactivation of nikA strongly reduced the activity of the nickel metalloenzyme urease, which was restored by addition of a nickel excess. Moreover, the nikA mutant of B. suis was functionally complemented with the E. coli nik gene cluster, leading to the recovery of urease activity. Reciprocally, an E. coli strain harboring a deleted nik operon recovered hydrogenase activity by heterologous complementation with the B. suis nik locus. Taking into account these results, we propose that the nik locus of B. suis encodes a nickel transport system. The results further suggest that nickel could enter B. suis via other transport systems. Intracellular growth rates of the B. suis wild-type and nikA mutant strains in human monocytes were similar, indicating that nikA was not essential for this step of infection. We discuss a possible role of nickel transport in maintaining enzymatic activities which could be crucial for survival of the bacteria under the environmental conditions encountered within the host.
The gram-negative bacteria Brucella spp. are the etiologic agents of brucellosis, a disease that is encountered worldwide and is endemic in many underdeveloped countries. Six distinct species have been identified in various mammalian hosts, including humans. In cattle, sheep, and goats, the disease still causes important economic losses. Among these species, Brucella melitensis, B. suis, and B. abortus are most frequently associated with pathogenicity in humans, characterized by undulant fever and other, less well-defined clinical symptoms. The infection can be asymptomatic, which may cause diagnostic difficulties and sometimes lead to chronic infections in bones, joints, and the central nervous system. Brucellae belong to the α-2 subdivision of the proteobacteria and are therefore phylogenetically related to the plant cell-associated species of the genera Rhizobium and Agrobacterium. Brucella spp. are facultative intracellular parasites that can survive within professional phagocytes. They are able to invade macrophages and to multiply inside acidified phagosomes (36). Among the mechanisms used by brucellae for intracellular survival are the inhibition of phagolysosomal fusion (35) and of tumor necrosis factor alpha production by macrophages (4), the activation of which under normal conditions is critical for the elimination of pathogens.
Despite intensive work, the mechanisms allowing Brucella to behave as an intracellular parasite have not been elucidated. To date, little is known about the bacterial factors contributing to the persistence and multiplication of this pathogen within human phagocytes. Upon infection of the macrophage by Brucella spp., certain stress proteins also induced at low pH and at high temperature were found to be expressed. Among those are the proteins HtrA, GroEL, and DnaK, the latter having been shown to be essential for replication of B. suis in human macrophage-like cell lines (23). Genes encoding these stress proteins (12, 17, 26) were frequently identified by the use of heterologous probes designed from previously characterized genes of other species. More recently, a two-component regulatory system (41) and a locus homologous to the VirB type IV secretion system (34) have been demonstrated to play important roles in intracellular survival. In an attempt to identify the bacterial genes expressed during multiplication of brucellae in host cells, a genetic tool was developed using the green fluorescent protein (GFP) gene as a reporter gene fused to randomly cloned B. suis promoters (22). Characterization of these genes, especially inactivation, should give an insight into the mechanisms allowing Brucella spp. to adapt to host macrophages. Several clones were selected on the basis of the inducible expression of GFP fluorescence after infection of a macrophage cell line. Here, we describe the identification of a gfp-fused B. suis gene highly homologous to nikA, the first gene of the Escherichia coli ATP-binding cassette (ABC) transport system specific for nickel (33). Uptake of nickel by the periplasmic binding-protein-dependent transport system encoded by the nikABCDE operon is required for the synthesis and activities of the E. coli hydrogenase isoenzymes under anaerobic conditions (46). Transcription of this operon is activated by the general anaerobic transcriptional factor Fnr and repressed by the nickel-responsive regulator NikR when intracellular nickel concentrations are high (9, 46, 47). Many microorganisms incorporate this metal ion into enzymes participating in important metabolic reactions of hydrogen metabolism, ureolysis, methane biogenesis, and acetogenesis (20). Hydrolysis of urea is catalyzed by the nickel-dependent urease, an enzyme produced by all Brucella species (7).
Our data show that B. suis possesses genes encoding counterparts highly similar to the five protein components of the E. coli NikABCDE permease and to the NikR regulator with a distinct genomic organization. We demonstrate that in vitro expression of the nikA promoter-gfp fusion is activated by low oxygen or nickel concentrations. We also show that inactivation of nikA severely alters the activity of the nickel metalloenzyme urease in this bacterium. In addition, successful reciprocal complementation of nik mutants of E. coli and B. suis with the nik operons supports the notion that the gene cluster of B. suis functions as a nickel transport system. Our data also suggest that, like E. coli and other bacteria, B. suis can use alternative ion transport systems for this metal when its concentration is high enough.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Brucella strains used in this study (B. suis 1330 [ATCC 23444] and derived mutants) were all grown in tryptic soy (TS) medium.For cultures carried out under microaerobic conditions, the brucellae were grown on TS agar plates placed in a jar with activated CampyPack Plus (Becton Dickinson, Baltimore, Md.) and palladium catalyst. The mutant strain containing the kanamycin resistance (Kanr) cassette was cultured in presence of 50 μg of the antibiotic ml−1 in the medium. DNA cloning for complementation studies in B. suis was performed using plasmid pBBR1MCS (24), a broad-host-range vector. Recombinant clones were selected on agar supplemented with chloramphenicol (25 μg ml−1), in combination, where appropriate, with kanamycin (50 μg ml−1). Construction of plasmid pBBR1-KGFP, designed for random cloning of B. suis DNA fragments fused to a promoterless gfp gene, and selection of the clone, 19A10, studied here were previously described (22).
E. coli strain DH5α was used as the recipient strain for cloning and was routinely grown in Luria-Bertani (LB) medium. The appropriate antibiotics (ampicillin, 50 μg ml−1; chloramphenicol and kanamycin as indicated above) were added when needed. E. coli strain MC4100 was used for hydrogenase assay. The deletion mutant HYD720 was derived from this strain by excision of the MudI (Apr lac) bacteriophage, randomly inserted in the E. coli chromosome. Plasmids pLW21 and pLW22 carried the entire E. coli nik operon cloned into two different vectors, pUC19 and pPH126, respectively (47).
The standard growth temperature for all bacterial strains was 37°C.
DNA manipulations and hybridization.
Plasmid DNA was isolated from B. suis and E. coli according to standard procedures (39). B. suis chromosomal DNA was prepared as previously described (2). DNA treatments with restriction and modification enzymes were performed according to the manufacturer's instructions. For labeling or subcloning, restriction fragments or PCR products were purified after separating bands on low-melting-point agarose gels (Life Technologies) by the Wizard DNA clean-up system (Promega, Madison, Wis.). The 975-bp DNA fragment isolated from plasmid pBBR1-KGFP of clone 19A10 was labeled with digoxigenin using a random prime kit (Boehringer, Mannheim, Germany), yielding a nonradioactive probe. Southern blotting (pocket blotting 8) and colony blotting were performed with Biodyne B nylon membranes (Pall, Port Washington, N.Y.). Under high-stringency conditions, the membranes were washed twice at 68°C for 15 min in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.0115 M sodium citrate) with 0.1% sodium dodecyl sulfate.
Cloning of the complete B. suis nik locus.
The 19A10 probe was hybridized to Southern blots of B. suis genomic DNA digested by various restriction enzymes. The probe bound to a single 6.4-kb EcoRI fragment, which was recloned into linearized and dephosphorylated plasmid pUC18 (Pharmacia Biotech). Recombinant E. coli clones were screened as described above. The positive clone used for this study was named pNIK-BS.
Nucleotide sequence and analysis.
The nucleotide sequence of the B. suis nik genes carried by pNIK-BS (EMBL accession number AJ278644) was determined with the universal M13 forward and reverse primers, along with specific internal sequencing primers based on the newly acquired sequences. Sequencing was performed by Applied Biosystems Dye deoxyterminator techniques according to the kit protocols (Perkin-Elmer, Roissy, France). Sequences were analyzed with an automated Applied Biosystems model 373A DNA sequencer. Primer synthesis and sequencing were carried out by Genome Express (Grenoble, France).
The DNA sequences obtained were translated into the six reading frames and compared to the proteins in the SwissProt database (Swiss Institute of Bioinformatics, Geneva, Switzerland) by using the program BLASTx (1) to identify similar sequences. B. suis and E. coli Nik amino acid sequences were aligned using the ALIGN program of the FASTA package (29). Prediction of transmembrane regions in the deduced proteins of B. suis was obtained by the TMHMM or HMMTOP method (42, 43).
Inactivation of B. suis nikA by homologous recombination.
The B. suis DNA fragment of 975 bp, which comprised 676 bp of the nikA open reading frame (ORF) present in plasmid pBBR1-KGFP of clone 19A10, was produced by PCR. Two oligonucleotides (5′-TGAGACACAACGTGGCTTTCC-3′ and 5′-CAAGAATTGGGACAACTCCAGTG-3′; Eurobio, Les Ulis, France) deduced from the 5′ sequences of the Kanr gene and the gfp gene of the vector, respectively, were used. The DNA amplification product was obtained after 30 cycles of a denaturation step at 95°C for 45 s, annealing at 62°C for 1 min, and elongation at 72°C for 2 min (5 min ending the last cycle). The PCR product was then cloned into pUC18.
The 1.2-kb blunted kanamycin resistance gene (kan) from plasmid pUC4K (Pharmacia Biotech, Orsay, France) was inserted into the unique ApaI site in the partial nikA gene (see Fig. 2), resulting in construct pUC18nikA::kan. The nikA-kan insert was excised as a 2.2-kb XbaI-SacI fragment and recloned into pCVD442 (10), containing sacB, coding for sucrose sensitivity (14, 15). B. suis was transformed with this suicide vector by electroporation as described previously (23). Mutants of B. suis that had integrated the inactivated nikA gene into the chromosome by double-recombination events were selected by their resistance to kanamycin and sucrose as reported elsewhere (11). Isolated clones were screened for the loss of the plasmidic ampicillin resistance marker.
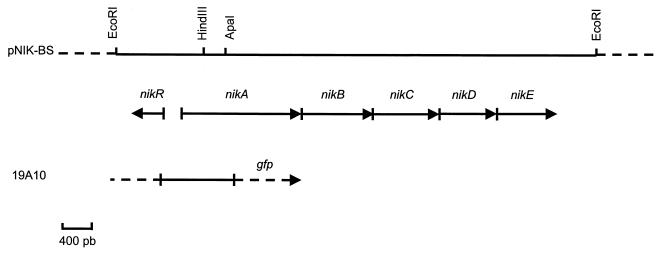
FIG. 2.
Physical map of the B. suis nik locus carried by clone pNIK-BS. The solid line delimits the insert DNA cloned into the EcoRI site of pUC18; arrows indicate the direction of transcription of the nikABCDER ORFs. The partial nikA gene and upstream sequence isolated in the initial clone 19A10 are shown below. The gfp reporter gene of plasmid pBBR1-KGFP is indicated by a dotted arrow.
Infection and intracellular growth of B. suis strains in murine J774A.1 macrophage-like cells and in human monocytes.
Infection of mouse J774.1 macrophage-like cells with recombinant Brucella strains containing plasmid pBBR1-KGFP, harboring a chromosomic DNA fragment fused to gfp was performed as described elsewhere (4, 19) in six-well plates (Falcon; Becton Dickinson, Meylan, France). Cells (2 × 106) cultured overnight in RPMI 1640 (Gibco BRL) supplemented with 10% heat-inactivated fetal calf serum (Gibco BRL) at 37°C and 5% CO2 were incubated for 45 min at 37°C with 1 ml of bacterial suspension, corresponding to a multiplicity of infection of 20. They were then washed three times with phosphate-buffered saline (PBS) to remove nonphagocytized bacteria. Infected cells were reincubated for 48 h in 4 ml of RPMI supplemented with 30 μg of gentamicin sulfate ml−1 to kill any remaining extracellular bacteria. The cells were washed with PBS and disrupted in water by incubation for 10 min on ice, scraped off, and centrifuged (353 × g [Heraeus Megafuge 1.0R], 5 min) to pellet the intact cells and nuclei. After a second centrifugation to eliminate cellular debris, the supernatant containing the bacteria was diluted in 2 ml of PBS for flow cytometry analysis.
Alternatively, for intracellular survival assay, human mononuclear cells were prepared from fresh blood by centrifugation with Ficoll-Hypaque (Sigma Chemical Co. Chimie), and monocytes were purified as described previously (4). Infection of 106 cells in 1 ml of RPMI was performed in 24-well plates as indicated above, at a multiplicity of infection of 20. At 1.5, 7, 24, and 48 h postinfection, cells were washed twice with PBS and lysed in 0.2% Triton X-100. CFU numbers were determined by plating serial dilutions on TS agar. Experiments were performed twice in duplicate.
Flow cytometry analysis.
Flow cytometry was carried out using a FACScalibur scanner (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). About 50,000 individual events were excited with a 488-nm argon-ion laser, and emission light was detected through a 530-nm bandpass filter. Cell Quest software (Becton Dickinson) was used for the quantitation of fluorescence.
Urease assay of B. suis strains.
Brucella strains were tested for urease activity by the phenol red spectrophotometric assay. A stationary-phase culture was diluted 1:100 in TS medium and grown for 24 h at 37°C with aeration in the presence of various concentrations of EDTA. Antibiotics were added when required. The optical density at 600 nm (OD600) was read at the end of the incubation period; high concentrations of EDTA reduced the growth rate. The bacterial samples were harvested and resuspended at equal densities in PBS. The urease assay was carried out directly on intact bacteria, using 500 μl of the suspension in 3 ml of urea test broth (32) containing 2% urea and 0.001% phenol red. A red-violet color after incubation at 37°C for 2.5 h indicated urease-positive cultures. The OD560 was measured against a blank of the reaction solution containing no urea.
Hydrogenase assay of E. coli strains.
E. coli strains MC4100 and HYD720 were grown in LB medium supplemented with 2 μM sodium selenite and 2 μM ammonium molybdate. Anaerobic growth was achieved in 250-ml bottles filled almost to the top, inoculated with 20 ml of overnight cultures, and tightly stoppered to maintain anaerobiosis. The bacterial samples were harvested by centrifugation at 5,000 × g for 10 min at 4°C. They were washed twice with 50 mM potassium buffer (pH 6.8) and resuspended in 1 to 4 ml of the same buffer. They were made permeable by addition of toluene (2% [vol/vol]) followed by vigorous shaking.
Hydrogenase (hydrogen:benzyl viologen oxidoreductase) was assayed spectrophotometrically at 600 nm and 30°C; 1.5-ml anaerobic cuvettes contained H2-saturated 100 mM potassium phosphate (pH 6.8) and 1.9 mM oxidized benzyl viologen. The reaction was initiated by addition of bacteria. One unit of activity is defined as 1 μmol of benzyl viologen reduced min−1 by using a molar extinction coefficient of 7,400 M−1 cm−1.
RESULTS
Isolation of an inducible gene from B. suis homologous to nikA.
A method was previously developed to identify genes which are preferentially expressed during the intracellular life of B. suis (22). The screening of such genes was based on the emission of fluorescence by J774 cells infected with B. suis containing plasmid constructs of a B. suis DNA library fused to a promoterless gfp gene. Clones harboring bacterial promoters induced in the host cell could therefore be visualized by fluorescence microscopy. Comparison with the fluorescence of free bacteria in culture medium allowed clones having promoters constitutively expressed to be distinguished from those exhibiting intracellularly inducible promoters; 24 such clones were selected for further analysis. Sequence analysis of B. suis DNA fragments fused to gfp and search for homologies to entries in the SwissProt database revealed a high degree of similarity (76%) of clone 19A10 with the NikA protein of E. coli over a stretch of 225 amino acids. This protein is the first component of the specific nickel transport system. While the role of this transporter in supplying nickel for hydrogenases activities is well documented in E. coli, its contribution to the in vitro and intracellular development of brucellae has not been studied. This gene was thus chosen for further investigation.
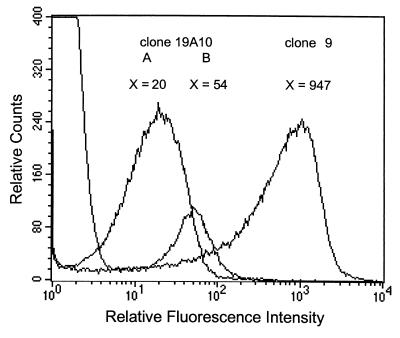
Activation of this promoter in J774 murine macrophage-like cells was controlled with a FACScalibur flow cytometer. The GFP-mediated fluorescence measured by analysis of clone 19A10 grown in TS culture medium was very low in comparison with that obtained by analysis of the constitutively expressed gfp gene in clone 9 cultured in the same conditions (Fig. 1). Fluorescence of the bacteria released after lysis of cells infected with clone 19A10 was 2.7-fold higher than that of free bacteria. No fluorescence was detectable with lysed uninfected cells.
FIG. 1.
Flow cytometry analysis of gfp expression by the B. suis nikA promoter isolated in clone 19A10. Fluorescence intensity of bacteria grown in TS culture medium (A) and of bacteria released after lysis of infected J774 macrophage-like cells (B) was compared to that of the constitutive clone 9, cultured in TS. X, mean relative fluorescence intensity of B. suis clones.
Cloning of genes homologous to the E. coli operon encoding the periplasmic binding-protein-dependent transport system for nickel.
Southern blots of B. suis chromosomal DNA showed that the 19A10 probe bound to a single EcoRI fragment (not shown). The EcoRI fragments of about 6 kb were therefore cloned into pUC18. The physical map of pNIK-BS, the selected recombinant clone, is shown in Fig. 2. Sequence analysis confirmed that B. suis possessed an ORF encoding a protein of 526 amino acids showing high (63%) identity to NikA of E. coli. Four ORFs were identified downstream of nikA and were predicted to be transcribed in the same direction (Fig. 2). Alignment of the proteins encoded by these ORFs with the different components of the E. coli nik operon (33) revealed that they were highly conserved in size and exhibited high (51 to 67%) identity (Table 1). The only motif predicted to form a stem-loop structure (−18 kcal mol−1 [45]) and followed by a run of T's was found 64 bp after the stop codon of nikE and therefore might potentially be recognized as a transcription terminator. These data thus suggested that these five genes are part of an operon which could represent the nik locus homolog in B. suis. A 399-bp ORF separated from nikA by 266 bp and potentially transcribed in the opposite direction (Fig. 2) was highly similar to NikR (Table 1), the repressor protein of the E. coli nik operon (9). The G+C content of the six Brucella genes was variable, as it is in their E. coli counterparts (Table 1).
TABLE 1.
Predicted ORFs homologous to the E. coli nik genes
| Gene | Size (bp) of B. suis gene | % G+C
|
No. of amino acids
|
% Amino acid identity | ||
|---|---|---|---|---|---|---|
| B. suis | E. coli | B. suis | E. coli | |||
| nikA | 1,581 | 54.9 | 53.3 | 526 | 524 | 63 |
| nikB | 945 | 59.2 | 58.4 | 314 | 314 | 67.3 |
| nikC | 873 | 60.5 | 60.9 | 290 | 277 | 62.4 |
| nikD | 768 | 59 | 58.4 | 255 | 253 | 52.7 |
| nikE | 801 | 57.3 | 56.2 | 266 | 268 | 51.1 |
| nikR | 399 | 53.4 | 57.2 | 132 | 133 | 49.3 |
Furthermore, a potential 24-amino-acid signal sequence suggested that B. suis NikA could be transported through the cytoplasmic membrane. This was confirmed by Western blot analysis using antibodies specific to E. coli NikA, which was found to be able to recognize the B. suis NikA protein in the periplasmic space (data not shown). The deduced nikB and nikC gene products were predicted to contain six and five transmembrane regions, respectively, and they possessed most of the histidines and cysteines potentially important for interaction of the homologous E. coli proteins with the metal ions (33). Both NikD and NikE proteins from B. suis possessed the two consensus sites A (GX2GXGKS) and B (DEX4LD) defining the binding motifs for ATP (44). These two proteins could represent the energy providers for the transport system. Structure prediction (16) of B. suis NikR detected the typical β-α-α pattern of its E. coli homolog (5) in the N-terminal region. Furthermore, the positions of the histidine and cysteine residues proposed to interact with nickel (5) were conserved in the C-terminal sequence of the B. suis protein.
Interestingly, palindromic sequences which could serve as binding sites for regulatory proteins were identified in the region between nikA and nikR. The putative Pribnow box (AAATAAT) found upstream of the first codon of nikA is surrounded by two imperfect inverted repeats, ATATGAN16TCATAC (not shown), which match the E. coli NikR operator site (6), with only one mutation in the 5′ half site. Another such motif, CGATCTGATN3ATCAGATCG, contains a DNA stretch strongly resembling to the consensus TTGATN4ATCAA Fnr target sequence.
In vitro induction of the B. suis nikA promoter.
Possible regulation of B. suis nikA expression depending on the oxygen level in the growth conditions used was then tested. GFP-mediated fluorescence under control of the nikA promoter cloned in the 19A10 construct was measured after culture under microaerobic conditions. On TS agar, B. suis strains grew microaerobically almost as well as aerobically (not shown). Comparison of fluorescence intensities detected by flow cytometry following growth of bacteria in different conditions showed that activity of this promoter increased 3.6-fold under microaerobic incubation (Fig. 3A). This induction was higher when bacteria were directly grown in the microaerobic jar than when they were preincubated for 24 h under normal, aerobic conditions. To control an eventual bias due to the production of CO2 by the microaerobic system used, the experiment was also performed in degassed liquid medium. The result (not shown) confirmed induction of the nikA promoter with decreasing oxygen concentration. In contrast, we detected no such effect on expression of the constitutively expressed promoter carried by clone 9 (not shown).
FIG. 3.
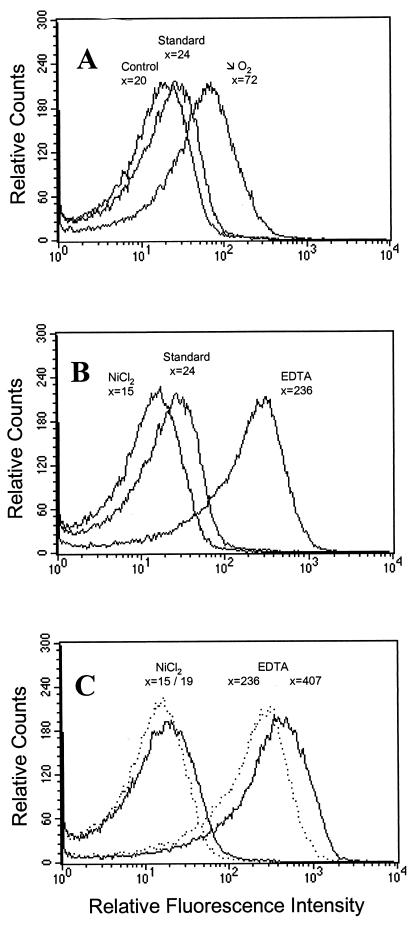
Flow cytometry analysis of the in vitro conditions regulating the expression of the nikA promoter-gfp fusion. x, mean relative fluorescence of the B. suis 19A10 clone. The standard curve represents the fluorescence of clone 19A10 grown under standard aerobic conditions. (A) Activation under microaerobic conditions. The control curve corresponds to bacteria transformed with native plasmid pBBR1-KGFP. (B) Stimulation by nickel deficiency. The right curve represents clone 19A10 grown in TS medium supplemented with 10 mM EDTA; the left curve represents clone 19A10 cultured in the presence of 500 μM NiCl2. (C) Additional effects of simultaneous nickel and oxygen deficiency on fluorescence increase. Shown are fluorescence intensities emitted by the 19A10 clone grown under standard aerobic (dotted curves) and microaerobic (solid curves) conditions in TS medium supplemented with a high concentration of EDTA (10 mM) or NiCl2 (500 μM).
A common feature of many metal transport systems is that their expression is stimulated by metal ion deficiency and inhibited by substrate excess. To verify whether the same was true for the promoter studied here, the 19A10 clone was grown in the presence of a high (10 mM) concentration of the metal ion chelator EDTA in the culture medium. The fluorescence emitted by the gfp fusion under these conditions was compared to that obtained under standard conditions (Fig. 3B). Fluorescence enhancement (approximately 10-fold) showed that the promoter cloned in 19A10 was strongly induced in the conditions of metal ion deficiency created during culture with 10 mM EDTA. In addition, when the medium was supplemented with 500 μM NiCl2, fluorescence intensity decreased below the standard, indicating that an increase in Ni2+ concentration repressed promoter activity. Cultures of clone 19A10 on minimal medium agar resulted in repression of the 19A10 promoter-gfp fusion (not shown). These results showed that under standard conditions, even minimal medium contained enough Ni2+ to repress the nikA promoter. Maximal activation of this promoter was obtained when effects of metal and O2 deficiency were combined. As shown in Fig. 3C, the fluorescence increase caused by lack of Ni2+ was 1.7-fold higher under microaerobic conditions. In contrast, inhibition of the promoter resulting from excess of Ni2+ remained stable.
Insertional inactivation of B. suis nikA.
A mutation in nikA resulting in insertional inactivation of the gene was constructed. The kan gene was inserted into the unique ApaI site within the partial nikA gene isolated from 19A10 (Fig. 2). This new construct, which contained 562 bp of nikA DNA upstream of the insertion site and 114 bp downstream, was introduced into the Brucella suicide plasmid pCVD442. After transformation of B. suis, mutants were obtained by allelic exchange between chromosomal nikA and the inactivated gene on the plasmid. Southern analysis was performed to verify that adequate allelic exchange took place. Chromosomal DNAs of the wild-type and nikA mutant strains of B. suis digested by HindIII were compared by probing with the 19A10 insert. The predicted 967-bp fragment, which originated from the presence of two HindIII sites in nikA and in the Kanr cassette, hybridized in the mutant DNA but was absent in the wild-type DNA (not shown).
Inhibition of urease activity.
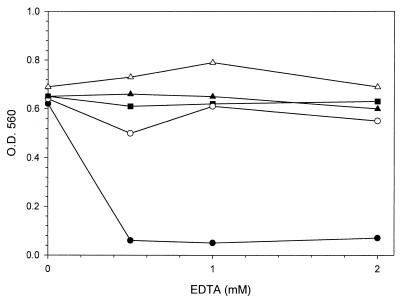
Since urease activity is dependent on nickel acquisition, we tested whether the nikA gene product is required for optimal activity of urease in B. suis. The inhibitory effect of EDTA as a divalent cation chelator on urease activity was examined in wild-type and nikA mutant strains. Bacteria were grown in medium containing various concentrations of EDTA. Urease activity was directly evaluated on washed culture aliquots by the phenol red-urea spectrophotometric assay (Fig. 4). The EDTA concentrations used in this experiment had no effect on the urease activity of the wild-type strain, but there was a marked drop in urease activity of the mutant strain. In the absence of EDTA, there was no difference between the parent and nikA mutant strains, indicating that the mutant retained urease activity. While NikA appeared to be required for full urease activity, other systems in B. suis seemed to supply urease with nickel.
FIG. 4.
Urease activity of B. suis wild type (■), nikA null mutant (●), nikA mutant under conditions of nickel excess (500 μM NiCl2) during the reaction (○), nikA null mutant complemented with the full-length nik locus of B. suis (▵), and nikA null mutant complemented with the nikABCDE genes of E. coli (▴). The inhibitory effect of a Ni2+ chelator was analyzed by addition of increasing EDTA concentrations to the TS growth medium. After culture for 24 h, the urease assay was performed by the phenol red-urea spectrophotometric method (OD560) on whole bacteria washed in PBS.
The nikA null mutant of B. suis was complemented in trans with the full-length nik locus from the parental strain. The 6.4-kb EcoRI fragment carrying nikABCDE and nikR genes from pNIK-BS (Fig. 2) was subcloned into plasmid pBBR1MCS. The complemented nikA mutant recovered maximal urease activity, slightly higher than that of the wild-type strain (Fig. 4). This result indicated that expression of the nikA gene in B. suis is essential for maintaining the optimal level of urease activity.
As depicted in Fig. 4, urease activity of the nikA mutant was stimulated by addition of 500 μM NiCl2 to the reaction. These findings demonstrate that the mutation of nikA results in a markedly reduced urease activity which is correlated to a lower Ni2+ content available for the enzyme. This effect on urease activity strongly supports a probable role for NikA in nickel acquisition by B. suis.
To verify this hypothesis, we tested the effect of heterologous trans complementation with the nik genes of E. coli on urease activity of the nikA mutant. To this end, the nik operon of E. coli, obtained from pLW22 (47), was truncated at the unique ApaLI restriction site, which eliminated 140 bp from the 3′ end of the nikR gene encoding the repressor protein. After subcloning into pBBR1MCS, the resulting plasmid was used to complement the nikA mutant strain of B. suis. The urease assay indicated that this complemented strain had enzymatic activity reaching levels similar to those obtained with the wild-type strain (Fig. 4). The compensatory effect of E. coli nik genes on nikA inactivation therefore confirms that nikA participates in the nickel processing in B. suis.
Functional complementation of a nik mutant of E. coli.
Previous biochemical and physiological studies demonstrated that all nik mutant strains of E. coli lacked hydrogenase activity (33, 46), which requires nickel in the active site of the enzyme. Heterologous complementation of strain HYD720 (Δnik 47) was performed with the integral nik gene cluster of B. suis into plasmid pNIK-BS. Hydrogenase activity of this complemented HYD720 strain was compared to that obtained by homologous complementation with pLW21 (47) bearing the entire nikA-R locus of E. coli. Genes from B. suis restored the hydrogenase activity to 25% of the level measured with the wild-type E. coli strain and mutant HYD720 complemented with pLW21 (Table 2). The B. suis nik system appeared to be able to replace the E. coli nik operon to ensure functional nickel transport and therefore to allow significant hydrogenase activity in an E. coli background.
TABLE 2.
Complementation of an E. coli nik deletion mutant by the B. suis nik genes
| E. coli strain | Plasmid | Genes carried by plasmid | Hydrogenase sp act (μmol of benzyl viologen reduced min−1 mg−1 [dry wt] of bacteria)a |
|---|---|---|---|
| MC4100 | 0.67 | ||
| HYD720 (Δnik)b | 0 | ||
| HYD720 (Δnik) | pLW21 | nikA–R of E. coli | 0.6 |
| HYD720 (Δnik) | pNIK-BS | nikR/A–E of B. suis | 0.17 |
Average of data obtained from two independent experiments.
Mutant HYD720 was derived from the wild-type strain MC4100 by deletion of the entire nik locus except the 5′ region of nikA (47).
Intracellular growth of the nikA mutant in human monocytes.
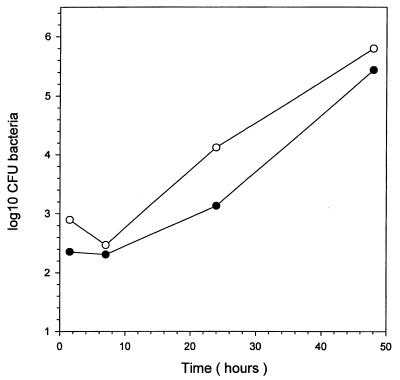
We investigated the effect of the nikA mutation on the intracellular survival of B. suis to assess the contribution of this gene to bacterial replication within its host cell. Multiplication rates of parental B. suis 1330 and of the nikA mutant were determined after infection of human monocytes by these strains. Although the isogenic nikA mutant replicated to a lesser extent than the parental strain at 24 h postinfection, the two strains displayed similar replication rates at 48 h (Fig. 5), both reaching a multiplication factor of approximately 1,000-fold. Infection of differentiated, human macrophage-like THP1 cells yielded similar intracellular growth curves (not shown). These data led us to conclude that in vitro nikA is not a critical factor for replication of B. suis within its host cell.
FIG. 5.
Growth of B. suis in human monocytes. Cells were infected with the wild-type strain (○) or the nikA null mutant (●).
DISCUSSION
The development of a GFP reporter gene fusion system adapted for the detection of B. suis genes induced in the host cell has led to the isolation of the B. suis homolog of the E. coli nikA gene. Further characterization of the nucleotide sequences surrounding this gene on the B. suis chromosome has revealed that this bacterium possesses a complete nik locus highly homologous to the E. coli operon (33). In the present study, several findings suggest a role of the B. suis nik system in nickel uptake. The structure of its components is highly similar to that of the five E. coli nik ORFs, included in a single transcription unit. Sequence comparison between the functionally related proteins has shown that the deduced B. suis Nik proteins, like their E. coli counterparts, belong to a binding-protein-dependent transport system. The first component NikA probably represents in B. suis the periplasmic binding protein as well. NikB and NikC could be membrane proteins analogous to transport system permeases and might function as metal transmitters. NikD and NikE exhibit typical ATP-binding domains of ABC transporters, suggesting their participation in energy production for the transport system. The high level of identity (57.6% on average) between the E. coli and B. suis proteins was rather unexpected for such distantly related organisms. To date, the newly identified nik genes in B. suis are the most homologous to those of E. coli.
Observations concerning the in vitro induction of the B. suis nikA promoter in the 19A10-gfp transcriptional fusion were consistent with a function in nickel transport. On one hand, it could be strongly activated as a result of divalent metal ion deficiency; on the other hand, it was repressed by an increase of the nickel concentration in the medium. Inhibition of nikA expression was observed for culture of 19A10 on minimal medium. As described for the E. coli nik operon, the B. suis nikA gene was suggested to be activated only at very low concentrations of nickel.
Because it is well documented that urease requires nickel ions in the active site to be functional (20, 28), the considerably lower urease activity of the B. suis nikA mutant than of the wild-type strain implicated this gene in nickel uptake. The finding that this apparent insufficiency of nickel could be overcome by addition of excess NiCl2 also demonstrated the involvement of nikA in nickel acquisition. Nevertheless, the enzyme of the mutated B. suis strain was not totally inactive. Despite the need for an intact nikA gene to produce fully active urease, the residual enzymatic activity of the nikA mutant indicated the existence of other Ni2+ uptake systems.
trans complementation of the B. suis nikA mutant and the E. coli nik null strain with the heterologous nik genes also showed that they were able to functionally replace the mutated genes in a heterologous background. In fact, the nikABCDE genes from E. coli fully operated in the mutant of B. suis which recovered optimal urease activity, whereas the intact nik gene cluster from B. suis significantly restored hydrogenase activity in a deletion mutant of E. coli. These results demonstrate that the B. suis nik locus mediates nickel transport.
While the structure of the B. suis nik gene cluster strongly resembles to that of the E. coli operon, the overall organization with respect to nikR was totally different. On the B. suis chromosome, this ORF was found in the vicinity of nikA, and in the opposite orientation of transcription, instead of being downstream of nikE and in the same orientation as in E. coli. Nevertheless, the activation of the B. suis nikA promoter was apparently regulated similarly to the E. coli homolog: induction by metal ion deficiency and low oxygen concentration, and inhibition by substrate excess.
Furthermore, in the promoter region of the B. suis nikA gene, the presence of a palindrome which might potentially serve as a NikR-binding site suggests that NikR could mediate repression of nikA expression by way of promoter occlusion, as described for E. coli (5, 6, 9). However, regulation of nikR gene activation is expected to be quite different in B. suis. Because of the genomic organization in B. suis, expression of nikR cannot be controlled through transcription from the nikA promoter region, in contrast to E. coli nikR (9), but might be driven only by its own promoter. A specific promoter was also identified upstream of E. coli nikR as being responsible for its constitutive transcription. Furthermore, as a consequence of transcriptional regulation at the level of the nik operon promoter, this gene is partially under the positive control of Fnr, the global regulator of anaerobic metabolism (9, 33). A sequence homologous to the FNR box was found in the region between the nikA and nikR genes of B. suis. Although this motif was not located at the proper spacing from the putative nikA promoter, it should be noted that its position, 43 bp upstream of the potential promoter of nikR, could correspond to that described for promoters positively controlled by Fnr. Because Brucella spp. are unable to grow anaerobically, the existence of such a global regulator in B. suis seems unlikely. The main question rising from this observation is whether the nik genes can undergo regulation depending on the oxygen level in the environment. While Brucella was classified among aerobic microorganisms, it can grow in vitro under standard microaerobic growth conditions. Our results have revealed the possible induction of the B. suis nikA promoter under microaerobiosis. We suggest that regulatory mechanisms could occur during the intracellular life of Brucella to enable the organism to cope with changes in the external oxygen concentration in the host. Many microorganisms previously described as obligate aerobes exhibited adaptation to fluctuations of the redox status (for a review see reference 40). For example, Bacillus subtilis was shown to possess a redox-sensing (ResDE) system which regulates expression of the B. subtilis fnr-like gene (30, 31). The requirement of an active nik gene cluster in this context could be viewed as a specific nickel provider for B. suis enzymes involved in adaptation to growth in a microaerobic environment. This is the case for E. coli hydrogenases (46), produced only under anaerobic conditions. Rhizobium species also express uptake hydrogenases, which may play a key role in improving the efficiency of N2 fixation in the microaerobic environment of the plant nodules. The Bradyrhizobium japonicum hydrogenase requires nickel for both enzyme activity and transcriptional regulation (21). These enzymes and the specific regulations required to adapt activities to the intracellular life-style remain to be discovered in Brucella spp.
Nickel is essential for other important enzymatic reactions, among which ureolysis has become of major interest since a relationship between the highly ureolytic pathogen H. pylori and the human peptic ulceration has been found (28). This organism has adopted a strategy of survival in the gastric mucosa that involves synthesis of urease, producing ammonia which in turn may neutralize the acidic pH in the local environment of the bacteria. Urease is absolutely essential for colonization of animal models by Helicobacter pylori (for a review reference 28).
We have shown that B. suis displayed maximal urease activity when the nik locus encoding a putative nickel transporter was intact. According to the above model of resistance to host defense, it might be speculated that Brucella, which is unable to multiply in strongly acidic medium (25), could be protected from acidification by the ammonia released from urea hydrolysis. Because the natural host infection by Brucella frequently occurs via the oral route, the bacterium has to tolerate low pH during its passage through the digestive tract. Recent work (18) reported that urease-negative mutants of Yersinia enterocolitica were less virulent after intragastric inoculation. Moreover, a previous study of the environmental conditions that B. suis encounters in the host cell has demonstrated that this bacterium survives and remains enclosed in phagosomes which are rapidly acidified. Early acidification was necessary for the multiplication of B. suis within macrophages (36). Ammonium chloride addition at 7 h postinfection did not affect the replication rate of intracellular bacteria, in contrast to early neutralization. This result suggested that the pH of the phagosome had already increased at this step of infection.
Rhizobium meliloti, phylogenetically closely related to Brucella, also harbors urease and hydrogenase, whose functions are important for nitrogen management. Interestingly, the same transcription unit of the R. meliloti chromosome contains the structural genes for urease, separated by ORFs also involved in determining hydrogenase activity (27).
In conclusion, B. suis seems to possess one or several enzymes whose full activity requires nickel ions. These activities could be necessary for adaptation to the environmental conditions encountered by the bacteria in the host (O2 level, acidic pH, etc.). In B. suis, as in many pathogenic microorganisms, the binding-protein-dependent transporter encoded by the E. coli nik operon homolog is probably not the only system supplying these enzymes with nickel. In addition to this specific high-affinity transporter, E. coli is able to use one, and perhaps two, nonspecific magnesium transport systems to accumulate nickel under conditions of substrate excess (33). Because urease activity is probably crucial for the virulence of H. pylori, its high-affinity nickel transport protein, NixA, is not the only ion transporter capable of nickel acquisition (3) and contributing to full catalytic activity.
For these reasons, it was not surprising to observe little difference between intracellular growth rates of the B. suis wild-type and nikA mutant strains. Nevertheless, it cannot be ruled out that this gene could participate in intracellular multiplication over the first 24 h of infection and that its inactivation in the mutant strain could be overcome by other systems expressed later. Results describing urease activity have indeed suggested that several systems could be apparently involved in nickel acquisition.
The nikA gene was previously selected on the basis of its induction within macrophages (22). However, the present results do not allow us to conclude that it is crucial for in vitro intracellular multiplication of B. suis. On the other hand, the presence of additional systems able to partially compensate for the inactivation of this gene suggested that it plays an important role in retaining enzyme activities which in turn could be critical for survival of brucellae under the environmental conditions of the host cell. Until now, no specific mechanism has been described as being involved in intracellular multiplication of Brucella spp. No liberation of toxin was detected, and few virulence factors (the stress proteins HF-I [38] and Lon [37], the VirB type IV secretion system [34], and a two-component regulatory system [41]) have been identified. Although analysis of the genes so far selected for their specific induction in macrophages is still in progress, the majority of the deduced proteins are components of transport systems, as is NikA (22). Therefore, we speculate that Brucella may be able to use enzymes involved in normal metabolic pathways to adapt to the conditions specific to the intracellular life-style of this bacterium. It was noteworthy that among B. suis mutants with attenuated virulence (13), most harbor mutations in genes encoding regulation factors and enzymes of the general metabolism. The ability of intracellular bacteria like Brucella species to sense changes in their environment and to rapidly modulate gene expression is essential for bacterial virulence. The search for global regulators, involved in sensing diverse stimuli such as oxygen and redox potential, could provide clues to the mechanisms contributing to adaptation of brucellae to the host cells.
ACKNOWLEDGMENTS
We thank S. Burkhardt for help with DNA cloning for complementation assays.
This work was supported in part by grant QLK2-CT-1999-00014 from the European Union.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 3.Bauerfeind P, Garner R M, Mobley H L T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron E, Gross A, Liautard J-P, Dornand J. Brucella species release a specific, protease-sensitive, inhibitor of TNF-α expression, active on human macrophage-like cells. J Immunol. 1996;156:2885–2893. [PubMed] [Google Scholar]
- 5.Chivers P T, Sauer R T. NikR is a ribbon-helix-helix DNA-binding protein. Protein Sci. 1999;8:2494–2500. doi: 10.1110/ps.8.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chivers P T, Sauer R T. Regulation of high affinity nickel uptake in bacteria. Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem. 2000;275:19735–19741. doi: 10.1074/jbc.M002232200. [DOI] [PubMed] [Google Scholar]
- 7.Corbel M J. Brucella. In: Parker M T, Collier L H, editors. Principles of bacteriology, virology, and immunity. 2. E. London, England: Arnold; 1990. pp. 339–353. [Google Scholar]
- 8.Cuny G, Veas F, Roizès G. “Pocket blotting”: a method for transferring nucleic acids onto nylon membranes. Anal Biochem. 1991;193:45–48. doi: 10.1016/0003-2697(91)90041-q. [DOI] [PubMed] [Google Scholar]
- 9.De Pina K, Desjardin V, Mandrand-Berthelot M-A, Giordano G, Wu L-F. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J Bacteriol. 1999;181:670–674. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekaza E, Guilloteau L, Teyssier J, Liautard J-P, Köhler S. Functional and analysis of the ClpATPase ClpA of Brucella suis, and persistence of a knockout mutant in BALB/c mice. Microbiology. 2000;146:1605–1616. doi: 10.1099/00221287-146-7-1605. [DOI] [PubMed] [Google Scholar]
- 12.Elzer P H, Phillips R W, Kovach M E, Peterson K M, Roop R M., II Characterization and genetic complemention of a Brucella abortus high-temperature requirement A (htrA) deletion mutant. Infect Immun. 1994;62:4135–4139. doi: 10.1128/iai.62.10.4135-4139.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foulongne V, Bourg G, Cazevieille C, Michaux-Charachon S, O'Callaghan D. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect Immun. 2000;68:1297–1303. doi: 10.1128/iai.68.3.1297-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gay P, Le Coq D, Steinmetz M, Ferrari E, Hoch J A. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geourjon C, Deléage G. SOPMA: significant improvement in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 17.Gor D, Mayfield J E. Cloning and nucleotide sequence of the Brucella abortus groE operon. Biochim Biophys Acta. 1992;1130:120–122. doi: 10.1016/0167-4781(92)90476-g. [DOI] [PubMed] [Google Scholar]
- 18.Gripenberg-Lerche C, Zhang L, Ahtonen P, Toivanen P, Skurnik M. Construction of urease-negative mutants of Yersinia enterocolitica serotypes O:3 and O:8: role of urease in virulence and arthritogenicity. Infect Immun. 2000;68:942–947. doi: 10.1128/iai.68.2.942-947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross A, Spiesser S, Terraza A, Liautard J-P, Caron E, Dornand J. Expression and bactericidal activity of nitric oxide synthase in Brucella suis-infected murine macrophages. Infect Immun. 1998;66:1309–1316. doi: 10.1128/iai.66.4.1309-1316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausinger R P. Nickel utilization by microorganisms. Microbiol Rev. 1987;51:22–42. doi: 10.1128/mr.51.1.22-42.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Maier R J. Transcriptional regulation of hydrogenase synthesis by nickel in Bradyrhizobium japonicum. J Biol Chem. 1990;265:18729–18732. [PubMed] [Google Scholar]
- 22.Köhler S, Ouahrani-Bettache S, Layssac M, Teyssier J, Liautard J-P. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect Immun. 1999;67:6695–6697. doi: 10.1128/iai.67.12.6695-6697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler S, Teyssier J, Cloeckaert A, Rouot B, Liautard J-P. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol Microbiol. 1996;20:701–712. doi: 10.1111/j.1365-2958.1996.tb02510.x. [DOI] [PubMed] [Google Scholar]
- 24.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 25.Kulakov Y K, Guigue-Talet P G, Ramuz M R, O'Callaghan D. Response of Brucella suis 1330 and B. canis RM6/66 to growth at acid pH and induction of an adaptive acid tolerance response. Res Microbiol. 1997;148:145–151. doi: 10.1016/S0923-2508(97)87645-0. [DOI] [PubMed] [Google Scholar]
- 26.Lin J, Adams L G, Ficht T A. Characterization of the heat shock response in Brucella abortus and isolation of the genes encoding the GroE heat shock proteins. Infect Immun. 1992;60:2425–2431. doi: 10.1128/iai.60.6.2425-2431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miksch G, Arnold W, Lentzsch P, Priefer U B, Pühler A. A 4.6 kb DNA region of Rhizobium meliloti involved in determining urease and hydrogenase activities carries the structural genes for urease (ureaA, ureB, ureC) interrupted by other open reading frames. Mol Gen Genet. 1994;242:539–550. doi: 10.1007/BF00285277. [DOI] [PubMed] [Google Scholar]
- 28.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers E W, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 30.Nakano M M, Hulett F M. Adaptation of Bacillus subtilis to oxygen limitation. FEMS Lett. 1997;157:1–7. doi: 10.1111/j.1574-6968.1997.tb12744.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakano M M, Zuper P, Glaser P, Danchin A, Hulett F M. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nash P, Krenz M M. Culture media. In: Balows A, et al., editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. p. 1280. [Google Scholar]
- 33.Navarro C, Wu L-F, Mandrand-Berthelot M-A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 34.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 35.Pizarro-Cerdá J, Moreno E, Sanguedolce V, Mege J-L, Gorvel J-P. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun. 1998;66:2387–2392. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porte F, Liautard J-P, Köhler S. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect Immun. 1999;67:4041–4047. doi: 10.1128/iai.67.8.4041-4047.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson G T, Kovach M E, Allen C A, Ficht T A, Roop R M., II The Brucella abortus Lon functions as a generalized stress response protease and is required for wild-type virulence in BALB/c mice. Mol Microbiol. 2000;35:577–588. doi: 10.1046/j.1365-2958.2000.01726.x. [DOI] [PubMed] [Google Scholar]
- 38.Robertson G T, Roop R M., II The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Sawers G. The aerobic/anaerobic interface. Curr Opin Microbiol. 1999;2:181–187. doi: 10.1016/S1369-5274(99)80032-0. [DOI] [PubMed] [Google Scholar]
- 41.Sola-Landa A, Pizarro-Cerda J, Grillo M-J, Moreno E, Moriyon I, Blasco J-M, Gorvel J-P, Lopez-Goni I. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol. 1998;29:125–138. doi: 10.1046/j.1365-2958.1998.00913.x. [DOI] [PubMed] [Google Scholar]
- 42.Sonnhammer E L L, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 43.Tusnády G E, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 44.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter A E, Turner D H, Kim J, Lyttle M H, Muller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L-F, Mandrand-Berthelot M-A, Waugh R, Edmonds C J, Holt S E, Boxer D H. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol Microbiol. 1989;3:1709–1718. doi: 10.1111/j.1365-2958.1989.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 47.Wu L-F, Navarro C, Mandrand-Berthelot M-A. The hydC region contains a multi-cistronic operon (nik) involved in nickel transport in Escherichia coli. Gene. 1991;107:37–42. doi: 10.1016/0378-1119(91)90294-l. [DOI] [PubMed] [Google Scholar]